Abstract
Tissue morphogenesis requires complex cellular interaction and communication. The sensory ray in the Caenorhabditis elegans male tail has a simple cellular make-up and a non-essential function, thus providing an ideal model for studying the mechanisms guiding morphogenesis. We present here the analysis of a novel gene, ram-5, mutations of which are characterized by abnormal lumpy rays in the male tail. Microscopic analysis and behavioral studies revealed that lumpy rays contain operational sensory neurons. However, abnormalities were observed in the hypodermis and structural cells as well as in appositions between these two cell types. Molecular cloning and expression studies revealed that the ram-5 gene encodes a transmembrane protein localized in sensory ray support cells, the structural cells. Expression of ram-5 in these cells is required for normal ray morphogenesis. ram-5-dependent cell–cell communication is implicated in organizing the structural cell and the hypodermis, potentially through adhesion at the structural cell–hypodermal cell border.
Keywords: cellular morphogenesis/ram-5/sensory ray
Introduction
Morphogenesis guided by a tightly regulated genetic program is important in defining the functional and structural characteristics of an organism (Poodry, 1992). For instance, the morphogenesis of Drosophila appendages requires interactions between cellular components through secreted ligand, receptor, transcription factors and adhesion molecules for the proper propagation of morphogenetic signals (von Kalm et al., 1995; Brabant et al., 1998). Localized expression of proteases in the imaginal disk cells allows tissue remodeling (Appel et al., 1993) while expression of adherens junction proteins and cytoskeletal molecules helps to re-establish cell–cell association and the formation of a new structural form (Geiger and Ayalon, 1992).
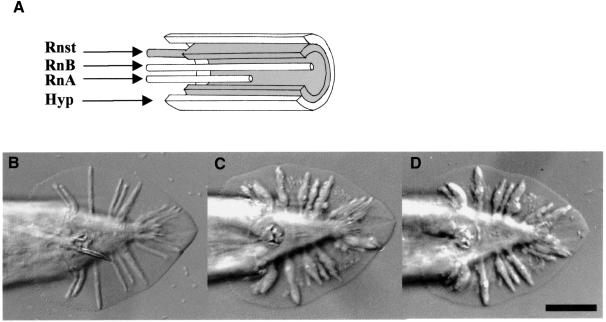
Caenorhabditis elegans has been used widely for developmental studies. Its well-documented cell lineage has facilitated the analysis of various developmental processes (Sulston and Horvitz, 1977; Kimble and Hirsh, 1979; Sulston et al., 1983), including morphogenesis of the male-specific sensory structures and the hermaphrodite vulva (Baird and Emmons, 1990; Herman et al., 1999; Newman et al., 1999). The sensory rays are organized bilaterally in a simple and specific pattern in the male tail and are used for active searching and locating the hermaphrodite vulva during mating (Liu and Sternberg, 1995). The ray precursor cells derive from the lateral seam cells. During the L4 stage, each precursor cell gives rise to four differentiated cells: a ray structural cell (Rnst), two ray neurons (RnA and RnB) and a hypodermal cell (hyp). Sulston and Horvitz (1977) noticed that when the Rnst cells were removed by laser microsurgery, rays failed to form. Thus, the structural cell is essential for ray formation. The hyp cells of rays 1–5 fuse together to form the tail seam (SET) and those of rays 6–9 fuse with the hyp7 compartment on the ventral surface (Sulston et al., 1980). During tail retraction, the somata of Rnst, RnA and RnB of each ray migrate anteriorly, leaving behind dendritic processes with the structural cell attached to the cuticle surface. This attachment is important for ray positioning and occurs early during papilla formation (Baird et al., 1991; Nguyen et al., 1999). The processes of RnA and RnB eventually detach from the cuticle and are wrapped by the Rnst process near the ray tip (Figure 1A) (Chow et al, 1995). Coincident with the completion of anterior migration of these cells, the hypodermis retracts dorsally and anteriorly, resulting in a thin hypodermal sheath covering the ray surface (Figure 1A) (Baird et al., 1991). Although the molecular mechanisms controlling ray morphology are largely unknown, proper synthesis of adhesion molecules is believed to facilitate the cellular association and movement of ray cells during morphogenesis.
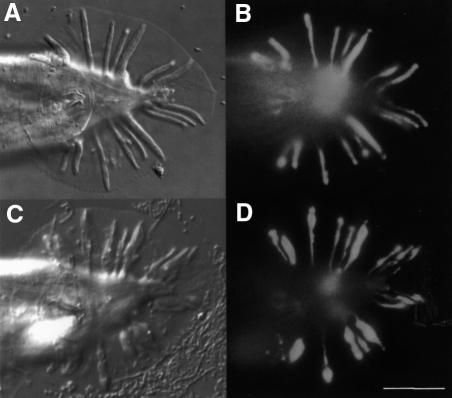
Fig. 1. (A) Schematic diagram showing a ray and its cellular constituents. (B–D) Morphology of wild-type and mutant male tails. Ventral view, anterior to the left with ray 1 at the most anterior position on each side of the fan. (B) him-5; (C) ram-5 (bx30); (D) ram-5 (bx81). Scale bar = 20 µm.
A number of genes have been shown to be necessary for male sensory ray morphogenesis (Hodgkin, 1983; Baird and Emmons, 1990; Baird et al., 1991). Among them are a group of ram genes, mutations of which result in lumpy sensory rays (Figure 1B, C and D). It has been postulated that Ram defects are caused by the failure of ray cell assembly (Baird and Emmons, 1990). Mutations in different ram genes failed to complement each other, which implies interactions among the ram gene products. Studies of a temperature-sensitive allele of ram-2 suggested that ram genes function at the final stage of ray cell morphogenesis rather than in cell fate determination steps (Baird and Emmons, 1990). In addition, tunicamycin treatment of wild-type males during the retraction period can result in the lumpy rays phenotype (Ko and Chow, 2000). In that particular study, four ram genes, including ram-5, have been suggested to act in a glycosylation-dependent pathway and interact with glycosylation-independent ram genes.
In this study, we explore ray morphogenesis by phenotypic characterization of ram-5 mutants at both the molecular and cellular level. ram-5 encodes a transmembrane protein, which is likely to be glycosylated, and interacts with other ram gene products to form an adhesive complex between ray cells.
