Abstract
Abnormalities in oligodendrocyte (OLG) differentiation and OLG gene expression deficit have been described in schizophrenia (SZ). Recent studies revealed a critical requirement for Disrupted-in-Schizophrenia 1 (DISC1) in neural development. Transgenic mice with forebrain restricted expression of mutant human DISC1 (ΔhDISC1) are characterized by neuroanatomical and behavioral abnormalities reminiscent of some features of SZ. We sought to determine whether the expression of ΔhDISC1 may influence the development of OLGs in this mouse model.
OLG- and cell cycle-associated gene and protein expression were characterized in the forebrain of ΔhDISC1 mice during different stages of neurodevelopment (E15 and P1 days) and in adulthood.
The results suggest that the expression of ΔhDISC1 exerts a significant influence on oligodendrocyte differentiation and function, evidenced by premature OLG differentiation and increased proliferation of their progenitors. Additional findings showed that neuregulin 1 and its receptors may be contributing factors to the observed upregulation of OLG genes.
Thus, OLG function may be perturbed by mutant hDISC1 in a model system that provides new avenues for studying aspects of the pathogenesis of SZ.
Keywords: Schizophrenia, disrupted-in schizophrenia 1, gene expression, oligodendrocyte, oligodendrogenesis, myelin, neuregulin
1. INTRODUCTION
Evidence for the involvement of oligodendrocytes (OLGs) and myelin in the pathophysiology of SZ has come from analysis of postmortem tissue from persons with schizophrenia (SZ) using gene (Hakak et al., 2001; Bahn and Jones, 2003; Tkachev et al., 2003; Aston et al., 2004; Katsel et al., 2005a; Katsel et al., 2005b; Iwamoto et al., 2005; Dracheva et al., 2006; Aberg et al., 2006; Barley et al., 2009) and protein expression studies (Flynn et al., 2003; Dracheva et al., 2006; Martins-de-Souza, 2010); light and electron microscopic studies (Miyakawa et al., 1972; Uranova et al., 2001; Uranova et al., 2004; Vostrikov et al., 2007), and in vivo neuroimaging studies (Buchsbaum et al., 1998; Lim et al., 1999; Foong et al., 2000; Foong et al., 2001; Bartzokis et al., 2003). In addition, morphometric studies of persons with SZ have shown reductions in glial cell densities in the anterior cingulate cortex (Cotter et al., 2001; Stark et al., 2004) as well as layer-specific reductions of glia in the dorsolateral prefrontal cortex (Hof et al., 2003; Rajkowska et al., 2002). We recently showed that the widespread OLGs/myelin abnormalities observed in SZ are associated with cell cycle abnormalities and incomplete differentiation of OLGs (Katsel et al., 2008; Kerns et al., 2010). Animal model studies have demonstrated that OLG-related cell cycle abnormalities can contribute to pervasive myelin deficits providing further insights into the potential neurobiological mechanisms of OLG-associated deficits in SZ (Katsel et al., 2008).
Disrupted-in schizophrenia 1 (DISC1) is a strong candidate gene for schizophrenia (SZ) and an association at the DISC1 locus has been demonstrated for a range of other mental disorders (Blackwood et al., 2001; Hennah et al., 2009; Macgregor et al., 2004), suggesting broad relevance of DISC1 for psychiatric illnesses. Detected in a large Scottish family (St Clair et al., 1990), the genomic translocation that disrupts DISC1 on chromosome 1q42 (Millar et al., 2000; Millar et al., 2001) may lead to haploinsufficiency and/or the production of a mutant truncated and presumably functionally deficient DISC1 protein. Studies of a number of DISC1 mutant or deficient mice have revealed behavioral and anatomical deficits (Shen et al., 2008; Clapcote et al., 2007; Hikida et al., 2007; Pletnikov et al., 2008) that can be related to psychiatric illness and have prompted a number of studies focusing on functions and pathways that may be affected by aberrant DISC1. Two recent studies in zebrafish have provided a potential linkage between OLG and DISC1. These studies suggest a critical role for DISC1 in oligodendroglial differentiation early in the development of the CNS and PNS (Wood et al., 2009; Drerup et al., 2009).
A recently developed transgenic mouse model with forebrain restricted inducible dominant negative expression of human mutant DISC1 showed the behavioral abnormalities and neuroanatomical changes reminiscent of SZ (Pletnikov et al., 2008), including enlargement of lateral ventricles and attenuated neurite outgrowth. Behavioral abnormalities included spontaneous hyperactivity and aggressive behaviors in male mutant mice and deficient spatial memory and elevated depression-like responses in female mutant mice (Pletnikov et al., 2008; Ayhan et al., 2010). The current study used this mouse model to evaluate the effects of mutant DISC1 on gene and protein expression of cell cycle proteins and OLG markers in several brain regions of ΔhDISC1 mice during different stages of neurodevelopment and adulthood.
The results demonstrate that forebrain restricted expression of mutant ΔhDISC1 is associated with upregulation of gene expression of markers of OLG precursor cells and mature myelinating OLGs in the forebrain during early development and adulthood. Mutant ΔhDISC1 expression is also associated with widespread abnormalities in the expression of cell cycle genes and oligodendroglial progenitor markers, suggesting expansion of glial progenitors in forebrain regions with a concurrent reduction of glial progenitor marker in the cerebellum. These findings support the hypothesis that the expression of ΔhDISC1 can exert a significant influence on oligodendrocyte differentiation, proliferation and function and that disturbance in DISC1 may at least partially contribute to oligodendroglia-associated abnormalities in SZ.
2. MATERIAL AND METHODS
2.1. Ethics Statement
Animal procedures were approved by the institutional review board of Johns Hopkins University School of Medicine.
2.2. Generation of ΔhDISC1 mouse
Inducible ΔhDISC1 mice were generated as described previously (Pletnikov et al., 2008). Inducible expression of mutant ΔhDISC1 (truncated – 64kD protein (Pletnikov et al., 2008)) was based on the Tet-off system. DISC1 transgenic mice B6;SJL-Tg(TRE-CMV-ΔhDISC1) were bred with single transgenic B6; CBA-Tg (Camk2a-tTA)1Mmay/j mice (The Jackson Laboratory) to generate double transgenic mice of the hybrid B6;SJL;CBA background. Forebrain-restricted inducible expression of mutant ΔhDISC1 protein was achieved because single transgenic B6 CBA-Tg(Camk2a-tTA)1Mmay/j mice express tTA under the control of the CAMKII promoter which yields a predominant expression of the transgene in the forebrain. tTA mice were used as controls.
In some experiments as described below, expression of ΔhDISC1 was controlled by doxycycline (DOX) added to mouse chow. DOX binds to tTA and blocks its biding to TRE, leading to suppression of transcription of ΔhDISC1. Delayed expression of ΔhDISC1 mice (only postnatal expression) were achieved by mating prospective parents on DOX enriched food that was continuously provided to pregnant dams until embryonic day 12 (E12). At E12, DOX enriched food was switched to regular food, and dams and their offspring were maintained on regular food until they were sacrificed (Ayhan et al., 2010).
2.3. Sample Information and Preparation of Total RNA
Genotypes were distinguished by PCR using conditions outlined previously (Pletnikov et al., 2008). Double transgenic ΔhDISC1 mice at embryonic day 15 (E15) (N=5); postnatal day 1 (P1; N=6); 12 month old (N=6), and single transgenic tTA mice at E15, P1 and 12 months of age were used as controls (N=5–6/group). Mice were sacrificed by decapitation and the brains were removed and immediately frozen before dissection. Each brain was divided midsagitally. The left halves of the brains were stored at −80°C. The right halves were dissected. Dissections of adult – 12 months old mice included the frontal cortex grey matter (from the frontal pole to the beginning of the parietal cortex (bregma −1.8 (Franklin and Paxinos, 1997)), white matter - the corpus callosum underlying the dissected frontal cortical grey matter, the hippocampus and cerebellum. The border of the frontal cortex and posterior cortical regions was identified as the coronal block where the middle cerebral artery bifurcates from the internal carotid artery (Zeman and Innes, 1963) and the dissections were limited to the cortical region dorsal to the rhinal fissure. Dissections of P1 day mice included the frontal cortex grey matter and cerebellum. Brains of E15 day mice were cut at the midbrain (coronal plane) resulting two parts: frontal (forebrain) and posterior (hindbrain). Aliquots (2–5 mg) from each dissected region were used for RNA isolation. Similarly, prepared aliquots from the frontal cortex (12 months old mice) or right frontal cortex (P1 day mice) were used in Western blot analyses. Brain samples from: frontal cortex, cerebellum and white matter (12 months old); frontal region and cerebellum (P1)/hindbrain (E15) were homogenized in 300 μl of lysis buffer and total RNA isolation was performed using a Maxwell 16 Nucleic Acid/Protein Isolation system (Promega Corp., Madison, WI) and Total RNA Purification Kits (Promega Corp). cDNA was generated from 2μg of total RNA using High-Capacity cDNA synthesis kits (Applied Biosystems, Foster City, CA).
2.4. RT-qPCR
The mRNA levels of selected neuronal, myelin and cell cycle genes were measured by qPCR using TaqMan® probes and primer sets (Supp. Table 1) using ABI Prism® 7900HT Sequence Detection System (all from Applied Biosystems). For relative quantification of mRNA expression, relative values of examined genes were calculated using the standard curve method which were further normalized to the geometric means (GMs) of endogenous control-genes as described previously.(Dracheva et al., 2005) Two housekeeping genes (GAPDH and PPIA) were used as the endogenous references.
