Abstract
Protein thermodynamic stability is a fundamental physical characteristic that determines biological function. Furthermore, alteration of thermodynamic stability by macromolecular interactions or biochemical modifications is a powerful tool for assessing the relationship between protein structure, stability, and biological function. High-throughput approaches for quantifying protein stability are beginning to emerge that enable thermodynamic measurements on small amounts of material, in short periods of time, and using readily accessible instrumentation. Here we present such a method, fast quantitative cysteine reactivity (fQCR), which exploits the linkage between protein stability, sidechain protection by protein structure, and structural dynamics to characterize the thermodynamic and kinetic properties of proteins. In this approach, the reaction of a protected cysteine and thiol-reactive fluorogenic indicator is monitored over a gradient of temperatures after a short incubation time. These labeling data can be used to determine the midpoint of thermal unfolding, measure the temperature dependence of protein stability, quantify ligand-binding affinity, and, under certain conditions, estimate folding rate constants. Here, we demonstrate the fQCR method by characterizing these thermodynamic and kinetic properties for variants of Staphylococcal nuclease and E. coli ribose-binding protein engineered to contain single, protected cysteines. These straightforward, information-rich experiments are likely to find applications in protein engineering and functional genomics.
Keywords: quantitative cysteine reactivity, thiol reactivity, protein thermodynamic stability, conformational free energy, protein folding kinetics, linkage analysis of protein stability, dissociation constants binding affinity, Staphylococcal nuclease, ribose-binding protein
Introduction
The genomic revolution has greatly influenced the scope and scale of quantitative experiments in biology1. High-throughput genomic and proteomic initiatives have explored transcription2, translation3, post-translational modification4-8, and protein degradation9,10 and turnover11 in response to changes in cellular physiology, and have expanded our knowledge of protein structure12-14. Less emphasis has been placed on developing methods that use small quantities of material to rapidly quantify protein thermodynamics and folding kinetics. Such advancements are essential to furthering our understanding of the relationship between protein sequence, structure, stability and function that underlies the molecular basis of disease and evolution. Here we present a miniaturized thermodynamic technique, fast determination of quantitative cysteine reactivity (fQCR), that measures protein stability (ΔGU), thermal stability (Tm), ligand-binding affinity (KD), and estimates (un)folding kinetics. By exploiting the fundamental linkage between protein thermodynamics and function, fQCR experiments can be used to identify and assess many aspects of biological function that affect protein conformational stability. These functions include the strength of biomolecular interactions15-18, the impact of deleterious mutations19,20, and the effects of post-translational modifications21-25.
Traditional approaches for measuring protein stability (e.g., temperature and chemical denaturation) and folding kinetics are low-throughput (requiring > 100 μg of protein) and time-consuming (taking hours to days). By contrast, the emergence of miniaturized techniques for determining Tm26-28, ΔGU26,29-31, and KD28,30,32,33 are beginning to enable the thermodynamic characterization of proteins using less than a few micrograms of purified material. One such technique, quantitative cysteine reactivity (QCR)26, uses cysteine protection to measure protein stability in a manner analogous to amide protection experiments34. Cysteine sidechains protected by burial in a folded protein structure are inaccessible to solvent and cannot be modified by a thiol-reactive probe. However, these residues can be modified upon exposure to solvent by transient unfolding reactions. Consequently, the kinetics of cysteine modification are coupled to the thermodynamic stability and (un)folding kinetics of the protein.
Here we present a development of the QCR technique that enables rapid determination of cysteine modification using fluorescence: fast determination of quantitative cysteine reactivity (fQCR). The fQCR approach was validated using variants of Staphylococcal nuclease (SN) and E. coli ribose-binding protein (RBP) engineered to contain single, buried cysteines. In fQCR experiments, the kinetics of cysteine labeling are determined using the fluorogenic thiol-probe, 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole (ABD). The fluorescence readout facilitates the rapid quantification of the degree of fractional cysteine labeling as a function of temperature. From these observations ΔGU, Tm, and, in some cases, folding kinetics can be determined. Additionally, fQCR experiments can be used to quantify the affinity of biomolecular interactions by measuring changes in ΔGU in the presence of a binding partner. This approach was demonstrated for SN and RBP by determining the binding affinities of these proteins for inhibitor and ribose respectively. Unlike traditional methods for measuring protein stability, folding kinetics, and ligand binding affinity, fQCR data can be acquired on non-specialized equipment within minutes, requires only micrograms of protein, and is not restricted to proteins that exhibit ideal two- or three-state unfolding behavior.
Materials and Methods
Protein Engineering and Purification
Linear DNA encoding SN.L36C was assembled from synthetic oligonucleotides35 and cloned into pET-21a (EMD Biosciences; 69740) at the XbaI and XhoI sites. A C-terminal GGSHHHHHHHK tag was introduced by QuickChange® (Stratagene; 200519).
Linear DNA fragments encoding RBP.L61C and RBP.A188C variants were assembled from synthetic oligonucleotides35 and cloned into pET-21a at NdeI and XhoI sites. These mutations were introduced into a variant of the wild-type E. coli ribose-binding protein in which rare codons have been replaced, the N-terminal signal sequence removed, and a C-terminal GGSHHHHHH tag added. All sequence-verified cloned variants were transformed into E. coli KRX strain (Promega L3002) and stored as glycerol stocks at -80°C.
Starter cultures from glycerol stocks were diluted 1:500 into auto-induction media ZYM-505236 supplemented with 0.04% L-Rhamnose for delayed induction of the KRX T7 expression system. Cells were grown for 12-16 hours at 30°C with shaking at 225-270 rpm and pelleted by centrifugation at 6,000 g. RBP variants were lysed by addition of 1 mL BugBuster® Master Mix (EMD Biosciences; 71456) for each 20 mL of culture. The SN.L36C culture was lysed on ice by sonication for two minutes after resuspension in TBS (20 mM Tris, 136 mM NaCl, pH 7.4). In all cases 5 mM β-mercaptoethanol was added to prevent disulfide bond formation during lysis. Lysed samples were centrifuged at 16,000 g for 10 min. The tagged recombinant proteins were purified by batch immobilized metal affinity chromatography by mixing the supernatant with an equal volume of binding buffer (15 mM imidazole, 20 mM MOPS, 500 mM NaCl, pH 7.5) before addition to His-Select Nickel affinity gel (Sigma P6611). Non-specifically bound protein was removed by four successive washes with 14 resin bed volumes of a 50:50 mixture of TBS and binding buffer supplemented with 5 mM β-mercaptoethanol. The tagged proteins were eluted in one bed volume of elution buffer (400 mM imidazole, 20 mM MOPS, 500 mM NaCl, pH 7.5), followed by buffer exchange on 10DG gel-filtration columns (Bio-Rad 732-2010) that were pre-equilibrated with 25 mM potassium phosphate, 100 mM KCl, pH 7. Samples were flash-frozen by dropwise addition (~20uL / drop) into liquid nitrogen, and stored as frozen beads at -80°C. No difference in stability (assessed by thermal melt) was observed between samples that underwent a single freeze-thaw cycle and aliquots stored overnight at 4°C. Protein concentration was measured spectroscopically using absorbance at 280 nm using extinction coefficients (εSN≈16,000 M-1cm-1, εRBP≈4,000 M-1cm-1) calculated from amino acid sequence37. Protein purity was assessed by SDS-PAGE.
Preparation of ABD stock solutions
A ~0.5 M stock solution was prepared by dissolving 200 mg of 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole (ABD; TCI America; A5597) in ~1.8 mLs of DMSO. The final concentration was determined spectroscopically (ε313nm=4,200 M-1cm-1)38. This stock solution was stored in 30 μL aliquots at –20°C, and was used over multiple freeze-thaw cycles. Working solutions of ABD were mixed immediately before an experiment by combining the appropriate volume of ABD stock solution and phosphate buffer (25 mM potassium phosphate and 100 mM KCl at pH 7).
Reaction of ABD with glutathione
The pseudo-first-order rate constants for the reaction of ABD with reduced glutathione (L-GSH; Sigma-Aldrich; G4251) were determined from full kinetic traces measured at combinations of four temperatures (20, 30, 40, and 50°C) and pH values (6, 7, 8, and 8.5) in buffers containing 25 mM potassium phosphate and 100 mM KCl. The progress curve of the reaction between 10 μM L-GSH and 2.5 mM ABD was monitored in a SLM Series 2 Luminescence Spectrometer by recording the emission intensity of the fluorescent ABD-L-GSH adduct (λex 389 nm; λem 513 nm; excitation bandwidth 8 nm; emission bandwidth 16 nm; PMT sensitivity 700). To ensure thermal equilibration, 1.45 mL of phosphate buffer was pre-incubated (with stirring) in a quartz cuvette for ten minutes in a temperature-controlled sample cell, after which 35.1 μL of 107 mM ABD was added (final concentration of 2.5 mM). The reaction was initiated after a further two minutes by addition of 15 μL of 1 mM L-GSH (final concentration of 10 μM).
