Abstract
In this study, the role of the double-stranded (ds) RNA-dependent protein kinase (PKR) in macrophage activation was examined. dsRNA [polyinosinic:polycytidylic acid (poly IC)]-stimulated inducible nitric oxide synthase, interleukin (IL)-1α and IL-1β mRNA expression, nitrite formation and IL-1 release are attenuated in RAW264.7 cells stably expressing dominant negative (dn) mutants of PKR. The transcriptional regulator nuclear factor (NF)-κB is activated by dsRNA, and appears to be required for dsRNA-induced macrophage activation. While dnPKR mutants prevent macrophage activation, they fail to attenuate dsRNA-induced IκB degradation or NF-κB nuclear localization. The inhibitory actions of dnPKR on dsRNA-induced macrophage activation can be overcome by treatment with interferon (IFN)-γ, an event associated with PKR degradation. Furthermore, dsRNA + IFN-γ stimulate inducible nitric oxide synthase expression, IκB degradation and NF-κB nuclear localization to similar levels in macrophages isolated from PKR–/– and PKR+/+ mice. These findings indicate that both NF-κB and PKR are required for dsRNA-induced macrophage activation; however, dsRNA-induced NF-κB activation occurs by PKR-independent mechanisms in macrophages. In addition, the PKR dependence of dsRNA-induced macrophage activation can be overcome by IFN-γ.
Keywords: dsRNA/IFN-γ/macrophage/PKR
Introduction
The antiviral response of infected cells includes the expression and release of type 1 interferons, which then stimulate the expression of the double-stranded (ds) RNA-dependent protein kinase (PKR; Meurs et al., 1990; Thomis et al., 1992; Tanaka and Samuel, 1994). PKR is a 68 kDa serine–threonine kinase that appears to play a primary role in mediating the antiviral activities of infected cells (Hovanessian, 1993; Gale and Katze, 1998). dsRNA produced during viral replication or viral RNA containing extended double-stranded secondary structure activates PKR (Hovanessian, 1993; Romano et al., 1995; Samuel et al., 1997; Gale and Katze, 1998). Binding to dsRNA induces PKR dimerization, autophosphorylation and activation (Wu and Kaufman, 1997). Once activated, PKR inhibits protein translation by phosphorylating the α-subunit of the initiation factor eIF2. Phosphorylation of eIF2α inhibits translation by sequestration of the guanine nucleotide exchange factor eIF2B. In addition, PKR has been shown to participate in transcriptional regulation of the E-selectin (Bandyopadhyay et al., 2000) and immunoglobulin κ light chain genes (Koromilas et al., 1995). Importantly, dsRNA-induced E-selectin expression is attenuated in endothelial cells isolated from PKR-deficient mice, suggesting that PKR is required for dsRNA-induced gene expression (Bandyopadhyay et al., 2000).
The transcriptional regulator nuclear factor κB (NF-κB) is activated by dsRNA, and this activation appears to be mediated by PKR. The NF-κB transcription factor family consists of c-Rel, p50 (NF-κB1), p52 (NF-κB2), p65 (RelA) and RelB. These factors are found in a variety of active homo- and heterodimers, the best characterized being the p50–p65 complex (Baeuerle and Henkel, 1994). NF-κB is found in the cytoplasm sequestered in a complex with inhibitor protein κB (IκB). Phosphorylation of IκB triggers ubiquitin-dependent degradation, and subsequent release of NF-κB (Liou and Baltimore, 1993). NF-κB is then free to translocate to the nucleus to activate gene transcription. IκB appears to be one substrate for PKR. In vitro phosphorylation studies have shown that PKR directly phosphorylates IκB (Kumar et al., 1994), and dsRNA-induced NF-κB nuclear localization is attenuated in mouse embryonic fibroblasts isolated from PKR-deficient mice (Yang et al., 1995; Kumar et al., 1997).
The antiviral response of macrophages includes the expression of inducible nitric oxide synthase (iNOS) and production of nitric oxide. Karupiah et al. (1993) first showed that interferon-γ (IFN-γ)-induced inhibition of viral replication requires macrophage production of nitric oxide, and viral infection has been shown to stimulate iNOS expression in both rodent and human macrophages (Bukrinsky et al., 1995; Kreil and Eibl, 1996). Macrophage expression of iNOS is dependent upon NF-κB activation. The 5′ untranslated region of the iNOS gene contains two NF-κB consensus sequences, and deletion of these sequences diminishes iNOS reporter activity in hepatocytes and macrophages (Diaz-Guerra et al., 1996; Kim et al., 1997). In addition, inhibition of NF-κB activation by treatment with the antioxidant pyrrolidinedithiocarbamate (PDTC) prevents macrophage expression of iNOS and production of nitric oxide in response to lipopolysaccharide (LPS) and LPS + IFN-γ (Mulsch et al., 1993). Recently, PDTC has been shown to prevent dsRNA and dsRNA + IFN-γ-induced iNOS expression, nitrite formation and interleukin (IL)-1 release by RAW264.7 cells and primary CD1 mouse peritoneal exudate cells (PEC), respectively (Heitmeier et al., 1998), suggesting that NF-κB may by required for viral-induced macrophage activation.
In this study, the role of PKR in dsRNA-induced macrophage activation has been examined. Using a dominant negative approach, evidence is presented indicating that dsRNA-induced iNOS and IL-1 expression by RAW264.7 macrophages is dependent on functional PKR. While dominant negative (dn) PKR expression prevents dsRNA-induced macrophage activation, it does not prevent dsRNA-induced IκB degradation or NF-κB nuclear localization. The inhibitory actions of dnPKR on dsRNA-induced macrophage activation can be overcome by co-treatment with IFN-γ. In addition, the genetic absence of PKR in primary macrophages does not affect the ability of dsRNA + IFN-γ to stimulate iNOS expression, IL-1 release or NF-κB activation. These findings indicate that dsRNA-induced RAW264.7 cell activation is PKR dependent, and that stimulation of a second signaling pathway, in response to IFN-γ, can overcome the inhibitory actions of dnPKR or the genetic absence of PKR on dsRNA-induced macrophage activation.
Results
Overexpression of dnPKR prevents dsRNA-induced iNOS expression by RAW264.7 macrophages
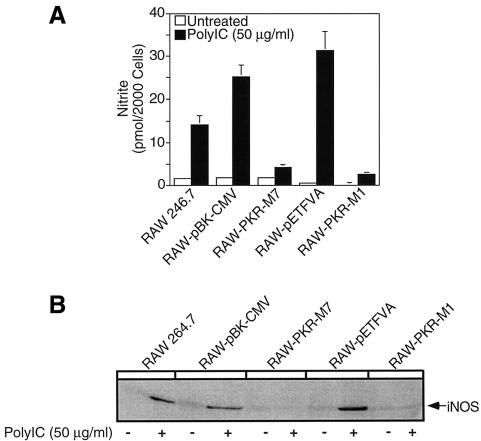
The role of PKR in dsRNA-induced macrophage activation was examined using RAW264.7 macrophages stably expressing the dn PKR mutants, PKR-M7, which lacks the first dsRNA-binding domain (Barber et al., 1992), and PKR-M1, which contains a K296P point mutation at the ATP-binding site (Wu and Kaufman, 1996). These well-characterized dnPKR mutants have been shown to prevent dsRNA-induced PKR activation in a number of cell types (Barber et al., 1992, 1995a, 1995b; Meurs et al., 1992, 1993; Lee et al., 1994; Wu and Kaufman, 1996, 1997; Srivastava et al., 1998). Incubation of RAW264.7 cells with 50 µg/ml polyinosinic:polycytidylic acid (poly IC) stimulates nitric oxide production and iNOS protein expression following a 24 h exposure, and iNOS mRNA accumulation following a 6 h incubation (Figure 1A–C). Poly IC fails to stimulate nitrite formation in RAW264.7 cells stably expressing the dnPKR mutants PKR-M1 or PKR-M7. Consistent with a lack of nitric oxide production, poly IC-induced iNOS protein expression and iNOS mRNA accumulation by RAW264.7 cells expressing PKR-M1 or PKR-M7 following 24 and 6 h incubations, respectively (Figure 1B and C), are attenuated as compared with the levels expressed in RAW264.7 cells. Also, poly IC stimulates iNOS mRNA accumulation, protein expression and nitric oxide production by RAW264.7 cells stably expressing the vector controls pBK-CMV and pETFVA (Figure 1A–C and data not shown). Western blot analysis was used to confirm a 2-fold increase in PKR-M1 expression by RAW-PKR-M1 cells as compared with RAW264.7 and vector control RAW-pETFVA cells (Figure 1D). Because the monoclonal antibody used to detect PKR and PKR-M1 protein expression failed to cross-react with the truncated form of human PKR, RT–PCR analysis was used to confirm expression of mutant PKR-M7 (Figure 1E).