2.5. Quantitative Western Blotting
Protein abundance was measured in the cortex and cerebellum from P1, P21 and 12 months old mice expressing ΔhDISC1 and controls using Western blotting. Tissue specimens (5mg) were homogenized in Tris/Triton solution: 250 mM sucrose, 50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 2 mM EGTA, 1% Triton X100 containing 1mM PMSF and supplemented with complete cocktails of proteinase/phosphatase inhibitors (Pierce Biotech Inc, Rockford, IL). Total protein concentration in the tissue homogenates was determined with a CBQCA Quantitation Kit (Molecular Probes Inc, Eugene, OR). Aliquot samples of 15μg of total protein in triplicate were loaded onto pre-cast 12% HEPES-glycine gels (Pierce Biotech Inc) under reduced conditions. A “standard-calibrator” (a mix of small aliquots of tissue from all samples) was used as a calibrator between the gels and run on each gel in triplicate. Blots were incubated with antibodies: mouse phosphodiesterase Cnp (CNP; 1:700 v/v) and mouse anti- platelet-derived growth factor receptor α (PDGFRA; 1:1000 v/v) – both from LifeSpan Biosciences (Seattle, WA); rabbit anti-mouse DISC1 (described previously(Pletnikov et al., 2008)); mouse anti-human GAPDH from Meridian Life Science, Inc. (Saco, ME) and rabbit anti-human TUBB (Novus Biologicals, Littleton, CO). Electrophoresis, blotting, immunostaining, and infrared (IR) fluorescence detection (IRDye 680 or 800 Goat Anti-appropriate species IgG, Li-Cor Biosciences, Lincoln, NE) were performed under standard conditions. Multiplex western blots were scanned on an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). The linearity of the dose responses for the antibodies used was established in preliminary experiments. Images were analyzed and quantitated with Odyssey software ver.3 (Li-Cor Biosciences, Lincoln, NE). To account for gel to gel variability, the relative expression value (REV) of analyzed proteins in each sample was calculated as a ratio between the averaged intensities of the band in the experimental sample and in the “standard-calibrator”. GAPDH and TUBB were used as loading controls.
2.6. Statistical Data Analysis
A two-tailed Student’s t-test was used to compare relative mRNA expression of analyzed genes in qPCR experiments and relative abundance of proteins in Western blots. Student’s t-test and correlation analyses were performed using Statistica (release 6.0).
3. RESULTS
3.1. Expression of oligodendrocyte lineage genes is strongly upregulated in frontal cortex and white matter, but not in the cerebellum of 12 months old ΔhDISC1 mice
qPCR analysis of 9 genes specific to OLG lineage cells included PDGFRA - platelet derived growth factor receptor alpha, CNP1 - phosphodiesterase Cnp, OLIG2 - oligodendrocyte transcription factor 2, SOX10 – SRY-box containing gene 10, QKI (5, 6 and 7 isoforms) – quaking homolog, KH domain RNA binding, MAG - myelin-associated glycoprotein, PLP - proteolipid protein 1 and CLDN11 – claudin 11 in frontal cortex, cerebellum and white matter of the 12 months old ΔhDISC1 mice. Marker selection was based on their temporal expression profile during maturation of myelinating OLGs (Miller and Reynolds, 2004).
The results of this study are shown in Figure 1 and showed significantly increased expression of eight genes: PDGFRA, CNP1, OLIG2, SOX10, QKI (5, 6 isoforms), MAG, PLP and CLDN11 (Table 1; all ps≤ 0.01) in frontal cortex and white matter (Table 1; all ps≤ 0.05), but not in cerebellum where only OLIG2 was significantly increased (Table 1; p≤ 0.001) in ΔhDISC1 mice compare to controls. Only isoform 7 of quaking homolog gene (QKI-7), which is more abundant during maturation and myelination stages of OLG development, was significantly decreased in frontal cortex and in cerebellum (Table 1; p≤ 0.001) of ΔhDISC1 mice, but it was unchanged in the white matter. In contrast to the cortex and white matter, gene expression of PDGFRA and PLP was significantly decreased (Table 1; p≤ 0.001) in the cerebellum of ΔhDISC1 mice compare to controls. Furthermore, expression of OLG lineage genes was highest in the white matter (4–10 folds higher than in frontal cortex) and smallest in the cerebellum confirming the fidelity of regional dissections.
Figure 1.
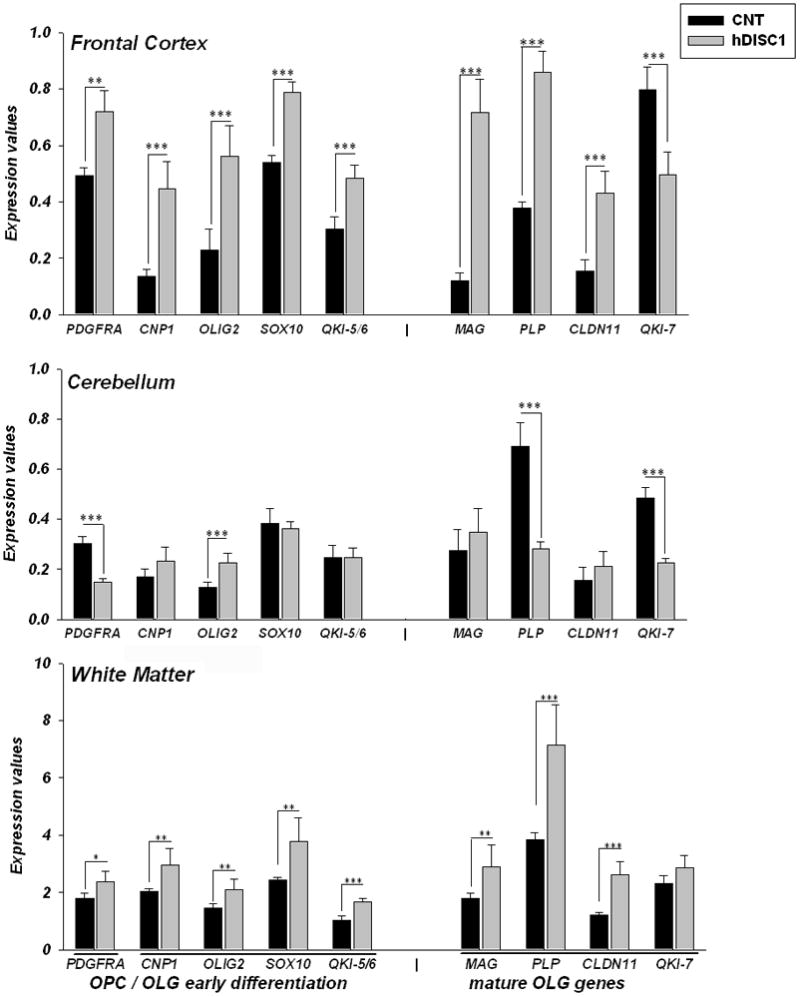
Gene expression levels of the markers of OLG lineage cells were significantly upregulated in the frontal cortex and white matter, but less so in the cerebellum of ΔhDISC1 (hDISC1) mice compared to control animals (CNT) at 12 months of age. Data are expressed as geometric means ±SEM of individual expression values normalized to the housekeeping genes as described in Methods. N=5–6/group. * p≤0.05; **p≤0.01; *** p≤0.001
Table 1.
Gene expression analysis of the oligodendrocyte lineage genes in frontal cortex (FrCtx), cerebellum (Crb) and the white matter (WM) in 12 months old mice.
| Gene | OLG Lineage marker | FC ratio FrCtx | p-val | FC ratio Crb | p-val | FC ratio WM | p-val |
|---|---|---|---|---|---|---|---|
| PDGFRA | OPC | 1.46 | 5E-04 | −2.00 | 1E-08 | 1.33 | 0.017 |
| CNP1 | OLG | 3.26 | 1E-05 | 1.37 | ns | 1.47 | 0.007 |
| OLIG2 | OLG | 2.45 | 2E-04 | 1.76 | 6E-04 | 1.44 | 0.011 |
| SOX10 | OLG | 1.46 | 2E-09 | −1.06 | ns | 1.56 | 0.006 |
| QKI-5/6 | OLG | 1.60 | 6E-05 | 1.00 | ns | 1.63 | 6E-06 |
| MAG | mature OLG | 6.01 | 1E-08 | 1.28 | ns | 1.62 | 0.015 |
| PLP | mature OLG | 2.27 | 1E-10 | −2.46 | 2E-07 | 1.87 | 4E-04 |
| CLDN11 | mature OLG | 2.79 | 9E-06 | 1.37 | ns | 2.20 | 1E-05 |
| QKI-7 | mature OLG | −1.60 | 8E-05 | −2.09 | 1E-09 | 1.23 | ns |
Gene expression from three brain regions were analyzed by qPCR (N=6/group). FC ratio-fold change ratios represent the ratio of means for each gene normalized to the geometric means of two housekeeping genes between controls and ΔhDISC1 mice. Significantly changed genes (t-test, p≤0.05) are highlighted in bold. ns- non-significant.
These data indicate that overexpression of ΔhDISC1 resulted in increased gene expression of markers of OLG progenitor cells, differentiated OLGs and myelinating OLGs in the frontal cortex and the forebrain white matter, but not in the cerebellum where no expression of ΔhDISC1 was present (Pletnikov et al., 2008).
3.2. Expression of oligodendrocyte related genes is strongly affected in the forebrain of ΔhDISC1 mice throughout development (E15 and P1)
As mutant hDISC1 has been found to be expressed as early as embryonic day 15 (E15) (Ayhan et al., 2010), we sought to determine whether ΔhDISC1expression had similar effects on the expression of OLG lineage genes during development, as it did in adulthood. We tested samples from ΔhDISC1 and control mice at E15 and after birth (postnatal day 1, P1) using qPCR.