Results
Phenotypic characterization of ram-5 mutation
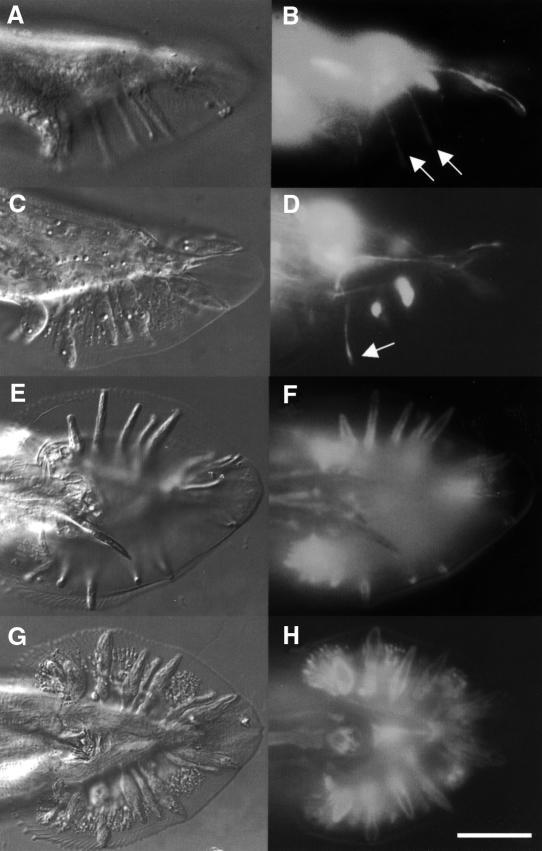
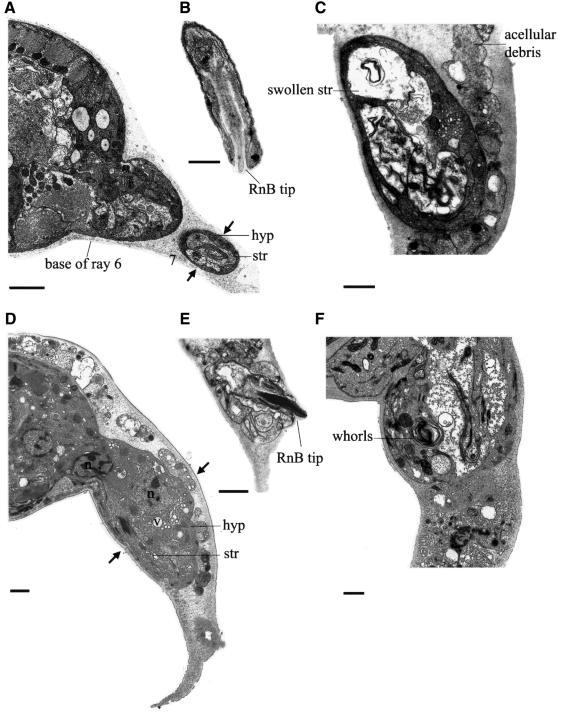
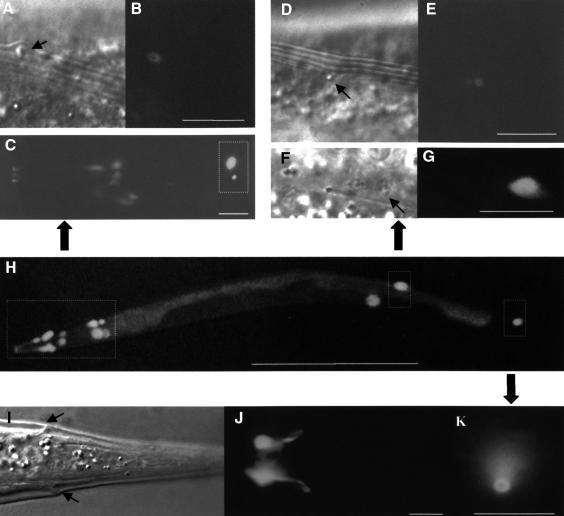
Animals with ram-5 mutations showed observable phenotype only in males and not in hermaphrodites. In ram-5 mutant males, all the rays are irregular (lumpy) in shape, but lie in their wild-type positions (Figure 1C and D). We used tissue-specific molecular markers and electron microscopy to examine the subcellular defect in ram-5 mutant animals. The promoter region from sek-1, the nematode homolog of the mammalian SEK-1 gene, was used to drive green fluorescent protein (GFP) expression in ray neurons (psek-1–GFP) (K.Matsumoto, personal communication). This GFP signal in both ray neurons, RnA and RnB, was used to monitor their morphological differentiation in ram-5 mutant males. Neuronal processes extend the full length of the rays in both mutant (Figure 2C and D) and wild type (Figure 2A and B). However, the expression of a hypodermal marker, pmab-21–GFP (S.S.H.Ho, G.M.K.So and K.L.Chow, in preparation), in ram-5 mutants revealed abnormal and irregular swelling in the hypodermal tissues (Figure 2G and H) as compared with that of wild-type rays (Figure 2E and F). Consistent with our observation by tissue-specific markers, transmission electron microscope (TEM) analysis also showed abnormal morphology of the hypodermal cell with entrapped nuclei, membranous whorls and cellular swelling (Figure 3C, D and F). The neuronal cells, RnA (not shown) and RnB (Figure 3B and E), appeared essentially normal and their distal ciliated tips were enclosed in a pocket of the structural cell process (not shown). In addition, acellular hypodermal debris, swollen structural cells with vacuoles and excessive membranous whorls were noted (Figure 3C and F). Hence, the lumpy ray phenotype must be due to abnormal differentiation of both structural and hypodermal cells but not gross abnormality in neurons.
Fig. 2. Mosaic animals showing ray neurons marked by psek-1–GFP in wild-type (A and B) and ram-5 (C and D) males. The hypodermal sheath of a ray is marked by pmab-21–GFP in wild-type (E and F) and ram-5 (G and H) animals. Scale bar = 20 µm. Arrows indicate normal neuronal extension.
Fig. 3. Subcellular anatomy of the rays in wild-type (A and B) and ram-5 (C–F) animals. (A) Transverse section of him-5 male tail showing rays 6 and 7 extending laterally to the right. Arrows indicate the girth of ray 7 at the midpoint, where a thin hypodermal (hyp) sheath wraps two neuron processes and the structural cell process (str). (B) A wild-type ray opens distally through a pore in the structural cell and cuticle to expose the tip of the RnB cilium. The ray is sheathed by a very thin layer of hyp. (C) A ram-5 ray is swollen at its midpoint with increased hyp volume and a very swollen str cell. The border between the str cell and the hyp is increased and disordered. Irregular membranes within the str process may be a disordered version of the tight spiral of membranes that is normally present within the str process [compare with (A)]. Acellular debris fills the cuticular fan, surrounding the ray. (D) Transverse section of a ram-5 male tail showing a fat ray at the same level as in (A). Note the increase in girth (between arrows) in comparison with wild type. The excess tissue is mostly hyp, including some vacuoles and a hyp nucleus, plus the nucleus of another cell at the base of the ray. The str cell looks normal here. (E) A ram-5 ray opens distally to expose the tip of the RnB cilium in a normal fashion [compare with (B)], but the boundary between hyp and str cell is disordered. (F) A ram-5 ray at midpoint is massively swollen by increased hyp volume, a very swollen str process and large whorls of membrane lying at the border, or perhaps within the hyp cytoplasm. Scale bars = 1 µm.
ram-5 males also displayed active mating engagement in mating tests and sired cross progeny efficiently (data not shown). While male searching behavior is dependent on the two ray neurons (Sulston et al., 1980; Liu and Sternberg, 1995), our observation suggests that ram-5 mutations have little or no effect on neuronal function of the rays.