The fQCR experiment
For a single fQCR experiment, 89 μL of 5-10 μM protein in phosphate buffer (25 mM potassium phosphate and 100 mM KCl at pH 7) was combined with 1 μL of 5 mM tris(2-carboxyethyl)phosphine (TCEP; Fluka; GA12963) and 10 μL of 10X ABD working stock solution (typically 1-20 mM). The final concentration of ABD was always at least 10-fold greater than the protein concentration to ensure that cysteine labeling followed pseudo-first-order kinetics. Following initial mixing of the 100 μL reaction, 7 μL aliquots were distributed into a PCR strip tube (12 tubes per strip) and incubated for 5 or 10 minutes (depending on the experiment) in a BioRad DNA Engine® Peltier gradient thermal cycler using a heated lid offset by +10°C. After labeling, the samples were immediately cooled in an aluminum block kept at room temperature. Average fluorescence and standard error values determined from three independent readings of 1.5 μL aliquots of each sample on a NanoDrop 3300 fluorospectrophotometer (Thermo Scientific).
The temperatures of the thermocycler gradient were calibrated using a digital thermometer (Cole-Parmer; 7001H) wired to a high-precision thermister (Measurement Specialties; 44033RC) embedded in a PCR strip tube using a high thermal conductance compound (Artic Silver Inc; artic silver II). The difference between the temperatures reported by the thermocycler software and the recorded temperature could vary by as much as 5°C, depending on the difference between the heat of the lid and the sample. Temperatures in each column of a gradient thermocycler were recorded after an equilibration time of one minute with a closed, heated lid (i.e. conditions identical to a fQCR incubation).
The fQCR experiments for RBP.L61C were modified to account for the relatively slow conformational equilibration of this protein over the experimental temperature range. To ensure conformational equilibrium was established for this variant, 9 μL aliquots of protein sample were transferred to PCR tubes and placed in the temperature gradient for 2 minutes prior to labeling. Following equilibration, 1 μL of ABD working stock solution was added to each aliquot using a multi-channel pipette to initiate the labeling reaction. After labeling for 5 minutes, the samples were processed as described above.
One fQCR data set (Fig. 6B) was collected using a Tecan Genios fluorescence plate reader. In this case, 40 μL samples (5 μM protein, 400 μM ABD, 50 μM TCEP, 25 mM potassium phosphate, 100 mM KCl, pH 7) were incubated in the gradient thermocycler for 10 min and 30 μL aliquots transferred to 384-well black plates (Corning; 3821) after labeling. Fluorescence was measured at 30°C using 405 nm and 465 nm excitation and emission filters respectively.
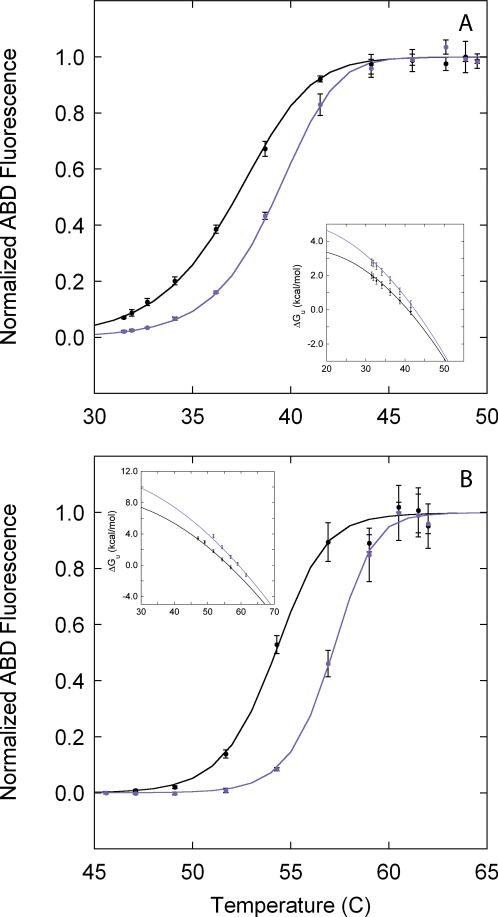
Figure 6. Ligand binding.
Temperature slices were collected in the absence (black) and presence (purple) of 100 μM pdTp and 20 μM ribose for SN.L36C (A) and RBP.A188C (B) respectively. The insets in panels A and B correspond to the free energy profiles derived from the primary fQCR data for SN.L36C in the absence (black, ΔHm = 83 ± 1 kcal/mol, Tm = 41 ± 1°C) and presence (purple, ΔHm = 94 ± 3 kcal/mol, Tm = 43 ± 1°C) of pdTp, and for RBP.A188C in the absence (black, ΔHm = 138 ± 1 kcal/mol, Tm = 57 ± 1°C) and presence (purple, ΔHm = 160 ± 8 kcal/mol, Tm = 59 ± 1°C) of ribose.
Thermal unfolding monitored by circular dichroism
Thermal unfolding experiments were carried out by preparing 3 mL of 3 μM protein in phosphate buffer (25 mM potassium phosphate and 100 mM KCl at pH 7) and monitoring the change in CD signal at 222 nm as a function of temperature in a Jasco J-815 CD spectrometer. Samples were heated at a rate of 1°C per minute and allowed to equilibrate for 2 minutes following each temperature change. The midpoint of thermal unfolding, Tm, was determined for each protein by fitting a two-state model of unfolding to the temperature dependence of normalized far-UV CD signal
| [1] |
where θN and θD are linear relationships that describe the native (N) and denatured (D) baselines (e.g., θN=mNT + bN, describe the native baseline), and ΔGGH(T) is the Gibbs-Helmholtz relationship (Eq. 11). In the process of using Eq. 1 to derive Tm, ΔCp was fixed at a value of 3 kcal mol-1 K-1.
Results and Discussion
Measuring protein stability by QCR
Cysteine sidechains are attractive targets for site-directed chemistry in proteins given the wide variety of commercially available compounds that preferentially label thiols (e.g., methanethiosulfonate (MTS), 5,5′-dithiobis(2-nitrobenzoate) (DTNB), iodoacetamide (IAM) and ABD). These reagents are most frequently used to covalently modify cysteine sidechains to introduce chromophores or fluorophores for determining protein concentration39,40, measuring binding affinity41,42, and assessing sidechain accessibility43,44. Such reagents can also be employed as site-specific probes of protein stability45-47 and folding kinetics48-51, but rarely are. Cysteine reactivity to characterize protein thermodynamics and function has been underutilized.
In the QCR method, protein stability is quantified by exploiting the relationship between conformational free energy and the reactivity of a buried cysteine26. In a folded protein (Closed-SH), a buried cysteine is concealed by protein structure and is inaccessible to bulk solvent. To be covalently modified by a thiol-reactive probe (P), the cysteine must become exposed to solvent (Open-SH) in an unfolding reaction
| [2] |
where kopen and kclose are the rate constants for opening (unfolding) and closing (folding), and kint is the rate constant for cysteine modification in the unprotected (open) state. Under the appropriate conditions (see below), the observed rate constant for labeling a buried cysteine (klabel) can expressed as52
| [3] |
If kclose >> kint, cysteine modification is said to proceed under EX2 conditions52 and kint can be eliminated from the denominator of Eq. 3, thereby directly relating observed cysteine reactivity with protein thermodynamic stability
| [4] |
where ΔGU is the conformational free energy of unfolding, ΔGU = RTln(kopen/kclose). Rearrangement of Eq. 4 yields
| [5] |
which is used to derive ΔGU from the labeling of single, buried cysteines in QCR-based experiments .
The mechanism of cysteine labeling
A protected cysteine can be exposed to bulk solvent by transient local or global unfolding events. Deeply buried sites are usually highly protected and only label as a consequence of global unfolding. Sites that are in proximity to the solvent-accessible surface of the protein can be labeled as a consequence of either local or global unfolding, depending on their structural context and experimental conditions. In the QCR approach, the effects of global unfolding are measured by manipulating the global thermodynamic stability to achieve a range of 99% to 1% native protein, typically by altering temperature. Under these conditions, which we refer to as the global unfolding window of observation (GUWO)26, global unfolding is the dominant mechanism for the exposure of protected cysteines, and measured values for klabel can be analyzed using Eq. 5 to give global protein thermodynamic stability. Outside of the GUWO, because the labeling of a buried cysteine can result from a variety of conformational fluctuations (i.e., transient local, subglobal or global unfolding events), ΔGU cannot be guaranteed to exclusively reflect the free energy of global unfolding.
