Abstract
The induction of immediate-early (IE) genes, including proto-oncogenes c-fos and c-jun, correlates well with a nucleosomal response, the phosphorylation of histone H3 and HMG-14 mediated via extracellular signal regulated kinase or p38 MAP kinase cascades. Phosphorylation is targeted to a minute fraction of histone H3, which is also especially susceptible to hyperacetylation. Here, we provide direct evidence that phosphorylation and acetylation of histone H3 occur on the same histone H3 tail on nucleosomes associated with active IE gene chromatin. Chromatin immunoprecipitation (ChIP) assays were performed using antibodies that specifically recognize the doubly-modified phosphoacetylated form of histone H3. Analysis of the associated DNA shows that histone H3 on c-fos- and c-jun-associated nucleosomes becomes doubly-modified, the same H3 tails becoming both phosphorylated and acetylated, only upon gene activation. This study reveals potential complications of occlusion when using site-specific antibodies against modified histones, and shows also that phosphorylated H3 is more sensitive to trichostatin A (TSA)-induced hyperacetylation than non-phosphorylated H3. Because MAP kinase-mediated gene induction is implicated in controlling diverse biological processes, histone H3 phosphoacetylation is likely to be of widespread significance.
Keywords: acetylation/histone H3/MAP kinase pathways/phosphorylation/proto-oncogene induction
Introduction
Transcription factors and co-activators such as GCN5 (Brownell et al., 1996), p300/CBP (Bannister and Kouzarides, 1996; Ogryzko et al., 1996), p/CAF (Yang et al., 1996), TAFII250 (Mizzen et al., 1996) and the nuclear hormone receptor co-activators ACTR (Chen et al., 1997) and SRC-1 (Spencer et al., 1997) have histone acetyltransferase (HAT) activity, a finding that implicates nucleosomal modifications in transcriptional regulation (reviewed in Imhof and Wolffe, 1998; Kuo and Allis, 1998; Struhl, 1998; Workman and Kingston, 1998; Grant and Berger, 1999; Johnson and Turner, 1999; Brown et al., 2000; Strahl and Allis, 2000). Histone acetylation was first observed by Vincent Allfrey and co-workers (Allfrey et al., 1964) who later, using a method of nucleosome subfractionation based on the accessibility of a cysteine residue on histone H3, provided the first indications of histone acetylation and alterations in nucleosome structure associated with the induction of two immediate-early (IE) genes, c-fos and c-myc (Allegra et al., 1987; Chen and Allfrey, 1987). These alterations were transient, correlating well with induction and superinduction of these genes and were reversed upon shut-off (Chen and Allfrey, 1987). More recently, Feng and Villeponteau (1990, 1992), who mapped the transient relaxation of chromatin that originated at the c-fos promoter and spread across the gene, argued that this occurred too rapidly to be the consequence of the passage of RNA polymerase II. Overall, these data suggest a model whereby alterations in chromatin and nucleosome structure, possibly involving histone acetylation, are a cause and not an effect of IE gene induction. Two recent findings support this model. First, p300/CBP, which by binding to the transcription factors ternary complex factor (TCF) (Janknecht and Nordheim, 1996a,b) and c-Jun (Arias et al., 1994; Bannister et al., 1995) is implicated in c-fos and c-jun induction, has intrinsic HAT activity (Bannister and Kouzarides, 1996; Ogryzko et al., 1996). Second, highly localized modulation of histone acetylation, spanning a few nucleosomes, has been demonstrated in vivo concomitant with gene induction (Kuo et al., 1998; Chen et al., 1999; Parekh and Maniatis, 1999) and repression (Kadosh and Struhl, 1998; Rundlett et al., 1998). The fact that p300/CBP is recruited by its interaction with sequence-specific transcription factors provides a long-sought mechanism by which localized nucleosomal alterations can be targeted to specific genes. Interference with the recruitment of p300/CBP to the human interferon-β (IFN-β) enhanceosome reduced transcription and suppressed the localized H3 and H4 hyperacetylation normally observed at the IFN-β promoter in response to viral infection (Parekh and Maniatis, 1999). Finally, proof that the upstream serum response element (SRE), which controls c-fos, can direct activation-dependent histone H4 acetylation of an associated reporter gene was recently provided by Treisman and colleagues (Alberts et al., 1998; see also O’Neill and Turner, 1995; discussed in Thomson et al., 1999a).
A second nucleosomal modification indissociable from IE gene transcription under diverse conditions of induction, superinduction (Mahadevan and Edwards, 1991; Barratt et al., 1994a,b; Cano et al., 1995) and inhibition (Hazzalin et al., 1996; Thomson et al., 1999b) is the phosphorylation of histone H3 on serine 10 (Mahadevan et al., 1991; Mizzen et al., 1998; Chadee et al., 1999; DeManno et al., 1999). In parallel, phosphorylation of the nucleosome-binding high mobility group protein HMG-14 on serine 6 is also invariably observed (Barratt et al., 1994a). MAP kinases, which control the induction of several IE genes by phosphorylating transcription factors such as TCFs, c-Jun, ATF-2 and RSRF/MEF2 (reviewed in Hunter and Karin, 1992; Cano and Mahadevan, 1995; Treisman, 1996), also mediate the phosphorylation of histone H3 and HMG-14 (Hazzalin et al., 1996; Thomson et al., 1999b). This occurs through the intermediary of the kinase MSK1 (Deak et al., 1998), which lies immediately downstream of extracellular signal regulated kinases (ERKs) and p38 (Deak et al., 1998; Thomson et al., 1999b). In addition, Rsk2, which lies downstream of ERKs, has also been implicated as a histone H3 kinase (Chen et al., 1992; Sassone-Corsi et al., 1999). The fact that the same signalling cascade mediates phosphorylation of transcription factors necessary for IE gene induction as well as the nucleosomal response adds to the case that the two are linked. Finally, we have shown by metabolic labelling studies that the fraction of histone H3 that becomes phosphorylated is extremely small and that these histone H3 molecules are especially susceptible to hyperacetylation; this implies that phosphorylation and acetylation are targeted to the same nucleosomes (Barratt et al., 1994b).
The model that emerges is one whereby IE genes, poised to transcribe by pre-association of transcription factors such as TCFs for c-fos and c-Jun or ATF-2 for c-jun, are triggered by MAP kinase-mediated phosphorylation of these factors (reviewed in Hunter and Karin, 1992; Treisman, 1996). They are then able to recruit HATs such as p300/CBP (Arias et al., 1994; Bannister et al., 1995; Janknecht and Nordheim, 1996a,b), providing a means for targeted acetylation of the adjacent IE gene nucleosomes. Our data suggest that histone H3 on these nucleosomes must also become phosphorylated by MSK1, which is activated either by ERK or by p38 MAP kinases, both of which are capable of forming stable complexes with transcription factors and could therefore produce localized activation of downstream kinases (reviewed in Thomson et al., 1999a). The missing piece of evidence that would validate this model is the direct demonstration that histone H3 on IE gene nucleosomes does indeed become both phosphorylated and acetylated upon gene activation. Here we provide this evidence. Antibodies were first raised against histone H3 phosphorylated at serine 10 to immunoselect nucleosomes for DNA analysis. This revealed a major complication in that the presence of acetylated lysines adjacent to the phosphoepitope prevented antibody recognition because these acetylated lysines occluded the serine 10 phosphoepitope. This problem was overcome by raising new antibodies that recognize only the doubly modified phosphoacetylated form of histone H3. These then allowed direct proof (i) that the phosphoacetyl epitope does occur in vivo upon stimulation of quiescent cells and (ii) that histone H3 on nucleosomes associated with c-fos and c-jun is both phosphorylated and acetylated upon transcriptional activation. These data prove for the first time that phosphoacetylation of H3 occurs on IE gene chromatin upon gene activation, suggesting its involvement in diverse biological instances where MAP kinase-mediated IE gene induction is observed.
Results
[32P]Phosphate-labelling and acetic acid–urea gel analysis of the nucleosomal response
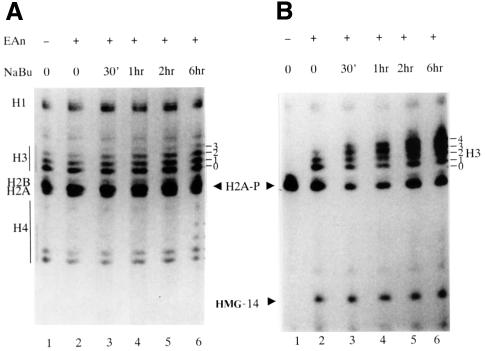
To show the relationship between H3 phosphorylation and acetylation, and to aid interpretation of acetic acid–urea gels and western blots using modification-specific antibodies, we first present data from a [32P]phosphate-labelling experiment. Hyperacetylation of histones in C3H 10T1/2 cells was induced by butyrate pretreatment for varying times (0–6 h) and histone H3 and HMG-14 phosphorylation elicited under superinducing conditions by stimulation for the last hour with a combination of epidermal growth factor (EGF) plus anisomycin (Edwards and Mahadevan, 1992; discussed in Hazzalin et al., 1998 and references therein). Acetic acid–urea gels were used in which successive modifications of histone H3 lead to incremental retardations, giving rise to a ladder of differentially modified H3 forms (see Coomassie-stained gel, Figure 1A; the H3 ladder is numbered according to the number of modifications). Previous experiments establish that phosphorylation and acetylation will both cause incremental retardations in this gel system (Barratt et al., 1994b). There are at least four acetylation sites (lysines 9, 14, 18 and 23), one proven site of inducible phosphorylation (serine 10) and other possible modifications such as a second potential site of phosphorylation (serine 28) (Goto et al., 1999) and methylation (Strahl et al., 1999; Strahl and Allis, 2000) that could affect H3 mobility on these gels. Each Coomassie-stained band above band 0 in the H3 ladder contains mixtures of modified H3. It is not possible at present to comprehensively assign complements of modifications at any position although metabolic labelling or modification-specific antibodies can prove the presence of particular modifications within each band (see Discussion).