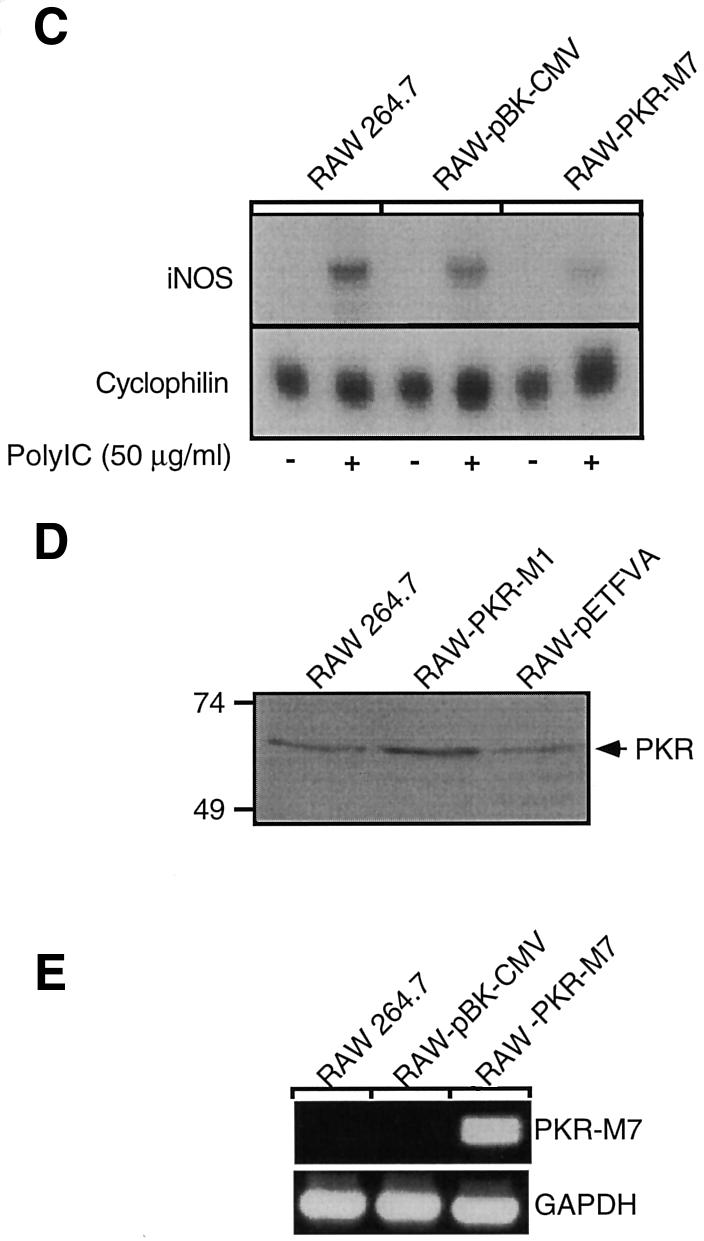
Fig. 1. Effects of dnPKR on poly IC-induced nitric oxide production and iNOS expression by RAW264.7 cells. RAW264.7, RAW-pBK-CMV and RAW-pETFVA (vector controls), or dnPKR-expressing RAW-PKR-M7 and RAW-PKR-M1 cells (4 × 105 cells/400 µl complete CMRL-1066) were treated with or without 50 µg/ml poly IC for 24 h at 37°C. Nitrite production was determined on the cell culture supernatant (A), and iNOS expression was examined on the isolated cells by western blot analysis (B). (C) Total RNA was isolated from RAW264.7, RAW-pBK-CMV or RAW-PKR-M7 cells (5 × 106 cells/3 ml complete CMRL-1066) following a 6 h incubation with or without poly IC, and iNOS mRNA accumulation was determined by northern blot analysis. To control for mRNA loading, cyclophilin mRNA accumulation is also shown. (D) SDS samples were prepared from RAW264.7, RAW-pETFVA and RAW-PKR-M1 cells, and dnPKR expression was examined by SDS gel electrophoresis and western blot analysis. Note that RAW-PKR-M1 cells express ∼2-fold higher levels of PKR (as determined by densitometry) as compared with vector control and wild-type RAW264.7 cells. (E) Total RNA isolated from RAW264.7, RAW-pBK-CMV and RAW-PKR-M7 was used to confirm dnPKR-M7 mRNA accumulation by RT–PCR. GAPDH mRNA accumulation is also shown. Results for nitrite production are the average ± SEM of three independent experiments; iNOS and PKR protein expression and mRNA accumulation are representative of three independent experiments.
Effects of dnPKR on poly IC-induced IL-1 expression and release
In an activated state macrophages express and release the proinflammatory cytokine IL-1. The effects of dnPKR on dsRNA-induced IL-1 expression and release were examined by northern blot analysis and the RINm5F cell IL-1 bioassay, respectively. Treatment of RAW264.7 cells or the vector control cells, RAW-pBK-CMV and RAW-pETFVA, for 24 h with 50 µg/ml poly IC results in an ∼16.5-fold increase in the release of IL-1 (Figure 2A). The release of IL-1 is significantly attenuated in cells expressing dnPKR, as poly IC fails to stimulate IL-1 release from RAW-PKR-M7 and RAW-PKR-M1 cells. The inhibitory actions of dnPKR on IL-1 release appear to reflect an inhibition of IL-1 mRNA accumulation. Poly IC-induced IL-1α and IL-1β mRNA accumulation is attenuated in RAW-PKR-M7 cells as compared with the levels that accumulate in RAW264.7 cells and the vector control RAW-pBK-CMV (Figure 2B). Similar results were obtained using RAW264.7 cells expressing the dnPKR point mutant PKR-M1 (data not shown). These findings suggest that PKR is required for poly IC-induced IL-1 expression and release by macrophages.