The results of this study are shown in Figure 2 and Table 2 and revealed that significant increases in the expression of eight genes: PDGFRA, CNP1, OLIG2, SOX10, QKI (5/6 and 7 isoforms), MAG and PLP (Table 2; all ps≤ 0.05) occurred as early as E15. The most striking effects of ΔhDISC1 expression were detected for the transcription factor - SOX10 (~11 fold increase, p<0.001) and for MAG (~45 fold increase, p<0.001) in the forebrain of ΔhDISC1 mice compare to control. Only four genes: PDGFRA, CNP, SOX10 and QKI-7 were significantly increased in E15 hindbrain (data not shown, all ps≤ 0.05). The magnitude of changes, however, was much lower than in the forebrain (from 1.3 to 2.2 fold change (FC)). The precise dissection of developing cerebellum at this stage of brain development is difficult. Therefore hindbrain dissections at E15 likely to included portions of the brain stem. OLG lineage genes were less affected right after birth (P1 day). Only four genes: PDGFRA, CNP, MAG and QKI-7 showed significant changes (Fig. 2 and Table 2, all ps≤ 0.05). As was the case in 12 months old mice, PDGFRA, CNP and MAG gene expression was upregulated (FC: 1.3, 2.2 and 2.3, respectively; all ps≤ 0.05) and QKI-7 was downregulated (FC= −1.5; all p= 0.02) in the forebrain of P1 ΔhDISC1 mice.
Figure 2.
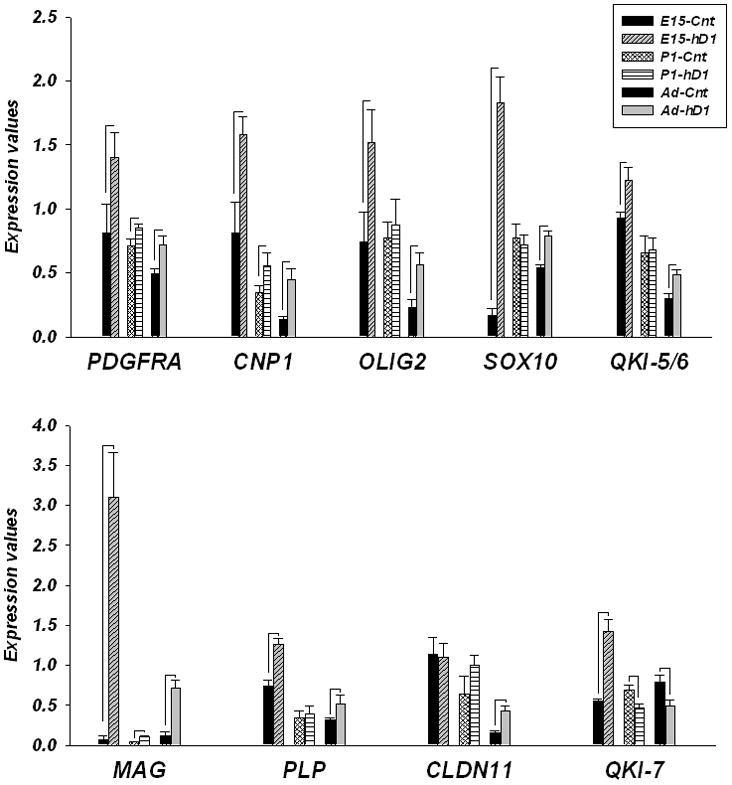
Gene expression levels of glial progenitor cells and OLG-related genes in forebrain regions of control (CNT) and ΔhDISC1 (hDISC1) mice throughout development (E15, P1) and at 12 months of age (Ad). N=5–6/group. Data is expressed as geometric means ± SEM. Significant changes (all ps<0.05) are marked by lines.
Table 2.
Gene expression analysis of the oligodendrocyte lineage genes in frontal cortex during development (E15, P1) and in 12 months old mice.
| Gene | OLG lineage marker | FC- E15 | p-val | FC- P1 | p-val | FC- 12mo | p-val |
|---|---|---|---|---|---|---|---|
| PDGFRA | OPC | 1.73 | 0.03 | 1.25 | 0.03 | 1.46 | 5E-04 |
| CNP1 | OLG | 1.94 | 0.01 | 2.19 | 2E-04 | 3.26 | 1E-05 |
| OLIG2 | OLG | 2.05 | 0.05 | 1.13 | ns | 2.45 | 2E-04 |
| SOX10 | OLG | 10.93 | 1E-04 | −1.07 | ns | 1.46 | 2E-09 |
| QKI-5/6 | OLG | 1.31 | 0.02 | −1.14 | ns | 1.60 | 6E-05 |
| MAG | mature OLG | 44.77 | 1E-05 | 2.34 | 0.02 | 6.01 | 1E-08 |
| PLP | mature OLG | 1.71 | 1E-03 | −1.10 | ns | 2.27 | 1E-10 |
| CLDN11 | mature OLG | −1.03 | ns | 1.57 | ns | 2.79 | 9E-06 |
| QKI-7 | mature OLG | 2.54 | 2E-04 | −1.47 | 0.02 | −1.60 | 8E-05 |
Gene expression from frontal cortex were analyzed by qPCR (N=6/group). FC ratio-fold change ratios represent the ratio of means for each gene normalized to the geometric means of two housekeeping genes between controls and ΔhDISC1 mice. Significantly changed genes (t-test, p≤0.05) are highlighted in bold. ns- non-significant.
These data indicate a significant and dramatic increase in expression of markers of OLG progenitor cells, immature OLGs and mature OLGs increases in the forebrain of ΔhDISC1 mice very early during prenatal development (E15), preceding the presumed ontogenetic course of normal active myelination postnatally.
3.3 Gene expression of cell cycle proteins associated with differentiation of OLG progenitor cells is altered in the forebrain, but not in the cerebellum of ΔhDISC1 mice throughout development and adulthood
Strong upregulation of OLG lineage genes as early as E15 suggested premature OLG differentiation. Therefore, mRNA levels of seven cell cycle proteins, expression of which is altered during the initial stages of OLG differentiation (Dugas et al., 2006), was measured in E15, P1 and 12 months old ΔhDISC1 and control mice. As shown in Fig. 3 (Upper panel) and Table 3 gene expression of CDK inhibitors, whose mounting abundances halt cell cycle progression, induces cell cycle exit and initiates OLG differentiation in progenitor cells, was significantly decreased at E15 (CDKN2C (p18Ink4) and CDKN1B (p27 Kip1), FC< −1.3; p<0.05). The expression of CDKN1C (p57Kip2) was unchanged. In contrast to E15, gene expression of p18Ink4 and p57Kip2 was significantly increased at P1 (FC>1.3; p<0.05) and in 12 months old mice (FC>2; p<0.001), while levels of p27 Kip1 remained significantly downregulated (FC< −1.3; p<0.01). These results suggest that at E15 OPCs proliferate but remain in an undifferentiated state and progress through G1/S phases of the cell cycle. While later in development (P1) and in adulthood, the upregulation of gene expression of p18Ink4 and p57Kip2 suggests enhanced differentiation (Fig.3; Upper panel). Contrasting directionality of gene expression changes for CCND2 (Fig.3; Lower panel), which is one of the main cyclins governing transition through the G1 phase of cell cycle, during development and in adult mice (FCs= −2.2 (E15); −1.4 (P1); −1.5 (12 months old), ps<0.05, Table 3), also supports the notion of enhanced differentiation. Three other cell cycle genes including CDC2A/CDK1, RAD51 and FOXN3/CHES1, whose expression is strongly downregulated after OPCs exit the cell cycle and during OLG differentiation [46], were upregulated in 12 months old mice (FC= 3.1, 1.6 and 1.3 respectively, all ps<0.05) as well as at P1 (FC= 1.5 (CDC2A) and 1.3 (FOXN3/CHES1); both ps<0.05; RAD51, p>0.05). Overall, gene expression changes of cell cycle markers in the white matter were similar to those observed for cortical grey matter (Table 3).
Figure 3.
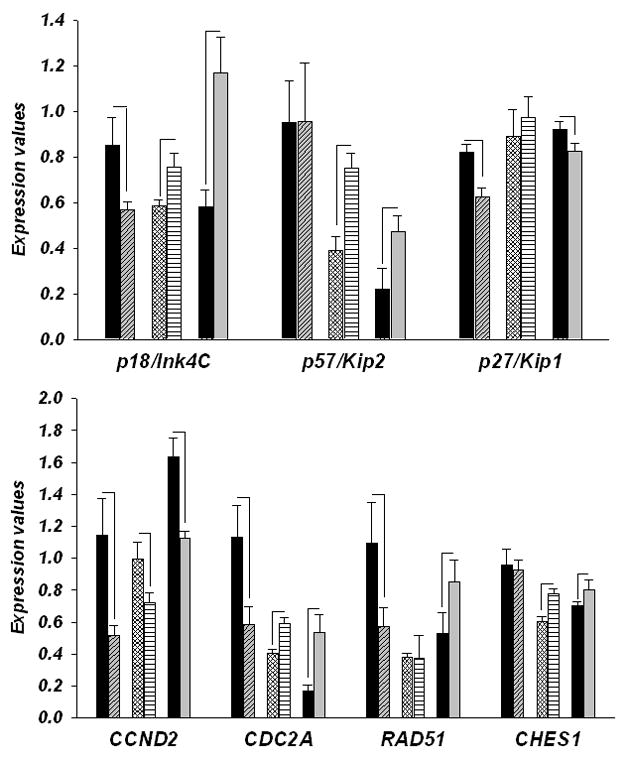
Gene expression of CDK inhibitors and cell cycle genes in forebrain regions of control (CNT) and ΔhDISC1 (hDISC1) mice throughout development (E15, P1) and at 12 months of age (Ad). N=5–6/group. Data is expressed as geometric means ± SEM. Significant changes (p<0.05) are marked by lines.
