Abstract
Calcineurin is the conserved target of the immunosuppressants cyclosporin A and FK506. Using the yeast two-hybrid system, we identified a novel calcineurin binding protein, CBP1, from the pathogenic fungus Cryptococcus neoformans. We show that CBP1 binds to calcineurin in vitro and in vivo, and FKBP12–FK506 inhibits CBP1 binding to calcineurin. Cryptococcus neoformans cbp1 mutant strains exhibit modest defects in growth under stress conditions and virulence, similar to but less severe than the phenotypes of calcineurin mutants. Saccharomyces cerevisiae mutants lacking the CBP1 homolog RCN1 are, like calcineurin mutants, sensitive to lithium cation stress. CBP1 shares a central peptide sequence motif, SPPxSPP, with related proteins in S.cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Caenorhabditis elegans and humans, and peptides containing this motif altered calcineurin activity in vitro. Interestingly, the human CBP1 homolog DSCR1 is encoded by the Down’s syndrome candidate region interval on chromosome 21, is highly expressed in the heart and central nervous system, and may play a role in calcineurin functions in heart development, neurite extension and memory.
Keywords: calcineurin/Cryptococcus neoformans/Down’s syndrome/pathogenic fungi/protein phosphatase
Introduction
Calcineurin is a Ca2+/calmodulin-activated serine/threonine-specific phosphatase that is structurally and functionally conserved from yeast to man (reviewed in Hemenway and Heitman, 1999). The enzyme is a heterodimer composed of a catalytic A subunit and a regulatory B subunit that has four EF hands and is structurally related to calmodulin. Calcineurin is a critical calcium sensor in cells. When Ca2+ levels are low, calcineurin is inactive because a C-terminal autoinhibitory domain is docked in the active site. When intracellular Ca2+ levels increase, Ca2+/calmodulin binds to a C-terminal calmodulin binding site on the calcineurin A subunit, causing conformational changes that dislodge the autoinhibitory domain to result in enzyme activation (Hashimoto et al., 1990). Introduction of a point mutation into the autoinhibitory domain or proteolytic cleavage of the autoinhibitory domain results in constitutive activation of calcineurin (Hubbard and Klee, 1989; Fruman et al., 1995).
The immunosuppressive drugs cyclosporin A (CsA) and FK506 inhibit calcineurin in association with highly conserved binding proteins, cyclophilin A and FKBP12, which are conserved from yeast to humans (Liu et al., 1991; Foor et al., 1992; Breuder et al., 1994; Odom et al., 1997; Cruz et al., 1999). CsA and FK506 inhibit the immune system by blocking signal transduction in the antigen response pathway of T-lymphocytes. Normally, antigen presentation to the T-cell receptor results in increased intracellular Ca2+ levels, and calcineurin is activated and regulates nuclear import of the transcription factor NF-AT (Shaw et al., 1995; Loh et al., 1996; Shibasaki et al., 1996; Wesselborg et al., 1996). In the yeast Saccharomyces cerevisiae, calcineurin is also a critical Ca2+ sensor and regulates nuclear import of a transcription factor, Tcn1/Crz1, which is distantly related to NF-AT and regulates the expression of genes whose products regulate cell wall biosynthesis and cation transport, including FKS2, PMR1 and PMC1 (Matheos et al., 1997; Stathopoulos and Cyert, 1997; Stathopoulos-Gerontides et al., 1999).
Activated calcineurin is a heterotrimer composed of the catalytic A subunit, the regulatory B subunit and calmodulin. In addition, calcineurin forms stable interactions with several other proteins. For example, calcineurin–NF-AT and calcineurin–Tcn1 complexes have been detected using affinity chromatography or the two-hybrid system (Wesselborg et al., 1996; Matheos et al., 1997). Calcineurin also physically interacts with the AKAP79 scaffold protein in a macromolecular complex that also contains cAMP-dependent protein kinase (Coghlan et al., 1995; Klauck et al., 1996). Finally, recent studies have identified a novel inhibitor of calcineurin, called Cabin 1 or Cain, which interacts with and inhibits calcineurin in a phosphorylation-dependent manner in mammalian cells (Lai et al., 1998; Sun et al., 1998).
Calcineurin is the conserved target for both the immunosuppressive and the antifungal actions of CsA and FK506 (Odom et al., 1997; Cardenas et al., 1998, 1999; Cruz et al., 2000). In the yeast S.cerevisiae, calcineurin regulates cation homeostasis, cell wall biosynthesis and responses to pheromone (Cyert and Thorner, 1992; Nakamura et al., 1993; Cunningham and Fink, 1994, 1996; Hemenway et al., 1995; Mazur et al., 1995; Moser et al., 1996). In the fission yeast Schizosaccharomyces pombe, calcineurin is required for growth at low temperature and for mating (Yoshida et al., 1994; Plochocka-Zulinska et al., 1995). Calcineurin is essential for cell cycle progression in Aspergillus nidulans (Rasmussen et al., 1994), and regulates hyphal elongation and is also essential for growth in Neurospora crassa (Prokisch et al., 1997; Kothe and Free, 1998). Although calcineurin is highly conserved from fungi to man, fungal homologs of the endogenous mammalian calcineurin inhibitor Cabin 1/Cain have not yet been identified.
In the studies described here we have identified a novel calcineurin binding protein, CBP1, from the pathogenic fungus Cryptococcus neoformans. Cryptococcus neoformans is an important opportunistic pathogen in immunocompromised patients, and previous studies have revealed that calcineurin is required for cation homeostasis, growth at 37°C and virulence of this pathogen (Odom et al., 1997; Cruz et al., 2000). Here a two-hybrid screen was conducted using the calcineurin A catalytic subunit to screen a C.neoformans two-hybrid library and identify the CBP1 protein. We show that calcineurin and CBP1 form a stable complex in vivo and in vitro and that FKBP12–FK506 inhibits CBP1 binding to calcineurin. The CBP1 gene was disrupted by transformation and homologous recombination. In contrast to calcineurin mutant strains, cbp1 mutants were viable at 37°C. The cbp1 mutant strain did exhibit a modest virulence defect, similar to but not as severe as calcineurin mutants, which are avirulent. Interestingly, CBP1 shares a small central region of homology with related proteins in S.cerevisiae and man, and is the most broadly conserved of all known calcineurin binding proteins with the exception of calmodulin. Saccharomyces cerevisiae mutants lacking the CBP1 homolog YKL159c were viable and sensitive to cation stress, similar to calcineurin mutants. Expression of the C.neoformans CBP1 protein in S.cerevisiae restored cation resistance in calcineurin mutant strains. In summary, our studies identify a novel, conserved calcineurin binding protein that regulates calcineurin signaling cascades in vivo. The presumptive human CBP1 homolog DSCR1 is the first gene contained in the Down’s syndrome candidate region. Both calcineurin and DSCR1 are highly expressed in the central nervous system (CNS) and heart, and could have shared functions that are perturbed in patients with Down’s syndrome (Fuentes et al., 1995; Miyazaki et al., 1996).
Results
Identification of the calcineurin binding protein CBP1
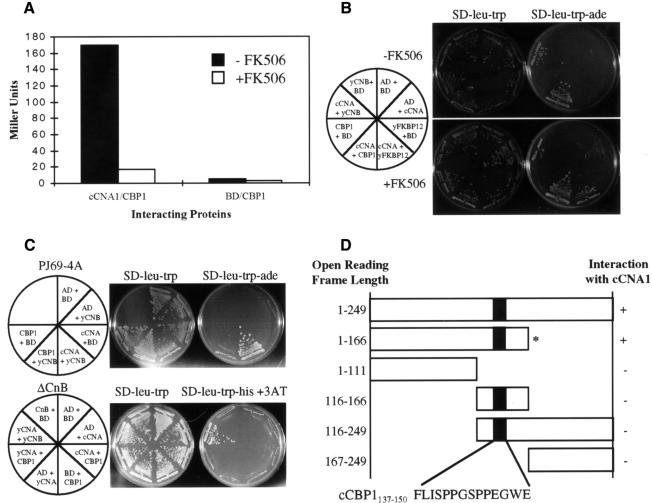
A two-hybrid screen was conducted to identify proteins that interact with calcineurin. The C.neoformans calcineurin A subunit was fused to the GAL4 DNA binding domain (GAL4-BD) and co-expressed in the yeast two-hybrid reporter strain PJ69-4A with a C.neoformans two-hybrid library consisting of cDNA fused to the GAL4 activation domain (GAL4-AD). A truncated form of calcineurin A lacking the C-terminal autoinhibitory domain was employed to allow binding to the calcineurin active site in the absence of Ca2+/calmodulin. From a screen of 5 million transformants, 24 Ade+ isolates were obtained (Figure 1A). All 24 isolates were also His+ and expressed β-galactosidase activity (Figure 1A), indicating that all three GAL reporter genes are expressed. The library plasmids were rescued in Escherichia coli, and all 24 activated the two-hybrid reporter genes when re-introduced with a plasmid expressing the GAL4–calcineurin fusion protein but not with a control vector (Figure 1A). Restriction mapping and sequencing revealed all isolates contained an identical ∼750 bp insert corresponding to a novel gene, which we named CBP1 for calcineurin binding protein 1.