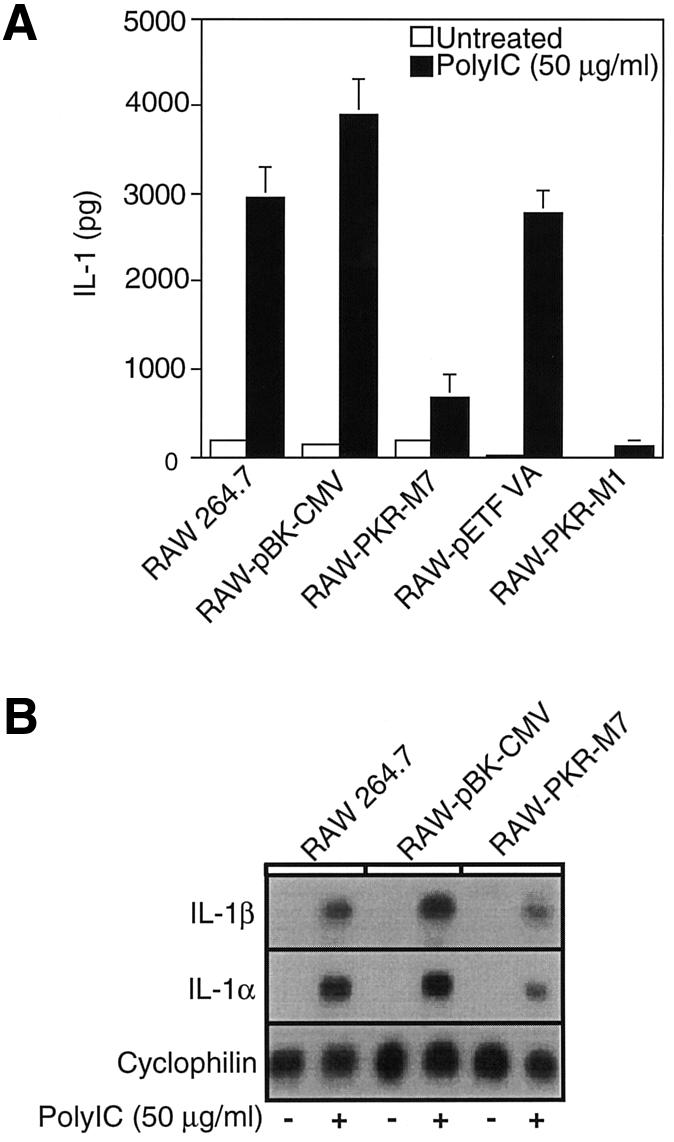
Fig. 2. Effects of dnPKR expression on poly IC-induced IL-1 expression and release from RAW264.7 cells. (A) Wild-type RAW264.7 cells, vector control RAW-pBK-CMV, RAW-pETFVA cells and dnPKR-expressing RAW-PKR-M7 and RAW-PKR-M1 cells (4 × 105 cells/400 µl complete CMRL-1066) were treated with or without 50 µg/ml poly IC for 24 h at 37°C. IL-1 released into the culture medium was determined using the RINm5F cell IL-1 bioassay. (B) RAW264.7, RAW-pBK-CMV or RAW-PKR-M7 cells (5 × 106 cells/3 ml complete CMRL-1066) were treated with or without 50 µg/ml poly IC for 6 h at 37°C, total RNA was isolated, and IL-1α and IL-1β mRNA accumulation was examined by northern blot analysis. Cyclophilin mRNA accumulation is shown as an internal RNA loading control. Results are the average ± SEM of three independent experiments for IL-1 release and representative of three independent experiments for IL-1 mRNA expression.
Effects of dnPKR on IκB degradation and NF-κB nuclear translocation
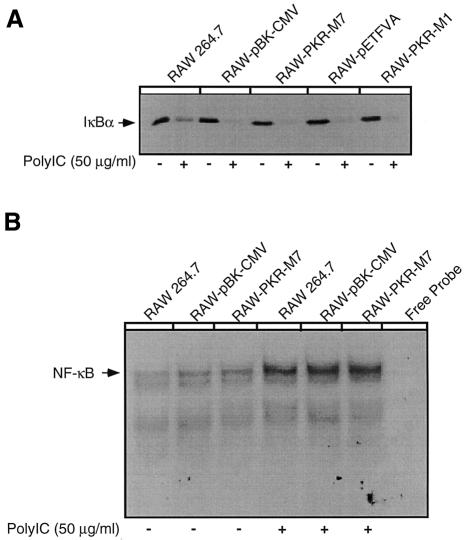
Since dsRNA-induced iNOS expression by macrophages requires NF-κB activation (Heitmeier et al., 1998), and IκB appears to be a substrate for PKR (Kumar et al., 1994, 1997; Yang et al., 1995), the effects of dnPKR expression on dsRNA-induced NF-κB nuclear localization and IκB degradation were examined. Consistent with previous studies (Heitmeier et al., 1998), treatment of RAW264.7 cells, or RAW cells expressing the vector controls pBK-CMV and pETFVA, with 50 µg/ml poly IC for 30 min results in IκB degradation (Figure 3A) and NF-κB nuclear translocation (Figure 3B). Surprisingly, poly IC-induced IκB degradation is not modulated by the expression of PKR-M1 or PKR-M7, as >85% (as determined by densitometry) of the cellular content of IκB is degraded in response to poly IC in both the vector control and dnPKR-expressing RAW264.7 cells. Also, expression of PKR-M7 does not impair poly IC-induced NF-κB nuclear localization (Figure 3B). Similar results were obtained using RAW-PKR-M1 cells (data not shown). These findings suggest that PKR is not required for dsRNA-induced NF-κB activation in RAW264.7 macrophages. To control for non-specific probe binding, excess (100-fold) cold NF-κB oligonucleotide inhibits the formation of the NF-κB–probe complex, and inclusion of antiserum specific for the p50 and p65 subunits of NF-κB reduces the mobility of the NF-κB–probe complex (supershift, data not shown).
Fig. 3. Effects of dnPKR expression on IκB degradation and NF-κB nuclear localization. (A) RAW264.7, vector control RAW-pBK-CMV and RAW-pETFVA, or dnPKR-expressing RAW-PKR-M7 and RAW-PKR-M1 cells (4 × 105 cells/400 µl complete CMRL-1066) were treated with or without 50 µg/ml poly IC for 30 min at 37°C. The cells were isolated and IκB levels were determined by western blot analysis. (B) RAW264.7, RAW-pBK-CMV and RAW-PKR-M7 cells (5 × 106 cells/3 ml complete CMRL-1066) were treated with or without 50 µg/ml poly IC for 30 min at 37°C. Nuclear extracts were isolated and gel shift analysis performed. Results are representative of seven independent experiments for IκB degradation (A) and three independent experiments for NF-κB nuclear localization (B).
IFN-γ overcomes the inhibitory effects of dnPKR on dsRNA-induced macrophage activation
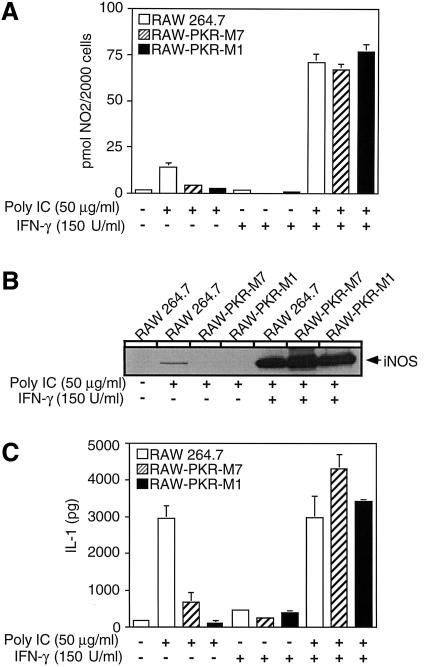
Alone, poly IC stimulates an ∼10-fold increase in nitrite production and iNOS expression by RAW264.7 cells. However, in the presence of IFN-γ (150 U/ml), the stimulatory effects of poly IC on macrophage production of nitrite are dramatically enhanced from ∼10- to ∼40-fold (Figure 4) (Heitmeier et al., 1998). IFN-γ also significantly enhances the level of iNOS protein expressed in response to poly IC (Figure 4B). Alone, IFN-γ fails to stimulate iNOS expression or nitric oxide formation by RAW264.7 cells (data not shown and Figure 4A, respectively). In contrast to the inhibitory actions of dnPKR on poly IC-induced macrophage activation, dnPKR does not prevent the stimulatory effects of poly IC + IFN-γ on iNOS expression and nitric oxide formation. Treatment of RAW-PKR-M1 and RAW-PKR-M7 cells with poly IC + IFN-γ results in the expression of iNOS and the production of nitrite to levels similar in magnitude to those produced by RAW264.7 cells.