Table 3.
Gene expression analysis of the cell cycle genes in frontal cortex during development (E15, P1) and 12 months old mice and in white matter of 12 months old mice.
| Gene | Cell cycle phase | FC- E15 FrCtx | p-val | FC- P1 FrCtx | p-val | FC- 12mo FrCtx | p-val | FC-12mo WM | p-val |
|---|---|---|---|---|---|---|---|---|---|
| PDGFRA | OPC | 1.73 | 0.03 | 1.25 | 0.03 | 1.46 | 5E-04 | 1.33 | 0.017 |
| CDKN2C/p18Ink4 | CDK inhibitor/G1 | −1.5 | 0.041 | 1.3 | 0.028 | 2 | 3E-05 | 1.3 | 0.02 |
| CDKN1C/p57Kip2 | CDK inhibitor/G1 | 1 | ns | 1.9 | 0.002 | 2.2 | 0.001 | 1.1 | ns |
| CDKN1B/p27Kip1 | CDK inhibitor/G1 | −1.3 | 0.004 | 1.1 | Ns | −1.3 | 0.01 | 1 | ns |
| CCND2 | G1 phase | −2.2 | 0.019 | −1.4 | 0.054 | −1.5 | 1E-05 | 1 | ns |
| CDC2A | G1/S/G2 phase | −1.9 | 0.028 | 1.5 | 0.003 | 3.1 | 1E-05 | 1 | ns |
| RAD51 | S phase | −1.9 | 0.048 | 1.1 | Ns | 1.6 | 0.054 | 1.5 | 0.008 |
| FOXN3 | G1/S checkpoint | −1 | ns | 1.3 | 0.014 | 1.3 | 0.026 | 1.5 | 3E-04 |
Gene expression from frontal cortex were analyzed by qPCR (N=6/group). FC ratio-fold change ratios represent the ratio of means for each gene normalized to the geometric means of two housekeeping genes between controls and ΔhDISC1 mice. Significantly changed genes (t-test, p≤0.05) are highlighted in bold. ns- Non-significant.
In the hindbrain (Table 4; Supp. Fig.1), cell cycle related genes showed a similar trend toward downregulation as in the forebrain at E15, where levels of CDC2A and RAD51 were significantly decreased (both FC< −1.9; ps≤0.05) and the level of PDGFRA was increased (FC=1.84, p=0.0004). In contrast to the forebrain, this downregulation trend persisted throughout the postnatal stage of development and become more pronounced in 12 months old ΔhDISC1 mice, where the levels of PDGFRA, p27Kip1 and RAD51 were significantly decreased (FC= −2.1, −1.3 and −2.1 respectively, all ps<0.001). The levels of CCND2 were modestly higher (FC=1.1, p=0.02).
Table 4.
Gene expression analysis of the cell cycle genes and OPC marker - PDGFRA during development in hindbrain (E15) and cerebellum of P1 and 12 months old mice.
| Gene | Cell cycle phase | FC- E15 | p-val | FC- P1 | p-val | FC- 12mo | p-val |
|---|---|---|---|---|---|---|---|
| PDGFRA | OPC marker | 1.84 | 4E-03 | −1.58 | 0.01 | −2.08 | 1E-08 |
| CDKN2C/p18Ink4 | CDK inhibitor/G1 | −1.22 | ns | 1 | ns | −1.03 | ns |
| CDKN1C/p57Kip2 | CDK inhibitor/G1 | 1 | ns | −1.45 | 0.03 | −1.21 | ns |
| CDKN1B/p27Kip1 | CDK inhibitor/G1 | 1.08 | ns | −1.15 | ns | −1.35 | 5E-07 |
| CCND2 | G1 phase | −1.19 | ns | −1.07 | ns | 1.1 | 0.02 |
| CDC2A | G1/S/G2 phase | −1.88 | 0.04 | −1.06 | ns | −1.2 | ns |
| RAD51 | S phase | −2.03 | 0.05 | −1.33 | ns | −2.06 | 1E-08 |
| FOXN3 | G1/S checkpoint | −1.07 | ns | −1.35 | 0.01 | −1.1 | Ns |
Gene expression from frontal cortex were analyzed by qPCR (N=6/group). FC ratio-fold change ratios represent the ratio of means for each gene normalized to the geometric means of two housekeeping genes between controls and ΔhDISC1 mice. Significantly changed genes (t-test, p≤0.05) are highlighted in bold. ns- Non-significant.
Decrease in the mRNA levels of CDK inhibitors and G1/S checkpoint proteins (RAD51 and CHES1) indicate that there is elevated cell cycle activity in the forebrain of E15 ΔhDISC1 mice suggestive of enhanced proliferation of multipotent progenitor cells. This conclusion is corroborated by the increased levels of the OPC marker – PDGFRA (Table 3). Contrasting directionality changes for the same genes during postnatal stages are suggestive of enhanced differentiation of OLG, which is also corroborated by expression of mature OLG markers (Table 2). At the same time data from cerebellum (Table 4) during postnatal period is suggestive of an opposite effect to the one observed in the forebrain.
3.4 Levels of the myelin specific protein, CNP, and the OPC marker protein, PDGFRA, are increased in parallel to gene expression in the frontal cortex, but not in the cerebellum of ΔhDISC1 mice
To determine whether protein expression changes parallel the upregulation of OLG-related gene expression in the forebrain of ΔhDISC1 mice we measured the levels of the myelin marker –CNP in the cortex of 12 months old ΔhDISC1 mice. As shown in Fig. 4A the levels of CNP protein were significantly increased in ΔhDISC1 mice compare to controls (p = 0.0003). We also measured levels of CNP in P1 mice. Consistent with the lack of myelogenesis at this early developmental stage (Scherer et al., 1994), no expression of CNP was found in control or ΔhDISC1 mice. In contrast to CNP, PDGFRA - an OPC specific protein- is easy detectable early in development. We measured levels of PDGFRA in the cortex of P1 mice. As shown in Fig. 5, the levels of PDGFRA protein was significantly increased in cortex (p= 0.026) of P1 ΔhDISC1 mice, but not in the cerebellum (not shown) of ΔhDISC1 mice compared to controls.
Figure 4.
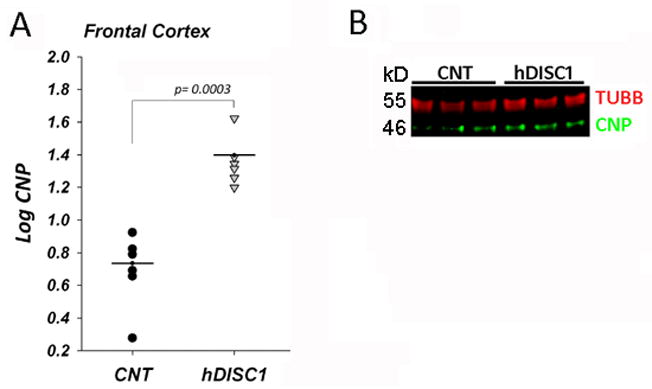
CNP protein levels in cortex of 12 months of age control (CNT) and ΔhDISC1 (hDISC1) mice (A). Multiplex blot (CNP and TUBB) for the samples from frontal cortex (B). Shown normalized log transformed intensity values for each sample. Means (N=5–6/group) are marked by horizontal lines. TUBB is a loading control.
Figure 5.
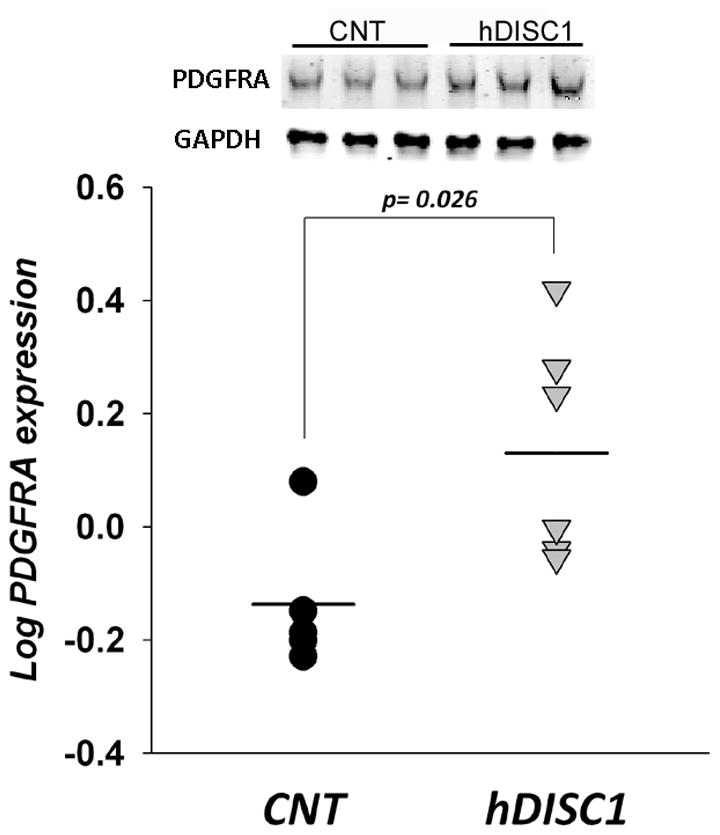
PDGFRA protein levels in forebrain of control (CNT) and ΔhDISC1 (hDISC1) mice at P1. N=5/group. Shown normalized log transformed intensity values of PDGFRA for each sample. Means are marked by horizontal lines. GAPDH is a loading control.