In a typical fQCR experiment, the GUWO comprises a 20-30 degree range of temperatures that span Tm. Under these conditions, ΔGU is highly dependent on temperature and the equilibrium fraction of folded and unfolded protein is variable. Consequently, the observed rate of cysteine labeling can be affected by kinetics of conformational equilibration (kconf = -(kopen + kclose) see SI-1) upon exposure to the temperature gradient. In cases where kint << kconf (steady-state exchange53), cysteine labeling is described by a single amplitude (As, which is equal to one) and a pseudo-first order rate constant (ks=klabel) (see SI-2)
| [6] |
at a given temperature T. When kint ≥ 0.1kconf (pre-steady-state exchange53), these conditions break down and cysteine labeling is described by two kinetic phases (see SI-2)
| [7] |
where the fast (amplitude Af, rate constant kf) and slow (amplitude As, rate constant ks) phases describe the kinetics of cysteine labeling in the unfolded and folded fraction of protein established at time t52,53. Under these conditions, neither amplitude is ever equal to one, and observed cysteine labeling is never pseudo-first order. This situation is identified experimentally by the EX2 test (see below). Equation 7 makes no assumptions regarding the kinetics of conformational equilibration, nor the relative magnitudes of kclose, kopen, and kint53. It does, however, assume that labeling of protected cysteines in the closed state is negligible.
Even though the possibility of multiple kinetic phases makes for a relatively complex analysis, under EX2 conditions klabel is pseudo-first order and can be extracted. To assist in the understanding of this process and to help identify appropriate experimental conditions (temperatures and label concentrations), we have developed some simulation tools (SI-2). By simulating Eq. 7, combinations of kclose, kopen and kint can be identified that eliminate the fast phase of the reaction. These simulations reveal that the fast phase is negligible under conditions where kint is an order of magnitude less than kclose (i.e., traditional EX2 conditions), regardless of the relative values of kclose and kopen. This generalization holds true even if the protein is 99% unfolded at equilibrium and kopen is >100 times kclose. Furthermore, these simulations demonstrate that QCR-based experiments can be designed to measure ΔGU at temperatures beyond Tm, providing EX2 conditions persist.
In the QCR method, the use of sidechain protection to measure protein stability at temperatures that promote unfolding is quite different from methods such as hydrogen-deuterium exchange (HDX) that exploit the differential reactivity of protected groups under native conditions (i.e. >99% folded, kclose > 100 kopen). HDX cannot be implemented at temperatures that promote unfolding because the long timescale and high protein concentration lead to protein aggregation54,55. QCR and HDX-like experiments can therefore be regarded as complementary ways for determining ΔGU under quite different experimental conditions.
Determinants of kint
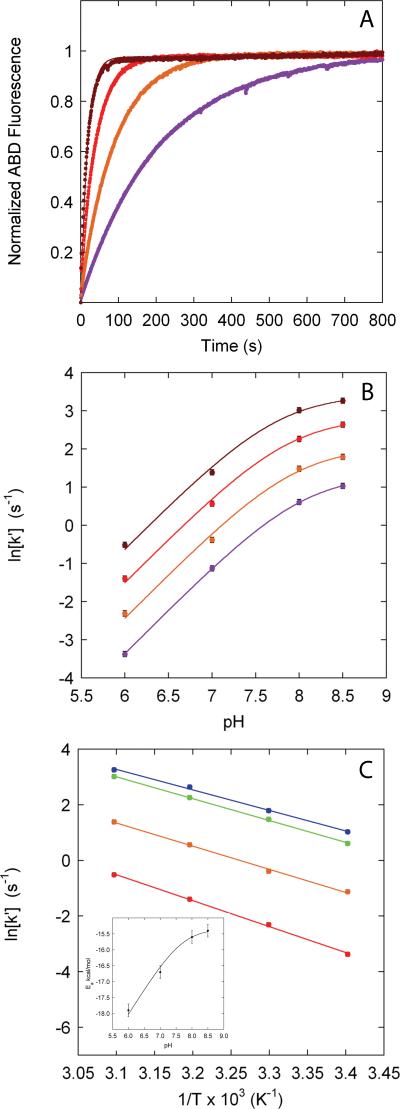
The reactivity of an unprotected cysteine, kint, can be affected by a number of factors including the cysteine pKa value in the unfolded state, solution pH, temperature, and probe concentration. To investigate the effects of pH and temperature on kint, we used the reaction between ABD and reduced glutathione (L-GSH), a tripeptide that serves as a model for the reaction of an unprotected cysteine (Fig. 1).
Figure 1. pH and temperature dependence of the reaction of ABD with L-GSH.
(A) Representative progress curves for the pseudo-first order reaction of 10 μM L-GSH with 2.5 mM ABD at pH 8.0 and ( ) 20°C, (
) 20°C, ( ) 30°C, (
) 30°C, ( ) 40°C and (
) 40°C and ( ) 50°C. Solid lines represent a fit of a first-order exponential function to obtain kint. Such progress curves were used to obtain the rate constants in B and C. (B) pH dependence of the second-order rate constants, k′=kint/[ABD], for the reaction of ABD with L-GSH at (
) 50°C. Solid lines represent a fit of a first-order exponential function to obtain kint. Such progress curves were used to obtain the rate constants in B and C. (B) pH dependence of the second-order rate constants, k′=kint/[ABD], for the reaction of ABD with L-GSH at ( ) 20°C, (
) 20°C, ( ) 30°C, (
) 30°C, ( ) 40°C and (
) 40°C and ( ) 50°C. Solid lines represent a fit of Eq. 8 to derive the apparent pKa of L-GSH as a function of temperature: (
) 50°C. Solid lines represent a fit of Eq. 8 to derive the apparent pKa of L-GSH as a function of temperature: ( , 8.03 ± 0.01), (
, 8.03 ± 0.01), ( , 7.9 ± 0.1), (
, 7.9 ± 0.1), ( , 7.88 ± 0.08), (
, 7.88 ± 0.08), ( , 7.75 ± 0.08). (C) Temperature dependence of k′ at pH (
, 7.75 ± 0.08). (C) Temperature dependence of k′ at pH ( ) 6, (
) 6, ( ) 7, (
) 7, ( ) 8 and (
) 8 and ( ) 8.5. Solid lines represent a fit of the Arrhenius relationship (Eq. 10) to obtain Ea/R as a function of pH: (
) 8.5. Solid lines represent a fit of the Arrhenius relationship (Eq. 10) to obtain Ea/R as a function of pH: ( , -9300 ± 200 K), (
, -9300 ± 200 K), ( , -8300 ± 200 K), (
, -8300 ± 200 K), ( , -7800 ± 200 K), (
, -7800 ± 200 K), ( , -7400 ± 200 K). The inset is a plot of the pH dependence of Ea derived from this data. lnA is independent of pH and temperature: (
, -7400 ± 200 K). The inset is a plot of the pH dependence of Ea derived from this data. lnA is independent of pH and temperature: ( , 28.4 ± 0.8), (
, 28.4 ± 0.8), ( , 27.1 ± 0.9), (
, 27.1 ± 0.9), ( , 27.3 ± 0.5), (
, 27.3 ± 0.5), ( , 26.2 ± 0.8). The average value and standard error of lnA (27.3 ± 0.3) is the same for any cysteine and was used in all fQCR simulations and data analysis.
, 26.2 ± 0.8). The average value and standard error of lnA (27.3 ± 0.3) is the same for any cysteine and was used in all fQCR simulations and data analysis.
The negatively charged form of a cysteine sidechain (thiolate) is the nucleophile that reacts with thiol-specific probes39. The second-order rate constant for labeling an unprotected cysteine (k′) is therefore pH dependent (Fig. 1B):
| [8] |
where k′ref is the pH-independent second-order rate constant measured over a pH range where cysteine is present in >99% thiolate form (in this case of L-GSH at pH values > 9).
The pKa values of unprotected cysteines can also be influenced by residual electrostatic interactions in the open (unfolded) state of proteins, and therefore may vary by location and protein. The pKa values of solvent-exposed ionizable sidechains are rarely perturbed from their model compound values by more than one pH unit56; the pKa values of most unprotected cysteines are expected to be within 8 ± 1. Any significant variation in the pKa of a cysteine in the unprotected state is observed directly in the fQCR experiment, and may report on residual structure and electrostatic interactions in the unfolded state57,58.