Fig. 4. IFN-γ overcomes the inhibitory actions of dnPKR on dsRNA-induced iNOS expression, nitric oxide production and IL-1 release by RAW264.7 macrophages. RAW264.7, RAW-PKR-M7 and RAW-PKR-M1 cells (4 × 105 cells/400 µl complete CMRL-1066) were treated with or without 50 µg/ml poly IC and/or 150 U/ml IFN-γ for 24 h at 37°C. Nitrite production was determined on culture supernatants (A) and the cells were isolated for western blot analysis of iNOS expression (B). (C) IL-1 released into the culture supernatant was determined using the RINm5F cell IL-1 bioassay. Results for nitrite production and IL-1 release are the average ± SEM of three independent experiments. Results for iNOS expression are representative of three independent experiments.
IFN-γ also prevents the inhibitory effects of dnPKR expression on dsRNA-induced IL-1 release by RAW264.7 cells. Poly IC alone stimulates IL-1 release by RAW264.7 cells, and this stimulatory action is attenuated in dnPKR-expressing RAW-PKR-M7 and RAW-PKR-M1 cells (Figure 4C). Alone, IFN-γ does not stimulate IL-1 release, nor does it enhance or reduce the levels of IL-1 released by RAW264.7 cells in response to poly IC. However, in the presence of IFN-γ, dsRNA stimulates the release of IL-1 to similar levels in RAW264.7 cells and RAW264.7 cells expressing PKR-M1 and PKR-M7. These findings indicate that IFN-γ overcomes the inhibitory actions of dnPKR on IL-1 release, nitric oxide production and iNOS expression.
Effects of IFN-γ on PKR expression by RAW264.7 cells
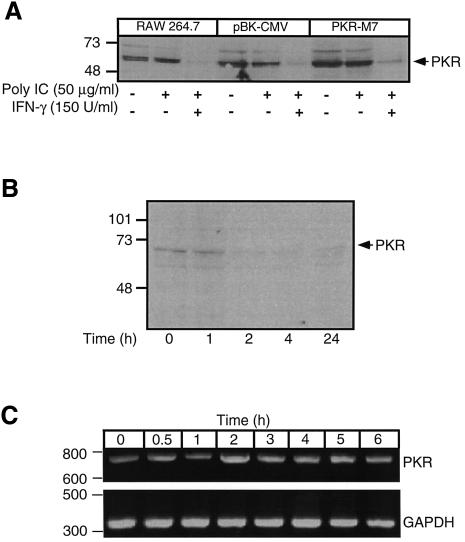
Previous studies have shown that IFNs stimulate PKR expression (Roberts et al., 1976; Galabru and Hovanessian, 1985; Tanaka and Samuel, 1994). Therefore, it is possible that the ability of IFN-γ to overcome the inhibitory actions of dnPKR on macrophage activation may be associated with increased expression of endogenous PKR by RAW264.7 macrophages. To examine this possibility, RAW264.7, RAW-pBK-CMV and RAW-PKR-M7 cells were treated for 24 h with 50 µg/ml poly IC alone or in combination with 150 U/ml IFN-γ. Surprisingly, treatment of each of these cell types with poly IC + IFN-γ results in the loss of PKR immunoreactivity (Figure 5A). IFN-γ appears to be the stimulus for the loss of PKR immunoreactivity, as poly IC alone does not alter PKR expression, nor does it prevent the loss of PKR immunoreactivity in the presence of IFN-γ. Alone, IFN-γ stimulates a >90% loss of PKR immunoreactivity that is first apparent and complete following a 2 h incubation (Figure 5B). PKR degradation is not associated with changes in PKR mRNA expression, as similar levels of PKR mRNA accumulate following treatment with or without IFN-γ (Figure 5C). These findings correlate the ability of IFN-γ to overcome the inhibitory actions of dnPKR on poly IC-induced iNOS expression, nitric oxide formation and IL-1 release with the degradation of PKR in RAW264.7 cells.
Fig. 5. IFN-γ stimulates PKR degradation in RAW264.7 cells. (A) RAW264.7, RAW-pBK-CMV or dnPKR-expressing RAW-PKR-M7 cells (4 × 105 cells/400 µl complete CMRL-1066) were treated with or without 50 µg/ml poly IC and/or 150 U/ml IFN-γ as indicated. The cells were isolated and PKR levels were evaluated by western blot analysis. (B) RAW264.7 cells (4 × 105 cells/400 µl complete CMRL-1066) were treated for the times indicated with 150 U/ml IFN-γ, the cells were isolated and PKR levels examined by western blot analysis. (C) RAW264.7 cells were incubated with 150 U/ml IFN-γ for the times indicated followed by RT–PCR analysis of PKR mRNA accumulation. Results for (A–C) are representative of three independent experiments.
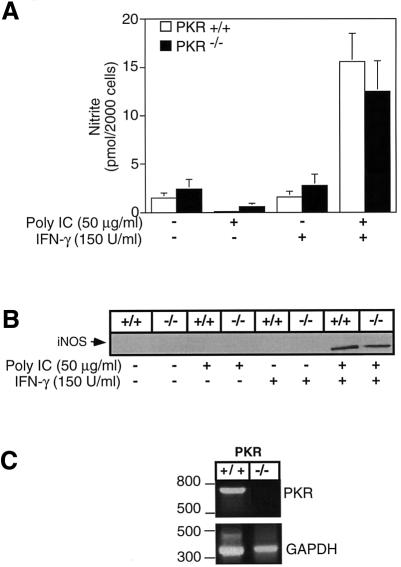
Effects of poly IC and IFN-γ on iNOS expression and nitric oxide production by peritoneal macrophages isolated from PKR–/– mice
While dsRNA alone is sufficient to stimulate iNOS expression, nitric oxide production and IL-1 release from the macrophage cell line RAW264.7, a combination of two proinflammatory signals is required to activate primary mouse peritoneal macrophages (Hibbs et al., 1990). Previous studies have shown that in combination, poly IC and IFN-γ are potent activators of CD-1 mouse PEC, stimulating iNOS expression, nitric oxide production and IL-1 release (Heitmeier et al., 1998). To determine if PKR is required for dsRNA + IFN-γ-stimulated iNOS expression, mouse macrophages were isolated from wild-type (PKR+/+) and PKR-deficient (PKR–/–) mice. Alone, neither poly IC nor IFN-γ stimulate nitrite formation or iNOS expression; however, in combination, poly IC + IFN-γ stimulates the expression of iNOS and production of nitrite to levels similar to those from macrophages isolated from PKR+/+ and PKR–/– mice (Figure 6A and B). RT–PCR and Southern blot analysis (Figure 6C and data not shown, respectively) were used to confirm the expression of PKR in PKR+/+ mice and the absence of PKR in PKR–/– mice.
Fig. 6. Effects of poly IC + IFN-γ on iNOS expression and nitric oxide production by peritoneal macrophages isolated from PKR–/– mice. Peritoneal macrophages (4 × 105 cells/400 µl complete CMRL-1066), isolated from PKR+/+ and PKR–/– mice, were treated with 50 µg/ml poly IC and/or 150 U/ml IFN-γ for 24 h at 37°C. Nitrite production was determined on the culture supernatants (A) and iNOS expression was determined by western blot analysis of the isolated cells (B). (C) PKR mRNA accumulation in macrophages isolated from PKR+/+ and PKR–/– mice was determined by RT–PCR. Results for nitrite production are the average ± SEM of three independent experiments, and for iNOS expression and PKR mRNA accumulation are representative of three independent experiments.