Fig. 1. Calcineurin and CBP1 interact in the two-hybrid assay. (A) The C.neoformans calcineurin A protein (cCNA1) specifically interacts with CBP1 and this interaction is inhibited by FK506. β-galactosidase assays were conducted in the presence and absence of 1 µg/ml FK506 as indicated. (B) FK506 inhibition of calcineurin–CBP1 binding requires FKBP12. An S.cerevisiae strain lacking FKBP12 (fpr1 strain SMY87-4) was transformed with plasmids expressing the GAL4 activation domain (AD) or DNA binding domain (BD) fused to the indicated proteins. To detect expression of the GAL-ADE2 reporter gene, cells were grown on synthetic medium minus leucine, tryptophan and adenine (SD-leu-trp-ade) for 7 days at 30°C. ‘y’ denotes S.cerevisiae proteins and ‘c’ denotes C.neoformans proteins. FK506 stimulated FKBP12 binding to calcineurin A as expected. FK506 did not inhibit CBP1 binding to calcineurin in these cells lacking endogenous FKBP12. Calcineurin A (cCNA1) binding to calcineurin B (yCNB) was not affected by FK506. (C) Saccharomyces cerevisiae calcineurin B interacts with CBP1 and is required for CBP1 binding to calcineurin A. Isogenic S.cerevisiae strains expressing (PJ69-4A) or lacking calcineurin B (ΔCnB, SMY3) were transformed with plasmids expressing the GAL4 AD or BD fused to the indicated proteins. Cells were grown on medium lacking adenine (SD-leu-trp-ade) to detect expression of the GAL-ADE2 reporter gene, and on medium lacking histidine (SD-leu-trp-his + 5 mM 3-AT) to detect expression of the GAL-HIS3 reporter gene. (D) Two-hybrid analysis of the binding of truncated forms of CBP1 to calcineurin A. The black box represents the highly conserved region of CBP1. Fragment length is indicated in amino acid residues. * indicates the GST–CBP1 fusion protein.
In two-hybrid assays, the C.neoformans CBP1 protein interacted with GAL4 fused to the C.neoformans, S.cerevisiae or murine calcineurin A catalytic subunits. The calcineurin inhibitor FK506 blocked CBP1–calcineurin interactions in the two-hybrid system (Figure 1A), and the effects of FK506 were dependent upon endogenous FKBP12 in the two-hybrid host strain (Figure 1B). The C.neoformans CBP1 protein also activated reporter gene expression when co-expressed with GAL4-BD–calcineurin B fusion protein in the two-hybrid assay, although the level of activation was reduced compared with CBP1 binding to calcineurin A, or calcineurin A binding to calcineurin B (Figure 1C, upper panel). CBP1 failed to interact with calcineurin A in a two-hybrid host strain lacking calcineurin B (Figure 1C, lower panel). Finally, a series of truncated forms of the CBP1 protein was assessed. This analysis revealed that residues 1–166 were sufficient for binding, whereas truncated proteins consisting of residues 1–111 or 116–249 failed to interact, although by western blotting we were unable to determine whether these proteins were stably expressed (Figure 1D and data not shown). These observations suggest that CBP1 interacts with both the calcineurin A and B subunits and may compete with FKBP12–FK506 binding to the calcineurin AB interface (Cardenas et al., 1995; Griffith et al., 1995; Kissinger et al., 1995).
Isolation of the CBP1 gene: CBP1 is conserved in yeasts, flies, worms and humans
The portion of the CBP1 gene obtained in the two-hybrid screen was used as a probe in Southern blot analysis and to clone genomic and cDNA clones corresponding to the CBP1 locus. Sequence analysis and comparison of cDNA and genomic clones revealed that the CBP1 gene encodes a 249 amino acid protein (Figure 2) and that the CBP1 gene contains a single intron. Moreover, this analysis revealed that, in the isolates obtained, the entire CBP1 protein was fused to the GAL4-AD.
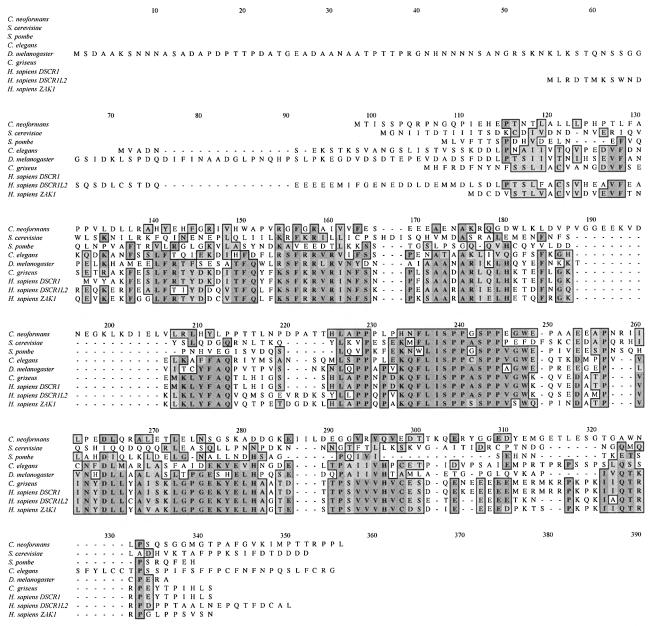
Fig. 2. Alignment of C.neoformans CBP1 protein homologs. An alignment of the C.neoformans, S.cerevisiae (DDBJ/EMBL/GenBank accession No. Z28159), S.pombe (accession No. Z67757), C.elegans (accession No. AF176115), D.melanogaster (accession No. AF147700), Cricetulus griseus (accession No. U60263) and human (Homo sapiens, accession Nos D83407, U28833 and AF176116) CBP1 proteins is depicted. Identical residues are in bold, darkly shaded and boxed; similar residues are lightly shaded and boxed.
Standard Blast searches with the CBP1 sequence (DDBJ/EMBL/GenBank accession No. AF230799) failed to identify any proteins with marked identity to the entire CBP1 protein. A nested BLAST search revealed a limited region of homology, spanning amino acids 125–160, shared between CBP1 and related proteins in S.cerevisiae, S.pombe, Drosophila melanogaster, Caenorhabditis elegans, hamster and humans, which are largely of unknown function (Figure 2). Interestingly, this region contains two conserved serine residues in the sequence SPPxSPP (Figure 2). These observations suggest that the C.neoformans CBP1 protein is a member of a family of broadly conserved calcineurin binding proteins.
CBP1 binds calcineurin in vitro and in vivo
We next sought to confirm that CBP1 interacts directly with calcineurin. A glutathione S-transferase (GST)–CBP1 fusion protein was expressed in bacteria, absorbed to glutathione Sepharose beads, and incubated in binding reactions with purified bovine calcineurin. Bound proteins were eluted, fractionated by SDS–PAGE, and the calcineurin A subunit was detected by western blotting. As shown in Figure 3A, GST–CBP1 bound to calcineurin A in the presence or absence of calmodulin. CBP1 binding to calcineurin A was completely inhibited by FKBP12–FK506, and was partially inhibited by EGTA (Figure 3A). No binding of calcineurin A was detected to GST alone (Figure 3A). These findings reveal that divalent cations are required for CBP1 binding to calcineurin and that CBP1 and FKBP12–FK506 compete for binding both in vitro and in the yeast two-hybrid system.