Cloning of the ram-5 locus
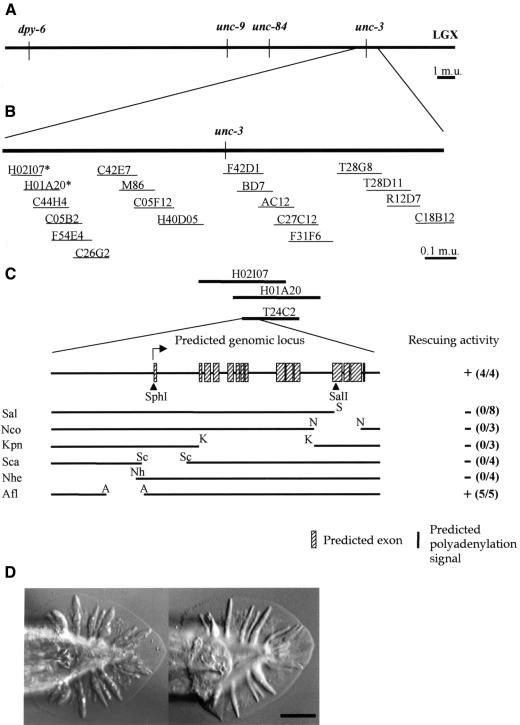
ram-5 was mapped on the right arm of chromosome X (Figure 4A and B). Rescue experiments with cosmids spanning this region identified three cosmids, H02I07, H01A20 and T24C2, which revert the mutant phenotype back to wild type (Figure 4D). Thus, the ram-5 locus must fall within their overlapping region. In these experiments, a swollen ray tip was occasionally observed when rescue was incomplete. Further deletion of T24C2 defined a minimal rescue DNA fragment of 9.5 kb (Figure 4C). Since animals carrying either bx30 or bx81 alleles of the ram-5 gene, obtained by ethylmethanesulfonate and di-epoxy-butane mutagenesis, respectively, could be rescued by this fragment, the mutations have probably occurred within this fragment.
Fig. 4. (A) Genetic map position of the ram-5 locus. (B) Arrangement of cosmids spanning through the mapped region. Asterisks mark the rescuing cosmids. (C) Predicted genomic organization of the ram-5 transcript and constructs for deletion-rescue analysis in a ram-5 mutant background. Independent transient lines were scored for the rescue activity (numbers in parentheses). Filled triangles are frameshift mutations. S, SalI; N, NcoI; K, KpnI; Sc, ScaI; Nh, NheI; A, AflII; (–), no rescue; (+), successful rescue. (D) Tail morphology of a ram-5 mutant male (left); animal transformed with 9.5 kb rescuing DNA fragment (right). Scale bar = 20 µm.
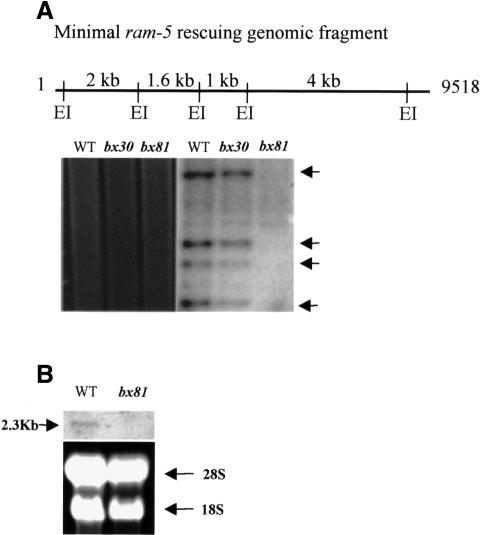
Using this 9.5 kb ram-5 genomic DNA as a probe, Southern hybridization was performed to map both ram-5 alleles. No hybridization signal was detected in the genomic DNA of ram-5 (bx81) worms, which indicates that the ram-5 locus was deleted in this allele (Figure 5A). Consistent with this finding, the northern analysis also failed to detect a 2.3 kb or a shorter ram-5-specific transcript in the bx81 allele (Figure 5B). Since this 9.5 kb fragment is sufficient to revert the mutant phenotype of bx81 allele to wild type, we conclude that the bx81 allele represents a null mutation of ram-5 and demonstrates the null phenotype. No chromosomal aberration was found in the bx30 allele. However, sequencing of the whole 9.5 kb fragment detected a nonsense mutation at Ala615 of the putative protein, changing it to an opal stop codon. Thus, bx30 allele produces a truncated non-functional RAM-5 protein that lacks a critical transmembrane (TM) domain (see below).
Fig. 5. (A) Southern blot analysis of genomic DNA from wild-type and two mutant alleles of the ram-5 gene, bx30 and bx81. Four EcoRI fragments spanning the ram-5 genomic region (arrows) were used as a probe to reveal the genomic aberration in the mutant animals. EI, EcoRI. (B) Northern blot analysis of wild-type and bx81 allele. A single ram-5 transcript was detected in RNA extracted from wild-type animals but not in the sample from a bx81-carrying strain. Ethidium bromide-stained gel showed equal loading of samples.
Characterization of ram-5 genomic organization and transcript
In terms of the genomic organization, both GENSCAN and GeneFinder programs suggested that there was only one probable transcript encoded on this 9.5 kb fragment (Burge and Karlin, 1997; Solovyev and Salamov, 1997). When a series of deletion constructs were made (Figure 4C), all deletions led to a loss of the rescuing activity except for an Afl deletion clone. Therefore, the ram-5 gene probably spans almost the entire 9.5 kb. In addition, a Nhe deletion clone without the putative promoter region from nucleotides 1 to 2438 failed to rescue the mutant phenotype, while a clone with an internal deletion from nucleotides 1630 to 2779 (Afl) could. The combined results showed that the 5′ promoter regulatory elements required for the rescuing activity are encompassed within the first 1630 nucleotides (Figure 4C).
To examine further the ram-5 coding region, frame-shift experiments were performed (Figure 4C). An SphI site at the putative translation start site at the predicted exon 1 was eliminated from this 9.5 kb fragment by T4 DNA polymerase modification. A frame-shift mutation at the SalI site in the predicted exon 12 was generated by a similar approach. Both mutations caused a loss of rescuing activity (SphI, 0/3 lines; SalI, 0/4 lines), which strongly implies that both sites fall within the coding region.