These data confirmed that protein levels of myelin marker CNP and OPC specific protein PDGFRA increased in parallel with gene expression in forebrain, but not in cerebellum of ΔhDISC1 mice.
3.5 Delayed postnatal expression of mutant ΔhDISC1 was associated with increased expression of OLG genes, but not OPC markers in the frontal cortex of adult mice
Upregulation of gene expression of PDGFRA and OLG specific genes in the forebrain of ΔhDISC1 mice as early as embryonic day 15 (E15) suggested increased proliferation of OLG precursor cells and their premature differentiation. To investigate the temporal relationship of the induction of mutant ΔhDISC1 expression to OLG and PDGFRA gene expression in the forebrain we compared gene expression of 12 months old ΔhDISC1 mice and similarly aged mice with exclusively postnatal expression of ΔhDISC1 (E12-ΔhDISC1) (Ayhan et al., 2010). Consequently, we had expected that embryonic OLG precursor cells would be less affected or remain unaltered during prenatal development. As shown in Fig. 1 and 6, levels of PDGFRA and OLG genes were significantly increased in the ΔhDISC1 mice in both the frontal cortex and the white matter at 12 months of age (ps≤ 0.02). Levels of OLG genes remained significantly upregulated (Table 5 and Fig. 6) also in E12-ΔhDISC1 mice (ps≤ 0.01) in both the frontal cortex and white matter, although for most of the genes the magnitude of the increase was lower in the frontal cortex compare to ΔhDISC1 mice. There were no significant changes for PDGFRA in either the frontal cortex or white matter of E12-ΔhDISC1 mice.
Figure 6.
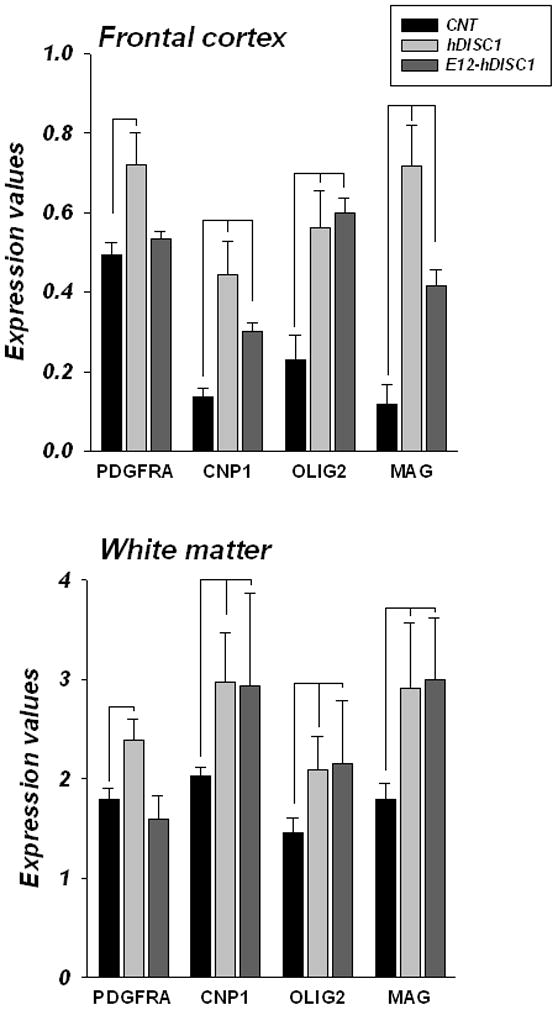
Gene expression levels of OLG-related genes in the frontal cortex and white matter of 12 month of age control (CNT) and ΔhDISC1 (hDISC1) mice and the same age mice with postnatal only expression of mutant DISC1 (E12-ΔhDISC1). N=5–6/group. Data is expressed as geometric means ± SEM. Significant (p<0.05) changes are marked by lines.
Table 5.
Gene expression analysis in frontal cortex and white matter of 12 months old ΔhDISC1 mice and the same age mice with delayed expression of mutant ΔhDISC1 at embryonic day 12 (E12/ΔhDISC1) mice as compared to controls.
| Gene | Cell marker | FC FrCtx ΔhDISC1/E12 | p-val ΔhDISC1/E12 | FC WM ΔhDISC1/E12 | p-val ΔhDISC1/E12 |
|---|---|---|---|---|---|
| PDGFRA | OPC | 1.5/1.1 | 5E-04/ns | 1.3/−1.1 | 0.02/ns |
| CNP | OLG | 3.3/2.2 | 1E-05/3E-08 | 1.5/1.5 | 0.007/0.03 |
| OLIG2 | OLG | 2.5/2.6 | 2E-04/1E-07 | 1.4/1.5 | 0.01/0.02 |
| MAG | mature OLG | 6.0/3.3 | 1E-08/5E-09 | 1.6/1.7 | 0.01/0.002 |
Gene expression from frontal cortex were analyzed by qPCR (N=6/group). FC ratio-fold change ratios represent the ratio of means for each gene normalized to the geometric means of two housekeeping genes between controls and ΔhDISC1 mice. Significantly changed genes (t-test, p≤0.05) are highlighted in bold. ns- Non-significant.
These data suggest that ΔhDISC1 can elicit its effect on OLG specific genes not only during embryonic development of OLG, but also postnatally, perhaps involving adult multipotent precursor cells and/or differentiated OLGs.
3.6 Forebrain restricted expression of mutant ΔhDISC1 affects neuregulin1 and its receptors
To address a possible link between DISC1 and NRG signaling, which is implicated in OLG differentiation and myelination in the CNS, the gene expression of NRG1 and its receptors known to be expressed by OLGs was assessed. Gene expression of NRG1, ErbB3 and ErbB4 was measured at E15 and P1 stages of development and in adult ΔhDISC1 mice and compared to controls. As shown on Fig. 7A the induction of ΔhDISC1 significantly affected the gene expression of NRG1 and its receptors in the forebrain and frontal cortex during development and in adulthood. NRG1 was upregulated in E15 and in 12 months old ΔhDISC1 mice (ps=0.01). ErbB3 was strongly upregulated in E15 (p=0.02) and in 12 months old ΔhDISC1 mice (p=1E-05), but downregulated in P1 mice (p=0.05) compare to age-matched controls. ErbB4 was upregulated in P1 (p=0.01) and in12 months old ΔhDISC1 mice (p=0.04). Similar upregulation of NRG1 and ErbB3 (p=0.04 and 0.01, respectively) was detected in the white matter of 12 months old ΔhDISC1 mice (Fig.7B). No significant changes in ErbB4 expression were detected in the white matter of 12 months old ΔhDISC1 mice (Fig.7B). No significant changes for NRG1 and its receptors were detected in cerebellum or the hindbrain during development. However, in contrast to the frontal cortex and white matter, NRG1 and ErbB4were significantly downregulated (p=8E-04 and p=3E-04, respectively) in the cerebellum of 12 months old ΔhDISC1 mice (Fig.7B).
Figure 7.
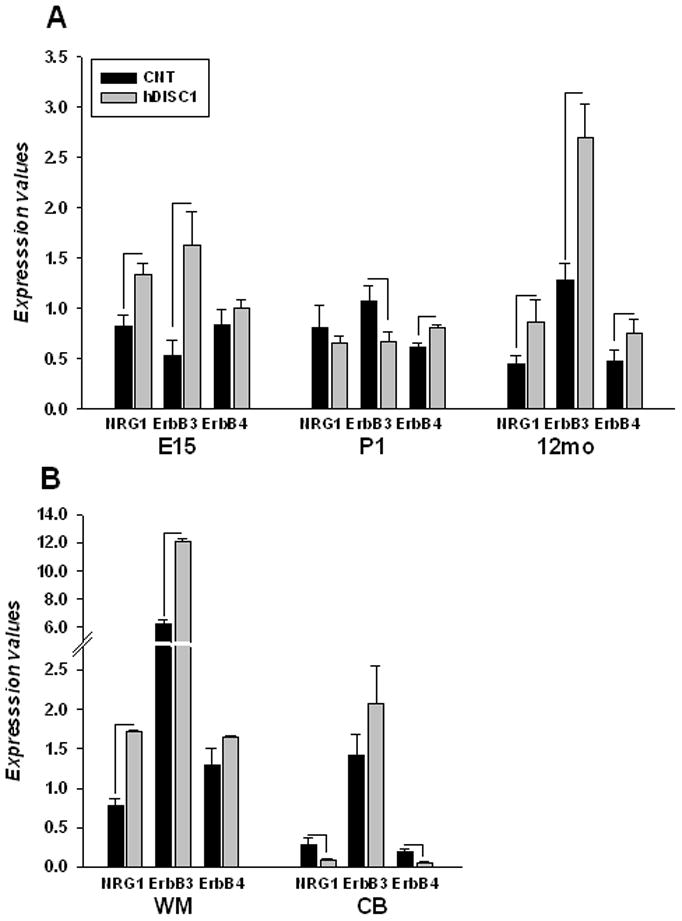
Gene expression of NRG1 and its receptors, ErbB3 and ErbB4, in control (CNT) and ΔhDISC1 (hDISC1) mice throughout development (E15, P1) and at 12-months of age in forebrain (A); or in the white matter (WM) and the cerebellum (CB) (B). N=5–6/group. Data is expressed as geometric means ±SEM. Significant changes (p<0.05) are marked by lines.