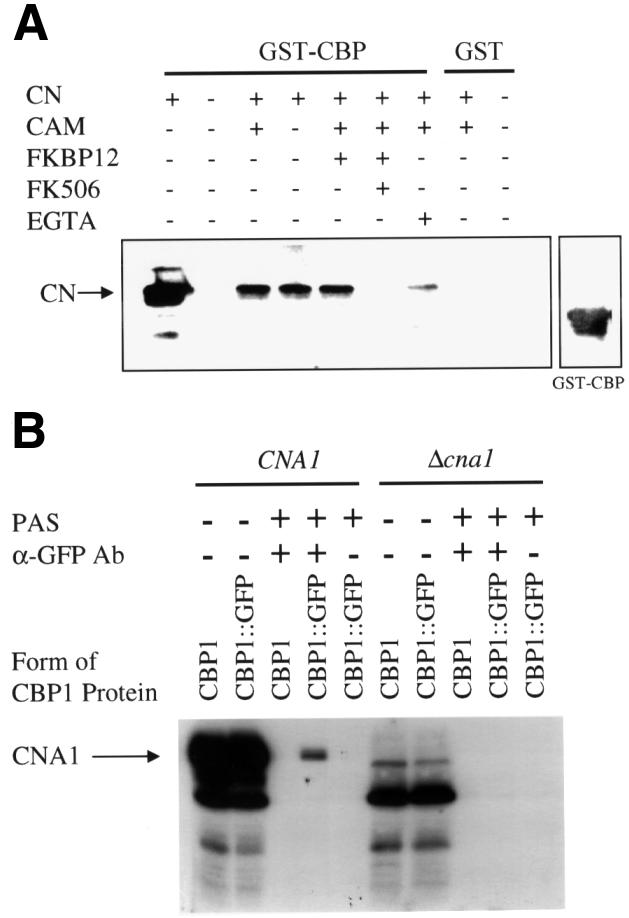
Fig. 3. CBP1 binds to calcineurin in vitro and in vivo. (A) Purified GST–CBP fusion protein bound to glutathione agarose was incu bated with bovine calcineurin in the presence (+) or absence (–) of calmodulin, FKBP12, FK506 or EGTA for 2 h at 4°C. Reactions were separated on 10% SDS–PAGE and transferred to PVDF membranes. Membranes were incubated with anti-calcineurin (bovine) antibody to detect binding of CBP to calcineurin or anti-GST antibody to detect the GST–CBP fusion protein. The arrow indicates the position of calcineurin. The panel on the right indicates the position of the GST–CBP fusion protein. (B) Wild-type strain H99 (CNA1) and the isogenic Δcna1 mutant strain expressing wild-type CBP1 (H99, AO4) or a CBP1–GFP fusion protein (JMC4, JMC6) were grown over night in rich medium, cells were mechanically disrupted, and immuno precipitation experiments were conducted with total cell extracts for 1 h at 4°C in the presence (+) and absence (–) of anti-GFP antisera (α-GFP Ab). Proteins bound to the antibody were subsequently precipitated with protein A–Sepharose (PAS) and separated on 12% SDS–PAGE. The calcineurin A protein (CNA1) was detected by incubating the western blots with [125I]calmodulin. The first two lanes of the right and left panels are total extract controls that were not incubated with the anti-GFP antisera.
To test whether CBP1 also binds calcineurin in vivo, the CBP1 gene was fused to the gene encoding the green fluorescent protein (GFP). The resulting CBP1–GFP fusion gene was transformed into C.neoformans cells, and the full-length fusion protein was detected by western blot analysis of whole-cell extracts with antisera against GFP (Figure 3B). Next, the CBP1–GFP fusion protein was immunoprecipitated with anti-GFP antibodies, and bound proteins were resolved by SDS–PAGE, transferred to nitrocellulose, and analyzed by overlay blotting with 125I-labeled calmodulin to detect calcineurin A. As shown in Figure 3B, the CBP1–GFP fusion protein co-immunoprecipitated with the calcineurin A subunit. In comparison, no interaction was detected with GFP alone or CBP1 in which the GFP epitope tag was omitted, there was no co-immunoprecipitating protein with extracts from a cna1 mutant strain lacking calcineurin A, and no CBP1–calcineurin complex was detected when the anti-GFP antibody was omitted (Figure 3B and data not shown). Thus, CBP1 and calcineurin A also interact in vivo.
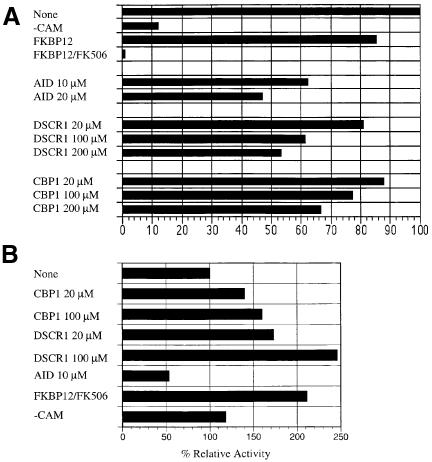
We tested whether CBP1 binding alters calcineurin enzymatic activity. Synthetic peptides corresponding to the conserved region of CBP1 or the human DSCR1 protein were found to inhibit the activity of bovine calcineurin towards a synthetic phosphopeptide derived from the RII subunit of cAMP-dependent protein kinase (Figure 4A). The magnitude of inhibition (∼50%) was less than that of FKBP12–FK506, but was similar to that observed with a peptide corresponding to the autoinhibitory domain of calcineurin. When calcineurin activity was measured towards the small substrate p-nitrophenylphosphate (pNPP), the CBP1 and DSCR1 peptides modestly stimulated activity (1.5- to 2.5-fold), similar to the effects of FKBP12–FK506, as has been previously reported (Liu et al., 1991). These findings provide additional evidence that CBP1 interacts with calcineurin and suggest that the conserved region of CBP1 and the CBP1 homolog DSCR1 can alter the activity of calcineurin by interacting with a site distinct from the active site.
Fig. 4. Effect of CBP1 peptides on calcineurin phosphatase activity. (A) Calcineurin phosphatase activity was determined by measurement of radiolabel released from [32P]phospho-RII peptide at 30°C in the presence or absence (–CAM) of calmodulin, and with the addition of FKBP12, FKBP12–FK506, autoinhibitory peptide (AID), or of the DSCR1 or CBP1 peptides at 20, 100 or 200 µM. (B) Calcineurin phosphatase activity was determined by spectrophotometric determination at 405 nm of the reaction product released from pNPP at 25°C. Values shown are % activities relative to uninhibited calcineurin control reactions. Each sample was performed in duplicate.
Calcineurin regulates the electrophoretic mobility of CBP1
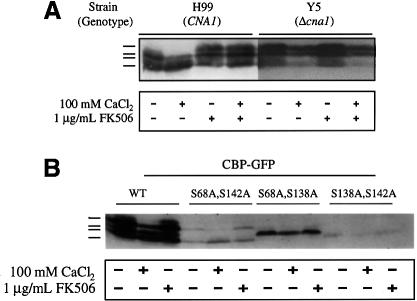
In western blot analysis, the CBP1–GFP fusion protein migrated as up to four distinct species with differing electrophoretic mobilities (Figure 5A). When cells were exposed to 100 mM extracellular Ca2+, the band with the slowest mobility disappeared and the relative abundance of the faster migrating species increased (Figure 5A). In contrast, when cells were exposed to the calcineurin inhibitor FK506, the relative abundance of the slowest migrating species increased, and FK506 blocked the effects of Ca2+ ions on CBP1 mobility (Figure 5A). To address further whether the effects of Ca2+ ions and FK506 were mediated via calcineurin, a similar analysis was conducted in a cna1 mutant strain lacking calcineurin A (Figure 5A). In this case, in control extracts the slowest migrating form of CBP1 was more abundant than in cells expressing calcineurin. Exposure to Ca2+ ions resulted in a decreased level of the faster migrating forms of CBP1. FK506 had no effect on CBP1 mobility in cells lacking calcineurin and did not block the effects of Ca2+. Finally, a series of mutant CBP1–GFP fusion proteins was generated in which the conserved serine residues in the FLISPPxSPP motif were replaced with alanines. In these cases, only one or two forms of the CBP1–GFP fusion protein were observed, and there was little or no effect of FK506 or Ca2+ (Figure 5B). In two cases protein stability may also be reduced. Taken together, these observations suggest that CBP1 may be a phosphoprotein and could be a substrate of both the Ca2+-activated phosphatase calcineurin and possibly also a Ca2+/calmodulin-dependent kinase.
Fig. 5. CBP1 electrophoretic mobility is regulated by calcineurin. (A) Strains H99 (CNA1) and Y5/AO4 (Δcna1) expressing a CBP1–GFP fusion protein were incubated for 2 h in the presence (+) or absence (–) of 100 mM CaCl2 or 1 µg/ml FK506. Total proteins were extracted, separated by 9% SDS–PAGE, transferred to nitrocellulose, and probed with anti-GFP antibody. (B) Strains expressing CBP1–GFP fusion proteins, either wild-type (CBP1) or mutant forms (CBP1S68A,S142A, CBP1S68A,S138A, CBP1S138A,S142A), were incubated for 2 h in the presence (+) or absence (–) of 100 mM CaCl2 or 1 µg/ml FK506. Total proteins were extracted, separated by SDS–PAGE, transferred to nitrocellulose, and probed with anti-GFP antibody.
Isolation and characterization of a C.neoformans cbp1 mutant strain
To disrupt the C.neoformans CBP1 gene, the CBP1 open reading frame from start to stop codon was replaced with the ADE2 gene and the cbp1Δ:ADE2 allele was transformed into the ade2 mutant strain M049 by biolistic transformation. Genomic DNA was isolated and analyzed by PCR and Southern blotting to identify isolates in which the CBP1 gene was replaced by the cbp1Δ::ADE2 allele by homologous recombination with no tandem or ectopic integrations. By Southern blotting, the CBP1 open reading frame was present in the CBP1 wild-type strain M049 and not in the cbp1Δ strain (not shown). Hybridization with the 3′ region of the CBP1 gene confirmed a single copy of the ADE2 gene was inserted. A cbp1 + CBP1 reconstituted strain was generated in which the CBP1–GFP gene was introduced into the cbp1Δ mutant. By northern blotting, the CBP1 gene was expressed in the wild-type strain H99 and in the cbp1 + CBP1 reconstituted strain, and was not expressed in the cbp1Δ::ADE2 mutant (not shown).