Fig. 1. EGF/anisomycin-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. (A) Coomassie-stained gel of histones resulting after sequential extraction of 32P-labelled cells pretreated with 5 mM sodium butyrate for various lengths of time before stimulation with EGF/anisomycin. Quiescent C3H 10T1/2 cells received no pretreatment (lanes 1 and 2) or were pretreated with sodium butyrate (NaBu, 5 mM) for 30 min (lane 3), 1 h (lane 4), 2 h (lane 5) or 6 h (lane 6). Cells were labelled with [32P]phosphate for the final 4 h and stimulated as indicated (lanes 2–6) with 50 ng/ml EGF plus 10 µg/ml anisomycin (EAn) for the last hour of labelling (lane 1; unstimulated cells). Acid extracts of the nuclear-enriched pellets containing histone and HMG proteins were electrophoresed on acid–urea gels and Coomassie stained. The portion of the gel containing proteins migrating more quickly than histone H1 is shown. The position of modified forms of histone H3 are numbered corresponding to the number of post-translational modifications visualized by Coomassie staining (0 = no modification). (B) Autoradiograph of stained gel in (A). The positions of the modified forms of histone H3 are numbered corresponding to the number of post-translational modifications, as in (A). The positions of phosphorylated H2A (H2A-P) and phosphorylated HMG-14 (HMG-14) are also marked.
In these cells, 4–6 h of treatment with deacetylase inhibitors produces maximal histone hyperacetylation (discussed in detail in Barratt et al., 1994b). The autoradiograph (Figure 1B) shows, firstly, that histone H3 and HMG-14 are both phosphorylated upon stimulation with EGF/anisomycin (Figure 1B; lanes 2–6). Relevant to western blotting data presented below (Figure 2), note that these phosphorylation events are not inhibited in the presence of butyrate. Secondly, there is clear H3 acetylation in untreated cells (three Coomassie-stainable bands in Figure 1A, lanes 1 and 2) that is only slightly augmented upon butyrate treatment (four stainable bands in Figure 1A, lanes 5 and 6) whereas the 32P-radiolabelled H3 bands are shifted to the top of this ladder by butyrate treatment, occupying positions where there is little or no Coomassie-stained protein (Figure 1B, lanes 5 and 6). This confirms our previous finding that the phosphorylation signal is targeted to a small fraction of histone H3 that is especially susceptible to hyperacetylation (discussed in detail in Barratt et al., 1994b).
Fig. 2. Anti-phospho-H3 antibodies recognize phosphorylated histone H3 but not histone H3 that is both phosphorylated and acetylated. Quiescent C3H 10T1/2 cells received no pretreatment (lanes 1–4) or were pretreated with 500 ng/ml TSA for 4 h (lanes 5–8) or 5 mM sodium butyrate for 5 h (NaBu, lanes 9–12). For each set, cells were stimulated with 50 ng/ml EGF plus 10 µg/ml anisomycin (EAn) for 30 min (lanes 2, 6 and 10), 45 min (lanes 3, 7 and 11) or 60 min (lanes 4, 8 and 12); lanes 1, 5 and 9 contain unstimulated cells. Acid-soluble nuclear proteins were resolved on 15% acid–urea gels, transferred to PVDF membrane and then analysed by western blotting using anti-acetyl-H3 antibodies (panel i), anti-phospho-H3 antibodies (panel ii) or anti-HMG-14 antibodies (panel iii). Coomassie-stained acid–urea gel is shown in panel iv. The positions of the modified forms of histone H3 are numbered corresponding to the number of post-translational modifications visualized by Coomassie staining of gels or Ponceau S staining of PVDF membrane after transfer (0 = no modification).
Anti-phospho-H3 antibodies recognize phosphorylated histone H3 but not histone H3 that is both phosphorylated and hyperacetylated
To prove by chromatin immunoprecipitation (ChIP) assays that the nucleosomal response is targeted to IE genes, we first raised phospho-specific antibodies against a synthetic phosphopeptide encompassing residues 7–20 of H3 within which serine 10 was phosphorylated (characterization described in Thomson et al., 1999b). Similar anti-phospho-H3 antibodies have been used to stain highly phosphorylated histone H3 on condensed mitotic chromosomes (Hendzel et al., 1997; Wei et al., 1999); this mode of widespread stable H3 phosphorylation on condensed chromosomes is distinct from events described here, which involves a minute fraction of histone H3 and is a rapid and transient response to diverse extracellular stimuli (Mahadevan et al., 1991; Barratt et al., 1994b).
The anti-phospho-H3 antibodies were used in western blotting analyses of acid–urea gels containing histones extracted from quiescent or stimulated cells, as well as from cells in which histone hyperacetylation was induced by pretreatment with the deacetylase inhibitors sodium butyrate or trichostatin A (TSA; Yoshida et al., 1990; reviewed in Yoshida et al., 1995). Anti-acetyl histone H3 antibodies that recognize hyperacetylated histone H3 (kindly given by Professor David Allis, University of Virginia, Charlottesville) were used to show that butyrate and TSA treatment both produced hyperacetylation of histone H3 (Figure 2, panel i, lanes 5–12), whereas EGF/anisomycin stimulation does not produce any significant increase of H3 acetylation detectable with these antibodies over that seen in quiescent cells (Figure 2, panel i, lanes 1–4). Probing these blots with the anti-phospho-H3 antibodies produced two interesting observations (Figure 2, panel ii). First, in 32P-labelling studies, a ladder of phosphorylated H3 is always seen (see Figure 1B) whereas the anti-phospho-H3 antibody picks up one band at position 1, presumably monophospho-H3 with no acetyl groups, and a trace of a second band above it (Figure 2, panel ii, lanes 1–4); the antibody does not recognize phospho-H3 demonstrable by 32P-labelling to be present in the upper bands of the H3 ladder (see Figure 1B). A clue to the cause of this arises from similar analyses performed in TSA- or butyrate-treated cells; in these lanes, recognition of phosphorylated histone H3 by the anti-phospho-H3 antibody is essentially lost (Figure 2, panel ii, lanes 5–12) although it is clear from the 32P-labelling studies described above that phospho-H3 in hyperacetylated forms is certainly present.
To show that the loss of anti-phospho-H3 signal is not due to inhibited signalling to nucleosomes, the blot was reprobed with antibodies raised against HMG-14, which reveals its phosphorylation by altered mobility on these gels (characterization described in Thomson et al., 1999b). This showed that EGF/anisomycin-stimulated phosphorylation of HMG-14 is not inhibited under any of these conditions (Figure 2, panel iii), confirming that signalling to nucleosomes is not affected by TSA or butyrate pretreatment.
Recognition of phosphoserine 10 by the anti-phospho-H3 antibody is prevented by acetylation of adjacent lysines
The marked difference between the anti-phospho-H3 immunostaining profile (Figure 2, panel ii) and the 32P-labelled histone H3 bands (Figure 1B) suggested that the presence of acetylated lysines might interfere with antibody recognition of the serine 10 phosphoepitope. This hypothesis was tested by analysing antibody recognition of a set of synthetic peptides (shown in Figure 3A) corresponding to differentially modified forms of the histone H3 tail. As expected, the anti-phospho-H3 antibody recognizes an H3 peptide with phosphate on serine 10 (Figure 3B, lane 2), whereas there is little or no binding to the non-phosphorylated peptide (Figure 3B, lane 1) or to a diacetylated peptide synthesized with acetyl groups on lysines 9 and 14 (Figure 3B, lane 3). These two lysines were chosen because they are closest to and most likely to occlude the serine 10 phosphoepitope. When a phosphodiacetylated peptide with phosphate at serine 10 and acetyl groups at lysines 9 and 14 was probed with the anti-phosphoserine-10 antibody, the signal was severely reduced (Figure 3B; compare lanes 4 and 2), suggesting that the acetyl groups at lysines 9 and/or 14 did interfere with antibody recognition of the serine 10 phosphoepitope. To prove directly that occlusion was due to the acetylated lysines, we used recombinant yeast histone deacetylase HOS3 (Rundlett et al., 1996; reviewed in Johnson and Turner, 1999; kindly given by Professor Peter Lewis; Toronto) to remove the acetyl groups from this peptide; deacetylation restored recognition of the serine 10 phosphoepitope by the anti-phospho-H3 antibody (lane 6). Note that the HOS3 preparation on its own did not yield a signal (lane 5) and HOS3 denatured by boiling was substantially ineffective in restoring antibody binding (lane 7). These data prove that our anti-phospho-H3 antibody recognizes histone H3 phosphorylated on serine 10 but that this epitope is occluded and is not recognized by the antibody when lysines 9 and/or 14 are acetylated (discussed further below).