These results confirmed that ΔhDISC1 expression may stimulate NRG1 expression and elicit signaling in OLGs that may, at least in part, account for augmented OLG gene expression.
4. DISCUSSION
Schizophrenia is a complex disease that has been associated with specific DISC1 and NRG1 haplotypes (Millar et al., 2001; Devon et al., 2001; Stefansson et al., 2002) and independently with regional deficit of expression of genes and proteins related to OLG function (Hakak et al., 2001; Martins-de-Souza, 2010; Katsel et al., 2008; Tkachev et al., 2003). The myelin associated expression deficits in SZ are due at least in part to a failure of execution of the normal cell-cycle arrest in postmitotic OLGs (Katsel et al., 2008; Kerns et al., 2010) that adversely affects the expression of OLG genes and myelin function. Two recent studies suggested that DISC1, in addition to its role in neuronal function, can affect oligodendroglial differentiation early in CNS and PNS development (Wood et al., 2009; Drerup et al., 2009). In this study, we used mutant DISC1 inducible mouse model to perturb functions of endogenous neuronal DISC1 to evaluate the region-specific oligodendroglial and myelin deficits observed in SZ.
This study analyzed gene expression of mature OLG markers and genes involved in differentiation of OLGs at different stages of development and the adulthood in transgenic mice with forebrain restricted expression of the mutant ΔhDISC1 gene. Expression of ΔhDISC1 was associated with a strong upregulation of gene and protein expression of markers of OLG precursor cells and mature myelinating OLGs in the frontal cortex and white matter, but not in the cerebellum of 12 month old mice. Similar to 12 month old ΔhDISC1 mice, markers of OLG precursor cells and differentiating OLGs were also upregulated during development (at E15 and P1 days) in the forebrain of ΔhDISC1 mice relative to controls. The greatest increase in expression of nine OLG specific genes, most strikingly SOX10 (11 fold) and MAG (45 fold), was observed at E15, coinciding with the highest level of mutant and endogenous DISC1 expression (Ayhan et al., 2010; Schurov et al., 2004) and with proliferation of multipotent glial progenitors. These results suggest accelerated differentiation of these cells into OLGs.
This remarkable increase in SOX10 is in agreement with a study of peripheral cranial glia migration and development in zebrafish in which endogenous DISC1 expression was disrupted (Drerup et al., 2009). Experimental evidence suggested that DISC1 functions as a repressor of expression of SOX10 and FOXD3- a possible activator of SOX10 expression in cranial neural crest cells. Knockdown of DISC1 resulted in an increased expression of SOX10 and expansion of peripheral glia in the zebrafish model. Consistently, forebrain restricted overexpression of dominant-negative mutant DISC1 was associated with decreased levels of endogenous mouse DISC1 ((Pletnikov et al., 2008) and Supp. Fig. 2) and increased expression of SOX10 in frontal regions, but not in the hindbrain or cerebellum. In contrast, Woods and others (Wood et al., 2009) have demonstrated that an interruption of translation of endogenous DISC1 that caused a loss of DISC1 protein resulted in hindbrain restricted localization of SOX10 positive cells and was associated with a deficit in the myelin markers (i.e., PLP1 and MBP) in zebrafish development.
We hypothesize that augmentation of gene expression of myelin-related genes in forebrain of ΔhDISC1 mice may be due to a decreased expression of endogenous mouse DISC1. However, we cannot rule out the possibility that the observed increase in gene expression might, at least in part, result from gain-of-function effects of mutant DISC1. Similarly, although the overexpression of MAG in E15 ΔhDISC1 mice indicates a pronounced dysregulation of oligodendroglial genes expression, there is no clear explanation for the inflated gene expression of this typically late myelination marker at a time when myelination would not have commenced in control mice. Future studies will elucidate the mechanisms of upregulation of OLG genes and its pathogenic significance during early stages of oligodendrogenesis.
The transcription factor Sox10 is restricted to OLG lineage in the CNS. It is expressed in proliferating OPC and continues to be expressed by mature, terminally differentiating OLGs (Stolt et al., 2002), thus it cannot be considered to be an exclusive marker of OPCs. Unlike SOX10 and OLIG2, PDGFRA, is abundantly expressed in OLG progenitor cells and rapidly downregulated when OPC differentiate into OLGs (Butt et al., 1997; Pringle and Richardson, 1993; Tekki-Kessaris et al., 2001). Therefore, PDGFRA is broadly considered to be a selective OPC marker (Rivers et al., 2008) and its upregulation in ΔhDISC1 mice provides strong evidence in favor of increased OPC proliferation. Widespread abnormalities in the expression of cell cycle genes and the upregulation of a OPC marker, PDGFRA, in the forebrain and its downregulation in the cerebellum suggests an expansion of glial progenitors toward frontal cortical regions, away from the cerebellum. Notably, a developmental disruption of DISC1 function results in altered development of neural cells and cell migration in zebrafish (Drerup et al., 2009; Wood et al., 2009; Meyer and Morris, 2008; Mao et al., 2009).
Cell cycle proteins orchestrate OLG differentiation (Raff et al., 2001; Casaccia-Bonnefil et al., 1999; Dugas et al., 2007) and modulate maturation of OLGs. For example, the levels of p27Kip1, a putative promoter of cell cycle arrest, progressively rise during transition from OPC to mature OLG (Miskimins et al., 2002). Studies of rodents OPC in vitro suggest that the differentiation process is likely governed by two opposing forces that either promote or inhibit cell cycle progression (Nguyen et al., 2006; Dugas et al., 2006). Three out of seven analyzed cell cycle genes that were altered in ΔhDISC1 mice belong to the family of CDK inhibitors (p18Ink4, p57Kip2 and p27 Kip1), whose increased expression halt cell cycle progression, induce cell cycle exit and initiates OLG differentiation (Nguyen et al., 2006; Dugas et al., 2007). Four others (CCND2, CDC2A, RAD51 and FOXN3) are critical for cell cycle progression. The CCND2 gene encodes cyclin, whose activity is required for cell cycle G1/S phase transition (Kozar et al., 2004). The protein encoded by the CDC2A (CDK1) gene is a catalytic subunit of the Ser/Thr protein kinase complex (a.k.a. the M-phase promoting factor) is essential for G1/S and G2/M phase transitions (Draetta et al., 1988). Rad51 and FOXN3/CHES1 are involved in the regulation of DNA damage-inducible cell cycle arrests at G1 and G2 checkpoints (Scott and Plon, 2005; Baumann and West, 1998). Our results demonstrate that ΔhDISC1 affects expression pattern of cell cycle genes in a complex way, suggesting increased proliferation in early development (E15), but accelerated oligodendroglial differentiation and maturation postnatally. One, albeit speculative, interpretation is that the expression changes in cell cycle genes may also indicate activation of cell cycle checkpoints that can lead to premature cell cycle exit and activation of apoptosis. Future studies will address these and other possibilities.
Profound alterations in gene expression of NRG1 and its receptors, ErbB3 and ErbB4, were associated with expression of ΔhDISC1 during development and in adult mice, suggesting that NRG1/ErbB signaling may contribute to the increased expression of OLG markers in the forebrain. The best understood function of NRG1 is to control myelination in PNS, where axonal NRG1 type III is required for the proliferation and differentiation of Schwann cells as well as induction of myelination (Michailov et al., 2004; Garratt et al., 2000; Taveggia et al., 2005). In the CNS, NRG1 signaling has been implicated in neuronal migration, axonal elongation and synaptic function (Flames et al., 2004; Lopez-Bendito et al., 2006; Mei and Xiong, 2008). The role for NRG1 in myelination by OLG has been debated. Myelination was not affected or delayed by conditional knockdown of NRG1 or its receptors, ErbB3/ErbB4, in the CNS during different stages of development, suggesting that NRG1 signaling is not obligatory for CNS myelination (Brinkmann et al., 2008). However, overexpression of either type 1 or type III NRG1, showed evidence for premature myelination and hypermyelination in mice (Brinkmann et al., 2008; Taveggia et al., 2008). Consistent with these findings, the PCR probe used in our experiments interrogated three major types of NRG1 and showed upregulation of NRG1 in frontal cortical regions of ΔhDISC1 mice. These results support the hypothesis that the effects of mutant ΔhDISC1 on OLG function could be mediated, at least in part, by NRG1/ErbB signaling.
Distinguishing between these various potential explanations for the current data will require considerable additional studies, however, given the associations and convergence of DISC1, NRG1 and OLGs and myelin with schizophrenia such studies are likely to be valuable to the understanding of the neurobiology of schizophrenia.
Given that expression of mutant DISC1 is regulated by the neuronal promoter, one could suggest that the effects of mutant DISC1 on OLG development and maturation may be mediated via neuron-glia interaction. This possibility has been demonstrated by a recent study where a neuron-specific mutation produced abnormal development and maturation of OLG (Stritt et al., 2009). We cannot completely rule out the possibility of ectopic expression of ΔhDISC1 in multipotent OPCs during development as two recent studies have demonstrated constitutive expression of CAMKII in several types of glia in the hippocampus(Suh et al., 2005) and the involvement of Ca2+-CaMK signaling in expression of myelin specific genes in differentiated OLGs(Marta et al., 2002).