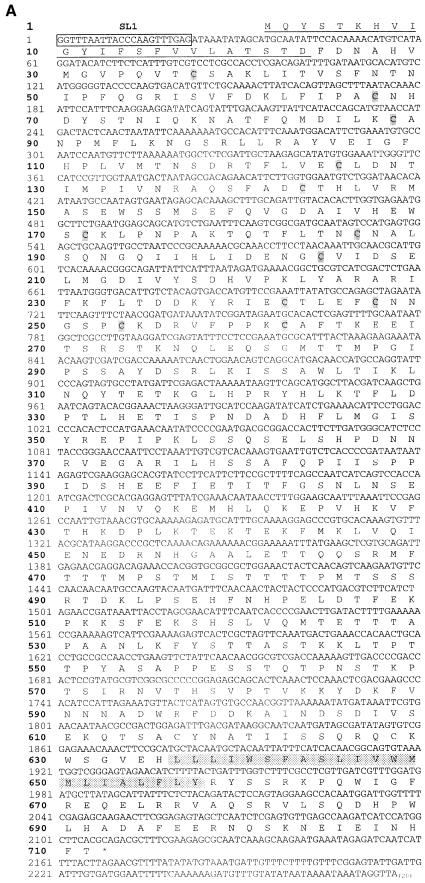
According to several prediction programs, the ram-5 gene has 14 exons (Figure 4C). Yet, no EST clone was identified based on the putative coding sequences. Neither did library screening yield any cDNA clone. We designed specific primers for 5′ RACE, 3′ RACE and RT–PCR to reconstruct a full-length ram-5 cDNA (Figure 6B). The cDNA obtained was ∼2.3 kb in length (Figure 6A), consistent with the transcript size detected by northern analysis (Figure 5B). It has 17 exons for the coding region and is transpliced to an SL1 leader at the 5′ end, thus representing a putative full-length cDNA (Figure 6B). Consensus signal sequences of splicing donor and acceptor sites at the exon–intron junctions were all confirmed in the genomic sequence.
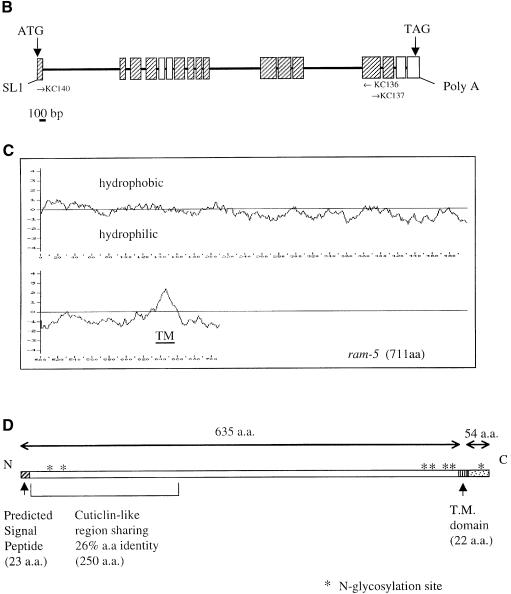
Fig. 6. Cloning and sequence analysis of ram-5 cDNA. (A) Nucleotide and predicted amino acid sequences of ram-5 cDNA. The 5′ end of cDNA contained trans-splice leader SL1. The predicted TM domain and stop codon are indicated by the shadowed box and by an asterisk, respectively. The underlined amino acids represents the predicted signal peptide. The 12 cysteine residues in the CUT-1-like domain are shadowed. (B) The genomic organization of the ram-5 gene. Hatched and white boxes indicate exons that are exactly identical to or different from the prediction, respectively. The position and orientation of PCR primers are indicated as arrows. (C) Hydrophobicity plot of the predicted RAM-5 amino acid sequence with the putative TM domain marked near the C-terminus. (D) Schematic organization of RAM-5 protein.
To test the functionality of this cDNA, double-stranded RNAs corresponding to regions spanning exons 1–4 and exons 9–12 were used for an interference study (RNAi) in wild-type animals (Fire et al., 1998). Male worms with lumpy rays and swollen ray tips were generated in both experiments, suggesting that the products derived from the PCR-based experiments correspond to portions of the ram-5 gene. When the putative full-length cDNA was put under the control of the ram-5 promoter, including all the genomic sequence 5′ to the translation start, it had full rescuing activity (bx30, 2/2 lines; bx81, 3/3 lines). Therefore, this cDNA encodes a functional RAM-5 protein.
Sequence analysis of ram-5
The predicted amino acid sequence of the RAM-5 protein obtained from the functional cDNA revealed a putative signal peptide at the N-terminus of RAM-5 (Figure 6A and D) (Nielsen et al., 1997). The N-terminal region shows a limited homology, 26% over 250 amino acids, to the CUT-1-like domain of cuticlin protein encoded by the C.elegans cut-1 gene (Figure 6D). However, while the CUT-1 domain contains 12 cysteine residues with conserved spacing, the 12 cysteines in RAM-5 have a slightly different spacing. In addition, a hydrophobic α-helical region between positions 636 and 657 represents a putative membrane-spanning domain of RAM-5 (Figure 6C). Because more positively charged amino acids reside in the C-terminus near the TM domain, the ‘positive inside rule’ predicts this protein to have a short cytoplasmic C-terminus and a large extracellular domain (Figure 6D) (Sipos and Von Heijne, 1993; Gafvelin et al., 1997). This prediction is in agreement with the position of the signal peptide.
RAM-5 protein has multiple potential glycosylation sites mostly on the extracellular domain (Figure 6D). It has a weak structural resemblance to cuticlin with a large extracellular domain and a short cytoplasmic tail (see above). Cuticlin was first identified as an insoluble non-collagenous component of nematode cuticle resistant to detergent and reducing agent treatment (Sebastiano et al., 1991). Although its biological function is unknown, it is expressed specifically in dauer larva forming an ∼2 µm wide ribbon along the lateral lines beneath the alae (Sebastiano et al., 1991) and may interact with the extracellular matrix in the dauer cuticle. The partial resemblance of RAM-5 to cuticlin suggests that it may interact with extracellular matrix components to dictate ray morphology.
Expression pattern of the ram-5 gene
To explore the expression pattern of ram-5, a GFP reporter was cloned into the genomic fragment at the putative translation start site at the SphI site (E1–GFP). Transgenic animals carrying this reporter gene were examined (Figures 7 and 8). Both hermaphrodites and males expressed GFP in a group of cells around the pharyngeal corpus (Figure 7C), which represented either the sheath or socket cells of the inner labial sensilla. Signal was also noted in the ADE socket cells (ADEso) (Figure 7C) and PDE socket cells (PDEso) (Figure 7F and G) in the midbody, and in phasmid socket cells (PHso2) (Figure 7I and J) in the tail, as defined by the presence of their openings, position and morphology. The openings of ADEso and PDEso lay beneath the lateral alae (Figure 7A, B and D, E), while those of Phso2 were near the tail tip (Figure 7K). GFP expression in these sensory support cells was detected soon after their birth and was maintained into adulthood.
Fig. 7. Common ram-5 expression pattern in wild-type males and hermaphrodites. ADE opening is displayed as a ring on the alae (A, arrow) with the ram-5 expression marked by fluorescence (B). (C) Labial neuron support cells around the pharyngeal corpus. The square box indicates the position of the ADE socket cell. PDE opening appears as a ring at the alae (D, arrow) with the fluorescent signal indicating ram-5 expression (E). The PDE socket cell (F, arrow) is also marked by GFP signal (G). (H) The overall expression pattern in a hermaphrodite with the magnified region marked by squares. (I) Phasmid socket cells (PhSo2) show GFP signal at the cell bodies, processes (J) and the opening (I, arrow; K). Scale bars = 10 µm [except (H), with scale bar = 0.5 mm].
