Abstract
Ovarian cancer is a complex and deadly disease that remains difficult to detect at an early curable stage. Furthermore, although some oncogenic (Kras, Pten/PI3K and Trp53) pathways that are frequently mutated, deleted or amplified in ovarian cancer are known, how these pathways initiate and drive specific morphological phenotypes and tumor outcomes remain unclear. We recently generated Pten fl/fl; KrasG12D;Amhr2-Cre mice to disrupt the Pten gene and express a stable mutant form of KrasG12D in ovarian surface epithelial (OSE) cells. Based on histopathologic criteria, the mutant mice developed low-grade ovarian serous papillary adenocarcinomas at an early age and with 100% penetrance. This highly reproducible phenotype provides the first mouse model in which to study this ovarian cancer subtype. OSE cells isolated from ovaries of mutant mice at 5 and 10 weeks of age exhibit temporal changes in the expression of specific Mullerian epithelial marker genes, grow in soft agar and develop ectopic invasive tumors in recipient mice, indicating that the cells are transformed. Gene profiling identified specific mRNAs and microRNAs differentially expressed in purified OSE cells derived from tumors of the mutant mice compared to WT OSE cells. Mapping of transcripts or genes between the mouse OSE mutant datasets, the Kras signature from human cancer cell lines and the human ovarian tumor array datasets, documented significant overlap, indicating that KRAS is a key driver of OSE transformation in this context. Two key hallmarks of the mutant OSE cells in these mice are the elevated expression of the tumor suppressorsTrp53 (p53) and its microRNA target, miR-34a-c. We propose that elevated TRP53 and miR-34a-c may exert negatively regulatory effects that reduce the proliferative potential of OSE cells leading to the low-grade serous adenocarcinoma phenotype.
Keywords: Kras, Pten, Trp53, ovarian cancer, serous
Introduction
Ovarian cancer is a complex disease that remains difficult to diagnose at early stages and hence is difficult to treat (Bast RC, Hennessy B, et al., 2009, Cho KR, 2009, Cho KR and Shih I-M, 2009). Epithelial ovarian cancers are subdivided into four major categories based on histological criteria and resemblance to epithelial components of the normal female reproductive tract (Cho KR and Shih I-M, 2009). Approximately 70% of ovarian carcinomas are categorized as serous tumors because their histoarchitecture recapitulates the epithelium lining the human fallopian tube. Other categories include endometrioid, mucinous and clear cell subtypes (Bast RC, Hennessy B, et al., 2009, Cho KR and Shih I-M, 2009). Ovarian carcinomas are also classified as either low-grade or high-grade tumors. Clinically, low and high-grade ovarian cancer behave very differently, with low-grade cancers being more resistant to cytotoxic chemotherapy (Cho KR and Shih I-M, 2009). Whereas low-grade tumors frequently exhibit alterations (mutations, deletions or amplifications) of genes in the Kras and Pten/PI3K pathways, high-grade tumors are associated frequently with mutations (or deletions) in the tumor repressor protein (Trp53; p53) gene (Cho KR and Shih I-M, 2009) and may be derived from fimbrial epithleial cells of the distal Fallopian tube (Kindelberger DW, Lee Y, et al., 2007, Lee Y, Miron A, et al., 2007) as well as from OSE cells (Auersperg N, Woo MMM, et al., 2008). High-grade tumors may also exhibit amplification of PIK3CA (Astanehe A, Arenillas D et al., 2008, Roh MH, Yassin Y, et al., 2010) suggesting a link between PTEN and p53. Recent gene profiling and sequencing databases have identified mRNAs and microRNAs relatively over-expressed or under-expressed in various ovarian cancer subtypes (Bast RC, Hennessy B, et al., 2009, Creighton CJ, Fountain Md, et al., 2010, Despierre E, Lambrechts D, et al., 2010, Kobel M, Kalloger SE, et al., 2008, Konstantinopoulos PA, Sppentzos D, et al., 2008, Page CL, Ouellet V, et al., 2006). However, our understanding this complex disease remains limited and continues to evolve as new information on the potential sites of origin and molecular pathways involved is obtained (Auersperg N, Woo MMM, et al., 2008, Crum CP, Drapkin R, et al., 2007, Karst AM and Drapkin R, 2010, Lee Y, Miron A, et al., 2007).
To help understand the etiology of human ovarian cancer, several mouse models of this disease have been devised in recent years using novel, pioneering strategies. Oncogenes introduced into dispersed Trp53 null ovarian cells using in vitro and in vivo approaches and avian retroviral receptors generated epithelial cell tumors (Orsulic S, Li Y, et al., 2002). Other investigators have injected adenoviral vectors expressing Cre recombinase under the ovarian bursa to generate Trp53fl/fl;Rbfl/fl (Flesken-Nikitin A, Choi K-C, et al., 2003), Ptenfl/fl; LSL-KrasG12D (Dinulescu DM, Ince TA, et al., 2005) and Ptenfl/fl;Apcfl/fl (Wu R, Hendrix-Lucas N, et al., 2007) mutations, characterized as poorly differentiated carcinomas, endometrioid-like adenocarcinomas, endometrioid adenocarcinomas, respectively. Although the adenoviral injection approach has been a major breakthrough, it depends on a highly technical intervention that can deliver the vectors to bursal and/or oviductal cells in addition to the OSE, thereby obscuring the identity of the actual transformed cell type(s) (Clark-Knowles KV, Senterman MK, et al., 2009). The anti-Mullerian hormone receptor (AMHR2) is expressed in multiple cells types, including Mullerian-derived ovarian surface epithelial (OSE) cells, stromal cells of the oviduct and uterus as well as ovarian granulosa cells (Fan HY, Shimada M, et al., 2008, Jamin SP, Arango NA, et al., 2002, Xing D, Scangas G, et al., 2009). When the Amhr2 promoter was used to drive expression of the SV40 T antigen, ovarian carcinomas developed with ~50% penetrance (Connolly DC, Bao R, et al., 2003) verifying expression of Amhr2-Cre in OSE cells. More recently, when the Amhr2-Cre mice were used to disrupt the Brca1 and p53 genes (Xing D, Scangas G, et al., 2009), uterine leiomyosarcomas developed rapidly indicating that uterine Amhr2-Cre expressing smooth muscle cells are acutely sensitive to these particular mutations. Importantly, activation of the PI3K pathway by loss of Pten (Ptenfl/fl;Amhr2Cre mice)(Fan HY, Liu Z, et al., 2009, Lague MN, Paquet M, et al., 2008) or over-expression of PI3K (Pik3ca;Amhr2Cre mice)(Liang S, Yang N, et al., 2009) or activation of the KRAS pathway by expression of stable mutant KRASG12D(KrasG12D;Amhr2Cre mice)(Fan HY, Liu Z, et al., 2009) alone does not lead to ovarian cancer but can enhance OSE cell proliferation and metaplasia (Liang S, Yang N, et al., 2009).