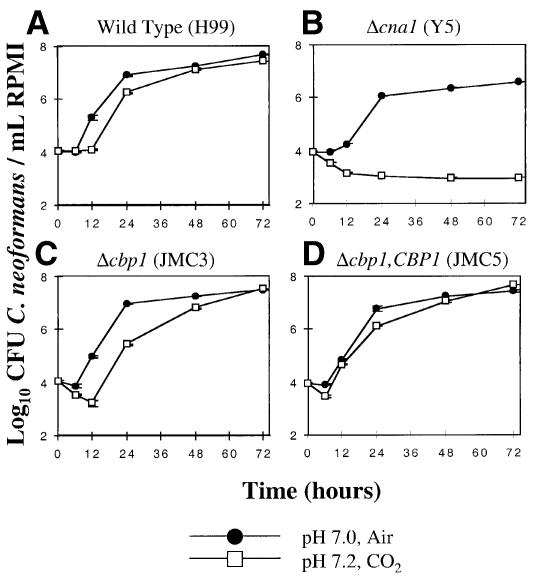
We next tested whether either the cbp1 mutant strain or the cbp1 + CBP1 reconstituted strain that overexpresses the CBP1 protein exhibited mutant phenotypes similar to isogenic cna1 mutants lacking calcineurin A. In contrast to cna1 mutants lacking calcineurin, both the cbp1 mutant and the cbp1 + CBP1 strain grew normally at 37 and 39°C (not shown) and were not sensitive to 50 mM LiCl or 1.5 M NaCl (not shown). The cbp1 mutant strain did exhibit a modest growth defect at pH 7.2/5% carbon dioxide (Figure 6C), which was complemented in the cbp1 + CBP1 reconstituted strain (Figure 6D), but this defect was less severe than in the isogenic calcineurin mutant (Figure 6B).
Fig. 6. cpb1 mutant strains are modestly sensitive to 5% CO2 and alkaline pH. The wild-type C.neoformans strain H99 (Wild Type) (A), the isogenic Δcna1 mutant strain (Y5/AO4) (B), Δcbp1 mutant strain (JMC3) (C) and the Δcbp1 mutant strain transformed with the CBP1 gene (JMC5) (D) were grown in triplicate in liquid RPMI medium at 30°C for 0–72 h in either an atmosphere of air at pH 7.0 (circles) or at 5% CO2 at pH 7.2 (squares) and the number of surviving colony-forming units (CFU) was measured by dilution and plating on YPD medium and the number of surviving colonies was averaged for triplicate cultures.
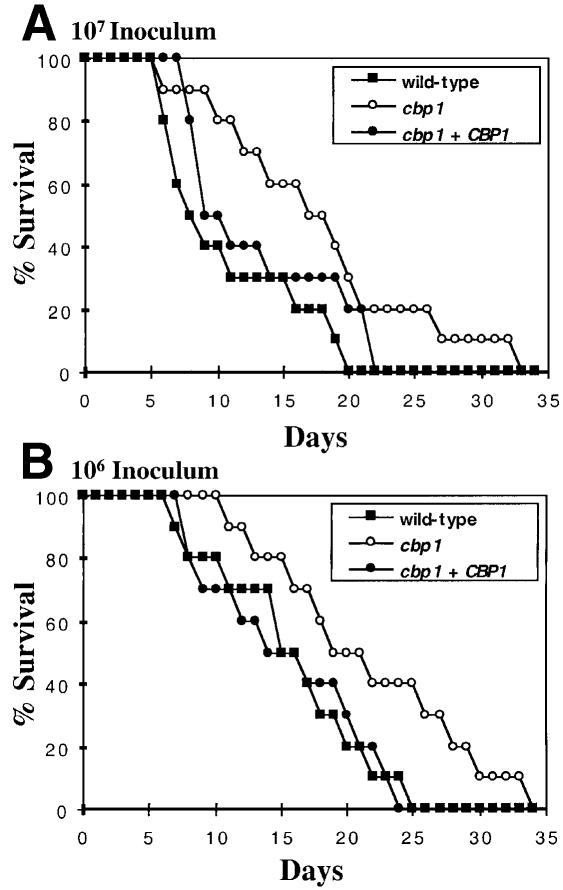
Calcineurin mutant strains are avirulent in both a rabbit model of cryptococcal meningitis and in a murine systemic infection model (Odom et al., 1997; Cruz et al., 2000). In contrast, persistence of the cbp1Δ mutant strain in the cerebrospinal fluid (CSF) of infected immunosuppressed rabbits was similar to the isogenic CBP1 wild-type strain, indicating that CBP1 is not required for survival in the CNS of this animal model (not shown), which is different from calcineurin mutants (Odom et al., 1997). However, in the murine model, virulence of the cbp1 mutant strain was modestly reduced compared with the wild-type strain. At an inoculum size of 107, 50% of mice infected with wild-type strain H99 survived to day 8 post-infection, compared with the cbp1 mutant in which 90% of animals were alive on days 8 and 9 and 50% survived until day 18 (Figure 7A). There was 100% mortality with the wild-type strain by day 20, whereas 100% mortality was delayed until day 33 with the cbp1 mutant (Figure 7A). Similar results were observed at an inoculum size of 106 cells (Figure 7B). Virulence of the cbp1 + CBP1 reconstituted strain was similar to the CBP1 wild type in both animal models (Figure 7 and data not shown). In comparison, the isogenic cna1 calcineurin A mutant was completely avirulent in this murine model (Cruz et al., 2000). Taken together, these findings indicate that the CBP1 protein plays a relative but not absolute role in some calcineurin functions in C.neoformans, possibly because it is redundant with other factors.
Fig. 7. CBP1 is required for full virulence of C.neoformans. Mice (10 each) were injected in the lateral tail vein with either 107 (A) or 106 (B) cells of the wild-type strain H99 (filled squares), the isogenic cbp1::ADE2 mutant strain (JMC3, open circles) or the Δcbp1 mutant strain transformed with the CBP1 gene (JMC5, filled circles). Survival was monitored and plotted with respect to time. P-values were gener ated using the Wilcoxon test. In (A), cbp1 (JMC3) versus CBP1 (H99) P <0.05, cbp1 (JMC3) versus cbp1 + CBP1 (JMC5) p >0.05. In (B), cbp1 (JMC3) versus CBP1 (H99) P = 0.05, cbp1 (JMC3) versus cbp1 + CBP1 (JMC5) p >0.05.
Analysis of S.cerevisiae CBP1 homolog YKL159c/RCN1
We next analyzed the functions of the S.cerevisiae CBP1-related protein YKL159c/RCN1. The YKL159c gene was replaced from start to stop codon with dominant selectable markers conferring G418 or hygromycin resistance by PCR-mediated gene disruption. ykl159c mutant strains were viable and had no growth defect on rich or minimal medium or at different temperatures (not shown). By western blotting, the calcineurin B subunit was stably expressed in ykl159c mutants, and thus CBP1 is not required for expression or stability of the CNB1 protein (not shown).
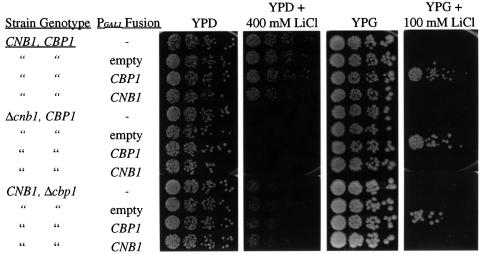
Several observations suggest that the presumptive CBP1 homolog YKL159c/RCN1 participates in some calcineurin functions in vivo. First, similar to S.cerevisiae calcineurin mutants, strains lacking YKL159c were moderately sensitive to Li+ cations (Figure 8). However, while growth of calcineurin mutants was completely blocked by 100 mM LiCl, growth of the cbp1 mutants was partially inhibited at 200 mM LiCl and more severely inhibited at 400 mM LiCl (Figure 8 and data not shown). Secondly, overexpression of the C.neoformans CBP1 protein in wild-type cells or a cnb1 mutant lacking calcineurin B conferred cation resistance on medium containing galactose (inducing) but not glucose (repressing). In contrast, calcineurin B overexpression did not restore Li+ resistance in cbp1 mutant cells (Figure 8 and data not shown). These findings suggest that CBP1 may function downstream of calcineurin.
Fig. 8. CBP1 functions in cation homeostasis in S.cerevisiae. The C.neoformans CBP1 or S.cerevisiae CNB1 genes were expressed from the S.cerevisiae GAL1 promoter on a 2µ plasmid. These plasmids were introduced into either wild-type (CNB1, CBP1), Δcnb1 or Δcbp1 mutant strains of S.cerevisiae. Strains with no plasmids (–) or harboring an empty vector (empty) were included as controls. Cells were grown in selective medium overnight and then serially diluted (1250, 250, 50 and 10 cells/spot from left to right) onto rich medium containing either glucose (YPD) or galactose (YPG) supplemented with 100 or 400 mM LiCl. Plates were incubated for 2 days (YPD, YPD + 400 mM LiCl) or 6 days (YPG, YPG + 100 mM LiCl) at 30°C.