Fig. 8. ram-5 (E1–GFP) expression in ray structural cells of the male tail in both wild type (A and B) and ram-5 mutant (C and D). Expressing cells in mutant tails display abnormal swelling of the structural cells. Scale bar = 20 µm.
More importantly, a GFP signal was observed in all the ray structural cells of the male tail (Figure 8A and B), but not in the hypodermal tissue. Expression in nine pairs of structural cells began just prior to the ray retraction period, peaked during the retraction and declined upon completion of ray extension. The same pattern of expression was also noted when a genomic fragment spanning nucleotides 1–1630 was used to drive the GFP reporter. This observation, in line with the deletion results, indicates that the regulatory cis-elements for ram-5 transcription activation reside within this 1.6 kb region.
ram-5 transcription is not dependent on expression of itself and other ram genes
With the E1–GFP reporter construct, we further examined the conditions for the transcriptional activation of the ram-5 gene in the ray structural cells. In a ram-5 null background, the transgene displayed identical spatial and temporal expression patterns to those in wild-type animals, suggesting that ram-5 gene transcription is independent of its own expression. Similarly, when the same transgene was crossed into five different ram mutants, ram-1 to ram-4 and mab-7, no change of expression was noticed (data not shown). It appears that the other ram genes are not required for tissue-specific expression of ram-5. Although these ram mutants have similar phenotypes and interact genetically, we conclude that mutations of them do not elicit the RAM phenotype through transcriptional regulation of the ram-5 gene.
Intriguingly, the ray structural cell morphology in ram-5 mutants, as revealed by this reporter, is swollen and vacuolated at regions where the structural cell contacts the hypodermal cell (n = 20) (Figure 8C and D). No other observable abnormality was noticed in other ram-5-expressing cells. In addition, structural cell bodies and processes within the animal body appear normal. This localized abnormality in the distal region of a structural cell implies that cell-specific expression of the ram-5 gene is required for normal differentiation of the sensory ray, but apparently does not affect the structural cell fate. Using this same E1–GFP reporter, the structural cell morphology in all the other ram mutants was shown to have similar abnormalities to different degrees (data not shown), suggesting that the structural cell is a common cellular target for all the ram mutations.
Localized expression of RAM-5 transmembrane protein is required for structural cell differentiation
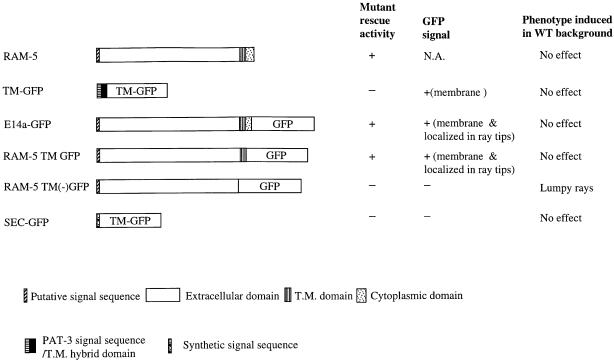
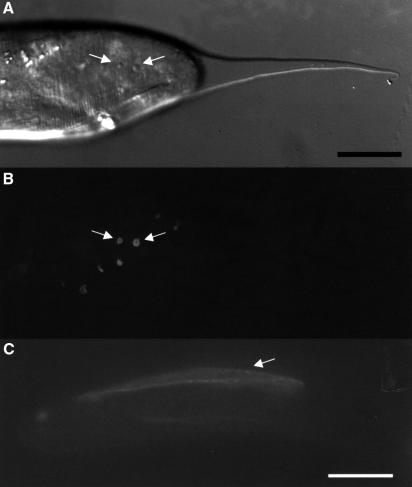
RAM-5 protein has a TM domain. To demonstrate its membrane-bound nature, E14a–GFP was made by in-frame fusion of GFP with RAM-5 after the predicted TM domain (Figure 10). When this fusion protein was driven by the ram-5 promoter, it behaved as a functional protein and rescued mutant ray phenotype (3/3 lines). The E14a–GFP transgene had an expression pattern identical to that of E1–GFP except that the fusion protein was detected on the structural cell surface and was highly localized near the distal pocket (Figure 9A and B). In contrast, the E1–GFP reporter presented an even distribution of GFP signal within the whole structural cell. This membrane association was further confirmed by ectopically expressing the fusion protein in the head hypodermal cells using a mab-21 promoter (Figure 9C). The localized RAM-5–GFP fusion protein (E14a–GFP) expression near the distal portion of the ray probably reflects a property of the RAM-5 protein but not the GFP moiety alone. When the transgene TM–GFP [including at its N-terminus a hybrid signal sequence and the PAT-3 protein TM domain (Gettner et al., 1995)] was expressed in the structural cells by the ram-5 promoter, the GFP signal was evenly distributed on the membrane of the entire structural cells including their processes (Figure 10). These results argue that domain(s) encoded by the first 15 exons of the ram-5 gene directs the subcellular localization of the protein at the ray tip.
Fig. 10. Summary of mutant rescuing activity, GFP signal detection and induced phenotype in wild-type background of different versions of RAM-5 fusion protein. All the fusion constructs were driven by a ram-5 promoter.
Fig. 9. Subcellular localization of RAM-5. (A) A lateral view of a retracting male tail with papillae (arrows) during the retraction period. (B) The same animal with E14a–GFP signal localized at the distal tip region of the ray structural cells. (C) E14a–GFP reporter driven by mab-21 promoter in hypodermis showed a membrane-bound signal. Scale bars = 20 µm.
RAM-5 cytoplasmic domain is dispensable
RAM-5 has a large N-terminal domain (635 amino acids) and only a short C-terminal tail (54 amino acids) (Figure 6D). In the E14a–GFP fusion construct, GFP was fused to RAM-5 protein after the TM domain with the last 24 amino acids deleted. These 24 amino acids appear dispensable for the RAM-5 biological function, and the construct could rescue the mutant ray phenotype. By site-directed mutagenesis, we introduced a SmaI site on the C-terminal side of the TM domain so that the entire cytoplasmic domain (54 amino acids) could be replaced by GFP (RAM-5 TM GFP) (Figure 10). This fusion protein was again localized to the ray tip and had full rescuing activity (2/2 lines). Hence, we conclude that the entire cytoplasmic domain of RAM-5 is dispensable for its normal function and its subcellular localization.
Another transgene, RAM TM(–) GFP, was produced without the TM domain, so that GFP was coupled only with the extracellular domain (Figure 10). Expression of this secreted form of fusion protein was not localized within specific cells and therefore not detectable. This fusion protein had no rescuing activity (0/4 lines). However, when these transgenes were introduced into wild-type animals, the secreted fusion protein could cause a lumpy ray phenotype at low penetrance while RAM-5 TM GFP or a secreted form of GFP (SEC–GFP) alone did not (Figure 10). These observations argue that the non-membrane-bound form of RAM-5 is expressed, and can interfere with normal RAM-5 function through its N-terminal extracellular domain. The lack of rescuing activity, on the other hand, highlights the importance of membrane anchorage of RAM-5 protein in normal ray morphogenesis.