Recent studies from our laboratory have shown that activation of the RAS, MEK1, and ERK1/2 pathway directs specific cell fate decisions of ovarian granulosa cells in growing and preovulatory follicles as well as in cells that comprise the ovarian surface epithelium (Fan HY, Liu Z, et al., 2009, Fan HY, Liu Z, et al., 2009). In preovulatory follicles activation of RAS and ERK1/2 by the LH surge is essential for ovulation and terminal differentiation of granulosa cells (Fan HY, Liu Z, et al., 2009). Conversely, premature expression of a mutant stable KRAS (KRASG12D) in granulosa cells of the KrasG12D;Amhr2Cre mice completely derailed early follicle development (Fan HY, Shimada M, et al., 2008). Granulosa cells ceased dividing, failed to undergo apoptosis and did not differentiate. Because KrasG12D mutant granulosa cells expressed high levels of the tumor suppressor PTEN, we further disrupted the Pten gene in the KrasG12D;Amhr2Cre mouse strain. Surprisingly, the fate of granulosa cells in the abnormal follicles was not markedly altered in the KrasG12D; Pten;Amhr2Cre double mutant mice. However, the ovarian surface epithelial (OSE) cells developed into low-grade serous papillary adenocarcinomas (as classified by expert mouse and human pathologists) with 100% penetrance and died within 4-6 months of age due to tumor volume (Fan HY, Liu Z, et al., 2009). Therefore, the Ptenfl/fl;KrasG12D;Amhr2-Cre mice provide the first evidence that granulosa cells are highly resistant to many oncogenic insults that profoundly impact OSE cells (Fan HY, Liu Z, et al., 2009, Fan HY, Shimada M, et al., 2008), thereby explaining why granulosa cell tumors have not been observed in the previous studies (Connolly DC, Bao R, et al., 2003, Fan HY, Liu Z, et al., 2009, Orsulic S, Li Y, et al., 2002).
Importantly, the low grade serous adenocarcinomas of the Ptenfl/fl;KrasG12D;Amhr2-Cre mutant mice represent the first mouse model of this specific ovarian cancer subtype (Fan HY, Liu Z, et al., 2009). The spontaneous and reproducible development of serous adenocarcinomas in these mice have lead us to determine what molecular pathways are altered in these cells and if this model can be used to understand the molecular events controlling the transformation of OSE cells and thereby provide some insights into this cancer subtype in women. In particular, this model, unlike other models, affords the opportunity to track the initiation of transformation of OSE cells at early stages in vivo by determining when the mutant OSE cells first exhibit altered morphology and functions and what signaling pathways are required to maintain transformation in the OSE cells in this context. The striking increases Mullerian cell markers in the OSE tumors indicate that these cells can differentiation into a Fallopian-like epithelium. In addition, the consistent increases in Trp53 expression and its target microRNA, miR-34a-c indicate that altered activation of the PI3K and RAS pathways is tightly linked to regulation of TRP53 levels and presumably function in these cells.
Results
OSE cell tumors are evident in the Ptenfl/fl; KrasG12D;Amhr2-cre mice as early as 5 weeks of age and express markers of human serous adenocarcinomas
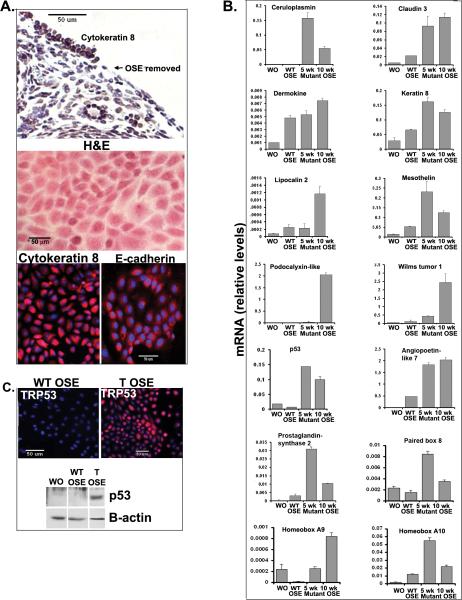
Histological sections prepared from ovaries of control mice at 5 and 10 weeks of age show that the OSE cell layer is comprised on a single layer of meso-epithelial cells as reported by others (Auersperg N, Wong AST, et al., 2001, Orsulic S, Li Y, et al., 2002). These cells stain for the epithelial cell markers cytokeratin 8 (Fan HY, Liu Z, et al., 2009) and for E-cadherin (CDH1) but not for vimentin (Figure 1A). In contrast, the OSE layer present in ovaries of the Ptenfl/fl;KrasG12D;Amhr2-Cre mice exhibits visible changes in morphology with obvious epithelial cell hyperplasia evident as early as 3 weeks of age in some ovaries and at 5 weeks in all ovaries. By 10 weeks of age, all mice exhibited low-grade serous, papillary-like adenocarcinomas as classified by expert pathologists (Fig. 1A). Of note, the OSE tumor cells stain for cytokeratin 8 (Fan HY, Liu Z, et al., 2009) and vimentin but not for E-cadherin (Figure 1A) suggesting that the cells have undergone an epithelial to mesenchymal type of transformation (Lee JM, Dedhar S, et al., 2006). Furthermore, the OSE tumor cells do not stain for calretinin, a marker of mesotheliomas and do not exhibit atypic nuclear morphology observed in high-grade adenocarcinomas (Supplemental Figure 1). However, the tumor OSE cells do stain positive for estrogen receptor alpha (ESR1), providing additional evidence that these tumors represent low-grade adenocarcinomas (Supplemental Figure 1) (Wong KK, Lu KH, et al., 2007).
Figure 1.

Temporal changes in OSE cell morphology and expression of specific genes associated with the serous adenocarcinoma phenotype in the Pten;KrasG12D;Amhr2-cre mice. A.) H&E and immuno-labeling of ovarian sections from WT and mutant mice at 5 and 10 weeks of age. B.) Gene expression patterns in whole ovaries of WT or tumor bearing (T) Pten;KrasG12D;Amhr2-cre mice at 5 and 10 weeks of age.