Fig. 3. Deacetylation of a synthetic phosphodiacetyl histone H3 peptide restores anti-phospho-H3 antibody binding. (A) Schematic summary of the phospho- (serine 10), diacetyl- (lysines 9 and 14) and phosphodiacetyl-H3 peptides synthesized for peptide screening and subsequent antibody generation. (B) The indicated mass of synthetic peptide was spotted onto Hybond-C in a volume of 1 µl. These correspond to histone H3 (lane 1), phospho-H3 (lane 2), diacetyl-H3 (lane 3) and phosphodiacetyl-H3 (lane 4). In vitro deacetylation of the synthetic phosphodiacetyl-H3 peptide was performed using recombinant yeast histone deacetylase HOS3. These peptide and enzyme samples were also spotted onto Hybond-C at the indicated mass in a volume of 1 µl. These correspond to HOS3 alone (lane 5), phosphodiacetyl-H3 plus HOS3 (lane 6) and phosphodiacetyl-H3 plus boiled HOS3 (lane 7). Spotted peptide and enzyme samples were air-dried and western analysis was performed using anti-phospho-H3 antibody.
Generation of antibodies against doubly modified phosphoacetyl-histone H3
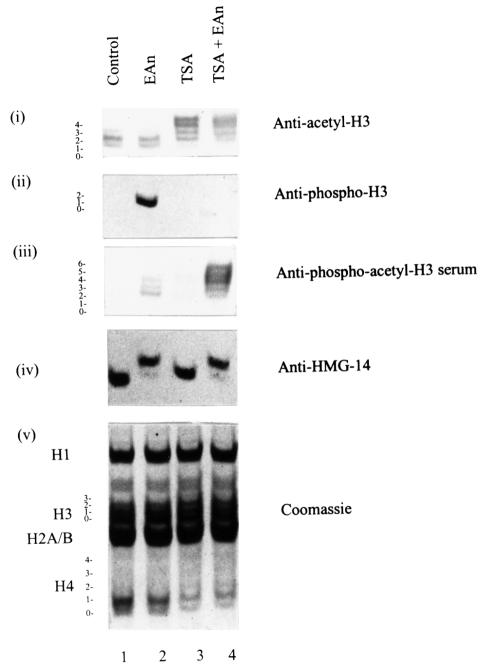
To investigate histone H3 phosphoacetylation further, we then raised antibodies against the phosphodiacetyl peptide shown in Figure 3A. The specificity of the resultant antibody was first verified using the peptides described above (Figure 3A; data not shown; see also Figure 5) and is confirmed by analyses of intact histones extracted from EGF/anisomycin- and TSA-treated cells (Figure 4). Nuclear extracts were prepared from control (Figure 4, lane 1) and EGF/anisomycin-stimulated cells (Figure 4, lanes 2 and 4) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of TSA to cause hyperacetylation, and resolved on acid–urea gels for western blotting analyses. Anti-acetyl-H3 antibodies were used to confirm hyperacetylation (Figure 4, panel i, lanes 3 and 4) and anti-HMG-14 antibodies to show signalling was unimpaired (Figure 4, panel iv, lanes 2 and 4); the Coomassie-stained gel (Figure 4, panel v) verifies equal loading in all lanes. Whereas the rabbit anti-phospho-H3 antibody only yields a signal from EGF/anisomycin-treated samples (Figure 4, panel ii, lane 2), the new sheep anti-phosphoacetyl-H3 antibody now recognizes the phosphorylated and acetylated histone H3 at the top of the H3 ladder (Figure 4, panel iii, lane 4). This antibody does not recognize H3 that is hyperacetylated but not phosphorylated (Figure 4, panel iii, lane 3). Further, it produces a weak signal in the EGF/anisomycin-treated cells (lane 2; see also Figure 5), showing that even in cells not treated with TSA to cause hyperacetylation, EGF/anisomycin stimulation does lead to some H3 becoming both phosphorylated and acetylated (see also Figure 8).
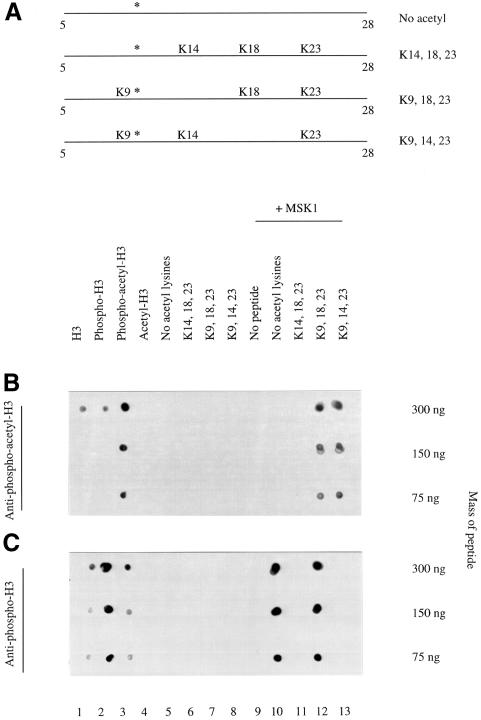
Fig. 5. Anti-phosphoacetyl-H3 antibodies specifically recognize acetyllysine 9 and phosphoserine 10. (A) Schematic diagram of the acetylated H3 peptides used in the in vitro kinase assays with MSK1. All peptides correspond to residues 5–28 of the H3 N-terminus, encompassing all the in vivo H3 acetylation sites. The first peptide contained no acetylated lysine residues, the second contained acetylated lysines 14, 18 and 23, the third contained acetylated lysines 9, 18 and 23 and the final peptide contained acetylated lysines 9, 14 and 23. The serine 10 residue that is phosphorylated by MSK1 in the kinase assay is indicated by an asterisk. (B) The indicated masses of the synthetic peptides were spotted onto Hybond-C in a volume of 1 µl. These were histone H3, phospho-H3, phosphodiacetyl-H3 and diacetyl H3 (lanes 1–4, respectively, as shown in Figure 3A), lanes 5–8 are the acetylated peptides shown in (A) and lanes 10–13 are the same peptides after in vitro phosphorylation with MSK1. Lane 9 contains MSK1 alone. Spotted peptide and enzyme samples were air dried and probed with the anti-phosphoacetyl-H3 antibody. (C) As in (B) but probed with anti-phospho-H3.
Fig. 4. Generation of antibodies against doubly modified phosphoacetyl histone H3. Acid-soluble nuclear proteins were extracted from quiescent C3H 10T1/2 cells (lane 1) or cells stimulated with 50 ng/ml EGF and 10 µg/ml anisomycin for 1 h (EAn, lane 2), pretreated with 500 ng/ml TSA for 4 h (lane 3) or pretreated with 500 ng/ml TSA for 4 h and then stimulated with EGF/anisomycin for the last hour (lane 4) and electrophoresed on 15% acid–urea gels. Proteins were transferred to PVDF membrane and analysed by western blotting using anti-acetyl-H3 antibodies (panel i), anti-phospho-H3 antibodies (panel ii), anti-phosphoacetyl-H3 antibodies (panel iii) or anti-HMG-14 antibodies (panel iv). Coomassie-stained gel is shown in panel v. The positions of the modified forms of histone H3 are numbered corresponding to the number of post-translational modifications visualized by Coomassie staining of gels or Ponceau S staining of PVDF membrane.
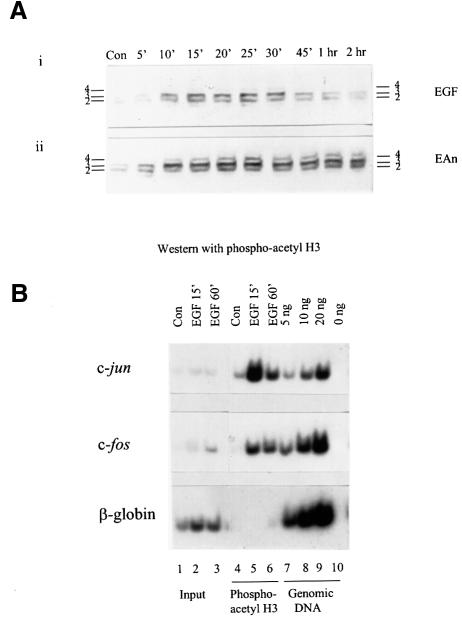
Fig. 8. Phosphoacetyl-H3 is associated with c-jun and c-fos chromatin upon physiological stimulation. (A) Quiescent control (Con) C3H 10T1/2 cells were stimulated with EGF (50 ng/ml) alone or with EGF (50 ng/ml) and anisomycin (10 µg/ml) for the times indicated. Acid-soluble nuclear proteins were resolved on 15% acid–urea gels, transferred to PVDF membrane and probed with the anti-phosphoacetyl-H3 antibody. (B) Formaldehyde cross-linked chromatin was prepared from quiescent (Con) and EGF (50 ng/ml)-stimulated mouse C3H 10T1/2 cells as described in Materials and methods, and immunoprecipitated with anti-phosphoacetyl-H3 antibodies (lanes 4–6). The recovered DNAs from the antibody-bound fractions, the total input DNA from released chromatin used for the ChIP assays (Input, lanes 1–3) and genomic DNA (lanes 7–9; lane 10 lacks template) were analysed for the presence of c-jun (upper panel), c-fos (middle panel) and β-globin (lower panel) sequences by PCR as described in the legend to Figure 7.