The yeast YKL159c/RCN1 protein does not play a role in all calcineurin functions in yeast. Mutants lacking YKL159c have no defect in response to or recovery from α-factor, whereas calcineurin mutations prevent recovery (not shown). Calcineurin mutations are synthetically lethal with vph6 or fks1 mutations (because calcineurin is required for cell wall biosynthesis or cation homeostasis in such mutants), whereas ykl159c vph6 and ykl159c fks1 double mutants were viable (not shown). Thus, in both S.cerevisiae and C.neoformans, CBP1 plays a role in some but not all calcineurin functions.
Discussion
We have identified a novel and conserved calcineurin binding protein, CBP1, from the pathogenic fungus C.neoformans. CBP1 interacts with both the calcineurin A and B subunits in the yeast two-hybrid assay, the calcineurin B subunit is required for formation of the calcineurin A–CBP1 complex, and FKBP12–FK506 inhibited CBP1 binding to calcineurin in the two-hybrid system. The interaction between CBP1 and calcineurin was confirmed by in vitro binding assays with purified proteins, revealing that CBP1 and calcineurin directly interact; calmodulin is not required for CBP1 binding to calcineurin. FKBP12–FK506 inhibited CBP1 binding to calcineurin in vitro, suggesting that CBP1 may bind to the interface between the calcineurin A and B subunits, which is known to be the case for FKBP12–FK506. This interpretation is consistent with the finding that CBP1 peptides activate calcineurin activity with the small substrate PNPP, and modestly inhibit activity towards larger phosphopeptide substrates. Finally, we showed that CBP1 and calcineurin can be co-immunoprecipitated from C.neoformans cell extracts, suggesting that CBP1 and calcineurin interact in vivo.
Our observations on the electrophoretic mobility of the CBP1 protein suggest that it could be a phosphoprotein substrate of calcineurin. Exposure of cells to FK506 or Ca2+ altered the mobility of CBP1, and site-directed mutagenesis of several conserved serine residues in the CBP1 protein resulted in a reduction or disappearance of altered mobility states of the protein. Because Ca2+ increased the mobility of CBP1 in cells expressing calcineurin, but had the opposite effect in calcineurin mutant cells, CBP1 could be a substrate of both calcineurin and a calcium/calmodulin regulated kinase. These findings may be analogous to other recent studies. For example, the Cot-1 serine/threonine kinase that regulates hyphal elongation in N.crassa is found in a physical complex with calcineurin (Gorovits et al., 1999). Similarly, the AKAP79 scaffolding protein forms complexes with both calcineurin and the cAMP-dependent protein kinase in mammalian cells (Coghlan et al., 1995; Klauck et al., 1996; Kashishian et al., 1998). Thus, both kinases and phosphatases may be present in common physical complexes with regulatory targets such as CBP1.
To determine the functions of CBP1, the CBP1 gene was disrupted by homologous recombination in C.neoformans. The cbp1 mutant strain was viable. In contrast to an isogenic calcineurin mutant, the cbp1 mutant was viable at 37°C and was not cation stress sensitive. The cbp1 mutant was modestly sensitive to other stress conditions that are toxic to calcineurin mutants (alkaline pH, CO2) and also exhibited a modest virulence defect in mice. Thus, CBP1 functions in some calcineurin-regulated pathways but not others, possibly because calcineurin has several targets, CBP1 is redundant with other factors, or CBP1 regulates only some calcineurin functions.
A small central region of the C.neoformans CBP1 protein shares identity with related proteins in S.cerevisiae, S.pombe, D.melanogaster, C.elegans and humans that have recently been identified by others and termed calcipressins (K.Cunningham and F.McKeon, personal communication; Rothermel et al., 2000). This conserved region contains the peptide sequence FLISPPxSPP, which could be involved in phosphorylation and calcineurin binding. This conserved peptide sequence is not present in the calcineurin binding sites of other calcineurin substrates or regulators, including NF-AT, Crz1/Tcn1, Cabin 1/Cain or AKAP79. We have disrupted the gene encoding the CBP1 homolog in S.cerevisiae (YKL159c). Mutants lacking the yeast CBP1 homolog are, like calcineurin mutants, cation sensitive. Overexpression of the C.neoformans CBP1 protein in wild-type or a cnb1 mutant strain conferred cation resistance, suggesting that CBP1 may function downstream of calcineurin when overexpressed. These observations reveal that the functions of CBP1 and calcineurin are related. CBP1 may be a target of the calcineurin signaling cascade, a regulator of calcineurin signaling functions, or both.
Most interestingly, the human CBP1 homolog DSCR1 is encoded on chromosome 21 and is the first gene in the Down’s syndrome candidate region interval (Fuentes et al., 1995; Miyazaki et al., 1996). Because Down’s syndrome results from trisomy for chromosome 21, DSCR1 may be relatively overexpressed in patients with this disorder. The most marked manifestations of Down’s syndrome include mental retardation and congenital heart defects. Both DSCR1 and calcineurin are highly expressed in the CNS and heart (Fuentes et al., 1995), and calcineurin and NF-AT are now known to play a prominent role in regulating cardiac development and hypertrophy (Molkentin et al., 1998; Ranger et al., 1998; Sussman et al., 1998). Calcineurin also regulates CNS functions, including neurite extension and both long-term memory and long-term potentiation (Ferreira et al., 1993; Mulkey et al., 1994; Chang et al., 1995; Mansuy et al., 1998; Winder et al., 1998). The role of calcineurin in neurite extension may be analogous to the known roles of calcium and calcineurin in regulating hyphal elongation in fungi (Jackson and Heath, 1993; Prokisch et al., 1997). These findings suggest an intriguing link between calcineurin, CBP1/DSCR1 and cell morphogenesis in both fungi and man.
In summary, our studies identify CBP1 as a highly conserved and ubiquitous calcineurin binding protein. CBP1 represents the first calcineurin binding protein, other than calmodulin, that is conserved from yeast and fungi to man. Our studies highlight the potential of molecular genetic studies in pathogenic fungi to identify conserved elements of signaling cascades, and may provide insights into the multiple roles of calcineurin in immunosuppressive drug action, cardiac and CNS functions, and fungal virulence.
Materials and methods
Yeast strains, media, genetic methods and compounds
Strains used in this study are listed in Table I. The S.cerevisiae strains PJ69-4A, JK9-3da, SMY3 and SMY87-4 used in this study were as described (Heitman et al., 1991; Cardenas et al., 1994; James et al., 1996; Arndt et al., 1999). The C.neoformans serotype A strain H99 and its derivative M049 were as described (Perfect et al., 1993). Both S.cerevisiae and C.neoformans strains were grown in complete (YPD) and minimal (SD) media as described (Sherman, 1991). Regeneration medium for biolistic transformation was prepared as described (Toffaletti et al., 1993). V8 mating-starvation medium was prepared as described (Odom et al., 1997). Immunosuppressant-containing medium was as described (Heitman et al., 1993). YPD was supplemented with salts as indicated. FK506 was from Fujisawa.
Table I. Strain list.
| S.cerevisiae strains | Genotype | Reference |
|---|---|---|
| PJ69-4A | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met::GAL7-lacZ | James et al. (1996) |
| SMY87-4 | PJ69-4A fpr1Δ::hisG | Arndt et al. (1999) |
| Y190 | MATa trp1-901 his3 leu2-3,112 ura3-52 ade2 gal4 gal80 URA3::GAL-lacZ LYS2::GAL-HIS3 | Harper et al. (1993) |
| SMY3 | Y190 TOR1-3 cnb1::ADE2 | Cardenas et al. (1994) |
| JK9-3da | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa | Heitman et al. (1991) |
| SCY78 |
JK9-3da ykl159cΔ::G418 |
this study |
|
C.neoformans strains |
Genotype (derived from strain) |
Reference |
| H99 | Perfect et al. (1993) | |
| M049 | ade2 (H99) | Perfect et al. (1993) |
| AO4 | cna1Δ::ADE2 ade2 (M049) | Odom et al. (1997) |
| AO10 | cna1Δ::ADE2 CNA1 ade2 (AO4) | Odom et al. (1997) |
| JMC3 | cbp1Δ::ADE2 ade2 (M049) | this study |
| JMC4 | CBP1::GFP Hygr ade2 (H99) | this study |
| JMC5 | cbp1Δ::ADE2 CBP1::GFP Hygrade2 (JMC3) | this study |
| JMC6 | cna1Δ::ADE2 CBP1::GFP Hygrade2 (AO4) | this study |
| MCC3 | MATa cna1Δ::ADE2 ade2 ura5 (serotype D, JEC156) | Cruz et al. (2000) |
| JMC7 | CBP1-S68A, S142A::GFP Hygr ade2 (H99) | this study |
| JMC8 | CBP1-S68A, S138A::GFP Hygr ade2 (H99) | this study |
| JMC9 | CBP1-S138A, S142A::GFP Hygr ade2 (H99) | this study |
Yeast two-hybrid system
Primers were designed to hybridize to regions of the C.neoformans CNA1 gene such that the autoinhibitory domain of the calcineurin A protein was omitted from the C-terminus of the protein. Using C.neoformans cDNA as a template, forward primer 5′-GCTTCCCCGGGCACTCAGACC (1420) and reverse primer 5′-GGCTCTGCAGCATTAAGAGGG (1422) were used to PCR amplify the 1.8 kb 5′ region of the CNA1 cDNA using standard PCR conditions. Primers 1420 and 1422 contain SmaI and PstI restriction sites, respectively. The PCR product was digested with both SmaI and PstI, gel purified, and ligated in-frame with the GAL4-BD in the two-hybrid vector pGBT9 to yield plasmid pJMM119.