To identify specific genes expressed in the tumor-bearing ovaries at 3 months of age, total RNA was extracted from ovaries of control (wild-type) and Pten fl/fl; KrasG12D;Amhr2-Cre mice and submitted for Microarray Analyses. Table 1 lists the top 25 most highly up-regulated genes in the Pten fl/fl; KrasG12D;Amhr2-Cre tumor-bearing ovaries (Supplementary Table S1). These include potential regulators of stem cells (Angtpl7) (Zhang CC, Kaba M, et al., 2006), markers of cancer cells (Nov, also known as Ccn3 (Bohlig L, Metzger R, et al., 2008), Mela and Ptgs2) (Wang D and DuBois RN, 2010), specific cytokines (Gkn1, Gnk2 ) and the acute phase factor (Hp) (Abdullah M, Schultz H, et al., 2009).
When we compared our microarray data derived from normal ovaries and tumor-bearing ovaries of Pten fl/fl; KrasG12D;Amhr2-Cre mice with data sets derived from human cancer cell lines expressing mutant or wild-type KRAS, 213 genes exhibited significantly over-lapping mutant Kras-related expression patterns (Figure 2 A and B) (Supplementary Table S2). Genes highly expressed in the human mutant Kras expressing cancer cells and the mouse ovary tumor samples include: Ptgs2, Mpzl2 (Eva1), Cldn3, Lcn2, Ccnd1, Adam8, Egfr, Btc, Podxl, Msln, Igfbp4 and Dusp6. These results indicate that the transformed mouse cells exhibit a mutant Kras signature similar to that observed in human cancer cell lines (GlaxoSmithKline GSK, 2008).
Figure 2.

Genes overlapping between the Pten;Kras;Amhr2-cre gene signature and relevant signatures from human cancers. (A) Overlaps between the set of human orthologs high (fold>2) in Pten;Kras;Amhr2-cre mice (compared to wild-type mice) and various sets of genes derived from human datasets: genes high/low (p<0.01, t-test) in k-ras mutant versus k-ras wild-type cell lines, and genes high/low (p<0.05) in serous, mucinous, or endometrioid ovarian cancers, as compared to normal ovary. Numbers of genes in each set indicated in parentheses. Numbers overlapping between sets were compared to chance expected. (B) Heat maps showing expression patterns for the Pten;Kras;Amhr2-cre gene signature, both in the Pten;Kras;Amhr2-cre mouse expression dataset and in an expression dataset of human cancer cell lines with or without mutations in kras (35). (C) Heat maps for the Pten;Kras;Amhr2-cre gene signature, both in the mouse dataset and in a dataset of human ovarian tumors (36). For parts B and C, genes represented in both heat maps are the same and have the same ordering, and genes not represented in the given human dataset are not shown.
The data sets from our tumor-bearing ovaries also exhibit overlapping expression with data sets from human ovarian cancers (as compared to normal ovary): 330 genes in human serous tumors and 331 in the mucinous tumors (Figure 2A and C) (Malpica A, Deavers MT, et al., 2004). Among the common genes are Cp, Lcn2, Dmkn, Msln, Cldn3, Podxl, Esr1 and Muc16 as well as specific Mullerian epithelial marker genes, Wt1, Pax8, Hoxa9 and Hoxa10. Genes encoding matrix components were also observed (Cheng W, Liu J, et al., 2005, Cho KR and Shih I-M, 2009, Despierre E, Lambrechts D, et al., 2010, Gava N, Clarke CL, et al., 281, Gorringe KL and IG, 2009, Kobel M, Kalloger SE, et al., 2008, Konstantinopoulos PA, Sppentzos D, et al., 2008, Malpica A, Deavers MT, et al., 2004, Santin AD, Zhan F, et al., 2004, Schwartz DR, Kardia SL, et al., 2002)(Supplementary Table S3). That many of the genes in the mouse and human ovarian cancer samples are common to the cancer cell Kras signature indicates that activation of Kras pathway (by mutations or other mechanisms) may be an underlying common feature of transformed OSE and Mullerian cells. Because a high percentage human ovarian cancer samples are likely to be high-grade serous adenocarcinomas, these tumors may harbor defects not only in the function of TP53 but also changes in the MET-RAS signaling pathway (Corney DC, Hwang C, et al., 2010, Zhao Z, Zuber J, et al., 2010).
Real-time RT-PCR results using RNA prepared from control and the tumor-bearing ovaries confirmed the Microarray data, documented age-dependent changes in gene expression profiles and verified up-regulation of specific genes known to be expressed in human ovarian cancers (Fig.1B). Cp, Podxl and Ptgs2 were expressed at elevated levels by 5 weeks of age whereas Cldn3, Dmkn, Krt8, Lcn2, Msln and Angptl were elevated at 10 weeks. Wt1, Pax8, Hoxa9 and Hoxa10 were also elevated in the mouse tumor tissue at 10 weeks of age indicating that these cells had acquired specific characteristics of Mullerian (Fallopian tube/endometrial) epithelial cells. Expression of the Trp53 (p53) was also elevated in the tumor cells providing further evidence that there are distinct temporal changes in gene expression patterns in the mutant OSE cells.
Most genes expressed at reduced levels in the tumor-bearing ovaries from the Pten fl/fl;KrasG12D;Amhr2-cre mice compared to controls are oocyte (*) or granulosa cell specific (Supplemental Table S4) and reflect the loss of oocytes and altered granulosa cell differentiation in the abnormal follicle structures (Supplemental Table S2) present in these ovaries (Fan HY, Shimada M, et al., 2008).
Tumor-related genes are selectively expressed in ovarian surface epithelial cells
To determine which of the genes associated with the tumor-bearing ovary at 5 and 10 weeks of age were expressed specifically in purified OSE cells, we isolated OSE cells from ovaries of WT and Pten fl/fl; KrasG12D;Amhr2-Cre mice. The WT and mutant OSE cells (cytokerain 8 positive) were removed from the epithelium by mild-trypsin digestion (Fig.3A, upper panel). The highly purified OSE cells from WT ovaries appear morphologically homogeneous in culture as indicated by the characteristic cuboidal-cell shape (Fig.3A, middle panel) and uniform immunolabeling of cytokeratin 8 and E-cadherin (Fig.3A, lower panel) as would be predicted from their presence in vivo (Fig. 1 and Fig.3A).
Figure 3.
Mutant OSE cells express genes associated with human serous adenocarcinomas in a time-dependent manner. A.) Cytokeratin 8 and E-cadherin-positive OSE cells removed from the ovarian surface exhibit a cobblestone appearance. B.) Expression of genes in RNA samples from whole ovaries and purified OSE cells isolated from ovaries of wild type and Pten;KrasG12D;Amhr2-cre mice at 5 and 10 weeks of age.