Figure 8 presents the scheme of the hypothetical signaling mechanisms whereby mutant hDISC1 may affect OLG proliferation and differentiation. However, there are many outstanding questions to be addressed in the future. What is the role of DISC1 in OLG lineage cells and what are the mechanisms through which DISC1 regulates OLG differentiation and maturation? What mechanisms mediate the effects of mutant DISC1 in the ΔhDISC1 mouse model? How does disruption of DISC1 functions produce the regional deficits in myelin markers in SZ? There is no direct explanation for apparent contradiction between the current mouse model data and gene expression profiling derived from postmortem studies. One speculative interpretation rests on the age and timing of mutant hDISC1 expression. The results suggests that in this model system the expression of mutant hDISC1 peaked during early development which may have affected migration, proliferation and differentiation of oligodendrocytes resulting in in overexpression of myelin markers in some regions and deficit in others. These and other questions can be experimentally addressed in future studies.
Figure 8.
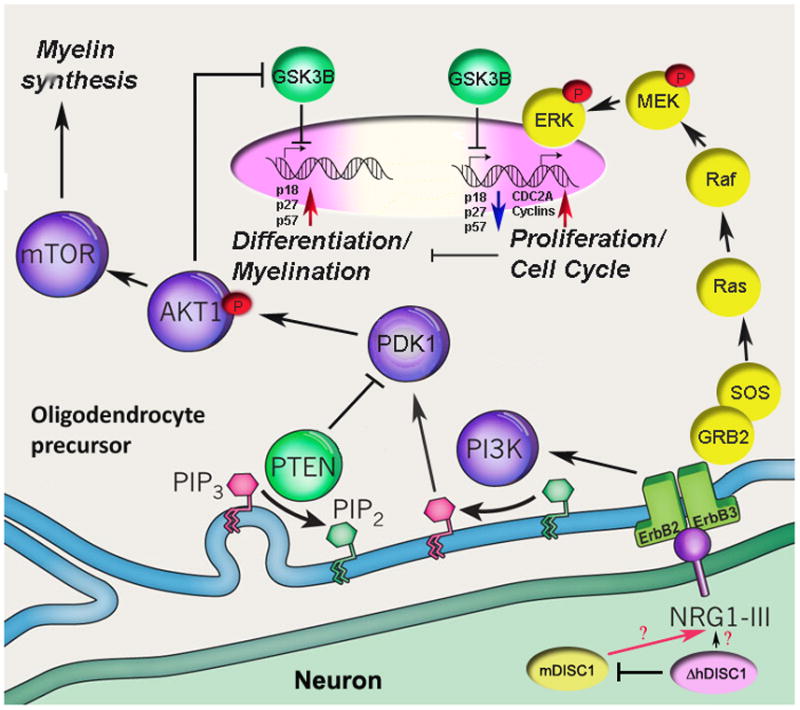
Schematic view of intracellular ErbB2/3 signaling cascade triggered by NRG1 and modulated by DISC1 (Adapted from (Nave, 2010)).
Signaling between neuronal membrane-bound NRG1(type III) and the ErbB family of proteins occurs at the glia–neuron interface and activates several second-messenger cascades, including phosphatidylinositol-3-kinase (PI3K)-Akt and mitogen-activated protein kinase/ERK pathways. Depicted here is a schematic representation of glial ErbB receptors and the sequential activation of PI3K– serine-threonine-specific protein kinase, AKT1– mammalian target of rapamycin (mTOR) kinases, leading to cell cycle arrest mediated by upregulation of CDK inhibitors (p18, p27 and p57) via inhibition of GSK3B activity and the activation of myelin-associated genes transcription. AKT1 signaling is tightly controlled and antagonized by the phosphatase and tensin homologue (PTEN) protein. NRG1- ErbB2, ErbB3 complex can also elicit MAPK/ERK signaling and activation of cell cycle and increased proliferation, which in turn suppress differentiation and expression of OLG genes. Neuronal NRG1expression is likely modulated by DISC1. Currently, it is not clear how expression of mutant hDISC1 (ΔhDISC1) can amend its effect on NRG1 expression. One of the possibilities is a dominant-negative suppression of expression of neuronal endogenous mouse DISC1 (mDISC1).
In conclusion, our results suggest that DISC1 may play a role in OLG proliferation and differentiation with relevance to myelination deficits in schizophrenia.
Supplementary Material
Abbreviations
- OLG
oligodendrocyte
- OPC
oligodendrocyte precursor cells
- SZ
schizophrenia
- DISC1
Disrupted-in-Schizophrenia 1
- CAMKII
calcium/calmodulin-dependent protein kinase II
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- Ayhan Y, Abazyan B, Nomura J, Kim R, Ladenheim B, Krasnova IN, Sawa A, Margolis RL, Cadet JL, Mori S, Vogel MW, Ross CA, Pletnikov MV. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol Psychiatry. 2010 doi: 10.1038/mp.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn S, Jones PB. Gene expression in schizophrenia and bipolar disorder. Schizo Res. 2003;60:60. [Google Scholar]
- Barley K, Dracheva S, Byne W. Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder. Schizophr Res. 2009;112:54–64. doi: 10.1016/j.schres.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Nuechterlein KH, Lu PH, Gitlin M, Rogers S, Mintz J. Dysregulated brain development in adult men with schizophrenia: a magnetic resonance imaging study. Biol Psychiatry. 2003;53:412–421. doi: 10.1016/s0006-3223(02)01835-8. [DOI] [PubMed] [Google Scholar]
- Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann BG, Agarwal A, Sereda MW, Garratt AN, Muller T, Wende H, Stassart RM, Nawaz S, Humml C, Velanac V, Radyushkin K, Goebbels S, Fischer TM, Franklin RJ, Lai C, Ehrenreich H, Birchmeier C, Schwab MH, Nave KA. Neuregulin-1/ErbB signaling serves distinct functions in myelination of the peripheral and central nervous system. Neuron. 2008;59:581–595. doi: 10.1016/j.neuron.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchsbaum MS, Tang CY, Peled S, Gudbjartsson H, Lu D, Hazlett EA, Downhill J, Haznedar M, Fallon JH, Atlas SW. MRI white matter diffusion anisotropy and PET metabolic rate in schizophrenia. NeuroReport. 1998;9:425–430. doi: 10.1097/00001756-199802160-00013. [DOI] [PubMed] [Google Scholar]
- Butt AM, Hornby MF, Ibrahim M, Kirvell S, Graham A, Berry M. PDGF-alpha receptor and myelin basic protein mRNAs are not coexpressed by oligodendrocytes in vivo: a double in situ hybridization study in the anterior medullary velum of the neonatal rat. Mol Cell Neurosci. 1997;8:311–322. doi: 10.1006/mcne.1996.0590. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Hardy RJ, Teng KK, Levine JM, Koff A, Chao MV. Loss of p27Kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 1999;126:4027–4037. doi: 10.1242/dev.126.18.4027. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, Kaneda H, Shiroishi T, Houslay MD, Henkelman RM, Sled JG, Gondo Y, Porteous DJ, Roder JC. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Cotter DR, Pariante CM, Everall IP. Glial cell abnormalities in major psychiatric disorders: the evidence and implications. Brain Res Bull. 2001;55:585–595. doi: 10.1016/s0361-9230(01)00527-5. [DOI] [PubMed] [Google Scholar]
- Devon RS, Anderson S, Teague PW, Burgess P, Kipari TM, Semple CA, Millar JK, Muir WJ, Murray V, Pelosi AJ, Blackwood DH, Porteous DJ. Identification of polymorphisms within Disrupted in Schizophrenia 1 and Disrupted in Schizophrenia 2, and an investigation of their association with schizophrenia and bipolar affective disorder. Psychiatr Genet. 2001;11:71–78. doi: 10.1097/00041444-200106000-00003. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Davis KL, Chin B, Woo DA, Schmeidler J, Haroutunian V. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in schizophrenia. Neurobiol Dis. 2006;79:157–173. doi: 10.1016/j.nbd.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Dracheva S, McGurk SR, Haroutunian V. mRNA expression of AMPA receptors and AMPA receptor binding proteins in the cerebral cortex of elderly schizophrenics. J Neurosci Res. 2005;79:868–878. doi: 10.1002/jnr.20423. [DOI] [PubMed] [Google Scholar]
- Draetta G, Piwnica-Worms H, Morrison D, Druker B, Roberts T, Beach D. Human cdc2 protein kinase is a major cell-cycle regulated tyrosine kinase substrate. Nature. 1988;336:738–744. doi: 10.1038/336738a0. [DOI] [PubMed] [Google Scholar]
- Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136:2623–2632. doi: 10.1242/dev.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Ibrahim A, Barres BA. A Crucial Role for p57Kip2 in the Intracellular Timer that Controls Oligodendrocyte Differentiation. The Journal of Neuroscience. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marin O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Flynn SW, Lang DJ, MacKay AL, Goghari V, Vavasour IM, Whittall KP, Smith GN, Arango V, Mann JJ, Dwork AJ, Falkai P, Honer WG. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and postmortem with analysis of oligodendrocyte proteins. Molecular Psychiatry. 2003;8:811–820. doi: 10.1038/sj.mp.4001337. [DOI] [PubMed] [Google Scholar]
- Foong J, Maier M, Barker GJ, Brocklehurst S, Miller DH, Ron MA. In vivo investigation of white matter pathology in schizophrenia with magnetisation transfer imaging. Journal of Neurology Neurosurgery and Psychiatry. 2000;68:70–74. doi: 10.1136/jnnp.68.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foong J, Symms MR, Barker GJ, Maier M, Woermann FG, Miller DH, Ron MA. Neuropathological abnormalities in schizophrenia: evidence from magnetization transfer imaging. Brain. 2001;124:882–892. doi: 10.1093/brain/124.5.882. [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. Academic Press; New York: 1997. [Google Scholar]