The S.cerevisiae reporter strain PJ69-4A (James et al., 1996) was cotransformed with plasmid pJMM119 and the C.neoformans two-hybrid library (Cruz et al., 1999) or with other control plasmids (Table II) by a high efficiency lithium acetate/heat shock method (Gietz et al., 1995). Double transformants were isolated on medium lacking tryptophan and leucine (to select for the introduced plasmids) and also lacking either adenine, or lacking histidine and containing 10 mM 3-aminotriazole to assay reporter gene expression. β-galactosidase assays were performed as described (Cardenas et al., 1994).
Table II. Plasmids.
| Plasmid | Description | Reference |
|---|---|---|
| pGAD424 | 2µ LEU2 GAL4(AD) | Bartel et al. (1993) |
| pGBT9 | 2µ TRP1 GAL4(BD) | Bartel et al. (1993) |
| pJMM119 | 2µ TRP1 GAL4(BD)::CnCNA1 | this study |
| pML59 | 2µ LEU2 GAL4(AD)::ScFKBP12 | Lorenz and Heitman (1995) |
| pYCBA | 2µ LEU2 GAL4(AD)::ScCNB1 | this study |
| pCH113BD | 2µ TRP1 GAL4(BD)::ScCMP1 | Cardenas et al. (1994) |
| pYDF2BD | 2µ TRP1 GAL4(BD)::MmCNA | Cardenas et al. (1994) |
| pmCnA | 2µ TRP1 GAL4(BD)::MmCNA (His-Glu) | this study |
| pYCBB | 2µ TRP1 GAL4(BD)::ScCNB1 | this study |
| pJMM131 | 2µ LEU2 GAL4(AD)::CnCNA1 | this study |
| pJMM160 | Δcbp1::ADE2 | this study |
| pJMM185 | Hph CBP1::GFP | this study |
| pJMM190 | Hph pGAL7::CBP1 | this study |
Cryptococcus neoformans cDNA library plasmids were rescued from strain PJ69-4A by transforming miniprep yeast DNA into E.coli strain HB101 and selecting for leucine prototrophy on M9 medium containing ampicillin. Three size classes of plasmids were identified. All plasmids were retransformed into strain PJ69-4A harboring plasmid pJMM119 to confirm the interaction. Only one class retained the ability to interact with calcineurin A. Several independent plasmids from this class were sequenced, revealing that all encoded the same protein, CBP1.
Protein truncations were generated by PCR amplification of the CBP1 gene using the two-hybrid library clone as a template and standard PCR conditions. Primer sequences were: 5′-GGTTGTCGGCTGCAGGTAGTGTAA (1915), 5′-GCAACGTCGACTGGAAACGC (1916), 5′-GCCGCGTCGACCATATCCC (1917), 5′-GGAATTCGCTGCAGCGTCGAC (1918), 5′-GTTTCCACTGCAGGTTGCTAATCCTCA (1919) and 5′-CCACCGTCGACTCAACCCCGACCC (1920). PCR products were digested with SalI and PstI, and ligated into SalI and PstI-digested plasmid pGAD424.
Isolation and characterization of the CBP1 genomic locus
Genomic DNA was isolated from C.neoformans serotype A strain H99 as described (Pitkin et al., 1996). DNA from the CBP1 two-hybrid library clone was gel-purified, random-primer labeled, and used to probe digested genomic DNA immobilized on Nytran+ membrane (Shleicher and Schuell) as described by the manufacturer. The CBP1 locus was mapped and a sub-genomic library of PstI-cleaved genomic DNA was size-selected, ligated into PstI-cleaved/Calf Intestine Alkaline Phosphatase-treated pBluescript SK, and screened by colony hybridization to isolate an 11.0 kb PstI fragment containing the entire CBP1 gene. One strongly hybridizing clone was isolated, mapped to confirm that it contained the CBP1 locus, and sequenced. Comparison of genomic sequence with the two-hybrid clone identified the intron– exon borders.
GST tagging of the CBP1 protein and calcineurin binding assays
A C-terminal truncation of the CBP1 coding region (residues 1–153) was obtained by amplification with primers CGCGTCGGATCCTATCCCCAGCAA (1782) and CTCCAGGAATTCCAGTGCACGTTG (2056) and cloned in-frame into the BamHI–EcoRI sites within the polylinker of the E.coli gene fusion vector pGEX-2TK (Pharmacia). The resulting GST–CBP1 fusion was expressed in TOP10 cells (Invitrogen).
Calcineurin binding assays with GST–CBP1 were performed using 50 µl of GST or GST–CBP1 bound to glutathione agarose per reaction in the presence or absence of 1 U (50 µg) of calcineurin (Sigma), 1 U (2 µg) of calmodulin (Sigma), 4 mM EGTA, 8 µg of FKBP12 (Sigma) or 20 µM FK506. Reactions were incubated for 1 h at 4°C in reaction buffer (50 mM Tris–HCl pH 7.4, 100 mM NaCl, 2 mM MgCl2, 0.2% Triton X-100, 7 mM CaCl2 and phenylmethylsulfonyl fluoride) prior to the addition of GST or GST–CBP1 substrate. Binding reactions were incubated at 4°C for 2 h, followed by two washes in reaction buffer. Samples were separated on 10% SDS–PAGE, transferred to PVDF and immunoblotted with polyclonal anti-calcineurin antibody (Upstate Biotechnology) or monoclonal anti-GST antibody (Santa Cruz Biotechnology) and visualized with ECL (Amersham).
Calcineurin phosphatase assays
Calcineurin phosphatase assays were performed with [32P]RII peptide as previously described (Liu et al., 1991; Heitman et al., 1993). Peptides were provided by the HHMI peptide synthesis facility at DUMC. Peptide sequences are as follows: [DSCR1 18] KQFLISPPASPPVGWKQV and [CBP1 18] HNFLISPPGSPPEGWEPA. The calcineurin autoinhibitory domain peptide [AID] ITSPEEAKGLDRINERMPPRRDAMP was obtained from Calbiochem. Prior to the addition of substrate, reactions containing calcineurin in the presence or absence of inhibitors were incubated at 30°C for 30 min. Reactions were terminated 20 min after the addition of substrate. Assays using pNPP were performed as described previously (Haddy et al., 1992).
In vivo tagging of the CBP1 protein
The genomic CBP1 locus was PCR amplified from the cloned 11.0 kb PstI fragment using forward primer 5′-TATTGTTGAGAAGTACTC (2387) and reverse primer 2041 and cloned into pCR2.1-TOPO (Invitrogen) resulting in plasmid pCn3gS. The open reading frame of GFP was amplified from a plasmid template (Del Poeta et al., 1999) using forward primer 5′-TATTAAAAGGCCTAAAGGT (2392) and reverse primer 5′-CTGAGGCCTTTTGTACAA (2465), digested with StuI and ligated into StuI-digested pCn3gS plasmid DNA. A clone with GFP in the correct orientation was digested with SpeI and NotI to release the CBP1::GFP gene fusion. This fragment was ligated into SpeI- and NotI-digested pHYG7-KB1 (Hua et al., 2000) to yield plasmid pCn3gS–GFP, which was transformed into C.neoformans strains H99, M049, JMC3 and AO4 (Y5) (Odom et al., 1997; Cruz et al., 2000) by biolistic transformation. Multiple hygromycin B resistant strains were isolated from each transformation event and were single colony purified. Cell extracts were prepared from at least two transformants of each strain (Odom et al., 1997), separated on 12% SDS–PAGE, transferred to nitrocellulose, and probed with Anti-Aquorea victoria GFP antibody. Subsequently an anti-rabbit IgG secondary antibody was applied and detected with the ECL detection system. All strains harboring the pCn3gS–GFP plasmid expressed a protein of ∼60 kDa that cross-reacted with the anti-GFP antibody; those lacking the plasmid had no cross-reacting protein. Highly conserved serine residues including those within the highly conserved SP domain of CBP1 were replaced with alanines by site-directed mutagenesis by PCR overlap.