Anti-phosphoacetyl-H3 antibodies are specific for acetyllysine-9 and phosphoserine-10
To define exactly which acetyl groups contribute to the phosphoacetyl epitope recognized by the new anti-phosphoacetyl-H3 antibody, synthetic H3 peptides acetylated at specific residues (Figure 5A; kindly provided by Professor Bryan Turner, Birmingham, UK) were phosphorylated in vitro to produce specifically phosphoacetylated histone H3 peptides for dot-blot analysis. The kinase used was recombinant MSK1 (kindly provided by Dr Dario Alessi, MRC Protein Phosphoryation Unit, Dundee, UK), which we recently showed is a potent kinase for histone H3 at serine 10 (Thomson et al., 1999b). The four peptides tested correspond to residues 5–28 with a C-terminal cysteine, encompassing all the in vivo H3 acetylation sites, either non-acetylated or with specific lysines acetylated as indicated in Figure 5A. That all these peptides can be phosphorylated by MSK1 was first confirmed using [32P]ATP and scintillation counting (Stuart Thomson and L.C.Mahadevan, data not shown) and is also shown by dot-blotting analyses discussed below.
Dot-blot analyses of these peptides performed with the new anti-phosphoacetyl antibodies (Figure 5B) showed that this antibody recognizes these peptides only after phosphorylation with MSK1, proving that the phosphate group at serine 10 is essential (Figure 5B, lanes 12 and 13). Most importantly, the antibody only recognizes the peptide when lysine 9 is acetylated and serine 10 phosphorylated (lanes 12 and 13); the combination acetyllysine 14 and phosphoserine 10 is not detectably recognized by the antibody (lane 11).
These phosphoacetyl-H3 peptides were also screened against our original anti-phospho-H3 antibody to determine which acetyl groups caused the occlusion of the serine 10 phosphoepitope (Figure 5C). This showed that whenever lysine 14 is acetylated (Figure 5C, lanes 11 and 13), the anti-phospho-H3 antibody failed to recognize the phosphoepitope at serine 10. By contrast, when lysine 9 was acetylated and lysine 14 was not (lane 12), the anti-phospho-H3 antibody remained able to recognize the phosphate at serine 10, suggesting that it is acetyllysine-14 that occludes the phosphate group at serine 10 from antibody recognition.
Thus, the new anti-phosphoacetyl-H3 antibody specifically recognizes an epitope comprising acetyllysine-9 and phosphoserine-10 on the H3 tail, whereas recognition of phosphoserine-10 by the original anti-phospho-H3 antibody is specifically impeded by acetyllysine-14. Investigation of the order of lysine modifications on the histone H3 tail showed that lysine 14 was the first to become acetylated (Thorne et al., 1990). The fact that this occludes the serine 10 phosphoepitope would explain why all upper bands of the H3 ladder are not recognized by the phospho-H3 antibody (Figures 2ii and 6ii). As might be expected, the acetylation status of the more distant lysines 18 and 23 on these peptides have no bearing on antibody recognition studied here.
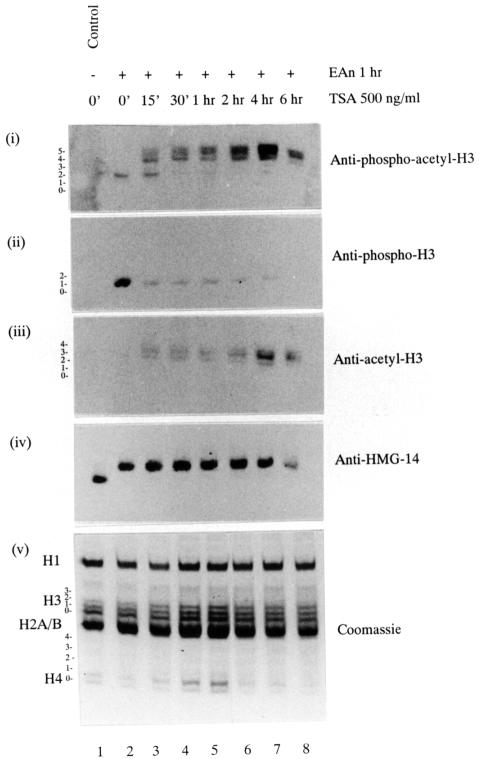
Fig. 6. Rapid increase in the phosphoacetyl-H3 epitope in response to TSA pretreatment. Quiescent C3H 10T1/2 cells received no pretreatment (lanes 1 and 2) or were pretreated with 500 ng/ml TSA for 15 min (lane 3), 30 min (lane 4), 1 h (lane 5), 2 h (lane 6), 4 h (lane 7) or 6 h (lane 8). Cells were stimulated with 50 ng/ml EGF plus 10 µg/ml anisomycin (EAn, lanes 2–8) for the last hour; lane 1 is an unstimulated control. Acid-soluble nuclear proteins were resolved on 15% acid–urea gels, transferred to PVDF membrane and analysed by western blotting using anti-phosphoacetyl-H3 antibodies (panel i), anti-phospho-H3 antibodies (panel ii), anti-acetyl-H3 antibodies (panel iii) or anti-HMG-14 antibodies (panel iv). Coomassie-stained gel is shown in panel v. The positions of the modified forms of histone H3 are numbered corresponding to the number of post-translational modifications visualized by Coomassie staining of gels or Ponceau S staining of PVDF membrane.
Phosphorylated histone H3 is exquisitely sensitive to TSA-induced hyperacetylation
We next asked if phosphoacetyl-H3 detected by the antibody in these experiments is identical to the phosphate-labelled histone H3 fraction in being especially sensitive to deacetylase inhibitors (shown in Figure 1). The anti-phospho-H3 and the new anti-phosphoacetyl-H3 antibodies were used in western blotting analyses of cells subjected to a time-course of TSA pretreatment (15 min to 6 h) to elicit hyperacetylation and EGF/anisomycin (1 h) to elicit H3 phosphorylation (Figure 6). The anti-phosphoacetyl-H3 antibody produces a weak signal in non-TSA-treated cells and an increasingly enhanced immunostaining profile of the upper bands with increasing TSA pretreatment (Figure 6, panel i), remarkably similar to the 32P-labelling profile in hyperacetylated cells (see Figure 1B). In contrast, the anti-phospho-H3 antibody signal is strong in non-TSA-treated cells (Figure 6ii, lane 2) but severely reduced upon TSA treatment (Figure 6ii, lanes 3–8). Note that even 15 min of pretreatment with TSA is sufficient to bring about virtually complete conversion of the epitope, attesting to the extreme sensitivity of phosphorylated histone H3 to TSA treatment. This is in striking contrast with the general state of histone modification as revealed by Coomassie staining (Figure 6, panel v), where no change is seen with 15 min of TSA pretreatment in either H3 or H4 ladders, or with the general appearance of hyperacetylated histone H3, which, as revealed by anti-acetyl-H3 antibody staining (Figure 6, panel iii), builds up gradually and peaks at 6 h. The anti-HMG-14 antibody was used to show that signalling to nucleosomes was unimpeded in all these conditions (Figure 6, panel iv).
These experiments prove conclusively that cells superinduced with EGF/anisomycin contain a small amount of phosphoacetyl-H3, but much more of phospho-H3 recognizable by our original anti-phospho-H3 antibody. Furthermore, upon treatment with deacetylase inhibitors such as butyrate or TSA, phosphorylated histone H3 is preferentially acetylated at a rate that far exceeds the modification of bulk histone H3 (Barratt et al., 1994b; discussed further below).
Phosphoacetyl-histone H3 is present on nucleosomes associated with c-fos and c-jun chromatin upon transcriptional activation
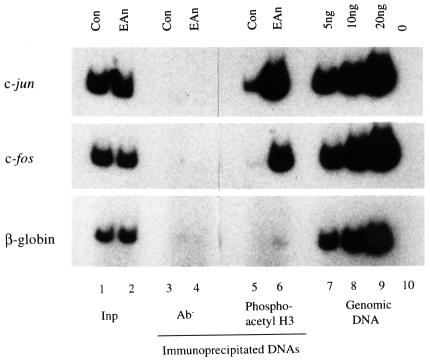
The final piece of evidence required to link phosphoacetyl-H3 to IE gene induction is the direct demonstration that nucleosomes associated with these genes carry the phosphoacetyl epitope in stimulated cells, but not in control cells. To this end, we used anti-phosphoacetyl-H3 antibodies to immunoprecipitate cross-linked chromatin fragments released by sonication (average size ∼2–3 kb) from control and EGF/anisomycin-stimulated cells. The precise conditions used (see Materials and methods) for ChIP assays and DNA analysis by PCR were derived by a series of preliminary experiments to optimize each step based on published methods (Braunstein et al., 1993; Orlando et al., 1997). Following chromatin immunoselection, DNA was prepared and analysed by PCR using primer pairs derived from the c-fos and c-jun promoters and from the β-globin gene, using [32P]dCTP to facilitate quantification by phosphorimaging of radiolabelled PCR products resolved on polyacrylamide gels (see Materials and methods; Figure 7). Each ChIP assay and analytical PCR reaction was internally controlled for quantification purposes (i) by analysis of the total cross-linked DNA used in each immunoselection (Figure 7; Input; lanes 1 and 2) and (ii) by analysis of standard masses (5, 10 and 20 ng) of genomic DNA to ensure that amplification reactions remain in the linear range (Figure 7; lanes 7–10). For all primer sets, comparison of the PCR signal using 5 ng of input DNA with the signal obtained using 5 ng of genomic DNA demonstrates that their representation in cross-linked released chromatin is approximately equivalent to that in the total genome. Mock immunoprecipitations were performed in the absence of antibody using cross-linked chromatin prepared from control and stimulated cells (lanes 3 and 4) to demonstrate the specificity of these ChIP assays.