Results presented in Figure 3 indicate that dnPKR expression does not prevent poly IC-induced IκB degradation by RAW264.7 cells. Consistent with these findings, poly IC also stimulates a similar time-dependent degradation of IκB in macrophages isolated from both PKR–/– and PKR+/+ mice (Figure 7A). Following a 15 min incubation with poly IC, >85% of IκB is degraded in macrophages isolated from both PKR–/– and PKR+/+ mice. The levels of immunoreactive IκB begin to return to basal levels following 3–4 h exposures to poly IC. In addition, following a 30 min incubation, poly IC stimulates NF-κB nuclear localization in macrophages isolated from PKR–/– and PKR+/+ mice (Figure 6B). These findings indicate that PKR does not appear to be required for dsRNA + IFN-γ-induced iNOS expression, NF-κB nuclear localization and IκB degradation in mouse peritoneal macrophages.
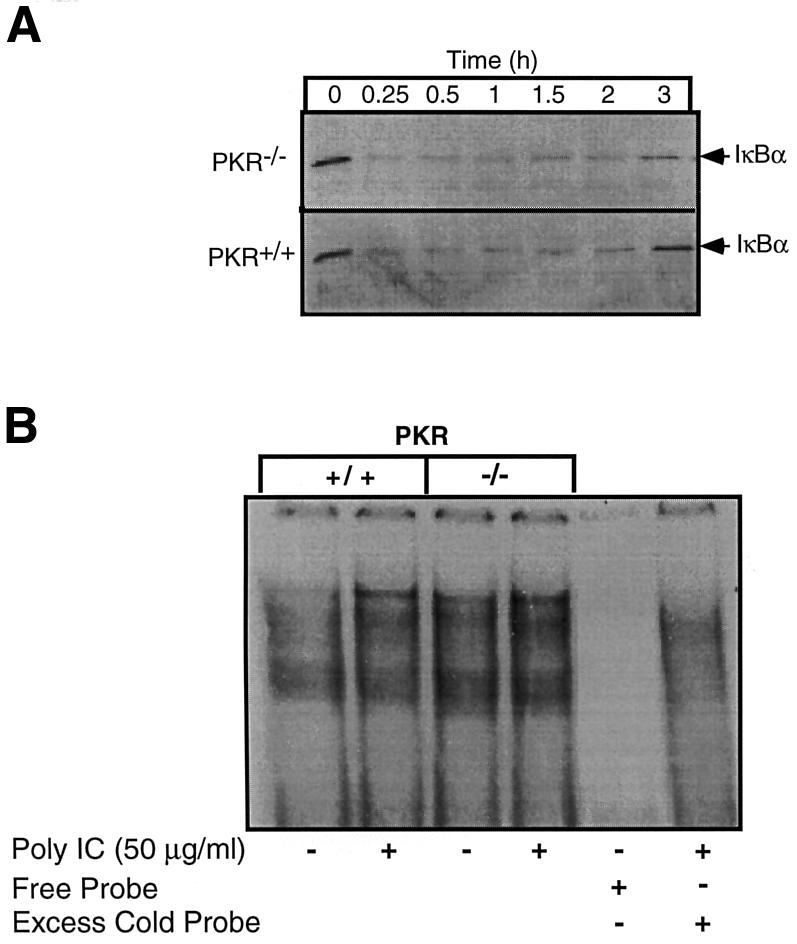
Fig. 7. Effects of poly IC on IκB degradation and NF-κB nuclear localization in macrophages isolated from PKR–/– mice. Peritoneal macrophages (4 × 105 cells/400 µl complete CMRL-1066) were isolated from PKR+/+ and PKR–/– mice and treated with 50 µg/ml poly IC for the times indicated. The cells were isolated and IκB degradation determined by western blot analysis (A). (B) Peritoneal macrophages (5 × 106 cells/400 µl complete CMRL-1066) isolated from PKR+/+ and PKR–/– mice were treated with 50 µg/ml poly IC for 30 min, nuclear extracts prepared, and NF-κB binding activity determined by gel shift analysis. Results for (A) and (B) are representative of two independent experiments.
Discussion
In this study, the role of PKR in dsRNA-induced macrophage activation was examined. Treatment of the macrophage cell line RAW264.7 with dsRNA (poly IC) results in iNOS and IL-1 expression, nitrite production and IL-1 release (Heitmeier et al., 1998). However, in two RAW264.7 cell lines stably expressing dominant negative forms of PKR (RAW-PKR-M1, K296P point mutant, and RAW-PKR-M7, first dsRNA-binding domain deletion mutant) dsRNA-induced IL-1α, IL-1β and iNOS mRNA accumulation, nitrite production and IL-1 release are attenuated. These findings support a role for PKR in the dsRNA-induced iNOS and IL-1 expression by RAW264.7 macrophages.
dsRNA-induced macrophage activation appears to require IκB degradation and NF-κB nuclear localization (Heitmeier et al., 1998). We have shown that PDTC prevents dsRNA-induced NF-κB nuclear localization, IκB degradation and iNOS expression by RAW264.7 cells and CD1 mouse macrophages (Heitmeier et al., 1998). PKR has been shown to activate NF-κB, either by directly phosphorylating IκB, as demonstrated by in vitro kinase assays (Kumar et al., 1994), or indirectly by activating IκB kinase (IKK) (Chu et al., 1999). In mouse embryonic fibroblasts and endothelial cells isolated from PKR–/– mice, dsRNA-induced NF-κB activation is attenuated or significantly delayed (Yang et al., 1995; Kumar et al., 1997). In contrast to these previous studies, we show that dsRNA stimulates a similar time-dependent degradation of IκB and the nuclear localization of NF-κB in RAW264.7 cells, and RAW264.7 cells expressing PKR-M1 and PKR-M7. This finding was confirmed using PKR-deficient macrophages, where dsRNA stimulates a similar time-dependent degradation of IκB, and NF-κB nuclear localization from PEC isolated from PKR+/+ and PKR–/– mice. These results suggest that dsRNA-induced NF-κB activation in macrophages does not require PKR, while dsRNA-induced iNOS and IL-1 expression appears to be NF-κB dependent. The reasons for the differences in dsRNA responsiveness of macrophages as compared with endothelial cells/fibroblasts isolated from PKR-deficient mice are unknown. It is possible, although unlikely, that NF-κB activation in RAW264.7 cells expressing the dnPKR point mutant M1 (K296P) may be associated with the direct activation of IKK by PKR-M1. Chu et al. (1999) have shown that the catalytically inactive dnPKR point mutant K296R can directly activate IKK in a concentration-dependent manner, where low levels of dnPKR expression fail to activate IKK, but high levels of dnPKR stimulate IKK activity. While Chu’s studies provide an alternative explanation for NF-κB activation in RAW264.7 cells expressing the dnPKR point mutant M1 (K296P), these studies are not consistent with our findings that dsRNA stimulates IκB degradation and NF-κB nuclear localization in macrophages isolated from PKR–/– mice. Our findings suggest that dsRNA activates macrophages by at least two pathways: (i) a PKR-dependent pathway that is required for IL-1 and iNOS expression and is independent of NF-κB activation; and (ii) a PKR-independent pathway resulting in the activation of NF-κB.