Several genes (Cldn3, Dmkn, Msln, Lcn2) were higher in the purified WT OSE cells compared to WT whole ovaries (Fig. 3B), suggesting that these genes are specific markers of OSE cells and function in the non-transformed epithelium. Furthermore, the expression of Wt1, Pax8, Hoxa9 and Hox10, specific markers of Mullerian-derived (Fallopian and endometrial) epithelia (Cheng W, Liu J, et al., 2005, Ko SY, Lengyel E, et al., 2010) were induced in the mutant mouse OSE cells (Figure 3B). These results document unequivocally that mouse OSE cells can acquire Mullerian duct markers during transformation by disrupting Pten and expressing mutant KrasG12D. Differential gene expression patterns in the mutant OSE cells were observed: some genes were elevated selectively in cells from 10 weeks (Hoxa9, Dmkn, Lcn2 and Podxl) whereas others were elevated at 5 and 10 weeks (Cp, Cldn3, Krt8, Msln, Ptgs2, Wt1, and Angptl7). A striking increase in Trp53 mRNA and protein were also observed in the mutant OSE cells compared to WT cells (Fig. 3C).
OSE cells isolated from ovaries of the Ptenfl/fl; KrasG12D;Amhr2-Cre mice are transformed
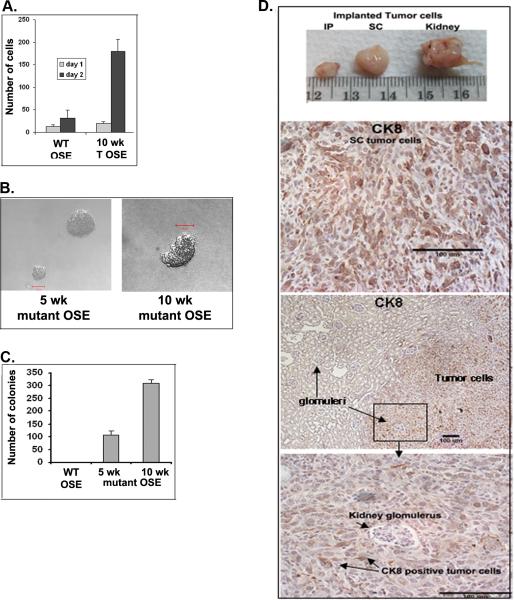
When OSE cells were isolated from WT or Pten fl/fl;KrasG12D;Amhr2-Cre mouse ovaries at 5 and 10 weeks and cultured for 2 days, the growth rate of the mutant cells far exceeded that of the WT cells (4A). The mutant, but not WT, OSE cells also generated stable cell lines. To further confirm that the mutant OSE cells were transformed, purified WT and mutant OSE cells were prepared and plated in soft agar. Mutant cells isolated from the Pten fl/fl;KrasG12D;Amhr2-Cre mice at 5 and 10 weeks formed colonies within 2 weeks whereas the WT cells did not (Figure 4B). Moreover, mutant OSE cells prepared at 10 weeks formed significantly more colonies than did cells collected at 5 weeks, providing additional evidence that temporal changes in the function of mutant OSE cells occur in vivo and are retained in culture (Figure 4C). Additionally, when the purified Pten fl/fl;KrasG12D;Amhr2-Cre OSE cells were injected into recipient mice in vivo, ectopic tumors positive for cytokeratin 8 developed rapidly (within 2 weeks) in the peritoneal cavity, at subcutaneous sites and under the kidney capsule where invasive activity into the kidney capsule was evident (Figure 4D). Thus, these transformed cells are highly proliferative and possess invasive activity.
Figure 4.
OSE cells isolated from ovaries of Pten;Kras;Amhr2-cre mice are transformed. A.) Mutant OSE cells proliferate faster than WT cells at 24 and 48h. B.) Mutant but not WT OSE cells grow in soft agar (C) and form ectopic tumors in the peritoneal cavity, subcutaneously and under the kidney capsule in vivo (D).
The PI3K/AKT signaling pathway and ERK1/2 impact in the functions of the mutant OSE cells
The disruption of Pten and expression of mutant KRASG12D are expected to increase the activity of the PI3K/AKT and RAS/MEK1/ERK1/2 pathways, respectively. However, there is also important cross-talk between these pathways and increasing evidence indicates that activation of the PI3K pathway is essential to maintain the growth promoting and transformation effects of KRAS (Miller KA, Yeager N, et al., 2009), in part, by blocking negative feedback regulatory loops (Wee S, Jagani Z, et al., 2009). Therefore, we determined if pharmacological disruption of either PI3K or MEK1/ERK1/2 activity would prevent colony formation and/or the expression of selected genes. As shown in Figure 5A, AKT and ERK1/2 are phosphorylated and hence activated in OSE cells isolated from ovaries of tumor-bearing mice at 10 weeks of age and this was blocked by blocked by inhibitors of PI3K (LY294002) and MEK1 (U0126), respectively. PTEN was undetectable and Foxo1 mRNA was markedly reduced in the mutant OSE cell (data not shown). When U0126 and/or LY294002 were added to the soft agar, they inhibited colony formation by approximately 70-90% (Fig. 5B).
Figure 5.
The MEK1/ERK1/2 and PI3K/AKT pathways drive gene expression and transformation of the mutant OSE cells. A.) Western blot of lysates prepared from mutant OSE cells cultured in media alone or with either the MEK1 inhibitor UO126 (10 □M) or the PI3K inhibitor LY294002 (10 □M) for 24 hrs. B.) Colony formation of mutant OSE cells grown in soft agar is inhibited by both UO126 and LY294002. C.) Gene expression is altered in cells cultured in media alone or with either UO126 or LY294002 for 24h. D.) TRP53 immunolabeling decreases in mutant cells cultured for 24 hours with or without 10 □M UO126 or LY294002 compared to media alone.
Loss of colony formation was associated with reduced expression of specific genes including Cp, Ptgs2, Dmnk, Lcn2 and Ptgs2 that were selectively down-regulated by inhibition of PI3K as well as Ccnd1,Cldn3, Krt8, Podxl and W1t that were reduced by either the PI3K/AKT or ERK1/2 signaling cascade inhibitors. Angptl7 was increased selectively by the PI3K inhibitor whereas Trp53 mRNA and protein were reduced equally by either LY294002 or UO126 (Figure 5C and 5D). Thus, the PI3K/AKT and MEK/ERK1/2 pathways are required to drive and maintain transformation of the mutant OSE cells.