Fig. 7. Phosphoacetyl-H3 is present on nucleosomes associated with c-jun and c-fos chromatin upon induction of these genes. Formaldehyde cross-linked chromatin was prepared from control (Con) quiescent and EGF/anisomycin (EAn)-stimulated mouse C3H 10T1/2 cells as described in Materials and methods, and immunoprecipitated with peptide affinity-purified anti-phosphoacetyl-H3 antibodies (lanes 5 and 6). The recovered DNAs from the antibody-bound fractions as well as the total input DNA from released chromatin used for the immunoprecipitations (Inp, lanes 1 and 2) were analysed for the presence of c-jun (upper panel), c-fos (middle panel) and β-globin (lower panel) sequences by PCR. Primers amplified –733 to –572 for c-jun, –131 to +93 for c-fos and +261 to +439 (exon 2) for β-globin (numbers are with respect to the transcription start sites). [32P]dCTP was incorporated into the amplified products, which were resolved on 6% acrylamide gels, visualized by autoradiography and quantified using a phosphorimager (see text). Input DNAs (5 ng) and 1/35 of the immunoprecipitated DNAs were used as template. Aliquots of genomic DNA (5, 10 and 20 ng, lanes 7–9) were always amplified alongside immunoprecipitated DNAs to verify that the PCR remained in the linear range (lane 10 lacks template). Similar results to those depicted here were obtained in at least two other independent sets of ChIPs.
The anti-phosphoacetyl-H3 antibody immunoselects nucleosomes associated with c-jun and c-fos from stimulated cells (Figure 7, lane 6), but produces a signal not much higher than the antibody-minus background from control cells (Figure 7, lane 5). In contrast, there is no selection of β-globin gene nucleosomes from stimulated or control cells (Figure 7, lanes 6 and 5). Note that these cells were not TSA treated to enhance acetylation; the phosphoacetyl-H3-containing nucleosomes immunoselected here occur normally during superinduction of these genes. The ratio of PCR signal from stimulated compared with control cells (stimulated:control ratio), as determined by phosphorimaging, was 13 for c-jun and 19 for c-fos. This stimulated:control ratio is representative of at least three independent experiments. Note, however, that there is some variation in these ratios between experiments, the main cause of which is the variation in background signal that shows up in the control lanes and is subject to inaccuracies associated with low signal levels. These data prove for the first time that nucleosomes associated with c-jun and c-fos, which we have used as model IE genes, contain phosphoacetyl-histone H3 upon transcriptional activation, whereas the β-globin gene, which is inactive in these cells, does not.
Note that we have also carried out ChIP assays using our anti-phospho-H3 antibody since it is clear that there is a population of nucleosomes with phospho-H3 in these cells whose levels of acetylation do not interfere with recognition by our antibody. PCR analyses of the resulting DNA suggests that there is, if any, a much smaller amount of c-fos and c-jun sequences immunoselected with the anti-phospho-H3 antibody compared with that bound to the anti-phosphoacetyl-H3 antibody (data not shown). This contrasts with recent work in which a different anti-phospho-H3 antibody was used in ChIP assays to immunoselect c-myc and c-fos gene sequences upon EGF or phorbol-12-myristate-13-acetate (TPA) stimulation (Chadee et al., 1999); this discrepancy may arise from differences in the antibodies used, especially in view of the sensitivity of our antibody to the acetylation status of the H3 tail.
Phosphoacetyl-H3 is associated with IE gene chromatin upon activation with a physiological stimulus
Thus far, we have studied phosphoacetylation of histone H3 either in TSA-treated cells or in cells superinduced with anisomycin, experimental conditions that greatly augment acetylation and phosphorylation of histone H3, respectively (Barratt et al., 1994b; Cano et al., 1995). Having established that H3 phosphoacetylation does occur under these conditions, we then asked whether it also occurs when normal physiological stimuli, such as EGF, are used. EGF-induced IE gene induction, activation of MAP kinases and phosphorylation of histone H3 and HMG-14 have previously been extensively characterized (Barratt et al., 1994a,b; Cano et al., 1995; Hazzalin et al., 1996).
Time-course analysis (Figure 8A) was first performed to determine the peak of EGF-induced histone H3 phosphoacetylation in order to pick an optimum time-point for ChIP assays. Western blotting analysis demonstrates that phosphoacetyl-H3 is detected after 5–10 min of EGF treatment (Figure 8A, panel i), increases to maximal levels at 15–25 min and decreases to much lower but detectable levels at 1 h. Thus, EGF stimulation produces transient histone H3 phosphoacetylation. By contrast, under superinducing conditions using EGF plus anisomycin, histone H3 phosphoacetylation is much stronger and more sustained (Figure 8A, panel ii), in complete accord with the fact that both kinase activation and IE gene transcription are stronger and sustained under these conditions (Edwards and Mahadevan, 1992).
We then performed ChIP assays using chromatin prepared from control and EGF-stimulated cells. Two time points of stimulation were selected, 15 min, at which time the level of H3 phosphoacetylation is maximal, and 60 min, when it is substantially reduced. DNA from chromatin immunoselected with the anti-phosphoacetyl-antibody was analysed for c-fos and c-jun sequences exactly as described above. Very low levels (if any) of c-jun, c-fos or β-globin sequences are immunoselected from control cells. After 15 min of treatment with EGF, both c-jun and c-fos sequences are associated with nucleosomes containing phosphoacetyl-H3, whereas the β-globin gene is not (Figure 8B, lanes 5). After 60 min of EGF stimulation, c-fos and c-jun sequences remain associated with phosphoacetyl-H3-containing nucleosomes, but at reduced levels. Taken together, these studies prove that phosphoacetyl-histone H3 occurs normally in vivo upon physiological stimulation and that it is associated with IE gene chromatin upon normal gene induction.
Discussion
We have shown that in one class of rapid transcriptional response, that involving the induction of IE genes typified by c-fos and c-jun, phosphorylation and acetylation of histone H3 occur as associated and highly targeted events co-localized to the same histone H3 tail. Phosphoacetylated histone H3 occurs on nucleosomes associated with the active but not the inactive c-fos and c-jun proto-oncogenes, or the inactive tissue-specific β-globin gene. These studies reveal an additional layer of complexity in the way that histone modifications are involved in this class of rapid transcriptional response.
Phosphoacetylation of histone H3 on IE gene nucleosomes
The modification-specific antibodies and western blotting analyses of H3 ladders resolved on acid–urea gels described above proves that the histone H3 on nucleosomes associated with active IE genes is phosphorylated on serine 10 and acetylated on lysine 9. However, it does not allow any conclusion about the type or extent of other modifications that may additionally be present on these tails. Given that each modification may be expected to cause an incremental shift up the histone H3 ladder (Barratt et al., 1994b), the acid–urea gels show that the anti-phosphoacetyl antibody picks up H3 tails that must by definition have two modifications, acetyllysine 9 and phosphoserine 10, but in addition, can have other modifications that cause it to migrate at higher positions on the H3 ladder, giving rise to multiple immunoreactive bands. Furthermore, the fact that our original anti-phospho-H3 antibody, which is specifically prevented from binding phosphoserine 10 by acetyllysine 14, does not substantially recover IE gene chromatin in ChIP assays, might suggest that lysine 14 is also acetylated at least on some of these nucleosomes, preventing its recovery with this antibody. However, there are other potential modifications such as methylation (lysine 4), further acetylation (possibly lysines 4 and 27) and phosphorylation (serine 28) of these nucleosomes, about which we cannot be conclusive. Note that the fact that the anti-phosphoacetyl antibody recognizes three bands in EGF-stimulated cells does suggest that, under physiological conditions, there are additional modifications on the H3 tail beyond phosphoserine 10 and acetyllysine 9. Since this study clearly shows that phosphorylated H3 can become preferentially acetylated, the simplest explanation is that other lysines on the same tail also become acetylated to account for these higher-migrating bands of phosphoacetylated H3.
More generally, these studies show that occlusion of epitopes by modification of adjacent amino acids can potentially be a serious problem in the use of antibodies and ChIP assays to analyse histone modifications associated with specific genes, an approach increasingly used now (Kuo and Allis, 1999; Orlando, 2000 and references therein). The most common post-translational modification of histones is acetylation, and the observation here that acetyl groups on adjacent lysines are sufficient to interfere with antibody recognition advocates care in the use of ChIP assays to study histone acetylation. The occlusion problem is probably minimal or non-existent in the use of general reagents such as antibodies against all acetylated lysines or against a particular hyperacetylated histone tail, but can potentially compromise experiments where site-specific anti-acetyllysine antibodies are used.