While dsRNA alone stimulates RAW264.7 cell expression of iNOS and production of nitric oxide, the levels produced are potentiated in the presence of IFN-γ. Importantly, in the presence of IFN-γ, dnPKR fails to prevent dsRNA-induced iNOS expression and nitric oxide production by RAW264.7 macrophages. Also, in the presence of IFN-γ, dnPKR fails to prevent dsRNA-induced IL-1 release by RAW264.7 cells. Consistent with our findings, Gusella et al. (1995) have shown that IFN-γ-stimulated tumoricidal activity of elicited mouse macrophages is not sensitive to the PKR inhibitor 2-aminopurine, while this inhibitor prevents LPS- and IFN-αβ-induced tumoricidal activity. We originally believed that the ability of IFN-γ to overcome the inhibitory actions of dnPKR on dsRNA-induced macrophage activation might be associated with an increase in the expression of endogenous PKR. However, IFN-γ treatment of RAW264.7 cells results in PKR degradation. IFN-γ-induced PKR degradation is not modulated by dsRNA, and dsRNA alone does not stimulate PKR degradation. Poliovirus infection has been shown to stimulate PKR degradation in HeLa cells by a mechanism that is independent of the poliovirus proteases 2A or 3C (Black et al., 1989, 1993). Also, poliovirus-stimulated PKR degradation is largely prevented by poly IC. Since poly IC does not modulate IFN-γ-induced PKR degradation, it is unlikely that the mechanisms for the loss of PKR in response to the two treatments are similar. Serine protease activation may be associated with IFN-γ-induced PKR degradation. In preliminary experiments we have shown that Nα-P-tyosyl-l-lysine chloromethyl ketone (a selective serine protease inhibitor) prevents IFN-γ-induced PKR degradation (unpublished observation). However, we have yet to identify this IFN-γ activated protease.
In contrast to RAW264.7 cells, activation of naive mouse peritoneal macrophages requires two signals, the classical stimuli being the combination of LPS and IFN-γ (Hibbs et al., 1990). Consistent with a requirement for two stimuli, alone neither dsRNA nor IFN-γ stimulates macrophage activation. However, in combination, dsRNA + IFN-γ stimulate iNOS expression, nitrite production and IL-1 release by primary mouse PEC (Heitmeier et al., 1998). Importantly, treatment of macrophages isolated from PKR–/– mice with dsRNA + IFN-γ results in the expression of iNOS, production of nitrite and release of IL-1 to levels comparable to those produced by macrophages isolated from PKR+/+ mice. These results are consistent with the ability of IFN-γ to overcome the inhibitory actions of dnPKR on dsRNA-induced activation of RAW264.7 cells, and indicate that dsRNA + IFN-γ-induced primary mouse macrophage activation may occur by mechanisms that are independent of PKR. These findings also dissociate the ability of IFN-γ to stimulate macrophage activation from the ability of this cytokine to stimulate PKR degradation, as PKR–/– mice fail to express PKR.
In summary, the findings presented in this study show that dsRNA-induced macrophage activation is dependent on PKR, but that the PKR dependence can be overcome by the addition of IFN-γ. dsRNA appears to activate macrophages by: (i) stimulating a PKR-dependent signaling pathway that is independent of NF-κB activation; and (ii) stimulating NF-κB activation that is independent of PKR. Also, by stimulating PKR degradation, IFN-γ may participate in the regulation of PKR-dependent antiviral activities of macrophages.
Materials and methods
Materials and animals
RAW264.7 cells, RINm5F cells and Dulbecco's modified Eagle (DME) media (DME containing 10% heat-inactivated fetal calf serum, 1× l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin) were obtained from Washington University Tissue Culture Support Center (St Louis, MO). CMRL-1066 tissue culture medium, l-glutamine, penicillin, streptomycin, mouse recombinant IFN-γ, Lipofectin reagent and geneticin (G418) were obtained from Gibco-BRL (Grand Island, NY). Poly IC was purchased from Sigma Chemical Co. (St Louis, MO). Poly IC was prepared by heating to 50°C in filter-sterilized Tris–EDTA buffer pH 7.4 containing 15 mM NaCl. Once solubilized, poly IC was slowly cooled to room temperature to allow for renaturation and then stored at –20°C prior to use. [α-32P]dCTP, [γ-32P]dATP and enhanced chemiluminescence (ECL) reagents were purchased from Amersham Pharmacia Biotech (Piscataway, NJ). The pBK-CMV mammalian expression vector was purchased from Stratagene (La Jolla, CA). NF-κB consensus oligonucleotide, rabbit anti-IκB, rabbit anti-p50 and goat anti-p65 antisera were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Horseradish peroxidase-conjugated donkey anti-rabbit IgG was purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Rabbit antiserum specific for the C-terminal 27 amino acids of mouse macrophage iNOS was a gift from Dr Thomas Misko (G.D.Searle, St Louis, MO). The dnPKR construct, PKR-M7, which lacks the first dsRNA-binding domain, was a gift from Dr M.G.Katze (University of Washington School of Medicine, Seattle, WA). The pETFVA–K296P dnPKR construct, which contains the K296P point mutation in the ATP binding site, rabbit anti-PKR antiserum, and PKR–/– mice [C57BL/6(J)×129/SV background] have been described (Yang et al., 1995; Wu and Kaufman, 1997). C57BL/6(J) wild-type mice (PKR+/+, 20–24 g) were purchased from Harlan (Indianapolis, IN). iNOS and cyclophilin cDNAs were gifts from Dr Charles Rodi (Monsanto Corporate Research, St Louis, MO) and Dr Steve Carroll (Department of Pathology, University of Alabama-Birmingham), respectively. IL-1α and IL-1β cDNAs were gifts from Dr Cliff Bellone (Saint Louis University, St Louis, MO). All other reagents were obtained from commercially available sources.
Transfections
RAW264.7 cells were transfected using Lipofectin reagent according to the manufacturer’s specifications creating four new cells lines: (i) RAW-pBK-CMV, which contains the pBK-CMV vector; (ii) RAW-PKR-M7, which contains the PKR-M7 mutant in the pBK-CMV expression vector; (iii) RAW-pETFVA, which contains the pETFVA expression vector and the pBK-CMV vector (required for stable selection); and (iv) RAW-PKR-M1, which contains the K296P mutant in the pETFVA expression vector and the pBK-CMV selection vector. Briefly, RAW264.7 cells were plated at 2 × 105 cells/3 ml complete CMRL-1066 (CMRL-1066 containing 2 mM l-glutamine, 10% heat-inactivated fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin) and allowed to grow to ∼50% confluence (∼24 h at 37°C under an atmosphere of 95% air/5% CO2). Plasmid DNA (2 µg), pBK-CMV, pBK-CMV-M7, or pETFVA and pETFVA–K296 in combination with 1 µg of pBK-CMV was incubated in 100 µl of CMRL-1066 (containing 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin) for 30 min at room temperature, 8 µg of Lipofectin were added and the mixture was incubated for an additional 15 min at room temperature followed by the addition of 800 µl of CMRL-1066. The cells were washed once with CMRL-1066, and the DNA–Lipofectin mixtures were added. Following a 24 h incubation at 37°C, the medium was replaced with DME and the RAW264.7 cells were cultured at 37°C for an additional 48 h. The cells were removed from growth flasks by treatment with 0.05% trypsin, 0.02% EDTA, and transferred to T-25 flasks in DME containing 400 µg/ml G418 for selection of stably transfected cells. The cells were maintained in DME containing 200 µg/ml G418.
Peritoneal macrophage isolation and cell culture
PEC were isolated from PKR-deficient C57BL/6(J) × 129/SV mice (PKR–/–) (Kumar et al., 1994; Yang et al., 1995; Der et al., 1997) and C57BL/6(J) wild-type mice by lavage as previously described (Beckerman et al., 1993). Following isolation, 4 × 105 cells/400 µl complete CMRL-1066 were incubated at 37°C under an atmosphere of 95%/air 5% CO2 for 3 h. Cells were washed three times with complete CMRL-1066 to remove non-adherent cells before treating with poly IC and/or IFN-γ as indicated in the figure legends. To control for potential strain differences, the effects of dsRNA + IFN-γ on iNOS expression and nitric oxide production were compared between macrophages isolated from C57BL/6 × 129 (Jackson Laboratories, Bar Harbor, ME) and C57BL/6 mice, and no differences were observed (data not shown).
RAW264.7 cells and RINm5F cells were removed from growth flasks by treatment with 0.05% trypsin, 0.02% EDTA at 37°C. Cells were washed twice with complete CMRL-1066, plated at the cell concentration indicated, and cultured for 2–3 h prior to initiation of experiments.