The temperature dependence of k′ is described by an Arrhenius relationship
| [9] |
where A is a pre-exponential factor and Ea is the activation energy for the reaction. Analysis of the temperature dependence of the reaction between L-GSH and ABD (Fig. 1C) shows that lnA (27.3 ± 0.3) is independent of both pH and temperature, and consequently remains fixed in the analysis of the temperature dependence of kint. However, Ea is dependent upon pH (Fig. 1C inset) and depends on the pKa of the specific cysteine under consideration. From this analysis, a general expression for the pseudo-first order rate constant kint at a given pH, temperature and probe concentration ([P]) can be derived
| [10] |
Measuring protein stability using fQCR
In the original implementation of the QCR experiment, klabel was derived for protected cysteines from full kinetic traces at a given temperature26. The experimental throughput of fQCR experiments is dramatically increased by determining klabel from single timed reaction endpoints multiplexed across a gradient of temperatures (“temperature slice”) and probe concentrations (“probe slice”). In a temperature slice, i parallel reactions are simultaneously initiated and incubated at different temperatures Ti in a gradient thermocycler, and quenched by rapid cooling or addition of acid after an incubation time, Δt. This procedure provides accurate pseudo-first order rate constants for klabel if cysteine labeling follows an EX2 mechanism (which is tested experimentally, see below). Under these conditions, the observed rate constant klabel is by definition pseudo-first order because kint and kconf are first order kinetic processes. In this case, values for klabel derived from fractional cysteine labeling at time t are an algebraic rearrangement of an integrated first-order rate equation (Eq. 13). Outside of EX2 conditions, klabel cannot be guaranteed to be pseudo-first order (Eq. 7), and accurate rate constants cannot be obtained from timed endpoint experiments. To demonstrate the procedure and accuracy of obtaining pseudo-first order rate constants from timed endpoint experiments, we have included a number of control experiments for the reaction of ABD with GSH as a function of pH (SI-6). These experiments show that rate constants derived from timed endpoint labeling experiments are equivalent to rate constants derived from full kinetic traces within experimental error.
In the fQCR experiment, the observed temperature dependence of cysteine labeling is an indirect measure of the temperature dependence of protein stability (Fig. 2 and SI-3), and can be analyzed in terms of the Gibbs-Helmholtz relationship
| [11] |
where ΔHm is the enthalpy of unfolding, ΔCp the change in heat capacity of unfolding, Tm the midpoint of thermal denaturation, and ΔGU,i is obtained experimentally from Eq. 5 as
| [12] |
with the pseudo-first order rate constant klabel,i given by
| [13] |
and the pseudo-first order rate constant kint,i given by
| [14] |
where fi is the fractional cysteine labeling observed at Δt and Ti, [P] is probe concentration, Ea is the activation energy of the labeling reaction, and k′i is the second-order rate constant for the labeling reaction at Ti (see Eq. 10). Combining 11-14 we obtain
| [15] |
which links the temperature dependence of fractional cysteine labeling at a set of constant endpoints to protein stability (Fig. 2). Rewriting Eq. 15 in terms of fluorescence measurements observed in an instrument gives
| [16] |
where RFUmax is the fluorescence value in arbitrary units of a fully labeled sample.
Figure 2. Simulation of the relationships between fractional labeling and protein stabilities at different temperatures in an fQCR experiment.
(A) Simulation of the Gibbs-Helmholtz relationship (Eq. 11) using a ΔCp, Tm and ΔHm of 3 kcal mol-1 K-1, 329 K (56°C) and 105 kcal/mol respectively. Colors in panels A, B and C indicate a set of temperatures within a typical GUWO: ( ) 60°C, (
) 60°C, ( ) 57°C, (
) 57°C, ( ) 55°C, (
) 55°C, ( ) 53°C, (
) 53°C, ( ) 51°C, (
) 51°C, ( ) 49°C, (
) 49°C, ( ) 47°C, (
) 47°C, ( ) 45°C and (
) 45°C and ( ) 43°C. (B) Simulated QCR kinetic traces calculated from klabel,i derived from ΔGU,i in panel A using Eq. 5. Values of kint were simulated using a lnA, Ea/R and ABD concentration of 27.3, -8300 K and 2.6 mM respectively. (C) Simulated temperature dependence of fractional labeling at 300 s (indicated by the arrow in panel B). The solid line is a simulation of Eq. 15 using a lnA, Ea/R and ABD concentration of 27.3, -8300 K and 2.6 mM respectively.
) 43°C. (B) Simulated QCR kinetic traces calculated from klabel,i derived from ΔGU,i in panel A using Eq. 5. Values of kint were simulated using a lnA, Ea/R and ABD concentration of 27.3, -8300 K and 2.6 mM respectively. (C) Simulated temperature dependence of fractional labeling at 300 s (indicated by the arrow in panel B). The solid line is a simulation of Eq. 15 using a lnA, Ea/R and ABD concentration of 27.3, -8300 K and 2.6 mM respectively.
The principal idea behind the fQCR approach is that the temperature dependence of ΔGU can be determined solely from experimentally measured values of fi and kint,i. Without further analysis, conformational free energies determined in this way quantify the temperature dependence of protein stability and can be used to measure ligand binding affinities. However, analysis of a single fQCR dataset by a least-squares fit of Eq. 16 requires the Gibbs-Helmholtz relationship (Eq. 11) to model the temperature dependence of protein stability. Robust determination of the thermodynamic parameters ΔHm and Tm require an accurate value of ΔCp, which is poorly determined by fQCR data collected over a 20-30 degree temperature interval that spans Tm59,60. To circumvent this issue, we fixed the value of ΔCp at an average value of 3, based on known values of ΔCp from structure-based calculations61 and experiments62-64. This procedure has no significant impact on the interpretation of the temperature dependence of ΔGU within the GUWO (see Fig. SI-7 and simulation SI-3).
EX2 conditions
Analysis of fQCR experiments by Eq. 15 and 16 is valid only under EX2 conditions, when kclose > 10kint (SI-2). A critical and simple diagnostic to identify whether cysteine labeling occurs under EX2 conditions is to measure the dependence of klabel on probe concentration (“probe slice”). At a given temperature Ti, if the corresponding klabel,i at different probe concentrations j exhibits a linear dependence (usually presented in a double log plot), EX2 conditions hold (Fig. 3). Furthermore, if EX2 conditions do not hold, the transformation of the Ti,fi relationship into Ti,ΔGUi (Eq. 15) no longer gives the shape characteristic of the Gibb-Helmholtz relationship (Fig. 2A), and instead “peels” off (Fig. 3B and SI-4). Visualization of these peeling regions indicates the combination of probe concentrations and temperatures where no inference can be drawn about protein stability. This behavior also indicates a range of conditions under which cysteine labeling is no longer a pseudo-first order kinetic process. At these temperatures, the observed fractional cysteine labeling at Δt cannot be converted into apparent rate constants using Eq. 13. However, because this observation indicates that kint ≥ 0.1kconf, it may be possible to extract information about folding kinetics in these regions.
Figure 3. Simulation of EX2 conditions.
(A) Dependence of log[klabel,i] on log[ABD] at ( ) 40°C, (
) 40°C, ( ) 45°C, (
) 45°C, ( ) 50°C, (
) 50°C, ( ) 55°C, (
) 55°C, ( ) 60°C and (
) 60°C and ( ) 70°C (refer to SI-4 for the details of this simulation). EX2 conditions (closed circles and solid lines) hold in the linear portion of these graphs (slope of 1.0 ± 0.2). (B) Apparent unfolding free energies derived by transforming the simulated rate constants in panel A using Eq. 5. The subset of free energy profiles (
) 70°C (refer to SI-4 for the details of this simulation). EX2 conditions (closed circles and solid lines) hold in the linear portion of these graphs (slope of 1.0 ± 0.2). (B) Apparent unfolding free energies derived by transforming the simulated rate constants in panel A using Eq. 5. The subset of free energy profiles ( ,
,  ,
,  ) that correspond to an EX2 mechanism give a coincident free energy profile. The subset of free energy profiles that correspond to a loss of EX2 conditions at higher ABD concentrations and temperatures (
) that correspond to an EX2 mechanism give a coincident free energy profile. The subset of free energy profiles that correspond to a loss of EX2 conditions at higher ABD concentrations and temperatures ( ,
,  ,
,  ) diverge from the coincident free energy profile “peeling”.
) diverge from the coincident free energy profile “peeling”.
Estimation of folding rate constants
Even though no thermodynamic inferences can be drawn in the absence of EX2 conditions, it is possible to obtain kinetic information in this regime under certain conditions. Under non-EX2 conditions, kint is similar to, or faster than kclose as temperature and probe concentration are increased. As discussed above, the situation is readily identifiable as nonlinearity in a plot of log[klabel,i] versus log[P] obtained from a probe slice (Fig. 3 and SI-4). In this nonlinear region, kint therefore serves as a proxy for protein (un)folding rate constants as the fQCR experiement transitions out of EX2 labeling conditions. Beyond this nonlinear region, klabel becomes independent of probe concentration and kint >> kclose.