To further characterize the specific effects of disrupting the PI3K pathway or activating the RAS pathway, we isolated and cultured OSE cells obtained from ovaries of Ptenfl/fl;Amhr2-Cre and KrasG12D;Amhr2-Cre mice as well as from the Pten fl/fl;KrasG12D;Amhr2-Cre mice and WT mice at 10 week of age. Cells from each genotype exhibited slightly different morphology (Fig 6A) and only cells from the Pten fl/fl;KrasG12D;Amhr2-Cre mice were transformed. Genes that were highly expressed in the OSE cells from the tumor (T) bearing ovaries of the Pten fl/fl;KrasG12D;Amhr2-Cre mice compared to WT were also elevated in OSE cells from the Ptenfl/fl;Amhr2-Cre mice. These include Podxl, markers of Mullerian epithelium (Wt1and Hoxa9), Trp53 and one of its target genes Cdkn1a (p21) as well as a marker of serous adenocarcinomas, Cldn3. These genes were less dramatically increased in the KrasG12D;Amhr2-Cre mice indicating that disrupting the PI3K pathway alone exerts the more potent response in this context (Fig 6B).
Figure 6.
Mutations in either Pten or Kras alone alter gene expression in purified OSE cells. A.) OSE cells were isolated from Pten;Amhr2-cre, Kras;Amhr2-cre, Pten;Kras;Amhr2-cre (tumor-bearing, T) and WT mice. The transformed cells (T) grow rapidly and eventually form clusters whereas the WT and KRAS OSE cells exhibit slower growth and well-defined boarders. OSE cells from the Pten null cells, like the transformed (T) cells, lack distinct borders but are not transformed. B.) The transformed cells and the Pten null cells exhibit striking similarities in the expression of specific mRNAs, including Trp53 and its target p21 (Cdkna1).
Specific microRNAs are regulated in the mutant OSE cells compared to WT OSE cells
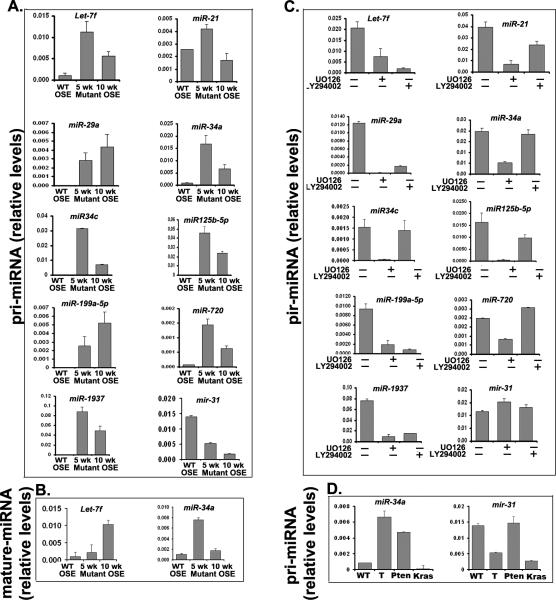
MicroRNAs comprise an extensive class of non-coding nucleic acids that govern broad gene regulatory pathways in development (Stafani G and Slack FJ, 2008). Specific microRNAs presumed to be linked to various cancers, including ovarian cancer, have been identified (Creighton CJ, Fountain Md, et al., 2010, Dahlya N, Sherman-Baust CA, et al., 2008, Iorio MV, Visone R, et al., 2007, Krichevsky AM and Gabriety G, 2009, Nam EJ, Yoon H, et al., 2008, Wyman SK, Parkin RK, et al., 2009, Yang H, Kong W, et al., 2008). Those highlighted most frequently are members of the Let-7 cluster that regulate (K/H)RAS and Hmga2, a potent regulator of proliferation (Roush S and Slack FJ, 2008), miR-21 that is transcriptionally controlled by STAT3, AP1 factors and/or TRP53 (Krichevsky AM and Gabriety G, 2009) and regulates such diverse processes as cell proliferation, invasion and migration (Krichevsky AM and Gabriety G, 2009), miR-34a/c that are targets of TRP53 and potent inhibitors of the cell growth regulators (Hermeking H, 2007, Ji Q, Hao X, et al., 2009), miR-29a that is a target of WNT/CTNNB1 (Kaplinas K, Kessler CB, et al., 2009) and regulates Trp53 (Park SY, Lee JH, et al., 2009), miR-125b that may target RAS (Rybak A, Fuchs H, et al., 2008) and miR-214 that impacts the level of mRNA encoding the tumor suppressor Pten (Yang H, Kong W, et al., 2008).
Based on these studies, we analyzed several microRNAs in our WT and mutant OSE cells in culture (Fig 7A). We show that primary transcripts of Let-7f and miR-21, miR-29a, miR-34a, miR-34c, miR-125b-5p, miR-199a-5p, miR-720 and miR-1937 were increased dramatically in the mutant cells isolated from tumors of mice at 5 and 10 weeks of age compared to WT cells whereas miR-31 was dramatically reduced. These results indicate that transcription of these microRNAs is being regulated in the mutant mouse OSE cells. The expression of the mature forms of these microRNAs was verified for Let-7f and miR-34a (Fig 7B). With the exception of miR-31, the expression of these microRNAs is regulated by inhibitors of PI3K (LY294002) or MEK1/ERK1/2 (U0126). Of particular interest is the down-regulation of miR-21, miR-29a, miR-34c, miR125b-5p, and miR-720 in response to U0126 indicating that ERK1/2 is a potent regulator of these microRNAs. Other microRNAs (miR-29a, miR-199a-5p, and miR-1937) were potently down-regulated by both inhibitors. Additionally, miR-34a, like Trp53, was increased in cells lacking Pten alone, whereas miR-31 was selectively suppressed in cells expressing KrasG12D but not those lacking Pten indicating that each pathway controls the expression of distinct miRNAs presumed involved in transformation.
Figure 7.
Specific miRNAs are expressed in the mutant compared to WT OSE cells and are regulated by inhibitors of MEK1/ERK1/2 and PI3K as shown by real-time RT-PCR. A and B.) Primary miRNA and mature transcripts are transcriptionally regulated in mutant OSE cells at 5 and 10 weeks of age. C.) Transcription of primary miRNAs is altered by UO126 or LY294002. D.) miR-34a is elevated in OSE cells from Pten;Kras;Amhr2-cre (tumor-bearing, T) mice and from Pten;Kras;Amhr2-cre mice (Pten) but not from Kras;Amhr2-cre mice (Kras). Conversely, the potential tumor suppressor miR-31 is down-regulated in the tumor OSE cells and Kras cells but not the Pten null cells, indicating differential regulation of these two pathways.