Gene replacement of the CBP1 open reading frame
The gene replacement construct was generated as follows. Forward primer 5′-TAGTTCGGAATTCCACAATC (2038) and reverse primer 5′-TGAAGGGATCCTGCGCGCGATATGG (2039) were used to amplify a 920 bp fragment of the promoter and 5′ untranslated region of the CBP1 gene. The PCR product was digested with EcoRI and BamHI and ligated into EcoRI- and BamHI-digested pUC119 to yield plasmid pJMM152. The 550 bp 3′ untranslated region and terminator of the CBP1 gene was amplified using forward primer 5′-TTTATGGTCGACCAGCGCGCGAGTCT (2040) and reverse primer 5′-GTATGCTTAAGCTTCTTCTCGG (2041), digested with SalI and HindIII, and ligated into plasmid pJMM152 digested with SalI and HindIII, yielding plasmid pJMM154. The ADE2 genomic locus was amplified from plasmid pCnade2ΔApaI (Sudarshan et al., 1999) using forward primer 5′-GGGTACCGCGCGCTTGAACGCC (1938) and reverse primer 5′-CAGGTGAAGTTGCGCGCGAAACGAAC (1939), digested with BssHII and ligated into plasmid pJMM154, yielding plasmid pJMM160. Approximately 10 µg of circular form pJMM160 plasmid DNA was transformed into the C.neoformans ade2 strain M049 by biolistic transformation (Toffaletti et al., 1993; Sudarshan et al., 1999). Transformants were isolated on synthetic regeneration medium lacking adenine. Approximately 530 transformants were isolated and screened by PCR amplification. Primers 5′-GGACGAGAGCAAGCAAGTCCG (1732) and 5′-CAGTTGAAGAGATGTTGCTGGGG (1612) amplify a 420 bp product from the CBP1 wild-type locus and no product from the cbp1Δ::ADE2 allele. Strains lacking the 420 bp PCR product were rescreened by Southern analysis to identify strains in which the CBP1 gene had been replaced by the ADE2 gene.
Northern blot analysis
Total RNA was isolated from C.neoformans strain H99 grown in 10 ml of YPD medium at 30°C as described (Chomczynski and Sacchi, 1987). Briefly, cells were centrifuged in a tabletop centrifuge and the cell pellet was frozen at –80°C and lyophilized overnight to complete dryness. The pellet was lysed by vortexing with 2 mm glass beads. To the pulverized cells was added 500 µl of RNA extraction buffer (guanidine thiocyanate, EDTA), 100 µl of 2 M sodium acetate pH 5.2, 200 µl of water-saturated phenol and 200 µl of chloroform, with vortexing after each addition. Cells were incubated on ice for 30 min and subsequently centrifuged at maximum speed in a tabletop centrifuge for 10 min at 4°C. The aqueous phase was transferred to an Eppendorf tube and an equal volume of isopropanol was added. RNA was precipitated on ice for 10 min and then centrifuged at maximum speed in a microfuge for 5 min. The pellet was washed with 70% ethanol, dried, and resuspended in ∼100 µl of water. RNA blot analysis was performed as described using Nytran+ membranes (Schleicher and Schuell).
Isolation of S.cerevisiae cbp1 mutant strain
The S.cerevisiae YKL159c gene was disrupted by PCR-mediated gene disruption with the KanMX gene cassette (Wach et al., 1994; Lorenz et al., 1995). Flanking primers with 40 bp of homology to the 5′ and 3′ regions of the YKL159c and 20 bp of homology to the KanMX cassette used for PCR amplification were: (2825) 5′-AAGCAATAAACCAACCGATATATAAAACACAGAACTGCAGCAGCTGAAGCTTCGTA CGC and (2826) 5′-GCATTTAAGTCTCTTAAGCCAACAAATCGCCTCGCCATCTGCATAGGCCACTAGTGGATCTG. The ykl159cΔ::G418 gene disruption PCR product was transformed into yeast strain JK9-3da, G418-resistant transformants were selected, and gene disruption was confirmed by isolation of genomic DNA and PCR analysis with the flanking primer (2827) CCATATTTACTTAGGTCA and primer 2825.
Acknowledgments
Acknowledgements
We thank Maria Cardenas for advice and discussions, Cristina Cruz for comments on the manuscript, Dena Toffaletti for technical assistance, Miguel Arevalo-Rodriguez for assistance with figures, Kyle Cunningham for communication of results prior to publication, and Jenny Lodge for the hygromycin resistance marker. These studies were supported by RO1 grants AI39115 and AI42159 from the NIAID (to J.R.P. and J.H.), 5T32 training grant AI07392 from the NIAID (to D.S.F.), and PO1 grant AI44975 from NIAID to the Duke University Mycology Research Unit. G.M.C. is a Burroughs Wellcome New Investigator in Molecular Pathogenic Mycology. J.H. is a Burroughs Wellcome Scholar in Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
References
- Arndt C., Cruz,M.C., Cardenas,M.E. and Heitman,J. (1999) Secretion of FK506/FK520 and rapamycin by Streptomyces inhibits the growth of competing Saccharomyces cerevisiae and Cryptococcus neoformans. Microbiology, 145, 1989–2000. [DOI] [PubMed] [Google Scholar]
- Bartel P., Chien,C.T., Sternglanz,R. and Fields,S. (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques, 14, 920–924. [PubMed] [Google Scholar]
- Breuder T., Hemenway,C.S., Movva,N.R., Cardenas,M.E. and Heitman,J. (1994) Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc. Natl Acad. Sci. USA, 91, 5372–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M.E., Hemenway,C., Muir,R.S., Ye,R., Fiorentino,D. and Heitman,J. (1994) Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. EMBO J., 13, 5944–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M.E., Muir,R.S., Breuder,T. and Heitman,J. (1995) Targets of immunophilin–immunosuppressant complexes are distinct highly conserved regions of calcineurin A. EMBO J., 14, 2772–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M.E., Sanfridson,A., Cutler,N.S. and Heitman,J. (1998) Signal-transduction cascades as targets for therapeutic intervention by natural products. Trends Biotechnol., 16, 427–433. [DOI] [PubMed] [Google Scholar]
- Cardenas M.E., Cruz,M.C., Del Poeta,M., Chung,N., Perfect,J.R. and Heitman,J. (1999) Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action. Clin. Microbiol. Rev., 12, 583–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.Y., Takei,K., Sydor,A.M., Born,T., Rusnak,F. and Jay,D.G. (1995) Asymmetric retraction of growth cone filopodia following focal inactivation of calcineurin. Nature, 376, 686–690. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Coghlan V.M., Perrino,B.A., Howard,M., Langeberg,L.K., Hicks,J.B., Gallatin,W.M. and Scott,J.D. (1995) Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science, 267, 108–111. [DOI] [PubMed] [Google Scholar]
- Cruz M.C., Cavallo,L.M., Görlach,J.M., Cox,G., Perfect,J.R., Cardenas,M.E. and Heitman,J. (1999) Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell. Biol., 19, 4101–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M.C., Sia,R.A.L., Olson,M., Cox,G.M. and Heitman,J. (2000) Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect. Immun., 68, 982–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K.W. and Fink,G.R. (1994) Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol., 124, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K.W. and Fink,G.R. (1996) Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2226–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M.S. and Thorner,J. (1992) Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol., 12, 3460–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Poeta M., Toffaletti,D.L., Rude,T.H., Sparks,S.D., Heitman,J. and Perfect,J.R. (1999) Cryptococcus neoformans differential gene expression detected in vitro and in vivo with green fluorescent protein. Infect. Immun., 67, 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A., Kincaid,R. and Kosik,K.S. (1993) Calcineurin is associated with the cytoskeleton of cultured neurons and has a role in the acquisition of polarity. Mol. Biol. Cell, 4, 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foor F., Parent,S.A., Morin,N., Dahl,A.M., Ramadan,N., Chrebet,G., Bostian,K.A. and Nielsen,J.B. (1992) Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from α-factor arrest in yeast. Nature, 360, 682–684. [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Pai,S.-Y., Burakoff,S.J. and Bierer,B.E. (1995) Characterization of a mutant calcineurin Aα gene expressed by EL4 lymphoma cells. Mol. Cell. Biol., 15, 3857–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes J.J., Pritchard,M.A., Planas,A.M., Bosch,A., Ferrer,I. and Estivill,X. (1995) A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum. Mol. Genet., 4, 1935–1944. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl,R.H., Willems,A. and Woods,R.A. (1995) Studies on the mechanism of high efficiency transformation of intact yeast cells. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Gorovits R., Propheta,O., Kolot,M., Dombradi,V. and Yarden,O. (1999) A mutation within the catalytic domain of COT1 kinase confers changes in the presence of two COT1 isoforms and in Ser/Thr protein kinase and phosphatase activities in Neurospora crassa. Fungal Genet. Biol., 27, 264–274. [DOI] [PubMed] [Google Scholar]
- Griffith J.P. et al. (1995) X-ray structure of calcineurin inhibited by the immunophilin–immunosuppressant FKBP12–FK506 complex. Cell, 82, 507–522. [DOI] [PubMed] [Google Scholar]