In order to access the folding kinetics of a particular protein, the intrinsic reaction rate of the probe has to match kclose within the range that is accessible by manipulation of probe concentration. Because protein folding rates vary over five orders of magnitude65, a given probe can access folding kinetics of only a subset of proteins. For instance, ABD can be used to measure folding kinetics for proteins with folding rate constants between ~3×10-4 s-1 and ~1 s-1.
Experimental fQCR results
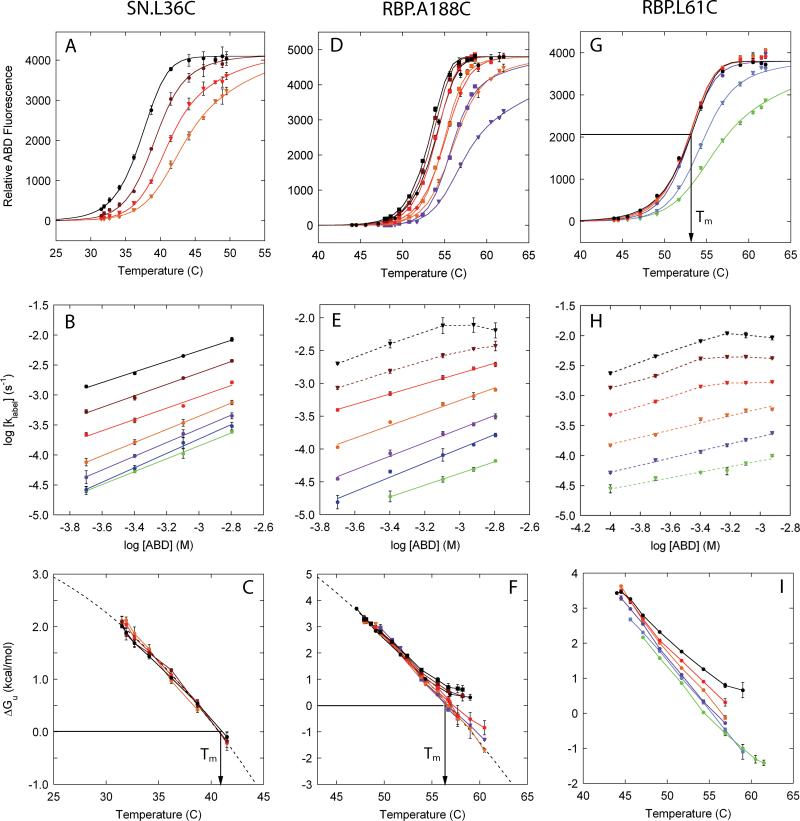
In a typical fQCR experiment, cysteine labeling is monitored as a function of temperature and probe concentration (Fig. 4A, D and G). Global analysis of these data using Eq. 16 provides kint, RFUmax, Tm, the temperature dependence of protein stability, estimates of folding rate constants, and confirms EX2 labeling conditions. Analysis of Eq. 16 (SI-3) using experimental parameters for ABD shows that kint,i is accurately determined under EX2 conditions at relatively low probe concentrations and short labeling times (typically 100-400 μM and 300 s). Outside of EX2 labeling conditions, kint can only be determined if kopen is much faster than kint (SI-2). To determine RFUmax in order to derive fi, relatively high probe concentrations need to be used (typically 400-2000 μM).
Figure 4. fQCR results for SN and RBP.
A, D, G: Temperature dependence of fractional labeling at single timed reaction endpoints monitored by ABD fluorescence fit with Eq. 16. Different curves represent different ABD concentrations or labeling times: SN.L36C, 300 s, ( ) 200 μM (
) 200 μM ( ) 400 μM, (
) 400 μM, ( ) 800 μM and (
) 800 μM and ( ) 1600 μM; RBP.A188C, 300 s (closed circles and triangles) or 600 s (closed squares), (
) 1600 μM; RBP.A188C, 300 s (closed circles and triangles) or 600 s (closed squares), ( ,
,  ) 200 μM (
) 200 μM ( ,
,  ) 400 μM, (
) 400 μM, ( ,
,  ) 800 μM, (
) 800 μM, ( ) 1200 μM, and (
) 1200 μM, and ( ,
,  ) 1600 μM; RBP.L61C, 300 s, (
) 1600 μM; RBP.L61C, 300 s, ( ) 100 μM (
) 100 μM ( ) 200 μM, (
) 200 μM, ( ) 400 μM, (
) 400 μM, ( ) 600 μM, (
) 600 μM, ( ) 800 μM and (
) 800 μM and ( ) 1200 μM. B, E, H: Dependence of labeling rate constant on ABD concentration to identify EX2 conditions: SN.L36C, (
) 1200 μM. B, E, H: Dependence of labeling rate constant on ABD concentration to identify EX2 conditions: SN.L36C, ( ) 31.5°C, (
) 31.5°C, ( ) 31.9°C, (
) 31.9°C, ( ) 32.7°C, (
) 32.7°C, ( ) 34.1°C, (
) 34.1°C, ( ) 36.2°C, (
) 36.2°C, ( ) 38.7°C and (
) 38.7°C and ( ) 41.5°C; RBP.A188C, (
) 41.5°C; RBP.A188C, ( ) 48.1°C, (
) 48.1°C, ( ) 49.6°C, (
) 49.6°C, ( ) 50.8°C, (
) 50.8°C, ( ) 52.4°C, (
) 52.4°C, ( ) 53.9°C, (
) 53.9°C, ( ) 55.6°C and (
) 55.6°C and ( ) 57.7°C; RBP.L61C, (
) 57.7°C; RBP.L61C, ( ) 45.6°C, (
) 45.6°C, ( ) 47.1°C, (
) 47.1°C, ( ) 49.1°C, (
) 49.1°C, ( ) 51.7°C, (
) 51.7°C, ( ) 54.3°C and (
) 54.3°C and ( ) 56.9°C. EX2 conditions (closed circles and solid lines) hold where the slope of the linear fit exceeds ~0.8. C, F, I: Transformation of fractional labeling to conformational free energy using Eqs. 13 and 5, and using the temperature dependence of kint obtained from Eq. 14. Gibbs-Helmholtz relationships (dashed line, Eq. 11 with ΔCp fixed at 3.0 kcal mol-1 K-1) can be fit to cases where EX2 conditions can be observed for at least some combinations of temperature and protein concentration (SN.L36C, RBP.A188C). Non-EX2 behavior is seen as peeling (F). The transformation of fi to ΔGU,i is done only for fi values between 0.02 and 0.95 to minimize artifacts associated with the experimental uncertainty encountered at very low and high levels of cysteine labeling.
) 56.9°C. EX2 conditions (closed circles and solid lines) hold where the slope of the linear fit exceeds ~0.8. C, F, I: Transformation of fractional labeling to conformational free energy using Eqs. 13 and 5, and using the temperature dependence of kint obtained from Eq. 14. Gibbs-Helmholtz relationships (dashed line, Eq. 11 with ΔCp fixed at 3.0 kcal mol-1 K-1) can be fit to cases where EX2 conditions can be observed for at least some combinations of temperature and protein concentration (SN.L36C, RBP.A188C). Non-EX2 behavior is seen as peeling (F). The transformation of fi to ΔGU,i is done only for fi values between 0.02 and 0.95 to minimize artifacts associated with the experimental uncertainty encountered at very low and high levels of cysteine labeling.
The fQCR experiments presented here used variants of SN and RBP that contain single, buried cysteines and exemplify three classes of behavior (Fig. 4): a system that is always in EX2 (SN.L36C, Fig. 4A-C); one that switches out of EX2 at some elevated probe concentrations and temperatures (RBP.A188C, Fig. 4D-F); and one that is never fully in EX2 (RBP.L61C, Fig. 4G-I). In the first system, only thermodynamic parameters can be determined; in the second, both thermodynamic parameters and estimates of folding rate constants can be obtained; and the third, estimates of folding rate constants and Tm can be made (Table I).
Table I.