- Garratt AN, Britsch S, Birchmeier C. Neuregulin, a factor with many functions in the life of a schwann cell. BioEssays. 2000;22:987–996. doi: 10.1002/1521-1878(200011)22:11<987::AID-BIES5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Thomson P, McQuillin A, Bass N, Loukola A, Anjorin A, Blackwood D, Curtis D, Deary IJ, Harris SE, Isometsa ET, Lawrence J, Lonnqvist J, Muir W, Palotie A, Partonen T, Paunio T, Pylkko E, Robinson M, Soronen P, Suominen K, Suvisaari J, Thirumalai S, St CD, Gurling H, Peltonen L, Porteous D. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andrade M, Tankou S, Mori S, Gallagher M, Ishizuka K, Pletnikov M, Kida S, Sawa A. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Haroutunian V, Friedrich VL, Jr, Byne W, Buitron C, Perl DP, Davis KL. Loss and altered spatial distribution of oligodendrocytes in the superior frontal gyrus in schizophrenia. Biol Psychiatry. 2003;53:1075–1085. doi: 10.1016/s0006-3223(03)00237-3. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, Kato T. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25:5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Gorman JM, Haroutunian V. Variations in differential gene expression patterns across multiple brain regions in schizophrenia. Schizo Res. 2005a;77:241–252. doi: 10.1016/j.schres.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions: A gene ontology study. Schizo Res. 2005b;79:157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Li C, Tan W, Greenstein E, Kleiner Hoffman LB, Haroutunian V. Abnormal Indices of Cell Cycle Activity in Schizophrenia and their Potential Association with Oligodendrocytes. Neuropsychopharmacology. 2008;33:2993–3009. doi: 10.1038/npp.2008.19. [DOI] [PubMed] [Google Scholar]
- Kerns DC, Vong GS, Barley K, Dracheva S, Katsel P, Casaccia P, Haroutunian V, Byne W. Gene expression abnormalities and oligodendrocyte deficits in the internal capsule in schizophrenia. Schizo Res. 2010;120:150–158. doi: 10.1016/j.schres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, Akashi K, Sicinski P. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Lim KO, Hedehus M, Moseley M, de Crespigny A, Sullivan EV, Pfefferbaum A. Compromised white matter tract integrity in schizophrenia inferred from diffusion tensor imaging. Archives of General psychiatry. 1999;56:367–374. doi: 10.1001/archpsyc.56.4.367. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Cautinat A, Sanchez JA, Bielle F, Flames N, Garratt AN, Talmage DA, Role LW, Charnay P, Marin O, Garel S. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125:127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Visscher PM, Knott SA, Thomson P, Porteous DJ, Millar JK, Devon RS, Blackwood D, Muir WJ. A genome scan and follow-up study identify a bipolar disorder susceptibility locus on chromosome 1q42. Mol Psychiatry. 2004;9:1083–1090. doi: 10.1038/sj.mp.4001544. [DOI] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, Biechele T, Petryshen TL, Moon RT, Haggarty SJ, Tsai LH. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marta CB, Davio C, Pasquini LA, Soto EF, Pasquini JM. Molecular mechanisms involved in the actions of apotransferrin upon the central nervous system: Role of the cytoskeleton and of second messengers. J Neurosci Res. 2002;69:488–496. doi: 10.1002/jnr.10317. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D. Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J Psychiatr Res. 2010;44:149–156. doi: 10.1016/j.jpsychires.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Morris JA. Immunohistochemical analysis of Disc1 expression in the developing and adult hippocampus. Gene Expr Patterns. 2008;8:494–501. doi: 10.1016/j.gep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Axonal Neuregulin-1 Regulates Myelin Sheath Thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- Millar JK, Christie S, Anderson S, Lawson D, Hsiao-Wei LD, Devon RS, Arveiler B, Muir WJ, Blackwood DH, Porteous DJ. Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry. 2001;6:173–178. doi: 10.1038/sj.mp.4000784. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Miller R, Reynolds R. Oligodendroglial Lineage. In: Lazzarini RA, editor. Myelin Biology and Disorders 1. Vol. 1. Elsevier Academic Press; San Diego, California: 2004. pp. 289–310. [Google Scholar]
- Miskimins R, Srinivasan R, Marin-Husstege M, Miskimins WK, Casaccia-Bonnefil P. p27(Kip1) enhances myelin basic protein gene promoter activity. J Neurosci Res. 2002;67:100–105. doi: 10.1002/jnr.10080. [DOI] [PubMed] [Google Scholar]
- Miyakawa T, Sumiyoshi S, Deshimaru M, Suzuki T, Tomonari H. Electron microscopic study on schizophrenia. Mechanism of pathological changes. Acta Neuropathol (Berl) 1972;20:67–77. doi: 10.1007/BF00687903. [DOI] [PubMed] [Google Scholar]
- Nave KA. Myelination and support of axonal integrity by glia. Nature. 2010;468:244–252. doi: 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Borgs L, Vandenbosch R, Mangin JM, Beukelaers P, Moonen G, Gallo V, Malgrange B, Belachew S. The Yin and Yang of cell cycle progression and differentiation in the oligodendroglial lineage. Ment Retard Dev Disabil Res Rev. 2006;12:85–96. doi: 10.1002/mrdd.20103. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, Mori S, Moran TH, Ross CA. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–86. 115. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- Raff M, Apperly J, Kondo T, Tokumoto Y, Tang D. Timing cell-cycle exit and differentiation in oligodendrocyte development. Novartis Found Symp. 2001;237:100–107. doi: 10.1002/0470846666.ch9. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Miguel-Hidalgo JJ, Makkos Z, Meltzer H, Overholser J, Stockmeier C. Layer-specific reductions in GFAP-reactive astroglia in the dorsolateral prefrontal cortex in schizophrenia. Schizophr Res. 2002;57:127–138. doi: 10.1016/s0920-9964(02)00339-0. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SS, Braun PE, Grinspan J, Collarini E, Wang DY, Kamholz J. Differential regulation of the 2′,3′-cyclic nucleotide 3′-phosphodiesterase gene during oligodendrocyte development. Neuron. 1994;12:1363–1375. doi: 10.1016/0896-6273(94)90451-0. [DOI] [PubMed] [Google Scholar]
- Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- Scott KL, Plon SE. CHES1/FOXN3 interacts with Ski-interacting protein and acts as a transcriptional repressor. Gene. 2005;359:119–126. doi: 10.1016/j.gene.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, Chatzi C, He S, Mackie I, Brandon NJ, Marquis KL, Day M, Hurko O, McCaig CD, Riedel G, St CD. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Stark AK, Uylings HB, Sanz-Arigita E, Pakkenberg B. Glial cell loss in the anterior cingulate cortex, a subregion of the prefrontal cortex, in subjects with schizophrenia. Am J Psychiatry. 2004;161:882–888. doi: 10.1176/appi.ajp.161.5.882. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Stern S, Harting K, Manke T, Sinske D, Schwarz H, Vingron M, Nordheim A, Knoll B. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat Neurosci. 2009;12:418–427. doi: 10.1038/nn.2280. [DOI] [PubMed] [Google Scholar]
- Suh HW, Lee HK, Seo YJ, Kwon MS, Shim EJ, Lee JY, Choi SS, Lee JH. Kainic acid (KA)-induced Ca2+/calmodulin-dependent protein kinase II (CaMK II) expression in the neurons, astrocytes and microglia of the mouse hippocampal CA3 region, and the phosphorylated CaMK II only in the hippocampal neurons. Neurosci Lett. 2005;381:223–227. doi: 10.1016/j.neulet.2005.01.089. [DOI] [PubMed] [Google Scholar]
- Taveggia C, Thaker P, Petrylak A, Caporaso GL, Toews A, Falls DL, Einheber S, Salzer JL. Type III neuregulin-1 promotes oligodendrocyte myelination. Glia. 2008;56:284–293. doi: 10.1002/glia.20612. [DOI] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, Yano H, Rosenbluth J, Einheber S, Xu X, Esper RM, Loeb JA, Shrager P, Chao MV, Falls DL, Role L, Salzer JL. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekki-Kessaris N, Woodruff R, Hall AC, Gaffield W, Kimura S, Stiles CD, Rowitch DH, Richardson WD. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, Starkey M, Webster MJ, Yolken RH, Bahn S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Uranova N, Orlovskaya D, Vikhreva O, Zimina I, Kolomeets N, Vostrikov V, Rachmanova V. Electron microscopy of oligodendroglia in severe mental illness. Brain Res Bull. 2001;55:597–610. doi: 10.1016/s0361-9230(01)00528-7. [DOI] [PubMed] [Google Scholar]
- Uranova NA, Vostrikov VM, Orlovskaya DD, Rachmanova VI. Oligodendroglial density in the prefrontal cortex in schizophrenia and mood disorders: a study from the Stanley Neuropathology Consortium. Schizo Res. 2004;67:269–275. doi: 10.1016/S0920-9964(03)00181-6. [DOI] [PubMed] [Google Scholar]
- Vostrikov VM, Uranova NA, Orlovskaya DD. Deficit of perineuronal oligodendrocytes in the prefrontal cortex in schizophrenia and mood disorders. Schizophr Res. 2007;94:273–280. doi: 10.1016/j.schres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 2009;18:391–404. doi: 10.1093/hmg/ddn361. [DOI] [PubMed] [Google Scholar]
- Zeman W, Innes JRM. Craigie’s neuroanatomy of the rat. Academic Press; New York: 1963. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