Discussion
These studies document that the Pten fl/fl;KrasG12D;Amhr2-Cre mice develop low-grade serous-type adenocarcinomas with 100% penetrance and at an early age. Detailed analyses of purified OSE cells obtained from the mutant mice provide unequivocal evidence that the cells are transformed, grow in soft agar and form ectopic invasive tumors when injected into recipient mice. That the tumors of Pten fl/fl;KrasG12D;Amhr2-Cre mice are the first to recapitulate diagnostic aspects of the human low-grade serous sub-type (Malpica A, Deavers MT, et al., 2004, Schwartz DR, Kardia SL, et al., 2002) is supported by several observations. Comparisons of microarray databases show highly significant overlap in genes expressed in the mouse and human tumor cells. Importantly, the Kras/Pten mutant cells but not WT OSE cells express Wt1, Hoxa9, Hox10 and Pax8 genes that are characteristic markers of Fallopian/Mullerian epithelia (Kurman RJ and Shih I-M, 2010). The expression of Hoxa9 is of particular interest because it is not only important for normal tissue development but is highly expressed in other cancer cell types, including leukemias (Wang Y, Krivtsov AV, et al., 2010) and astrocytomas (Costa BM, Smith JS, et al., 2010) where it is thought to maintain the survival of progenitor cells and to be regulated by PI3K (Costa BM, Smith JS, et al., 2010). Other genes that are up-regulated in the mutant mouse OSE cells are similar to ones that are enhanced in human ovarian serous adenocarcinomas (Cho KR and Shih I-M, 2009, Despierre E, Lambrechts D, et al., 2010, Gava N, Clarke CL, et al., 281, Gorringe KL and IG, 2009, Kobel M, Kalloger SE, et al., 2008, Konstantinopoulos PA, Sppentzos D, et al., 2008, Malpica A, Deavers MT, et al., 2004, Santin AD, Zhan F, et al., 2004, Schwartz DR, Kardia SL, et al., 2002). These include Cldn3 and Msln that are increased preferentially in serous adenocarcinomas expressing Hoxa9 (Cheng W, Liu J, et al., 2005). Thus, although recent evidence suggests that some ovarian cancers, especially high-grade serous adenocarcinomas, may be derived from mutant Fallopian tube or endometrial epithelial cells that migrate to the ovarian surface (Crum CP, Drapkin R, et al., 2007, Dubeau L, 2008, Karst AM and Drapkin R, 2010, Kurman RJ and Shih I-M, 2010, Lee Y, Miron A, et al., 2007, Roh MH, Yassin Y, et al., 2010), our results and those of others (Connolly DC, Bao R, et al., 2003, Dinulescu DM, Ince TA, et al., 2005, Flesken-Nikitin A, Choi K-C, et al., 2003, Orsulic S, Li Y, et al., 2002, Wu R, Hendrix-Lucas N, et al., 2007) document that mouse OSE cells can transform if exposed to specific oncogenic insults and that they may have some characteristics of multi-potent cells (Bowen NJ, Walker LD, et al., 2009) (Auersperg N, Woo MMM, et al., 2008).
Many microRNAs expressed in the mutant mouse OSE tumor cells are also present in human ovarian cancer. Of particular relevance are members of the Let-7 cluster (Roush S and Slack FJ, 2008) that regulate Kras, miR-21 (Krichevsky AM and Gabriety G, 2009) that is regulated by Kras, miR-34a/c that are transcriptional targets of TRP53 and potent inhibitors of the cell growth and therefore appear to act as tumor suppressors (Hermeking H, 2007, Ji Q, Hao X, et al., 2009), miR-29a (Kaplinas K, Kessler CB, et al., 2009) that regulates Trp53 (Park SY, Lee JH, et al., 2009), miR-125b that may target RAS (Rybak A, Fuchs H, et al., 2008) and miR-214 that impacts the level of mRNA encoding the tumor suppressor Pten (Yang H, Kong W, et al., 2008). That these miRNAs have been detected in various human ovarian cancer samples underscores their potential regulatory roles in the initiation and/or maintenance of the transformed phenotype. That many miRNAs were highest in mutant OSE cells isolated from ovaries of mice at 5 weeks indicates that the molecular activities as well as the growth rate of the mutant OSE cells change with time and tumor growth.
The molecular basis of the different histological phenotypes of ovarian cancer subtypes remains puzzling. For example, the serous adenocarcinomas of the Pten fl/fl;KrasG12D;Amhr2-Cre mice described herein (Fan HY, Liu Z, et al., 2009) differ in their histological architecture from the endometrioid-like OSE tumors in the mice reported by Dinulescu et al (Dinulescu DM, Ince TA, et al., 2005). Although the same mutant Pten and KrasG12D mouse strains were used, the tumors in each model were generated by different approaches and in OSE cells at different stages of differentiation. In our model OSE cells are transformed by endogenous Amhr-2Cre expressed directly in these cells in vivo and in mice prior to puberty; hence prior to increases in ovarian steroid production. Conversely, the Kras and Pten mutations in the Dinulescu model were generated by adenoviral Cre injections into the ovarian bursa of mice primed with gonadotropin (Dinulescu DM, Ince TA, et al., 2005) and therefore have been exposed to steroids. Because phenotypic outcome in each model appears to be highly reproducible, one plausible explanation for the distinct histological features is that the stage of OSE cell differentiation when they are exposed to the oncogenes determines the response.
Impressively, many genes selectively expressed in the Pten fl/fl; KrasG12D;Amhr2-Cre mouse tumor cells as well as in serous and mucinous human tumors share a common “Kras signature” observed in other cancer cell types (Figure 2), suggesting that activation of Kras pathway, by mutation or other mechanisms provide a mechanism that yields a common transformed outcome. Although a “Pten signature” was not significant, selective activation of the PI3K pathway by loss of Pten in OSE cell of the Pten fl/fl;Amhr2-Cre mice leads to specific and marked changes in mRNA and miRNA profiles in the OSE cells. Of note, Trp53, Cdkna1, Cldn3 and miR-34a are increased suggesting that components of the Trp53 pathway are key targets of the PI3K pathway. Conversely, Trp53, miR-34a and miR-31 are expressed but at lower levels in the tumor cells of Pten fl/fl; KrasG12D;Amhr2-Cre mice and OSE cells of KrasG12D;Amhr2-Cre mice indicating that activation of the KRAS pathway alters the effects of the PI3K pathway.
Moreover, when inhibitors of the PI3K and RAS pathways were added in culture to OSE cells already transformed, they blocked mutant OSE cell growth in soft agar and selectively altered the expression of key marker genes, indicating that both pathways are critical to maintain the transformed phenotype. Because the inhibition of PI3K was more effective than inhibition of MEK1/ERK1/2 on cell growth indicates that the PI3K pathway may be a more potent driver of proliferation, perhaps involving the regulatory effects of Hoxa9 and Meis1 (Costa BM, Smith JS, et al., 2010). However, the observed potency of PI3K may also be related to the ability of PI3K to activate the RAS/MEK1/ERK1/2 pathway, thereby, stimulating both signaling cascades. Conversely, the lesser effects of U0126 indicate that blocking MEK1 and ERK1/2 disrupts only one arm of the RAS signaling cascade but will not block the affects of KRAS on PI3K or other pathways in these cells (Wee S, Jagani Z, et al., 2009). Thus, the critical role of KRAS is underscored most impressively by the extensive number of human mutant Kras target genes (~213) that are also expressed in the murine and human OSE tumors cells (Figure 2). Collectively, these results provide strong evidence that the Pten fl/fl;KrasG12D;Amhr2-Cre mouse model is relevant to a subset of the human ovarian cancers and for determining the molecular events by which these two oncogenic pathways intersect to initiate transformation and also control the TRP53 pathway.