Experimentally derived fQCR parameters for SN and RBP
| Protein | Δt s | [ABD] μM | Ea/R K | RFUmax | ΔHm kcal/mol | Tm °C | EX2 | ~kclose at Tm s-1 |
|---|---|---|---|---|---|---|---|---|
| SN.L36C | 300 | 200 | -7760 ± 8 | 41 ± 1 | Yes | >>1 × 10-2d | ||
| 300 | 400 | -7875 ± 7 | 42 ± 1 | Yes | >>1 × 10-2d | |||
| 300 | 800 | 4100 ± 30 | 80 ± 3 | 41 ± 1 | Yes | >>1 × 10-2d | ||
| 300 | 1600 | 4100 ± 20 | 78 ± 1 | 41 ± 1 | Yes | >>1 × 10-2d | ||
| -7820a | 4100a | 83 ± 1b | 41 ± 1b | |||||
| RBP.A188C | 300 | 200 | -8163 ± 6 | 56 ± 1 | Yes | |||
| 300 | 400 | -8110 ± 14 | 57 ± 1 | Yes | ||||
| 300 | 800 | 4800 ± 40 | 121 ± 4 | 57 ± 1 | Yes | |||
| 300 | 1600 | 4700 ± 50 | 117 ± 8 | 57 ± 1 | No | 4 × 10-2 | ||
| 600 | 200 | 56 ± 1 | Yes | |||||
| 600 | 400 | 56 ± 1 | Yes | |||||
| 600 | 800 | 4800 ± 40 | 128 ± 3 | 57 ± 1 | Yes | |||
| 600 | 1200 | 4700 ± 40 | 125 ± 5 | 58 ± 1 | No | 4 × 10-2 | ||
| 600 | 1600 | 4700 ± 40 | 121 ± 4 | 58 ± 1 | No | 4 × 10-2 | ||
| 300f | 500f | -8133 ± 53f | 123 ± 16f | 56 ± 1f | Yes | |||
| 300f | 500f | -8173 ± 19f | 112 ± 5f | 56 ± 1f | Yes | |||
| -8140a | 4740a | 138 ± 1b | 57 ± 1b | |||||
| RBP.L61C | 300 | 100 | No | < 2 × 10-2e | ||||
| 300 | 200 | No | < 2 × 10-2e | |||||
| 300 | 400 | 3926 ± 40 | 53 ± 1c | No | < 2 × 10-2e | |||
| 300 | 600 | 3930 ± 40 | 53 ± 1c | No | < 2 × 10-2e | |||
| 300 | 800 | 3845 ± 30 | 53 ± 1c | No | < 2 × 10-2e | |||
| 300 | 1200 | 3824 ± 30 | 53 ± 1c | No | < 2 × 10-2e | |||
| 3881a | 53 ± 1c |
Average values for these parameters
Thermodynamic parameters derived from a fit of the coincident free energy profiles in Fig. 4C and 4F using the Gibbs-Helmholtz relationship (Eq. 11)
Quantified graphically
The folding rate constant is at least faster than the greatest value of kint,i encountered over this range of temperatures and probe concentrations
In the absence of EX2 conditions, kint represents the upper limit of kclose
These values were derived from the data collected using the PHEREstar FS microplate reader (Fig. 7)
Tm values
At Tm, ΔGU,i is zero and Eq. 5 reduces to kint,i=2klabel,i. Under EX2 conditions, Tm can therefore be identified unambiguously as the temperature at which this relation is true. Alternatively, Tm can be derived from fQCR datasets collected under EX2 conditions by identifying the temperature at which the fitted Gibbs-Helmholtz relationship is zero (Fig. 4C and 4F). If cysteine labeling never conforms to an EX2 mechanism (Fig. 4G), the determination of Tm is more complex. In these cases, under the appropriate combination of labeling time, probe concentration, and where kopen < kint, the midpoint of a single fQCR curve approximates Tm (Fig. 4G). These situations can only be identified by simulation (SI-5). In the case of RBP.L61C (Fig. 4G), a labeling time of 300 s, probe concentrations between 400-1200 μM, and folding rate constants between 10-2-10-3 give a set of fQCR curves with midpoints that estimate Tm. The Tm values reported by these different classes of fQCR data (Table I) are in agreement with those reported by unfolding monitored by CD (Fig. 5 and Table), and are consistent with previous results derived from full kinetic traces26.
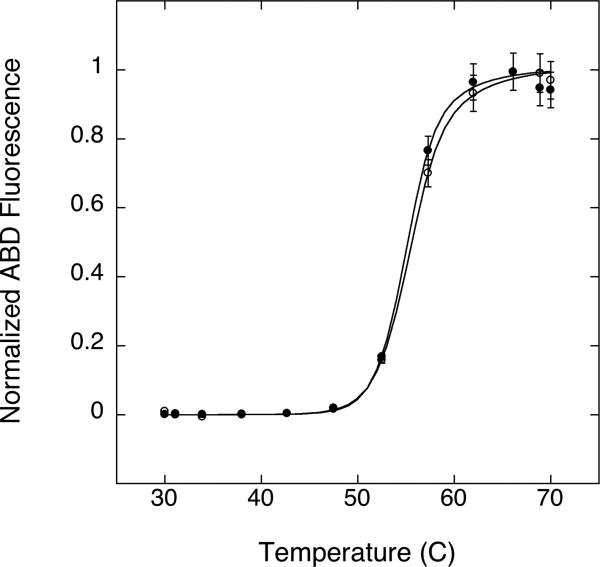
Figure 5. Thermal unfolding of SN and RBP.
Temperature unfolding of SN.L36C ( ), RBP.L61C (
), RBP.L61C ( ) and RBP.A188C (
) and RBP.A188C ( ) at pH 7 monitored by CD signal at 222 nm. A fit of Eq. 1 to the data (solid lines) gives apparent Tm values of 40 ± 1°C, 53 ± 1°C and 56 ± 1°C respectively.
) at pH 7 monitored by CD signal at 222 nm. A fit of Eq. 1 to the data (solid lines) gives apparent Tm values of 40 ± 1°C, 53 ± 1°C and 56 ± 1°C respectively.
Assessing protein function by fQCR
Biomolecular function is the consequence of proteins interacting with other macromolecules (e.g., nucleic acids and other proteins) and small molecules (e.g., ligands, substrates and inhibitors)15,16,18. All of these interactions affect protein stability, typically by stabilizing the folded state17. Consequently, stability measurements in the absence and presence of a binding partner serve as a general tool for identifying and quantifying protein function. Fig. 6A and 6B demonstrate how fQCR experiments can be used to assess protein function in terms of binding interactions. Temperature slices were acquired for SN.L36C (Fig. 6A) and RBP.A188C (Fig. 6B) in the absence (Apo) and presence of binding partner: 100 μM nucleotide inhibitor (SN) and 20 μM D-ribose (RBP). In both cases, addition of binding partner increases protein stability and shifts the endpoint labeling curves to higher temperatures. The free energy profiles in the absence (ΔapoGU) and presence (ΔLGU) of binding partner (see insets of Fig. 6A and 6B), and the relative difference in these free energies (i.e., relative stability, ΔΔGU) can be interpreted in terms of the linkage relationship
| [17] |
to obtain the equilibrium constant of binding (KD) as function of temperature and free ligand concentration ([L]) within a GUWO (Table II). These values are consistent with previous results derived from full kinetic traces26.
Table II.
KD of SN and RBP as a function of temperature
| Protein | T °C | ΔapoGU kcal/mol | ΔLGU kcal/mol | KD μM |
|---|---|---|---|---|
| SN.L36C | 40 | 0.2 | 0.8 | 65 |
| 38 | 0.7 | 1.3 | 54 | |
| 36 | 1.1 | 1.9 | 45 | |
| 34 | 1.6 | 2.3 | 38 | |
| 32 | 1.9 | 2.8 | 32 | |
| 30 | 2.3 | 3.2 | 27 | |
| RBP.A188C | 56 | 0.1 | 1.5 | 2.7 |
| 54 | 0.9 | 2.4 | 2.3 | |
| 52 | 1.6 | 3.2 | 2.0 | |
| 50 | 2.3 | 4.0 | 1.7 | |
| 48 | 3.0 | 4.8 | 1.4 | |
| 46 | 3.7 | 5.5 | 1.2 |
Quantitation and sensitivity of the fQCR method
One advantage of the fQCR approach is that it is adaptable. A number of thiol-reatives probes can be employed66, and cysteine labeling can be quantified in a variety of ways: electrophoresis (SDS-PAGE), fluorescence, mass spectrometry (MS), etc. The combination of probe and readout determines the amount of material and time required in an fQCR experiment. In the work presented here, we elected to implement the fQCR method using fluorescence because such an approach is readily accessible to a broad scientific community. Moreover, the constant improvement of fluorescence instrumentation has steadily reduced the amount of material and time required for multiplexed labeling experiments. For example, using a sensitive microplate reader we were able measure ABD-labeling quickly with relatively little material (Fig. 7): the experiment can be set up, executed, and quantified in minutes using as few as 30 nanograms (i.e., 60 uL of 0.01 uM protein) of protein. This is three orders of magnitude less material than is required for traditional CD thermal melts or high-throughput differential scanning calorimetry (DSC) experiments.