Co-targeting of histone H3 kinases and histone acetyltransferases to IE genes
IE gene induction is elicited by physiological (growth factors, cytokines), pharmacological (TPA, okadaic acid, anisomycin) and stress stimuli (hyperosmolarity, heavy metals, UV radiation), all of which also elicit the nucleosomal response (Mahadevan et al., 1991; Edwards and Mahadevan, 1992; Cano et al., 1995; Hazzalin et al., 1996). Despite their extreme diversity, all these agents share the ability to activate MAP kinase cascades (Cano et al., 1995; Hazzalin et al., 1996, 1997; reviewed in Cano and Mahadevan, 1995), which is essential for delivery of signals across the nuclear membrane and IE gene induction (Hazzalin et al., 1996, 1997; Thomson et al., 1999b). However, these stimuli differentially activate ERK, JNK/SAPK and p38 MAP kinase subtypes, and the strength and latency of activation of each subtype is a stable and reproducible characteristic of each stimulus (discussed in Cano et al., 1995; Hazzalin et al., 1996; Thomson et al., 1999b). We recently reported that ERK and p38 MAP kinase cascades can mediate the nucleosomal response, and that MSK1, a kinase that lies downstream of either ERKs or p38 (Deak et al., 1998), is the best candidate for the nuclear kinase that phosphorylates histone H3 and HMG-14 (Thomson et al., 1999b). Rsk-2, which lies downstream of ERKs, has also been proposed to be a mitogen-stimulated histone H3 kinase (Chen et al., 1992; Mizzen et al., 1998; Sassone-Corsi et al., 1999). MSK1 is reported to be a nuclear kinase (Deak et al., 1998; see also Jin et al., 1999) and the fact that its upstream activators can be translocated to specific genes through its ability to form stable complexes with transcription factors provides a potential mechanism by which histone H3 and HMG-14 phosphorylation can be highly targeted to nucleosomes in the vicinity of these transcription factors; these would include nucleosomes associated with the c-fos and c-jun genes.
Transcription factors that form complexes with, and are substrates for, MAP kinases can also bind to co-activators such as p300/CBP (Arias et al., 1994; Bannister et al., 1995; Janknecht and Nordheim, 1996a,b) and thereby p/CAF, all of which possess intrinsic HAT activity (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Yang et al., 1996). Studies on the IFN-β enhanceosome shows that interference with the recruitment of CBP/p300 affects transcriptional induction as well as localized histone hyperacetylation (Parekh and Maniatis, 1999). In vitro studies on peptides and isolated histones suggest that lysine 14 in the H3 tail is a preferred acetylation site for purified HATs (Kuo et al., 1996; Tse et al., 1998). However, an expanded specificity that targets lysine 14 and 18 equally well and includes lysine 9 is seen when HATs present in large native complexes such as the ADA or SAGA (Spt-Ada-Gen5 acetyltransferase) complexes are used to acetylate nucleosomes (Grant et al., 1999; Schiltz et al., 1999), arguably a more physiological assay. Thus, existing precedents and mechanisms could potentially explain how MAP kinase cascades and CBP/p300 can both be targeted through the same transcription factors to elicit phosphorylation and acetylation of the same histone H3 tail on nucleosomes asociated with IE genes.
The extent and distribution of phosphoacetylated histone H3 along IE gene chromatin in stimulated cells
Under superinducing conditions (EGF plus anisomycin; Edwards and Mahadevan, 1992; Hazzalin et al., 1998), extremely strong signalling responses, for example the phosphorylation of every molecule of HMG-14 in the nucleus, are observed (see Figure 2). With physiological stimuli such as growth factors or cytokines, quantitative phosphorylation of HMG-14 is never observed and histone H3 phosphorylation is likewise much weaker (Mahadevan et al., 1991; Barratt et al., 1994b; and data not shown); IE gene induction is also weaker under these conditions (Hazzalin et al., 1998). We have shown here (Figure 8) that histone H3 phosphoacetylation is associated with active c-fos and c-jun chromatin under physiological conditions using EGF as a stimulus. The timing and extent of H3 phosphoacetylation reflects the more transient nature of the gene responses observed under these conditions (Hazzalin et al., 1998), and it would now be useful to understand the origin and spread of the double modification under these physiological conditions. All current models predict that phosphorylation and acetylation directed by transcription factors would originate around regulatory elements in the enhancers of these genes. The question then arises as to how far the double modification extends along each gene and whether or not there is directionality in this response (see Feng and Villeponteau, 1990, 1992). The average length of the cross-linked sonicated chromatin fragments used for the experiments presented here is 2–3 kb, which prevents higher-resolution analyses of the association of these nucleosomes with specific regions of c-fos and c-jun. The next phase of this work will be to use cross-linked chromatin fragments prepared from physiologically stimulated cells and subfractionated so that they are of a defined size, possibly encompassing a maximum of three to four nucleosomes, in conjunction with more comprehensive sets of PCR primers along the genes to investigate the precise extent and spreading of these modifications along each gene.
Phosphoacetylation of histone H3 is restricted to a specific subset of rapid transcriptional responses
Regulatory modifications involving histone acetyltransferases and deacetylases are implicated in diverse types of transcriptional control including rapid gene induction (Chen and Allfrey, 1987; Alberts et al., 1998; Chen et al., 1999), telomeric and mating-type silencing in yeast (reviewed in Grunstein, 1998), DNA methylation-linked transcriptional control (reviewed in Ng and Bird, 1999), domain-linked histone acetylation (Hebbes et al., 1994), and various types of derepression (Kadosh and Struhl, 1998; Rundlett et al., 1998). Furthermore, CBP/p300 is known to be able to interact with many other transcription factors and is implicated in diverse physiological as well as viral-induced pathological modes of transcriptional control (Parekh and Maniatis, 1999; reviewed in Janknecht and Hunter, 1996a; Shikama et al., 1997). However, rapidly inducible H3 phosphorylation and phosphoacetylation is so far strictly limited to MAP kinase-mediated gene induction, observed in response to diverse stimuli but restricted to the control of a subset of inducible genes (Mahadevan et al., 1991; Chadee et al., 1999; DeManno et al., 1999; reviewed in Thomson et al., 1999a). The histone H3 and HMG-14 kinase MSK1 is only activated by agents that activate ERK and/or p38 MAP kinases (Hazzalin et al., 1996; Thomson et al., 1999b); these cascades also produce phosphorylation of CREB (Deak et al., 1998 and references therein) and in many instances of MAP kinase-targeted transcription factors, CBP/p300 is strongly implicated as a co-activator (reviewed in Hunter and Karin, 1992; Janknecht and Hunter, 1996a,b).
However, it is worth noting that there are other equally rapid transcriptional responses, for example involving signal transducers and activators of transcription (STATs) (reviewed in Darnell, 1997) or NF-κB (reviewed in Baeuerle and Baltimore, 1996), where MAP kinase activation is not obligately required. In these cases, there is at present no indication that phosphoacetyl-H3 might be involved with gene activation, although it is important to note that this cannot be excluded as yet because many stimuli that activate these signalling systems also activate MAP kinases in parallel. Finally, it is also worth noting that serine 10 on histone H3, equivalents of the ERK and p38 MAP kinases and some of their transcription factor substrates, are all conserved in Saccharomyces cerevisiae, but homologues to the downstream kinase MSK1 that mediates histone H3/HMG-14 phosphorylation, to the histone acetyltransferase CBP implicated here, and to HMG-14 itself are all missing in S.cerevisiae, suggesting that the complexity of transcriptional control described here represents an evolutionarily more recent elaboration of simpler modes of transcriptional control seen in yeast.
Materials and methods
Materials
Peptides used for antibody production and peptide screening were synthesized by Dr G.Bloomberg, University of Bristol, UK. TSA was kindly given by Professor Minoru Yoshida, University of Tokyo, Japan. Anti-acetyl-H3 antibody was kindly provided by Professor David Allis, University of Virginia, USA. Yeast histone deacetylase HOS3 was kindly given by Professor Peter Lewis, University of Toronto, Canada. Acetylated H3 peptides were kindly provided by Professor Bryan Turner, Birmingham, UK. Anisomycin was purchased from Sigma.
Cell culture and stimulation of C3H 10T1/2 cells
C3H 10T1/2 mouse fibroblasts were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% (v/v) fetal calf serum (FCS) and 2 mM glutamine. Confluent cultures were rendered quiescent by incubation in DMEM containing 0.5% (v/v) FCS and 2 mM glutamine for 24–36 h. Cells were pretreated as indicated with the histone deacetylase inhibitor TSA (500 ng/ml) and then stimulated as indicated with 50 ng/ml EGF and 10 µg/ml anisomycin. Medium was aspirated and cells harvested as described below at the times indicated after stimulation.
Extraction of histones from cell nuclei
Quiescent and stimulated cells were lysed directly in culture plates by the addition of 0.5 ml ice-cold cell lysis buffer [0.2% Triton X-100, 10 mM HEPES pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 100 µM sodium orthovanadate, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 0.1 µM okadaic acid and protease inhibitor cocktails A and B (Barratt et al., 1994a)] and left on ice for a minimum of 10 min. Extracts were centrifuged [3000 r.p.m., 3 min, 4°C in a Biofuge fresco microfuge (Heraeus)] and nuclear pellets were washed in 0.5 ml of the same buffer. Histones and other basic proteins were extracted by the addition of 150 µl 0.4 M HCl for 1 h on ice. Extracts were centrifuged [13 000 r.p.m., 15 min, 4°C in a Biofuge fresco microfuge (Heraeus)] and the resulting supernatant was acetone-precipitated overnight at –20°C. Precipitated proteins were collected by centrifugation [13 000 r.p.m., 15 min, 4°C in a Biofuge fresco microfuge (Heraeus)], washed with acetone three times and dried.