More aggressive ovarian cancer is associated with frequent mutations or deletions of TRP53 (Cho KR and Shih I-M, 2009). Strikingly, the mutant OSE cells from the Pten fl/fl;KrasG12D;Amhr2-Cre mice as well as in the Pten fl/fl;Amhr2-Cre mice express increased levels of Trp53 mRNA and protein. As a potent tumor suppressor (Connolly DC, Bao R, et al., 2003, Flesken-Nikitin A, Choi K-C, et al., 2003, Orsulic S, Li Y, et al., 2002), TRP53 may exert negative regulatory effects in the mutant Pten fl/fl;Amhr2-Cre and Pten fl/fl;KrasG12D;Amhr2-Cre cells to prevent more aggressive proliferation and metastatic activity (Astanehe A, Arenillas D et al., 2008). TRP53 appears to be active in the mutant cells because known targets are elevated selectively in the mutant OSE cells, including microRNAs, miR34a and miR34c, that have also been shown to exhibit tumor suppressor activity, genes encoding regulators of cell proliferation such as p21CIP (Corney DC, Flesken-Nikitin A, et al., 2007, Corney DC, Hwang C, et al., 2010, Hermeking H, 2007) and CCN3 that regulates cell adhesion (Bohlig L, Metzger R, et al., 2008). Thus, TRP53 and miR-34a in mouse and human low-grade adenocarcinomas may serve as an underlying negative molecular regulatory network that controls the low-grade tumorgenicity and perhaps cytotoxic resistance of these cells.
In summary, murine OSE cells respond to known oncogenic insults, Pten deletion and Kras activation (KrasG12D), and undergo transformation, leading to the formation of low-grade serous adenocarcinomas spontaneously in vivo. The mutant mouse OSE cells acquire morphological and biochemical characteristics of Mullerian derived epithelial cells. Moreover, these “differentiated” transformed cells express specific mRNA and miRNAs known to be expressed in human ovarian cancers indicating that this mouse model has relevance for understanding the specific effects of these two oncogenic factors in this cellular context and their interactions alone and together on the TRP53 pathway.
Materials and Methods
Animal procedures
LSL-KrasG12D;Amhr2-Cre, Ptenfl/fl;Amhr2-Cre; LSL-KrasG12D;Ptenfl/fl;Amhr2-Cre, mice were derived and genotyped as previously described (Fan HY, Liu Z, et al., 2009, Fan HY, Shimada M, et al., 2008). Trp53fl/fl;Amhr2-cre mice were derived from previously described Amhr2-Cre and Trp53fl/fl mice (obtained from the Mouse Models of Human Cancer Consortium, MMHCC, NCI-Frederick, MD (Raimondi AR, Molinolo A, et al., 2009). Animals were housed under a 16-h light/8-h dark schedule in the Center for Comparative Medicine at Baylor College of Medicine and provided food and water ad libitum. Animals were treated in accordance with the NIH Guide for the Care and Use of Laboratory Animals, as approved by the Animal care and Use Committee at the Baylor College of Medicine.
Histology and Immunohistochemistry
Ovaries were collected and fixed in 4% parformaldehyde, embedded in paraffin and processed by routine procedures for immunohistochemisrty (Fan HY, Liu Z, et al., 2009) of Cytokeratin 8 (ab59400 Abcam, Cambridge, MA).
Immunofluorescence
Ovaries were fixed in 4% parformaldehyde, embedded in optimal cutting temperature (OCT) compound (Sakura Finetek Inc., USA) and stored at -70° C. 7uM sections were immunostained (Fan HY, Liu Z, et al., 2009) with antibodies against: cytokeratin 8 (as above), E-cadherin (24E10), Vimentin (R28) from Cell Signaling (Danvers, MA) and TRP53 (p53) (Sc-6243) from Santa Cruz Biotechnology (Santa Cruz, CA).
Western blots
Whole cell extracts were prepared by lysing ovaries in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Roche, Nutley, NJ). Western blot analyses was performed using 30ug of lysate protein for each sample. Cell extracts were prepared from cultured OSE cells by lysis 1X sodium dodecyl sulfate (SDS) sample buffer at 100F for 5 min and 35 ul of each sample analyzed by Western blot using antibodies, AKT and phospho-AKT (9272, 4058L), and ERK and phospho-ERK (9102, 9101S) from Cell Signaling (Danvers, MA) and TRP53 (p53) (Sc-6243) from Santa Cruz Biotechnology (Santa Cruz, CA).
Isolation and culture of primary OSE cells
Ovaries were harvested from mice of indicated ages and gently washed once with serum free DMEM-F12 media. The OSE cells were released by mild-trypsin digestion (Flesken-Nikitin A, Choi K-C, et al., 2003) with modifications. The ovaries from 1-5 mice were then placed in a 15 ml conical tube containing 5 mls of room temperature (20-24 °C) buffer (0.25% w/v trypsin –EDTA)( Gibco/Invitrogen, Carlsbad, CA) for 30 minutes in a 37° incubator with 5%CO2. After 30 minutes the tube was gently tipped back and forth 10 times and the supernatant containing the OSE cells was transferred to a new tube and the cells collected by centrifugation at 1,000xg for 10 minutes. The cells from each tube were resuspended in DMEM-F12 growth media (10% FBS, 5% Insulin-Transferrin-selenium-A [Gibco/Invitrogen, Carlsbad, CA]) and 5% Penicillin-Streptomycin [Gibco/Invitrogen, Carlsbad, CA]) and plated in separate wells of a 24-well tissue culture plate. Fresh media was added every 2-3 days and the cells harvested when 90-100% confluent (7-10 days).
Innoculation of cells into mice
100,000 cultured OSE cells isolated from a 10 week Ptenfl/fl;KrasG12D;Amhr2-Cre mouse were grafted along with collagen under the renal capsule of wild type mice with the same genetic background. 100,000 cells were also injected subcutaneously along with Matrigel (BC Biosciences) according to the manufacturer's protocol. In addition, mice were injected itraperitoneally with 100,000 cells. Tumors were harvested and fixed in 4% paraformaldehyde after 20 days.