Expression of ram-5 in structural cells can restore normal ray morphology
ram-5 mutants have both structural and hypodermal cell defects. To address the tissue-specific expression and requirement of ram-5 activity, we ectopically expressed the functional RAM-5–GFP fusion, E14a–GFP, in the other ray cells. Hypodermis-specific promoters from dpy-11 (F.C.F.Ko and K.L.Chow, in preparation) and a ray neuron-specific promoter from the sek-1 gene were chosen to express the functional GFP-tagged RAM-5 protein in hypodermal and neuronal cells, respectively, during the ray retraction period in mutant ram-5 worms. Both constructs failed to rescue the mutant phenotype, although membrane-bound GFP signal was observed in the targeted tissues at the correct temporal windows (pdpy-11-ram-5, 0/4 lines; psek-1-ram-5, 0/3 lines). Hence, RAM-5 proteins must function by anchoring on the structural cell but not other neighboring cell types.
Discussion
ram-5 product is required during the ray retraction
ram-5 mutant worms grow well with a normal life cycle. Embryonic lethality has not been noticed even in the null allele, bx81, and there is no apparent defect in hermaphrodites. Obviously, ram-5 is not an essential gene for normal development except for establishing ray morphology in male tails. Since lumpy rays do not interfere with mating success, ram-5 is required only for ray morphogenesis but not their sensory function. Therefore, this gene represents a true ‘morphogenesis gene’.
Temporal expression of ram-5, as revealed by the GFP reporter E1–GFP in the structural cells, peaks at the beginning of retraction, but is absent earlier when ray cell identity or cell fate are determined. Its expression window is consistent with that defined by heat shock phenocopy and temperature-sensitive allele analysis on ram gene function (Baird and Emmons, 1990; Chow and Chan, 1999). Expression of ram-5 declines substantially after the proper ray association is established. Adult male animals present little or no E14a–GFP expression near the ray tips, suggesting that ram-5 products are quickly turned over after ray morphogenesis completes. This observation (and heat shock analysis) indicates that ram-5 products are required only in actively differentiating ray cells but not for maintaining the structural features of a ray once the morphology is established.
Level of ram-5 activity is important for ray morphogenesis
In a previous study, ram-1 to -4 mutations were shown to be semidominant or haploinsufficient. Heterozygous animals showed variably swollen ray tips, whereas homozygotes had lumpy rays (Baird and Emmons, 1990). Since ram-5 resides on the X chromosome, haploinsufficiency or semidominance was not tested in the previous study. Our rescue and RNAi experiments offer clues that the ram-5 expression level is also critical to its normal function. Swollen ray tips were occasionally observed in rescued animals, and were also found in dsRNA-injected animals. Two hypotheses may account for this observation: (i) mutant RAM-5 protein can act semidominantly (gene interference) or (ii) normal ram-5 function is dependent on its expression level. Since the rescue experiment was performed in a null background (bx81) with a wild-type gene fragment, the swollen ray tips cannot be explained by the interference of mutant proteins. Neither could they be caused by overexpression of RAM-5, because multiple copies of rescuing transgene in wild-type animals generated no abnormality. In fact, the absence of a mutant RAM-5 protein in the bx81 rescue experiments favors the latter hypothesis, consistent with the results from RNAi experiments where wild-type protein synthesis was reduced and no mutant protein was introduced at all. Hence, the swollen ray tips phenotype can be considered to be a result of lowering the wild-type RAM-5 protein level below a certain threshold.
ram-5 is expressed exclusively in the sensory support cells
All the cells expressing ram-5 are neuronal support cells, including the sheath and socket cells of other sensilla. Except for the rays, none of these cells was abnormal in the mutant animals as determined by molecular markers and light microscopic examination. The ADEso, PDEso, Phso2 and labial support cells in the pharyngeal region each extend fine, morphologically normal processes for their corresponding sensilla in ram-5 mutant animals. Hence, ram-5 expression may not be required for morphogenesis of any other sensilla except male rays. Since no sensilla in the head or midbody of ram-5 mutants were examined by TEM, it remains possible that subtle defects went undetected in ram-5 mutants, or functionally redundant genes do exist for the differentiation of other socket and sheath cells.
The structural cell defects in the ram-5 mutants are distinctive. The swollen structural cell morphology seen by light microscopy and in EM thin sections is limited to the ray process and is strongest at the distal region, where it contacts with the hypodermal cell and forms a special pocket to envelop the cilia. Both these cells showed local swelling, vacuolation and membranous whorls in the mutants. The remaining portion including the cell bodies and the proximal processes within the worm body appeared normal. Consistently, the E14a–GFP fusion construct showed localized GFP signal near the structural cell pocket at the ray tip during ray extension. Based on the ram-5 expression profile and the specific requirement of RAM-5 protein in the structural cell, we conclude that ram-5 is needed for structural cell differentiation primarily at the structural cell–hypodermis junction during the ray retraction period. In this regard, it is noteworthy that the distal pocket does form in ram-5 engulfing the ciliated dendrites at the tip.
The ram-5 reporter construct (E1–GFP) also helped to uncover a similar cellular abnormality in all different ram mutants. The observation reveals the importance of structural cells in establishing the form and shape of sensory rays, echoing the original laser ablation results of Sulston and Horvitz (1977). This shared phenotype among many different ram mutants reflects an interactive mechanism for ray morphogenesis, where the Ram phenotype in other ram mutants did not result from the failure of transcriptional activation of the ram-5 gene, but was more likely due to failure of regulatory and interactive events occurring at the post-transcriptional level.
ram-5 is required by both structural and hypodermal cells
When cellular and functional abnormalities in ram-5 rays were examined, both the hypodermis and the structural cell were swollen. Excess tissue, swelling, membranous whorls and vacuoles were often found in both mutant hypodermis and structural cells. Retained nuclei [of hyp, structural cell process (str) and other cells] were common in the mutant tail fan as was acellular debris, suggesting that the retraction phase of morphogenesis requires RAM-5. Disruption of the cell border between the hypodermis and structural cell was also observed. Based on the structure of RAM-5 and its localized expression, we hypothesize that RAM-5 and other ram gene products participate in forming an adhesive complex at the borders between these two cells at a brief critical period of male tail retraction. We predict that specific cell surface molecules expressed on the hypodermis would bind avidly to the extracellular domain of the RAM-5 proteins. This notion is supported by the observation that normal RAM-5 function is subject to interference by a non-membrane-bound RAM-5 protein. This non-functional truncated RAM-5 at high level can act as a dominant-negative mutant molecule and competes with the wild-type RAM-5 protein for its interacting partners, impairs the cell–cell association and gives rise to a mutant phenotype.