Antibody generation, characterization and purification
Antibodies were raised at the Scottish Antibody Production Unit (SAPU), Lanarkshire, UK. Characterization of anti-HMG-14 and anti-phospho-H3 antibodies has been described previously (Thomson et al., 1999b). The anti-phosphoacetyl-H3 antibody was raised in sheep. The sequence of the phosphodiacetyl-H3 peptide [ARTKGTARK(acetyl)S(phospho)TGGK(acetyl)APRKQLC, where ‘phospho’ indicates that the preceding residue is phosphorylated and ‘acetyl’ that the preceding residue is acetylated] corresponds to residues 1–20 of mouse histone H3 with the addition of a cysteine residue at the C-terminus for coupling. The peptide was conjugated to keyhole limpet haemocyanin (KLH) and injected into two sheep. Antibodies were purified from serum using peptide affinity columns (Pierce).
Acid–urea polyacrylamide gel electrophoresis and immunoblotting of protein samples
Proteins were resolved by acid–urea polyacrylamide gel electrophoresis as described previously (Lennox and Cohen, 1989; Barratt et al., 1994b). After separation, gels were washed in 5% acetic acid/10% methanol for 30 min followed by 5 min in transfer buffer (0.1% acetic acid/10% methanol). Proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Immobilon-P, Millipore) using transfer buffer. Membranes were incubated with blocking buffer (5% powdered skimmed milk in phosphate-buffered saline) for 1 h and then incubated at room temperature with the indicated primary antibody diluted in blocking buffer with 0.1% Tween-20 for 2 h. The membranes were washed in four changes of wash buffer (PBS, 0.1% Triton X-100), then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody in blocking buffer with 0.1% Tween-20 for 1 h. Membranes were washed in five changes of wash buffer and proteins were detected using the enhanced chemiluminescence reagent (Amersham). The anti-HMG-14, anti-phospho-H3 and anti-phosphoacetyl-H3 antibodies were used at a 1:1000 dilution. The anti-acetyl-H3 antibody was used at a 1:5000 dilution.
Deacetylation of phosphodiacetyl peptide by HOS3
Phosphodiacetyl peptide (0.5 mg/ml) was incubated overnight at 30°C with 10 µl recombinant yeast histone deacetylase HOS3 in NBQ buffer (10 mM sodium phosphate pH 7.5, 1 mM EDTA, 5 mM β–mercaptoethanol). Peptides were spotted at the concentrations indicated on Hybond-C membrane (Amersham). Immunoblotting was performed as described above using anti-phospho-H3 antibody as the primary antibody.
Formaldehyde cross-linked chromatin preparation
Confluent plates of quiescent, EGF/anisomycin- or EGF-stimulated C3H 10T1/2 cells were formaldehyde cross-linked for 10 min at room temperature by adding 0.1 vol of cross-linking solution (11% formaldehyde, 50 mM HEPES pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) directly to the culture medium in the plates. Cross-linking was stopped by the addition of glycine to a final concentration of 125 mM. Cells were washed twice with ice-cold PBS, then harvested in lysis buffer (10 mM Tris–HCl pH 8.0, 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM sodium butyrate, 100 mM sodium chloride, 20 mM β-glycerophosphate, 100 µM sodium orthovanadate and protease inhibitors A and B) and left on ice for 10 min. Cells were centrifuged at 3000 r.p.m. for 4 min at 4°C. Nuclei were resuspended in wash buffer (10 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 µM sodium orthovanadate and protease inhibitors A and B) then centrifuged as above. Nuclear pellets were resuspended in sonication buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 µM sodium orthovanadate and protease inhibitors A and B) and sonicated using an MSE Soniprep 150 sonicator or a Sonics Vibracell 150 W sonicator. Sodium dodecyl sulfate (SDS) was added to a final concentration of 1% and the chromatin solutions were rotated at room temperature for 1 h. Insoluble material was removed by centrifugation at full speed in a microcentrifuge at room temperature for 15–20 min. The soluble supernatant chromatin was diluted 10-fold to reduce the concentration of SDS to 0.1%, then concentrated using Vivaspin (Vivascience) concentrators. The resulting chromatin solutions were used for immunoprecipitations.
Chromatin immunoprecipitation
ChIPs were carried out as described previously (Braunstein et al., 1993; Orlando et al., 1997) with a few modifications. Cross-linked released chromatin fractions were adjusted to RIPA buffer (10 mM Tris–HCl pH 8.0, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 µM sodium orthovanadate and protease inhibitors A and B). For immunoprecipitation, chromatin from a 140 mm dish of cells in a volume of 500–700 µl was mixed with 50 µl of affinity purified anti-phosphoacetyl-H3 antibodies (equivalent to ∼65 µl of serum) and incubated with rotation for 2 h at 4°C. Aliquots of control, EGF–anisomycin and EGF chromatin were also used for antibody-minus controls and input samples. The chromatin/antibody solutions were adjusted to 0.02% bovine serum albumin (BSA) followed by addition of 5 µg of sonicated phage lambda DNA and protein A– or G–Sepharose beads in RIPA buffer containing 0.02% BSA and incubated with rotation at 4°C for a further 2 h. The beads were harvested by centrifugation and the unbound supernatant chromatin was retained. The beads were washed sequentially with 3× 1 ml RIPA buffer, 1–3× 1 ml RIPA, 500 mM NaCl or 1–3× 1 ml RIPA, 1 M NaCl, 1–3× 1 ml 0.25 M LiCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 20 mM β-glycerophosphate, 10 mM sodium butyrate, 100 µM sodium orthovanadate and protease inhibitors A and B and 2–3× 1 ml TE pH 7.5, 10 mM sodium butyrate, 20 mM β-glycerophosphate and protease inhibitors A and B. The antibody-bound chromatin was eluted from the beads with 200 µl of TE pH 7.5, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 30 mM NaCl containing 1.5% SDS followed by elution with 200 µl of the same buffer containing 0.5% SDS. The input, unbound and antibody bound chromatins were incubated at 65°C for ≥6 h to reverse the formaldehyde cross-links. The DNA was ethanol-precipitated overnight at –20°C, resuspended in water and treated with RNase A (50 µg/ml) for 1 h at 37°C followed by proteinase K treatment (100 µg/ml) in the presence of 0.25% SDS at 37°C overnight. The DNAs were extracted with phenol–chloroform and chloroform, precipitated with ethanol and dissolved in water. Input DNAs were quantified by UV spectrophotometry. Immunoprecipitated DNAs were analysed for c-jun, c-fos and β-globin gene sequences by PCR.
PCR analyses of immunoprecipitated DNA
32P-labelled dCTP was incorporated into the PCR-amplified products for visualization and quantification. All PCR reactions were performed using a Perkin–Elmer GeneAmp 2400 thermal cycler and AmpliTaq Gold DNA polymerase. The linear range for all primer pairs was determined empirically using different amounts of C3H 10T1/2 genomic DNA. Subsequent PCR analyses were carried out using the optimum cycle number determined for each primer set. Genomic DNA control reactions were always carried out alongside the immunoprecipitated DNA samples. Generally 5 ng of input DNAs and 1/35 of the total immunoprecipitated DNAs were used as templates. All primer pairs amplify fragments with sizes ranging from 158 to 224 bp (see legend to Figure 7 for details). PCR products were resolved on 6% polyacrylamide–TAE gels, dried, visualized by autoradiography and quantified using a PhosphorImager (Molecular Dynamics).
Acknowledgments
Acknowledgements
We thank Professor David Allis (University of Virginia, Charlottesville, USA) for the anti-acetyl-H3 antibody and for helpful discussions, Dr G.Bloomberg (University of Bristol, UK) for peptide synthesis, Professor P.Lewis (University of Toronto, Canada) for the yeast histone deacetylase HOS3, Professor Bryan Turner (Birmingham, UK) for the acetylated H3 peptides and Dr Dario Alessi (Dundee, UK) for recombinant MSK1. We thank Dr Heike Weber and Dr Soledad Lopez for advice on PCR and all members of the Nuclear Signalling Laboratory for discussions and comments on the manuscript. This work is funded principally by the Wellcome Trust (A.L.C.). S.R. is funded by an MRC PhD Studentship.