Polymerase chain reaction
Total RNA (2.5 µg), isolated from RAW264.7, RAW-pBK-CMV and RAW-PKR-M7 cells using the Qiagen RNeasy kit according to the manufacturer’s instructions, was used to prepare a first-strand cDNA library using the Superscript Preamplification System from Gibco-BRL (Grand Island, NY) according to the manufacturer’s specifications. A standard 25 µl PCR was performed as previously described (Arnush et al., 1998). PCR primers for human PKR were forward primer, 5′-GGTACAGGTTCTACTAAACAGG-3′, reverse primer, 5′-GAAAACTTGGCCAAATCCACC-3′ (PCR product size = 413 bp); and murine PKR, forward primer, 5′-GCCAGATGCACGGAGTAGCC-3′, reverse primer, 5′-GAAAACTTGGCCAAATCCACC-3′ (PCR product size = 722 bp). The human and murine PKR PCR products were confirmed by restriction digestion with BglII and PstI (Promega, Madison, WI), respectively. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers have been described previously (Arnush et al., 1998), and yield a PCR product of 343 bp. The PCRs used the following profile: an initial denaturing step of 94°C for 2 min, 28 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 1 min 15 s. The samples were then incubated at 30°C for 2 min. Two microliters of 10× PCRs loading dye (25% Ficoll 400, 0.5% xylene cyanole) were added and 20 µl of each sample were separated on 1.5% agarose gels containing 0.05 µg/ml ethidium bromide and products were visualized by UV exposure.
Nitrite and IL-1 determination
Nitrite production was determined by mixing 50 µl of culture medium with 50 µl of Greiss reagent as described previously (Green et al., 1982). The absorbance at 540 nm was measured and nitrite concentrations were calculated from a sodium nitrite standard curve. IL-1 release was determined using the RINm5F cell bioassay as described previously (Hill et al., 1996).
Western blot analysis
Protein samples were separated by SDS–PAGE and transferred to Nitrocell nitrocellulose membranes (Amersham Pharmacia Biotech, Piscataway, NJ) under semidry transfer conditions as previously described (Laemmli, 1970; Heitmeier et al., 1997). Antibody dilutions were: rabbit anti-mouse iNOS, 1:2000; rabbit anti-human IκB, 1:1000; murine anti-human PKR, 1:1000; horseradish peroxidase-conjugated donkey anti-rabbit, 1:7000; and donkey anti-mouse, 1:5000. Antigen was detected by ECL according to the manufacturer’s specifications.
Northern blot analysis
Total RNA (5 µg) was denatured and fractionated by electrophoresis on 1% agarose gels containing 2.2 M formaldehyde. RNA was transferred by capillary action in 20× SSC (3 M NaCl, 0.3 M sodium citrate pH 7.0) to Duralon UV nylon membranes (Stratagene, La Jolla, CA), and the membranes were hybridized to 32P-labeled probes specific for iNOS, IL-1α, IL-1β and cyclophilin. The DNA probes were radiolabeled with [α-32P]dCTP by random priming using the Prime-a-Gene nick translation system from Promega (Madison, WI). The iNOS, IL-1α and IL-1β cDNA probes have been described previously (Godambe et al., 1993; Heitmeier et al., 1998). Cyclophilin was used as an internal RNA loading control. Hybridization and autoradiography were performed as described previously (Heitmeier et al., 1998).
Nuclear protein isolation and gel shift analysis
Nuclear extracts were prepared from RAW264.7, RAW-pBK-CMV, RAW-PKR-M7 cells and PEC isolated from PKR–/– and PKR+/+ mice as described previously (Heitmeier et al., 1998). Gel shift analysis of NF-κB binding to an end-labeled radiolabeled NF-κB consensus DNA oligonucleotide (5′-AGTTGAGGGGACTTTCCCAGGC-3′) was performed as described previously (Heitmeier et al., 1998).
Densitometry and image analysis
Autoradiograms were scanned into NIH Image version 1.59 using a COHU high performance CCD camera (Brookfield, WI) and densities were determined using NIH Image version 1.59 software.
Acknowledgments
Acknowledgements
We thank Jessica Gorman, Joe Martin and Tracey Baird for expert technical assistance and Dr Michael Moxley for critical review of this manuscript. This work was supported by National Institutes of Health Grants DK52194 and AI44458 (J.A.C.), AI42394 (R.J.K.) and a Career Development Award from the Juvenile Diabetes Foundation International (to J.A.C.).
References
- Arnush M., Scarim,A.L., Heitmeier,M.R., Kelly,C.B. and Corbett,J.A. (1998) Potential role of resident islet macrophage activation in the initiation of autoimmune diabetes. J. Immunol., 160, 2684–2691. [PubMed] [Google Scholar]
- Baeuerle P.A. and Henkel,T. (1994) Function and activation of NF-κB in the immune system. Annu. Rev. Immunol., 12, 141–179. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S.K., de La Motte,C.A. and Williams,B.R. (2000) Induction of E-selectin expression by double-stranded RNA and TNF-α is attenuated in murine aortic endothelial cells derived from double-stranded RNA-activated kinase (PKR)-null mice. J. Immunol., 164, 2077–2083. [DOI] [PubMed] [Google Scholar]
- Barber G.N., Tomita,J., Garfinkel,M.S., Meurs,E., Hovanessian,A. and Katze,M.G. (1992) Detection of protein kinase homologues and viral RNA-binding domains utilizing polyclonal antiserum prepared against a baculovirus-expressed ds RNA-activated 68 000-Da protein kinase. Virology, 191, 670–679. [DOI] [PubMed] [Google Scholar]
- Barber G.N., Jagus,R., Meurs,E.F., Hovanessian,A.G. and Katze,M.G. (1995a) Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J. Biol. Chem., 270, 17423–17428. [DOI] [PubMed] [Google Scholar]
- Barber G.N., Wambach,M., Thompson,S., Jagus,R. and Katze,M.G. (1995b) Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol. Cell. Biol., 15, 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman K.P., Rogers,H.W., Corbett,J.A., Schreiber,R.D., McDaniel,M.L. and Unanue,E.R. (1993) Release of nitric oxide during the T cell-independent pathway of macrophage activation. Its role in resistance to Listeria monocytogenes. J. Immunol., 150, 888–895. [PubMed] [Google Scholar]
- Black T.L., Safer,B., Hovanessian,A. and Katze,M.G. (1989) The cellular 68 000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J. Virol., 63, 2244–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black T.L., Barber,G.N. and Katze,M.G. (1993) Degradation of the interferon-induced 68 000-Mr protein kinase by poliovirus requires RNA. J. Virol., 67, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I., Nottet,H.S., Schmidtmayerova,H., Dubrovsky,L., Flanagan,C.R., Mullins,M.E., Lipton,S.A. and Gendelman,H.E. (1995) Regulation of nitric oxide synthase activity in human immunodeficiency virus type 1 (HIV-1)-infected monocytes: implications for HIV-associated neurological disease. J. Exp. Med., 181, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.M., Ostertag,D., Li,Z.W., Chang,L., Chen,Y., Hu,Y., Williams,B., Perrault,J. and Karin,M. (1999) JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity, 11, 721–731. [DOI] [PubMed] [Google Scholar]
- Der S.D., Yang,Y.L., Weissmann,C. and Williams,B.R. (1997) A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl Acad. Sci. USA, 94, 3279–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Guerra M.J.M., Velasco,M., Martin-Sanz,P. and Bosca,L. (1996) Evidence for common mechanisms in the transcriptional control of type II nitric oxide synthase in isolated hepatocytes. Requirement of NF-κB activation after stimulation with bacterial cell wall products and phorbol esters. J. Biol. Chem., 271, 30114–30120. [DOI] [PubMed] [Google Scholar]