The presence of cellular defects in both hypodermal and structural cells also indicates that RAM-5 may be required for more than an adhesion function and may facilitate a reciprocal signaling process between the expressing cell and its neighbor within a differentiating ray. Although the exact mechanism is not clear, abnormality found in both cell types in mutants implies that RAM-5 functions early in the reciprocal exchange of signals. Were ram-5 responsible for processing a secondary (later) signal, only one of the two cell types would be affected. Interestingly, such reciprocal information exchange is evident for several other morphogenetic steps in C.elegans development, including early steps of male tail morphogenesis, and may act through adhesive intercellular junctions (Simske et al., 1996; Nguyen et al., 1999). These junctional complexes, including small adherens junctions and gap junctions, do link the structural cell pocket to the hypodermis at the distal portion of a ray (D.H.Hall, unpublished). Although the RAM-5 TM GFP fusion protein does not show the discrete punctate localization typical for either class of cell junctions, its localized expression on the distal membrane of structural cells may represent a novel cellular communication complex acting in concert with other junctional components. In addition, according to this signaling model, some of the ram gene products must form links to the cytoskeleton in each cell type to foster cell shape changes during retraction.
In this context, it is intriguing that the C-terminal tail of the RAM-5 protein is dispensable for its function. Should signaling events occur between the ray cells, RAM-5 may simply elicit an initiation signal to the neighboring cells through its extracellular domain alone, or it may transduce an external signal into the cytoplasm of the structural cell through an associated molecule spanning the cell membrane. In either case, RAM-5 protein must interact with other cell surface molecules, probably modulated by glycosylation of its extracellular domain. The identification of these interacting components, likely to be encoded by other ram genes, will help to reveal the mechanism of ray morphogenesis.
Materials and methods
Culture of nematodes
Caenorhabditis elegans strains were maintained by standard laboratory procedures (Brenner, 1974). All strains in this study carried a him-5 (e1490) mutation, which gives a high incidence of male progeny (Hodgkin, 1983). Other worm strains used include: CB4088: him-5 (e1490)V; EM139: ram-1 (bx34)I; him-5 (e1490)V; EM268: ram-2 (bx76)II; him-5 (e1490)V; EM97: ram-3 (bx32)II; him-5 (e1490)V; EM68: ram-4 (bx25)IV; him-5 (e1490)V; EM81: him-5 (e1490)V; ram-5 (bx30)X; EM309: him-5 (e1490)V; ram-5 (bx81)X; and KC64: him-5 (e1490)V; mab-7 (e1599)X.
Electron microscopy
Fixation, thin sectioning and TEM were performed based on standard procedures (Hall, 1995). Healthy adult ram-5 mutant males were picked, washed in M9 buffer and fixed in 0.1 M HEPES buffer containing 2.5% glutaraldehyde, 1% formaldehyde for 90 min at room temperature. Tails were cut open in the primary fixative with a razor blade to improve tissue access. After buffer washes, samples were re-fixed in 0.1 M HEPES with 1% osmium tetroxide, 0.5% KFe(CN)6 for 60 min at room temperature, washed in buffer and stained in 1% uranyl acetate, 0.1 M sodium acetate. After embedding in 3% agarose to position the tails in small groups, specimens were embedded in Scipoxy resin (Energy Beam Sciences) and heat cured. Thin sections were collected transverse to the body axis, post-stained with uranyl acetate and lead citrate, and examined in a Philips CM10 electron microscope.
Engineering of deletion and reporter constructs
All constructs were made following standard molecular procedures described in Sambrook et al. (1989). SalI–BglII mab-21 promoter fragments (2.3 kb) and HindIII–XbaI sek-1 promoter fragments (2.2 kb) were subcloned into pPD95.77 to generate hypodermal and neuronal markers for ray analysis. Plasmid carrying a 9.5 kb ram-5 genomic fragment was used as the parental vector for expression study. E1–GFP and SEC–GFP were generated by subcloning the GFP NLS– from pPD95.81 or GFP with a synthetic signal sequence from pPD95.85, respectively, downstream of the translation start at the SphI site. E14a–GFP was constructed by in-frame blunt-end ligation of GFP NLS– from pPD95.77 into ram-5 cDNA at the NcoI site, located in exon 14a, after T4 DNA polymerase treatment. The ram-5 promoter in E14a–GFP was replaced with a 2.3 kb SalI–BglII mab-21 promoter, 2.1 kb XhoI–BsmI dpy-11 promoter and 2.2 kb HindIII–XbaI sek-1 promoter to generate pmab-21-ram-5, psek-1-ram-5 and pdpy-11-ram-5, respectively, for ectopic expression study. The TM–GFP reporter construct was prepared by cloning of TM–GFP, from pPD122.39, under the control of the ram-5 promoter at the SphI site.
Mutagenesis was performed on ram-5 cDNA to introduce a SmaI site before and after the TM domain at positions 1935 and 2007 of the cDNA, respectively. Mutagenic PCR was performed with KC172 (5′-GGTCGGGAGTAGAACCCGGGCATCTTTTACTGATTTGG-3′)/KC173 (5′-CCAAATCAGTAAAAGATGCCCGGGTTCTACTCCCGACC-3′) and KC174 (5′-GCATTATTTCTCTACAGACCCGGGTACTCCAGTAGGAAGCC-3′)/KC175 (5′-GGCTTCCTACTGGAGTACCCGGGTCTGTAGAGAAATAATGC-3′) as described by Vallejo et al. (1995). GFP from pPD95.79 was then cloned into the cDNA at the SmaI site before and after the TM domain to generate the RAM-5 TM(–) GFP and RAM-5 TM GFP, respectively.
Rescue experiment
Microinjection of DNA into gonadal syncytium of hermaphrodites was carried out as described by Mello et al. (1991). Tested cosmids or plasmids were co-injected with a dominant rol-6 marker at a concentration of 100–150 ng/µl. Multiple transgenic lines were scored for each transformation experiment.
Mutation mapping
Genomic DNA from wild-type and mutant worms was isolated as described by Sulston and Hodgkin (Wood, 1988). Five micrograms of genomic DNA were digested with EcoRI, run on a 0.7% agarose gel and transferred onto nitrocellulose membrane for Southern analysis as described (Sambrook et al., 1989). Radiolabeled probe was prepared by random primer labeling of the 9.5 kb ram-5 genomic fragment to detect aberration of the bx81 allele. In the case of the bx30 allele, nine sets of gene-specific primers were designed for amplification of exons including its splice sites from the mutant genomic DNA. At least two independent PCRs were performed for each set of primers. The amplified genomic fragments were PCR sequenced to map the point mutation.
Extraction of total RNA, cloning of cDNA and northern analysis
Total RNA was extracted by a modified guanidinium thiocyanate–cesium chloride method (Sambrook et al., 1989). RT–PCR experiments were performed using a pair of gene-specific primers, KC136 (5′-GGTCAACTTTTTGGTCGACG-3′) and KC140 (5′-CTTCTCATTTGTCGTCCTCG-3′), or SL1 primer (5′-GGTTTAATTACCCAAGTTTGAG-3′) with KC136 for amplification. A 1.6 kb cDNA fragment, which was identical to the predicted cDNA, except that two additional exons, Ad1 and Ad2, had been inserted between the predicted exons 4 and 5 (Figure 6B). Both 5′ RACE RT–PCR experiments with KC136 and an anchor primer or with KC136 and SL1 primers resulted in a 1.6 kb amplified product with SL1 sequence on the 5′ end. Sequence analysis confirmed that the SL1 leader was trans-spliced to the 5′ end of cDNA at position –11 with a 3′ splice acceptor consensus (Figure 6A). 3′ RACE experiment with KC137 (5′-GAAGTTCTATTCAACAACGG-3′) and an anchor primer amplified a 0.7 kb fragment corresponding to the predicted 3′ end with an additional intron splitting the predicted exon 14 into exons 14a and 14b (Figure 6B). Northern analysis was performed with RNA samples separated on a 1% formaldehyde–agarose gel, transferred onto nitrocellulose membrane and hybridized with a radiolabeled ram-5 cDNA probe prepared by random labeling (Sambrook et al., 1989).