Figure 7. The sensitivity of the fQCR experiment.
fQCR data collected in a PHERAstar FS plate reader for variant RBP.A188C at 1 μM (closed circles) and 0.01 μM (open circles) concentrations. 10 μL of protein sample was labeled for 300 seconds at each temperature in the gradient and loaded into a 384-well microplate for quantitation. Lines represent a fit of Eq. 15, and error bars correspond to the propagated error of RFUmax in each experiment.
Currently, ABD is the most useful, commercial available thiol-reactive probe for the fQCR experiment. It is resistant to temperature-dependent hydrolysis, especially on the short timescale of an experiment (typically 5 minutes)67. Most importantly, ABD is highly fluorogenic, which presents an enormous advantage over other fluorescence probes that are fluorescent in both their reacted and unreacted forms. The use of nonfluorogenic probes, such as IAM- and MTS-fluorescein, requires that samples containing very dilute protein be desalted or precipitated to remove unreacted probe, which presents a major technical limitation that we have found difficult to overcome. Other fluorogenic probes can present other serious practical limitations. For example, we have found that thiol-reactive probe N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) suffers from limited solubility and a high rate of spontaneous hydrolysis that is strongly temperature dependent (data not shown). The latter issue prevents the quantitation of small amounts of labeled protein due to relatively high values of background fluorescence.
Another advantage to using ABD is that it reacts relatively slowly. Although this limits the range of folding rate constants that can be measured, it is a great advantage for measuring protein thermodynamics and assessing function in a wide variety of proteins. Due to the relatively slow speed of the ABD labeling reaction, labeling reactions are done in minutes in thermocycler, and can be quenched by cooling on ice or by addition of acid. Probes that react faster, such MTS and DTNB, require specialized instrumentation to initiate and quench labeling50,51.
Conclusions
In the 1950's Linderstrøm-Lang demonstrated how protein stability and folding kinetics are inextricably linked68. It is therefore possible to determine protein stability from reaction rates of probes with groups that are differentially accessible in the native and denatured states34. Here we show that these general principles can be applied to cysteine reactivity using ABD, a probe that becomes fluorescent only upon reaction with a thiolate. This fast determination of quantitative cysteine reactivity technique (fQCR) enables protein stability and folding kinetic properties to be determined in a short period of time with readily accessible instrumentation, using up to 1000 times less material than CD temperature melts or high-throughput DSC. In an fQCR experiment, data is acquired as fluorescence intensities obtained at single timed reaction endpoints following addition of probe. A single protein sample is multiplexed into a two-dimensional set of parallel points, in which the first dimension samples different temperatures using a gradient PCR thermocycler, and the second dimension alters probe concentration. From this block of data we can extract probe labeling rate constants, and identify different exchange regimes within which Tm, ΔGU, and folding kinetics can be determined in various combinations. In this format, data can be rapidly acquired (within minutes) at low protein concentrations (submicromolar) to minimize potential complications from irreversible unfolding or aggregation. For systems fully in EX2, Tm and ΔGU can be determined; for systems that switch out of EX2 under some conditions, folding rate constants can additionally be extracted; for systems that never attain EX2, it may be possible to estimate folding rate constants and Tm. In all cases, biological function as defined by the effects of molecular interactions can be evaluated by virtue of thermodynamic linkage between binding, stability, and folding kinetics. These relatively straightforward experiments can provide a remarkable amount of information regarding protein thermodynamic stability, folding kinetics, and biological function.
Acknowledgements
This work was supported by the NIH Director's Pioneer Award (5DPI OD000122) and the Homeland Security Advanced Research Projects Agency (HSH ODC-08-C-00099).
Footnotes
The institution at which the work was performed: Duke University, Department of Biochemistry
Contributor Information
Daniel G. Isom, Duke University, Department of Biochemistry
Philippe R. Marguet, Duke University, Department of Biochemistry
Terrence G. Oas, Duke University, Department of Biochemistry
Homme W. Hellinga, Duke University, Department of Biochemistry
References
- 1.Snyder M, Gallagher JEG. Systems biology from a yeast omics perspective. FEBS Letters. 2009;583:3895–3899. doi: 10.1016/j.febslet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeRisi J, Iyer V, Brown P. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean R, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 5.Kung LA, Tao S-C, Qian J, Smith MG, Snyder M, Zhu H. Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Molecular Systems Biology. 2009;5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nature Biotechnology. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 7.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee S-J, Mah AS, Meng L, Stark MJR, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 8.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR. Global analysis of protein sumoylation in Saccharomyces cerevisiae. The Journal of Biological Chemistry. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 9.Agard NJ, Wells JA. Methods for the proteomic identification of protease substrates. Current Opinion in Chemical Biology. 2009;13:503–509. doi: 10.1016/j.cbpa.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucet A, Butler GS, Rodríguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Molecular & Cellular Proteomics. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome.. Proceedings of the National Academy of Sciences of the United States of America; 2006. pp. 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews BW. Protein Structure Initiative: getting into gear. Nature Structural & Molecular Biology. 2007;14:459–460. doi: 10.1038/nsmb0607-459. [DOI] [PubMed] [Google Scholar]
- 13.Weigelt J, McBroom-Cerajewski LDB, Schapira M, Zhao Y, Arrowsmith CH. Structural genomics and drug discovery: all in the family. Current Opinion in Chemical Biology. 2008;12:32–39. doi: 10.1016/j.cbpa.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Thiele I, Weekes D, Li Z, Jaroszewski L, Ginalski K, Deacon AM, Wooley J, Lesley SA, Wilson IA, Palsson B, Osterman A, Godzik A. Three-dimensional structural view of the central metabolic network of Thermatoga maritima Science. 2009;325:1544–1549. doi: 10.1126/science.1174671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pace C, McGrath T. Substrate stabilization of lysozyme to thermal and guanidinehydrochloride denaturation. Journal of Biological Chemistry. 1980;255:3862–3865. [PubMed] [Google Scholar]
- 16.Schellman J. Macromolecular Binding. Biopolymers. 1975;14:999–1018. [Google Scholar]
- 17.Waldron T, Murphy K. Stabilization of proteins by ligand binding: Application to drug screening and determination of unfolding energetics. Biochemistry. 2003;42:5058–5064. doi: 10.1021/bi034212v. [DOI] [PubMed] [Google Scholar]
- 18.Xie D, Gulnik S, Erickson J. Dissection of binding energy with native and ligand-bound protein stabilities: Determining the affinity of ultratight-binding inhibitors of HIV-1 protease and its drug-resistance mutants. Journal of the American Chemical Society. 2000;122:11533–11534. [Google Scholar]
- 19.Boeckler F, Joerger A, Jaggi G, Rutherford T, Veprintsev D, Fersht A. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug.. Proceedings of the National Academy of Sciences of the United States of America; 2008. pp. 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutherford K, Alphandéry E, McMillan A, Daggett V, Parson WW. The V108M mutation decreases the structural stability of catechol O-methyltransferase. Biochimica et Biophysica Acta. 2008;1784:1098–1105. doi: 10.1016/j.bbapap.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Hagihara Y, Tan Y, Goto Y. Comparison of the conformational stability of the molten globule and native states of horse cytochrome c. Effects of acetylation, heat, urea and guanidine-hydrochloride. Journal of Molecular Biology. 1994;237:336–348. doi: 10.1006/jmbi.1994.1234. [DOI] [PubMed] [Google Scholar]
- 22.Nosworthy NJ, Peterkofsky A, König S, Seok YJ, Szczepanowski RH, Ginsburg A. Phosphorylation destabilizes the amino-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. Biochemistry. 1998;37:6718–6726. doi: 10.1021/bi980126x. [DOI] [PubMed] [Google Scholar]
- 23.Parsell D, Sauer R. The structural stability of a protein is an important determinant of its proteolytic susceptibility in escherichia coli. Journal of Biological Chemistry. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 24.Sinha S, Surolia A. Attributes of glycosylation in the establishment of the unfolding pathway of soybean agglutinin. Biophysical Journal. 2007;92:208–216. doi: 10.1529/biophysj.106.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toska K, Kleppe R, Armstrong C, Morrice N, Cohen P, Haavik J. Regulation of tyrosine hydroxylase by stress-activated protein kinases. Journal of Neurochemistry. 2002;83:775–783. doi: 10.1046/j.1471-4159.2002.01172.x. [DOI] [PubMed] [Google Scholar]