- Galabru J. and Hovanessian,A.G. (1985) Two interferon-induced proteins are involved in the protein kinase complex dependent on double-stranded RNA. Cell, 43, 685–694. [DOI] [PubMed] [Google Scholar]
- Gale M. Jr and Katze,M.G. (1998) Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther., 78, 29–46. [DOI] [PubMed] [Google Scholar]
- Godambe S.A., Chaplin,D.D. and Bellone,C.J. (1993) Regulation of IL-1 gene expression: differential responsiveness of murine macrophage lines. Cytokine, 5, 327–335. [DOI] [PubMed] [Google Scholar]
- Green L.C., Wagner,D.A., Glogowski,J., Skipper,P.L., Wishnok,J.S. and Tannenbaum,S.R. (1982) Analysis of nitrate, nitrite and [15N]nitrate in biological fluids. Anal. Biochem., 126, 131–138. [DOI] [PubMed] [Google Scholar]
- Gusella G., Musso,T., Rottschafer,S.E., Puulkki,K. and Varesio,L. (1995) Potential requirement of a functional double-stranded RNA-dependent protein kinase (PKR) for the tumoricidal activation of macrophages by lipopolysaccharide or IFN-αβ, but not IFN-γ. J. Immunol., 154, 345–354. [PubMed] [Google Scholar]
- Heitmeier M.R., Scarim,A.L. and Corbett,J.A. (1997) Interferon-γ increases the sensitivity of islets of Langerhans for inducible nitric-oxide synthase expression induced by interleukin 1. J. Biol. Chem., 272, 13697–13704. [DOI] [PubMed] [Google Scholar]
- Heitmeier M.R., Scarim,A.L. and Corbett,J.A. (1998) Double-stranded RNA-induced inducible nitric-oxide synthase expression and interleukin-1 release by murine macrophages requires NF-κB activation. J. Biol. Chem., 273, 15301–15307. [DOI] [PubMed] [Google Scholar]
- Hibbs J.B. Jr, Taintor,R.R., Vavrin,Z., Granger,D.L., Drapier,J.-C., Amber,I.J. and Lancaster,J.R.,Jr (1990) Synthesis of nitric oxide from a terminal guanidino nitrogen atom of l-arginine: a molecular mechanism regulating cellular proliferation that targets intracellular iron. In Moncada,S. and Higgs,E.A. (eds), Nitric Oxide from L-arginine: A Bioregulatory System. Elsevier Science Publishing Co., New York, NY, pp.189–223. [Google Scholar]
- Hill J.R., Corbett,J.A., Baldwin,A.C. and McDaniel,M.L. (1996) Nitric oxide production by the rat insulinoma cell line, RINm5F, is specific for IL-1: a spectrophotometric IL-1 bioassay. Anal. Biochem., 236, 14–19. [DOI] [PubMed] [Google Scholar]
- Hovanessian A.G. (1993) Interferon-induced and double-stranded RNA-activated proteins as key enzymes regulating protein synthesis. In Ilan,J. (ed.), Translational Regulation of Gene Expression. Plenum Press, New York, NY, pp. 163–185. [Google Scholar]
- Karupiah G., Xie,Q.W., Buller,R.M., Nathan,C., Duarte,C. and MacMicking,J.D. (1993) Inhibition of viral replication by interferon-γ-induced nitric oxide synthase. Science, 261, 1445–1448. [DOI] [PubMed] [Google Scholar]
- Kim Y.M., Lee,B.S., Yi,K.Y. and Paik,S.G. (1997) Upstream NF-κB site is required for the maximal expression of mouse inducible nitric oxide synthase gene in interferon-γ plus lipopolysaccharide-induced RAW264.7 macrophages. Biochem. Biophys. Res. Commun., 236, 655–660. [DOI] [PubMed] [Google Scholar]
- Koromilas A.E., Cantin,C., Craig,A.W., Jagus,R., Hiscott,J. and Sonenberg,N. (1995) The interferon-inducible protein kinase PKR modulates the transcriptional activation of immunoglobulin κ gene. J. Biol. Chem., 270, 25426–25434. [DOI] [PubMed] [Google Scholar]
- Kreil T.R. and Eibl,M.M. (1996) Nitric oxide and viral infection: NO antiviral activity against a flavivirus in vitro and evidence for contribution to pathogenesis in experimental infection in vivo. Virology, 219, 304–306. [DOI] [PubMed] [Google Scholar]
- Kumar A., Haque,J., Lacoste,J., Hiscott,J. and Williams,B.R. (1994) Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc. Natl Acad. Sci. USA, 91, 6288–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. (1997) Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J., 16, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lee S.B., Green,S.R., Mathews,M.B. and Esteban,M. (1994) Activation of the double-stranded RNA (dsRNA)-activated human protein kinase in vivo in the absence of its dsRNA binding domain. Proc. Natl Acad. Sci. USA, 91, 10551–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou H.C. and Baltimore,D. (1993) Regulation of the NF-κB/rel transcription factor and IκB inhibitor system. Curr. Opin. Cell Biol., 5, 477–487. [DOI] [PubMed] [Google Scholar]
- Meurs E., Chong,K., Galabru,J., Thomas,N.S., Kerr,I.M., Williams,B.R. and Hovanessian,A.G. (1990) Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell, 62, 379–390. [DOI] [PubMed] [Google Scholar]
- Meurs E.F., Watanabe,Y., Kadereit,S., Barber,G.N., Katze,M.G., Chong,K., Williams,B.R. and Hovanessian,A.G. (1992) Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J. Virol., 66, 5804–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E.F., Galabru,J., Barber,G.N., Katze,M.G. and Hovanessian,A.G. (1993) Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl Acad. Sci. USA, 90, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsch A., Schray-Utz,B., Mordvintcev,P.I., Hauschildt,S. and Busse,R. (1993) Diethyldithiocarbamate inhibits induction of macrophage NO synthase. FEBS Lett., 321, 215–218. [DOI] [PubMed] [Google Scholar]
- Roberts W.K., Hovanessian,A., Brown,R.E., Clemens,M.J. and Kerr,I.M. (1976) Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature, 264, 477–480. [DOI] [PubMed] [Google Scholar]
- Romano P.R., Green,S.R., Barber,G.N., Mathews,M.B. and Hinnebusch,A.G. (1995) Structural requirements for double-stranded RNA binding, dimerization and activation of the human eIF-2α kinase DAI in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E., Kuhen,K.L., George,C.X., Ortega,L.G., Rende-Fournier,R. and Tanaka,H. (1997) The PKR protein kinase—an interferon-inducible regulator of cell growth and differentiation. Int. J. Hematol., 65, 227–237. [DOI] [PubMed] [Google Scholar]
- Srivastava S.P., Kumar,K.U. and Kaufman,R.J. (1998) Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem., 273, 2416–2423. [DOI] [PubMed] [Google Scholar]
- Tanaka H. and Samuel,C.E. (1994) Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc. Natl Acad. Sci. USA, 91, 7995–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomis D.C., Doohan,J.P. and Samuel,C.E. (1992) Mechanism of interferon action: cDNA structure, expression and regulation of the interferon-induced, RNA-dependent P1/eIF-2α protein kinase from human cells. Virology, 188, 33–46. [DOI] [PubMed] [Google Scholar]
- Wu S. and Kaufman,R.J. (1996) Double-stranded (ds) RNA binding and not dimerization correlates with the activation of the dsRNA-dependent protein kinase (PKR). J. Biol. Chem., 271, 1756–1763. [DOI] [PubMed] [Google Scholar]
- Wu S. and Kaufman,R.J. (1997) A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem., 272, 1291–1296. [DOI] [PubMed] [Google Scholar]
- Yang Y.L., Reis,L.F., Pavlovic,J., Aguzzi,A., Schafer,R., Kumar,A., Williams,B.R., Aguet,M. and Weissmann,C. (1995) Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J., 14, 6095–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]