- Haddy A., Swanson,S.K.-H., Born,T.L. and Rusnak,F. (1992) Inhibition of calcineurin by cyclosporin A–cyclophilin requires calcineurin B. FEBS Lett., 314, 37–40. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Perrino,B.A. and Soderling,T.R. (1990) Identification of an autoinhibitory domain in calcineurin. J. Biol. Chem., 265, 1924–1927. [PubMed] [Google Scholar]
- Heitman J., Movva,N.R., Hiestand,P.C. and Hall,M.N. (1991) FK506-binding protein proline rotamase is a target for the immunosuppressive agent FK506 in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 88, 1948–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Koller,A., Cardenas,M.E. and Hall,M.N. (1993) Identification of immunosuppressive drug targets in yeast. Methods, 5, 176–187. [Google Scholar]
- Hemenway C.S. and Heitman,J. (1999) Calcineurin: structure, function and inhibition. Cell Biochem. Biophys., 30, 115–151. [DOI] [PubMed] [Google Scholar]
- Hemenway C.S., Dolinski,K., Cardenas,M.E., Hiller,M.A., Jones,E.W. and Heitman,J. (1995) vph6 mutants of Saccharomyces cerevisiae require calcineurin for growth and are defective in vacuolar H+-ATPase assembly. Genetics, 141, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Meyer,J.D. and Lodge,J.K. (2000) Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol., 7, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard M.J. and Klee,C.B. (1989) Functional domain structure of calcineurin A: mapping by limited proteolysis. Biochemistry, 28, 1868–1874. [DOI] [PubMed] [Google Scholar]
- Jackson S.L. and Heath,I.B. (1993) Roles of calcium ions in hyphal tip growth. Microbiol. Rev., 57, 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashishian A., Howard,M., Loh,C., Gallatin,W.M., Hoekstra,M.F. and Lai,Y. (1998) AKAP79 inhibits calcineurin through a site distinct from the immunophilin-binding region. J. Biol. Chem., 273, 27412–27419. [DOI] [PubMed] [Google Scholar]
- Kissinger C.R. et al. (1995) Crystal structures of human calcineurin and the human FKBP12–FK506–calcineurin complex. Nature, 378, 641–644. [DOI] [PubMed] [Google Scholar]
- Klauck T.M., Faux,M.C., Labudda,K., Langeberg,L.K., Jaken,S. and Scott,J.D. (1996) Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science, 271, 1589–1592. [DOI] [PubMed] [Google Scholar]
- Kothe G.O. and Free,S.J. (1998) Calcineurin subunit B is required for normal vegetative growth in Neurospora crassa. Fungal Genet. Biol., 23, 248–258. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Burnett,P.E., Wolosker,H., Blackshaw,S. and Snyder,S.H. (1998) Cain, a novel physiologic protein inhibitor of calcineurin. J. Biol. Chem., 273, 18325–18331. [DOI] [PubMed] [Google Scholar]
- Liu J., Farmer,J.D., Lane,W.S., Friedman,J., Weissman,I. and Schreiber,S.L. (1991) Calcineurin is a common target of cyclophilin–cyclosporin A and FKBP–FK506 complexes. Cell, 66, 807–815. [DOI] [PubMed] [Google Scholar]
- Loh C., Shaw,K.T.-Y., Carew,J., Viola,J.P.B., Luo,C., Perrino,B.A. and Rao,A. (1996) Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J. Biol. Chem., 271, 10884–10891. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C. and Heitman,J. (1995) TOR mutations confer rapamycin resistance by preventing interaction with FKBP12–rapamycin. J. Biol. Chem., 270, 27531–27537. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Muir,R.S., Lim,E., McElver,J., Weber,S.C. and Heitman,J. (1995) Gene disruption with PCR products in Saccharomyces cerevisiae.Gene, 158, 113–117. [DOI] [PubMed] [Google Scholar]
- Mansuy I.M., Mayford,M., Jacob,B., Kandel,E.R. and Bach,M.E. (1998) Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell, 92, 39–49. [DOI] [PubMed] [Google Scholar]
- Matheos D.P., Kingsbury,T.J., Ahsan,U.S. and Cunningham,K.W. (1997) Tcn1p/Crzlp, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev., 11, 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P., Morin,N., Baginsky,W., El-Sherbeini,M., Clemas,J.A., Nielsen,J.B. and Foor,F. (1995) Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol., 15, 5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T., Kanou,Y., Murata,U., Ohmori,S., Niwa,T., Maeda,K., Yamamura,H. and Seo,H. (1996) Molecular cloning of a novel thyroid hormone-responsive gene, ZAKI-4, in human skin fibroblasts. J. Biol. Chem., 271, 14567–14571. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Lu,J.R., Antos,C.L., Markham,B., Richardson,J., Robbins,J., Grant,S.R. and Olson,E.N. (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell, 93, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Geiser,J.R. and Davis,T.N. (1996) Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol., 16, 4824–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkey R.M., Endo,S., Shenolikar,S. and Malenka,R.C. (1994) Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature, 369, 486–488. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Liu,Y., Hirata,D., Namba,H., Harada,S.-i., Hirokawa,T. and Miyakawa,T. (1993) Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J., 12, 4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A., Muir,S., Lim,E., Toffaletti,D.L., Perfect,J. and Heitman,J. (1997) Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J., 16, 2576–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect J.R., Toffaletti,D.L. and Rude,T.H. (1993) The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun., 61, 4446–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkin J.W., Panaccione,D.G. and Walton,J.D. (1996) A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology, 142, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Plochocka-Zulinska D., Rasmussen,G. and Rasmussen,C. (1995) Regulation of calcineurin gene expression in Schizosaccharomyces pombe. J. Biol. Chem., 270, 24794–24799. [DOI] [PubMed] [Google Scholar]
- Prokisch H., Yarden,O., Dieminger,M., Tropschug,M. and Barthelmess,I.B. (1997) Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol. Gen. Genet., 256, 104–114. [DOI] [PubMed] [Google Scholar]
- Ranger A.M., Grusby,M.J., Hodge,M.R., Gravallese,E.M., de la Brousse,F.C., Hoey,T., Mickanin,C., Baldwin,H.S. and Glimcher,L.H. (1998) The transcription factor NF-ATc is essential for cardiac valve formation. Nature, 392, 186–190. [DOI] [PubMed] [Google Scholar]
- Rasmussen C., Garen,C., Brining,S., Kincaid,R.L., Means,R.L. and Means,A.R. (1994) The calmodulin-dependent protein phosphatase catalytic subunit (calcineurin A) is an essential gene in Aspergillus nidulans. EMBO J., 13, 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel B., Vega,R.B., Yang,J., Wu,H., Bassel-Duby,R. and Williams,R.S. (2000) A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem., 275, 8719–8725. [DOI] [PubMed] [Google Scholar]
- Shaw K.T.-Y., Ho,A.M., Raghavan,A., Kim,J., Jain,J., Park,J., Sharma,S., Rao,A. and Hogan,P.G. (1995) Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc. Natl Acad. Sci. USA, 92, 11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Shibasaki F., Price,E.R., Milan,D. and McKeon,F. (1996) Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature, 382, 370–373. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A.M. and Cyert,M.S. (1997) Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev., 11, 3432–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A., Guo,J.J. and Cyert,M.S. (1999) Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev., 13, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshan S., Davidson,R.C., Heitman,J. and Alspaugh,J.A. (1999) Molecular analysis of the Cryptococcus neoformans ADE2 gene, a selectable marker for transformation and gene disruption. Fungal Genet. Biol., 27, 36–48. [DOI] [PubMed] [Google Scholar]
- Sun L., Youn,H.-D., Loh,C., Stolow,M., He,W. and Liu,J.O. (1998) Cabin 1, a negative regulator for calcineurin signaling in T lymphocytes. Immunity, 8, 1–20. [DOI] [PubMed] [Google Scholar]
- Sussman M.A. et al. (1998) Prevention of cardiac hypertrophy in mice by calcineurin inhibition. Science, 281, 1690–1693. [DOI] [PubMed] [Google Scholar]
- Toffaletti D.L., Rude,T.H., Johnston,S.A., Durack,D.T. and Perfect,J.R. (1993) Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol., 175, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wesselborg S., Furman,D.A., Sagoo,J.K., Bierer,B.E. and Burakoff,S.J. (1996) Identification of a physical interaction between calcineurin and nuclear factor of activated T cells (NFATp). J. Biol. Chem., 271, 1274–1277. [DOI] [PubMed] [Google Scholar]
- Winder D.G., Mansuy,I.M., Osman,M., Moallem,T.M. and Kandel,E.R. (1998) Genetic and pharmacological evidence for a novel, intermediate phase of long-term potentiation suppressed by calcineurin. Cell, 92, 25–37. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Toda,T. and Yanagida,M. (1994) A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J. Cell Sci., 107, 1725–1735. [DOI] [PubMed] [Google Scholar]