- 26.Isom D, Vardy E, Oas T, Hellinga H. Picomole-scale characterization of protein stability and function by quantitative cysteine reactivity.. Proceedings of the National Academy of Sciences of the United States of America; 2010. pp. 4908–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 28.Pantoliano M, Petrella E, Kwasnoski J, Lobanov V, Myslik J, Graf E, Carver T, Asel E, Springer B, Lane P, Salemme F. High-density miniaturized thermal shift assays as a general strategy for drug discovery. Journal of Biomolecular Screening. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 29.Ghaemmaghami S, Fitzgerald M, Oas T. A quantitative, high-throughput screen for protein stability.. Proceedings of the National Academy of Sciences of the United States of America; 2000. pp. 8296–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park C, Marqusee S. Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding. Nature Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 31.West GM, Tang L, Fitzgerald MC. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Analytical Chemistry. 2008;80:4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 32.Cimmperman P, Baranauskiene L, Jachimoviciūte S, Jachno J, Torresan J, Michailoviene V, Matuliene J, Sereikaite J, Bumelis V, Matulis D. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophysical Journal. 2008;95:3222–3231. doi: 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell KD, Fitzgerald MC. High-throughput screening assay for the tunable selection of protein ligands. Journal of Combinatorial Chemistry. 2004;6:262–269. doi: 10.1021/cc034051e. [DOI] [PubMed] [Google Scholar]
- 34.Huyghues-Despointes BM, Pace CN, Englander SW, Scholtz JM. Measuring the conformational stability of a protein by hydrogen exchange. Methods in Molecular Biology. 2001;168:69–92. doi: 10.1385/1-59259-193-0:069. [DOI] [PubMed] [Google Scholar]
- 35.Cox JC, Lape J, Sayed MA, Hellinga HW. Protein fabrication automation. Protein Science. 2007;16:379–390. doi: 10.1110/ps.062591607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier F. Protein production by auto-induction in high-density shaking cultures. Protein Expression and Purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Gill S, von Hippel P. Calculation of protein extinction coefficients from amino-acid sequence data. Analytical Biochemistry. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 38.Toyo'oka T, Imai K. New fluorogenic reagent having halogenobenzofurazan structure for thiols: 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole. Analytical Chemistry. 1984;56:2461–2464. [Google Scholar]
- 39.Bulaj G, Kortemme T, Goldenberg D. Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 1998;37:8965–8972. doi: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- 40.Ellman G. Tissue sulfhydryl groups. Archives of Biochemistry and Biophysics. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 41.Dattelbaum JD, Looger LL, Benson DE, Sali KM, Thompson RB, Hellinga HW. Analysis of allosteric signal transduction mechanisms in an engineered fluorescent maltose biosensor. Protein Science. 2005;14:284–291. doi: 10.1110/ps.041146005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Lorimier RM, Smith JJ, Dwyer MA, Looger LL, Sali KM, Paavola CD, Rizk SS, Sadigov S, Conrad DW, Loew L, Hellinga HW. Construction of a fluorescent biosensor family. Protein Science. 2002;11:2655–2675. doi: 10.1110/ps.021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gross A, Columbus L, Hideg K, Altenbach C, Hubbell W. Structure of the KcsA potassium channel from Streptomyces lividans: A site-directed spin labeling study of the second transmembrane segment. Biochemistry. 1999;38:10324–10335. doi: 10.1021/bi990856k. [DOI] [PubMed] [Google Scholar]
- 44.Shuck K, Lamb RA, Pinto LH. Analysis of the pore structure of the influenza A virus M(2) ion channel by the substituted-cysteine accessibility method. Journal of Virology. 2000;74:7755–7761. doi: 10.1128/jvi.74.17.7755-7761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Z, Butler M, Alam S, Loh S. On the nature of conformational openings: Native and unfolded-state hydrogen and thiol-disulfide exchange studies of ferric aquomyoglobin. Journal of Molecular Biology. 2001;314:153–166. doi: 10.1006/jmbi.2001.5117. [DOI] [PubMed] [Google Scholar]
- 46.Silverman JA, Harbury PB. Rapid mapping of protein structure, interactions, and ligand binding by misincorporation proton-alkyl exchange. The Journal of Biological Chemistry. 2002;277:30968–30975. doi: 10.1074/jbc.M203172200. [DOI] [PubMed] [Google Scholar]
- 47.Silverman JA, Harbury PB. The equilibrium unfolding pathway of a (beta/alpha)8 barrel. Journal of Molecular Biology. 2002;324:1031–1040. doi: 10.1016/s0022-2836(02)01100-2. [DOI] [PubMed] [Google Scholar]
- 48.Ha J, Loh S. Changes in side chain packing during apomyoglobin folding characterized by pulsed thiol-disulfide exchange. Nature Structural Biology. 1998;5:730–737. doi: 10.1038/1436. [DOI] [PubMed] [Google Scholar]
- 49.Sridevi K, Udgaonkar JB. Unfolding rates of barstar determined in native and low denaturant conditions indicate the presence of intermediates. Biochemistry. 2002;41:1568–1578. doi: 10.1021/bi011494v. [DOI] [PubMed] [Google Scholar]
- 50.Jha SK, Udgaonkar JB. Exploring the cooperativity of the fast folding reaction of a small protein using pulsed thiol labeling and mass spectrometry. The Journal of Biological Chemistry. 2007;282:37479–37491. doi: 10.1074/jbc.M706714200. [DOI] [PubMed] [Google Scholar]
- 51.Ramachandran S, Rami BR, Udgaonkar JB. Measurements of cysteine reactivity during protein unfolding suggest the presence of competing pathways. Journal of Molecular Biology. 2000;297:733–745. doi: 10.1006/jmbi.2000.3605. [DOI] [PubMed] [Google Scholar]
- 52.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Advances in Protein Chemistry. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 53.Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Itzhaki LS, Neira JL, Fersht AR. Hydrogen exchange in chymotrypsin inhibitor 2 probed by denaturants and temperature. Journal of Molecular Biology. 1997;270:89–98. doi: 10.1006/jmbi.1997.1049. [DOI] [PubMed] [Google Scholar]
- 55.Neira JL, Mateu MG. Hydrogen exchange of the tetramerization domain of the human tumour suppressor p53 probed by denaturants and temperature. European Journal of Biochemistry. 2001;268:4868–4877. doi: 10.1046/j.1432-1327.2001.02414.x. [DOI] [PubMed] [Google Scholar]
- 56.Schutz C, Warshel A. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins-Structure, Function and Genetics. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- 57.Fitzkee NC, García-Moreno EB. Electrostatic effects in unfolded staphylococcal nuclease. Protein Science. 2008;17:216–227. doi: 10.1110/ps.073081708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhlman B, Luisi DL, Young P, Raleigh DP. pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry. 1999;38:4896–4903. doi: 10.1021/bi982931h. [DOI] [PubMed] [Google Scholar]
- 59.Chaires J. Possible origin of differences between van't Hoff and calorimetric enthalpy estimates. Biophysical Chemistry. 1997;64:15–23. doi: 10.1016/s0301-4622(96)02205-3. [DOI] [PubMed] [Google Scholar]
- 60.Prabhu NV, Sharp KA. Heat capacity in proteins. Annual Review of Physical Chemistry. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 61.Myers J, Pace N, Scholtz J. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Science. 1995;4(10):2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomez J, Hilser V, Xie D, Freire E. The heat-capacity of proteins. Proteins-Structure, Function and Genetics. 1995;22:404–412. doi: 10.1002/prot.340220410. [DOI] [PubMed] [Google Scholar]
- 63.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Science. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rees D, Robertson A. Some thermodynamic implications for the thermostability of proteins. Protein Science. 2001;10:1187–1194. doi: 10.1110/ps.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maxwell KL, Wildes D, Zarrine-Afsar A, De Los Rios MA, Brown AG, Friel CT, Hedberg L, Horng J-C, Bona D, Miller EJ, Vallée-Bélisle A, Main ERG, Bemporad F, Qiu L, Teilum K, Vu N-D, Edwards AM, Ruczinski I, Poulsen FM, Kragelund BB, Michnick SW, Chiti F, Bai Y, Hagen SJ, Serrano L, Oliveberg M, Raleigh DP, Wittung-Stafshede P, Radford SE, Jackson SE, Sosnick TR, Marqusee S, Davidson AR, Plaxco KW. Protein folding: defining a “standard” set of experimental conditions and a preliminary kinetic data set of two-state proteins. Protein Science. 2005;14:602–616. doi: 10.1110/ps.041205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, Zhou Y, Peng X, Yoon J. Fluorescent and colorimetric probes for detection of thiols. Chemical Society Reviews. 2010;39:2120–2135. doi: 10.1039/b925092a. [DOI] [PubMed] [Google Scholar]
- 67.Toyo'oka T, Imai K. Isolation and characterization of cysteine-containing regions of proteins using 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole and high-performance liquid chromatography. Analytical Chemistry. 1985;57:1931–1937. doi: 10.1021/ac00286a032. [DOI] [PubMed] [Google Scholar]
- 68.Englander SW, Mayne L, Bai Y, Sosnick TR. Hydrogen exchange: the modern legacy of Linderstrøm-Lang. Protein Science. 1997;6:1101–1109. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]