References
- Alberts A.S., Geneste,O. and Treisman,R. (1998) Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell, 92, 475–487. [DOI] [PubMed] [Google Scholar]
- Allegra A.P., Sterner,R., Clayton,R.F. and Allfrey,V.G. (1987) Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J. Mol. Biol., 196, 379–388. [DOI] [PubMed] [Google Scholar]
- Allfrey V.G., Faulkner,R. and Mirsky,A.E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias J., Alberts,A.S., Brindle,P., Claret,F.X., Smeal,T., Karin,M., Feramisco,J. and Montminy,M. (1994) Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature, 370, 226–229. [DOI] [PubMed] [Google Scholar]
- Baeuerle P.A. and Baltimore,D. (1996) NF-κB: ten years after. Cell, 87, 13–20. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Oehler,T., Wilhelm,D., Angel,P. and Kouzarides,T. (1995) Stimulation of c-Jun activity by CBP: c-Jun residues Ser 63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene, 11, 2509–2514. [PubMed] [Google Scholar]
- Barratt M.J., Hazzalin,C.A., Zhelev,N. and Mahadevan,L.C. (1994a) A mitogen-stimulated and anisomycin-stimulated kinase phosphorylates HMG-14 in its basic amino-terminal domain in vivo and on isolated mononucleosomes. EMBO J., 13, 4524–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt M.J., Hazzalin,C.A., Cano,E. and Mahadevan,L.C. (1994b) Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc. Natl Acad. Sci. USA, 91, 4781–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Brownell J.E., Zhou,J.X., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 4, 843–851. [DOI] [PubMed] [Google Scholar]
- Cano E. and Mahadevan,L.C. (1995) Parallel signal processing among mammalian MAPKs. Trends Biochem. Sci., 20, 117–122. [DOI] [PubMed] [Google Scholar]
- Cano E., Hazzalin,C.A., Kardalinou,E., Buckle,R.S. and Mahadevan,L.C. (1995) Neither ERK nor JNK/SAPK MAP kinase subtypes are essential for histone H3/HMG-14 phosphorylation or c-fos and c-jun induction. J. Cell Sci., 108, 3599–3609. [DOI] [PubMed] [Google Scholar]
- Chadee D.N., Hendzel,M.J., Tylipski,C.P., Allis,C.D., Bazett-Jones,D.P., Wright,J.M. and Davie,J.R. (1999) Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem., 274, 24914–24920. [DOI] [PubMed] [Google Scholar]
- Chen T.A. and Allfrey,V.G. (1987) Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc. Natl Acad. Sci. USA, 84, 5252–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Sarnecki,C. and Blenis,J. (1992) Nuclear localisation and regulation of Erk- and Rsk-encoded protein kinases. Mol. Cell. Biol., 12, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Xie,W., Wilpiz,D. and Evans,R.M. (1999) Regulation of hormone induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. (1997) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Deak M., Clifton,A.D., Lucocq,L.M. and Allessi,D.R. (1998) Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J., 17, 4426–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Manno D.A., Cottom,J.E., Kline,M.P., Peters,C.A., Maizels,E.T. and Hunzicker-Dunn, M. (1999) Follicle-stimulating hormone promotes histone H3 phosphorylation on serine-10. Mol. Endocrinol., 13, 91–105. [DOI] [PubMed] [Google Scholar]
- Edwards D.R. and Mahadevan,L.C. (1992) Protein synthesis inhibitors differentially superinduce c-fos and c-jun by three distinct mechanisms: lack of evidence for labile repressors. EMBO J., 11, 2415–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.L. and Villeponteau,B. (1990) Serum stimulation of the c-fos enhancer induces reversible changes in c-fos chromatin structure. Mol. Cell. Biol., 10, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. and Villeponteau,B. (1992) High-resolution analysis of c-fos chromatin accessibility using a novel DNase I-PCR assay. Biochim. Biophys. Acta, 1130, 253–258. [DOI] [PubMed] [Google Scholar]
- Goto H. et al. (1999) Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem., 274, 25543–25549. [DOI] [PubMed] [Google Scholar]
- Grant P.A. and Berger,S.L. (1999) Histone acetyltransferase complexes. Semin. Cell Dev. Biol., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Eberharter,A., John,S., Cook,R.G., Turner,B.M. and Workman,J.L. (1999) Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem., 274, 5895–5900. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1998) Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell, 93, 325–328. [DOI] [PubMed] [Google Scholar]
- Hazzalin C.A., Cano,E., Cuenda,A., Barratt,M.J., Cohen,P. and Mahadevan,L.C. (1996) p38/RK is essential for stress-induced nuclear responses: JNK/SAPKs and c-Jun/ATF-2 phosphorylation are insufficient. Curr. Biol., 6, 1028–1031. [DOI] [PubMed] [Google Scholar]
- Hazzalin C.A., Cuenda,A., Cano,E., Cohen,P. and Mahadevan,L.C. (1997) Effects of the inhibition of p38/RK MAP kinase on induction of five fos and jun genes by diverse stimuli. Oncogene, 15, 2321–2331. [DOI] [PubMed] [Google Scholar]
- Hazzalin C.A., LePanse,R., Cano,E. and Mahadevan,L.C. (1998) Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol. Cell. Biol., 18, 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase-I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei,Y., Mancini,M.A., VanHooser,A., Ranalli,T., Brinkley,B.R., Bazett-Jones,D.P. and Allis,C.D. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentric heterochromatin and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma, 106, 348–360. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Karin,M. (1992) The regulation of transcription by phosphorylation. Cell, 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Imhof A. and Wolffe,A.P. (1998) Transcription: gene control by targeted histone acetylation. Curr. Biol., 8, R422–424. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Hunter,T. (1996a) Transcriptional control: versatile molecular glue. Curr. Biol., 6, 951–954. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Hunter,T. (1996b) Transcription—a growing co-activator network. Nature, 383, 22–23. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Nordheim,A. (1996a) MAP kinase dependent transcriptional co-activation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun., 228, 831–837. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Nordheim,A. (1996b) Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene, 12, 1961–1969. [PubMed] [Google Scholar]
- Jin Y., Wang,Y., Walker,D.L., Dong,H., Conley,C., Johansen,J. and Johansen,K.M. (1999) JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell, 4, 129–135. [DOI] [PubMed] [Google Scholar]
- Johnson C.A. and Turner,B.M. (1999) Histone deacetylases: complex transducers of nuclear signals. Semin. Cell Dev. Biol., 10, 179–188. [DOI] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1998) Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localised domain of repressed chromatin in vivo. Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1998) Roles of histone acetylation and deacetylation in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Zhou,J.X., Churchill,M.E.A. and Allis,C.D. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H., Brownell,J.E., Sobel,R.E., Ranalli,T.A., Cook,R.G., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature, 383, 269–272. [DOI] [PubMed] [Google Scholar]
- Lennox R.W. and Cohen,L.H. (1989) Analysis of histone subtypes and their modified forms by polyacrylamide gel electrophoresis. Methods Enzymol., 170, 532–549. [DOI] [PubMed] [Google Scholar]
- Mahadevan L.C. and Edwards,D.R. (1991) Signalling and superinduction. Nature, 349, 747–748. [DOI] [PubMed] [Google Scholar]
- Mahadevan L.C., Willis,A.C. and Barratt,M.J. (1991) Rapid histone H3 phosphorylation in response to growth-factors, phorbol esters, okadaic acid, and protein-synthesis inhibitors. Cell, 65, 775–783. [DOI] [PubMed] [Google Scholar]
- Mizzen C.A. et al. (1996) The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Mizzen C. et al. (1998) Signaling to chromatin through histone modifications: how clear is the signal? Cold Spring Harbor Symp. Quant. Biol., 63, 469–481. [DOI] [PubMed] [Google Scholar]
- Ng H.H. and Bird,A. (1999) DNA methylation and chromatin modification. Curr. Opin. Genet. Dev., 9, 158–163. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- O’Neill L.P. and Turner,B.M. (1995) Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J., 14, 3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. (2000) Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci., 25, 99–104. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Parekh B.S. and Maniatis,T. (1999) Virus infection leads to localised hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell, 3, 125–129. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Kabayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Mizzen,C.A., Cheung,P., Crosio,C., Monaco,L., Jacquot,S., Hanauer,A. and Allis,C.D. (1999) Requirements of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science, 285, 886–891. [DOI] [PubMed] [Google Scholar]
- Schiltz R.L., Mizzen,C.A., Vassilev,A., Cook,R.G., Allis,C.D. and Nakatani,Y. (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem., 274, 1189–1192. [DOI] [PubMed] [Google Scholar]
- Shikama N., Lyon,J. and La Thangue,N.B. (1997) The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol., 7, 230–236. [DOI] [PubMed] [Google Scholar]
- Spencer T.E. et al. (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature, 389, 194–198. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Ohba,R., Cook,R.G. and Allis,C.D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA, 96, 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Thomson S., Mahadevan,L.C. and Clayton,A.L. (1999a) MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin. Cell Dev. Biol., 10, 205–214. [DOI] [PubMed] [Google Scholar]
- Thomson S., Clayton,A.L., Hazzalin,C.A., Rose,S., Barratt,M.J. and Mahadevan,L.C. (1999b) The nucleosomal response associated with immediate early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J., 18, 4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne A.W., Kmiciek,D.K., Mitchelson,K., Sautiere,P. and Crane-Robinson,C. (1990) Patterns of histone acetylation. Eur. J. Biochem., 193, 701–713. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1996) Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol., 8, 205–215. [DOI] [PubMed] [Google Scholar]
- Tse C., Georgieva,E.I., Ruiz-Garcia,A.B., Sendra,R. and Hansen,J.C. (1998) Gcn5, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J. Biol. Chem., 273, 32388–32392. [DOI] [PubMed] [Google Scholar]
- Wei Y., Yu,L., Bowen,J., Gorovsky,M.A. and Allis,C.D. (1999) Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell, 97, 99–109. [DOI] [PubMed] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Kijima,M., Akita,M. and Beppu,T. (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem., 265, 17174–17179. [PubMed] [Google Scholar]
- Yoshida M., Horinouchi,S. and Beppu,T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays, 17, 423–430. [DOI] [PubMed] [Google Scholar]