GenBank accession number
The DDBJ/EMBL/GenBank accession No. is AF218866.
Acknowledgments
Acknowledgements
We thank K.Matsumoto for the sek-1 genomic DNA. We thank K.C.Chow, V.Unkefer and members of our laboratory for critical comments on this manuscript. This research was supported by Hong Kong Research Grants Council grants to K.L.C. and NIH Grant RR12596 (NCRR) to D.H.H. for the Center for C.elegans Anatomy. R.Y.L.Y. is a Croucher Foundation Graduate Fellow. Some nematode strains used in this work were provided by Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources.
References
- Appel L.F., Prout,M., Abu-Shumays,R., Hammonds,A., Garbe,J.C., Fristrom,D. and Fristrom,J. (1993) The Drosophila Stubble-stubbloid gene encodes an apparent transmembrane serine protease required for epithelial morphogenesis. Proc. Natl Acad. Sci. USA, 90, 4937–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S.E. and Emmons,S.W. (1990) Properties of a class of genes required for ray morphogenesis in Caenorhabditis elegans. Genetics, 126, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S.E., Fitch,D.H.A., Kassem,I.A.A. and Emmons,S.W. (1991) Pattern formation in the nematode epidermis: determination of the arrangement of peripheral sense organs in the C.elegans mail tail. Development, 113, 515–526. [DOI] [PubMed] [Google Scholar]
- Brabant M.C., Fristrom,D., Bunch,T.A., Baker,S.E., Brower,D.L. (1998) The PS integrins are required for a regulatory event during Drosophila wing morphogenesis. Ann. N Y Acad. Sci., 23, 99–109. [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Chow K.L. and Chan,K.W. (1999) Stress induced phenocopy of C.elegans defines functional steps of sensory organ differentiation. Dev. Growth Differ., 41, 629–637. [DOI] [PubMed] [Google Scholar]
- Chow K.L., Hall,D.H. and Emmons,S.W. (1995) The mab-21 gene of C.elegans encodes a novel protein required for choice of alternate cell fates. Development, 121, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gafvelin G., Sakaguchi,M., Andersson,H. and von Heijne,G. (1997) Topological rules for membrane protein assembly in eukaryotic cells. J. Biol. Chem., 272, 6119–6127. [DOI] [PubMed] [Google Scholar]
- Geiger B. and Ayalon,O. (1992) Cadherins. Annu. Rev. Cell Biol., 8, 307–332. [DOI] [PubMed] [Google Scholar]
- Gettner S.N., Kenyon,C. and Reichardt,L.F. (1995) Characterization of β pat-3 heterodimers, a family of essential integrin receptors in C.elegans. J. Cell Biol., 129, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.H. (1995) Electron microscopy and three dimensional image reconstruction. Methods Cell Biol., 48, 395–436. [DOI] [PubMed] [Google Scholar]
- Herman T., Hartwieg,E. and Horvitz,H.R. (1999) sqv mutants of Caenorhabditis elegans are defective in vulval epithelial invagination. Proc. Natl Acad. Sci. USA, 96, 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. (1983) Male phenotypes and mating efficiency in C.elegans. Genetics, 103, 43–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. and Hirsh,D. (1979) Post-embryonic cell lineages of the hermaphrodite and male gonads in C.elegans. Dev. Biol., 70, 396–417. [DOI] [PubMed] [Google Scholar]
- Ko F.C.F. and Chow,K.L. (2000) Synthesis of glycoprotein is important for molting and male tail sensory ray morphogenesis in Caenorhabditis elegans. Dev. Growth Differ., 42, 69–78. [DOI] [PubMed] [Google Scholar]
- Liu K.S. and Sternberg,P.W. (1995) Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron, 14, 79–89. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.P., Acton,G.Z., Hartwieg,E., Horvitz,H.R. and Sternberg,P.W. (1999) The lin-11 LIM domain transcription factor is necessary for morphogenesis of C.elegans uterine cells. Development, 126, 5319–5326. [DOI] [PubMed] [Google Scholar]
- Nguyen C.Q., Hall,D.H., Yang,Y. and Fitch,D.H. (1999) Morphogenesis of the Caenorhabditis elegans male tail tip. Dev. Biol., 207, 86–106. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Poodry C.A. (1992) Morphogenesis of Drosophila. In Alexander,S. and Rossomando,E.F. (eds), Morphogenesis: An Analysis of the Development of Biological Form. Marcel Dekker, New York, NY, pp. 143–188. [Google Scholar]
- Sambrook J., Fritsch.E.F. and Maniatis.T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sebastiano M., Lassandro,F. and Bazzicalupo,P. (1991) cut-1 a Caenorhabditis elegans gene coding for a dauer-specific non-collagenous component of the cuticle. Dev. Biol., 146, 519–530. [DOI] [PubMed] [Google Scholar]
- Simske J.S., Kaech,S.M., Harp,S.A. and Kim,S.K. (1996) LET-23 receptor localization by the cell junction protein LIN-7 during C.elegans vulval induction. Cell, 85, 195–204. [DOI] [PubMed] [Google Scholar]
- Sipos L. and von Heijne,G. (1993) Predicting the topology of eukaryotic membrane proteins. Eur. J. Biochem., 213, 1333–1340. [DOI] [PubMed] [Google Scholar]
- Solovyev V.A. and Salamov,A.A. (1997) The Gene-Finder computer tools for analysis of human and model organisms genome sequences. In Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 294–302. [PubMed] [Google Scholar]
- Sulston J.E. and Horvitz,H.R. (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol., 56, 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Albertson,D.G. and Thomson,J.N. (1980) The C.elegans male: postembryonic development of nongonadal structures. Dev. Biol., 78, 542–576. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg,E., White,J.G. and Thomson,J.N. (1983) The embryonic cell lineage of the nematode C.elegans. Dev. Biol., 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Vallejo A.N., Pogulis,J.R. and Pease,L.R. (1995) Mutagenesis and synthesis of novel recombinant genes using PCR. In Dieffenbach,C.W. and Dveksler,G.S. (eds), PCR Primer: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 603–612. [Google Scholar]
- von Kalm L., Fristrom,D. and Fristrom,J. (1995) The making of a fly leg: a model for epithelial morphogenesis. BioEssays, 17, 693–702. [DOI] [PubMed] [Google Scholar]
- Wood W.B. (1988) The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 604–605. [Google Scholar]