Real time RT-PCR for mRNAs, primary(pri)-miRNAs and mature miRNAs
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Germantown, MD) and treated with DNase I (DNA-free™, Ambion) according to the manufacturer's instructions. cDNA was synthesized with a Taqman reverse transcriptase reagent kit (Applied Biosystems, Foster City, CA) primed with random hexamers. Real-time PCR was performed using the Light Cycler DNA Master SYBR Green I kit (Roche Applied Sciences, Nutley, NJ). Primers (Supplementary Table S5) were used at a concentration of 0.5 μM and MgCl2 at 2.4 mM. Samples were denatured for 10 min at 95 °C and then 40 cycles of 95 °C for 20 s, 60 °C for 20 s and 72 °C for 20s as previously(Fan HY, Liu Z, et al., 2009). Data were normalized to L19 using the comparative Ct method. Data are presented as the mean +/- SEM of a representative of at least 3 experiments performed in triplicate.
Small RNAs required for detecting and measuring mature miRNAs were extracted using the mirVANA miRNA Isolation Kit (Ambion, Inc. Austin, TX) according to manufacturer's instructions. Reverse transcription was performed as described above. For quantification of mature miRNAs, the TaqMan MicroRNA assay kit (Applied Biosystems, Foster City, CA) was used according to the manufacturer's instructions.
Microarray
Total RNA was prepared using the RNeasy Mini Kit (Qiagen, Germantown, MD from ovaries of control and Ptenfl/fl;KrasG12D;Amhr2-cre mice at 3 months of age when ovaries of the mutant mice contained a substantial amount of tumor. The quality of the RNA was verified in the MicroArray Core Facility at Baylor College of Medicine and hybridized in duplicate to Mouse 420.3 Affymetrix Chips using routine procedures.
Gene Expression Analysis
After scanning and low-level quantification using Microarray Suite (Affymetrix), DNA Chip (dChip) analyzer (www.dchip.org) was used to estimate expression values. Fold changes between control and Ptenfl/fl;KrasG12D;Amhr2-cre mice were estimated, using the ratio of expression values. Expression profiles of human cell lines were obtained from Glaxo (GlaxoSmithKline GSK, 2008) and expression profiles of human ovarian tumors and normal ovary were obtained from GEO (GSE6008). Two-sided t-tests using log-transformed data determined significant differences in mean gene mRNA levels between sample groups. The mapping of transcripts or genes between the mouse signature and the human tumor array datasets was made on the Entrez Gene identifier (using the human orthologs from the mouse dataset); where multiple human array probe sets referenced the same gene, the probe set with the highest variation was used to represent the gene. One-sided Fisher's exact tests determined the significance of overlap between the mouse and human gene sets (using the 15092 unique human gene orthologs represented on the mouse array as the population). Expression patterns were visualized as color maps using the JavaTree Software (Saldanha AJ, 2004).
Supplementary Material
Acknowledgements
The authors thank Alan J. Herron D.V.M., Professor and Head of the Comparative Pathology Laboratory for his advice and insights into the mouse ovarian tumor phenotype, Yuet Lo and Azam Zariff for technical assistance and the Microscopy Core at Baylor College of Medicine for their expertise. We also thank the Immunohistochemistry Laboratory at The University of Texas MD Anderson Cancer Center for performing the calretinin and ESR1 immunostaining. Supported in part by NIH-HD-16229 (JSR), NRSA (LM), a Program Project Development Grant from the Ovarian Cancer Research Fund (CJC, PG, MA) and The University of Texas MD Anderson Cancer Center Specialized Program of Research Excellence in Ovarian Cancer (P50 CA08369)(KKW).
Footnotes
The authors have nothing to declare.
Conflict of Interest: There is no conflict of interest
References
- Abdullah M, Schultz H, K. D, Branscheid D, Dalhoff K, Zabel P, et al. Expression of the acute phase protein haptoglobin in human lung cancer and tumor-free lung tissues. Pathol Res Pract. 2009;205:639–647. doi: 10.1016/j.prp.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Auersperg N, Wong AST, Choi K-C, Kang SK, Leung PCK. Ovarian surface epithelium: biology, endocrinology and pathology. Endocrine Rev. 2001;22:255–288. doi: 10.1210/edrv.22.2.0422. [DOI] [PubMed] [Google Scholar]
- Auersperg N, Woo MMM, Gilks CB. The origin of ovarian carcinomas: a developmental view. Gynecol Oncol. 2008;110:452–454. doi: 10.1016/j.ygyno.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Bast RC, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nature Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlig L, Metzger R, Rother K, Till H, Engeland K. The CCN3 gene coding an extracellular adhesion-related protein is transcriptionaly activated by the p53 tumor suppressor. Cell Cycle. 2008;7:1254–1261. doi: 10.4161/cc.7.9.5812. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Walker LD, Matyunina L, Logani S, Totten KA, Benigno BB, et al. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med Genomics. 2009;2:71. doi: 10.1186/1755-8794-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancer is controll by HOX genes that specify regional identity in the reproductive tract. Nature Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- Cho KR. Ovarian Cancer Update: Lessons from morphology, molecules and mice. Arch Pathol Lab Med. 2009;133:1775–1781. doi: 10.5858/133.11.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KR, Shih I-M. Ovarian Cancer. Ann Rev Path. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Knowles KV, Senterman MK, Collins O, Vanderhyden BC. Conditional inactivation of Brca1, p53 and Rb in mouse ovaries results in the development of leiomyosarcomas. PLoS One. 2009;4:e8534. doi: 10.1371/journal.pone.0008534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly DC, Bao R, Nikitin Y, Stephens KC, Poole TW, Hua X, et al. Female mice chimeric for expression of the Simian virus 40T Ag under the control of the MISRIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–1397. [PubMed] [Google Scholar]
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Corney DC, Hwang C, Matoso A, Vogt M, Fleskin-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa BM, Smith JS, chen Y, Chen J, Phillips HS, aldape KD, et al. Reversing HOXO9 oncogene actvation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453–462. doi: 10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Fountain Md, Yu Z, Nagaraja AK, Zhu H, Khan M, et al. Molecular-profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum CP, Drapkin R, Kindelberger D, Medeiros F, Miran A, Lee Y. Lessons from BRCA: The tubal fimbria emerges as an origin for pelvic serous cancer. Clin Med & Research. 2007;5:35–44. doi: 10.3121/cmr.2007.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlya N, Sherman-Baust CA, Wang T-L, Davidson B, Shih I-M, Zhang Y, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:1–11. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despierre E, Lambrechts D, Neven P, Amant F, Lambrechts S, Vergote L. The molecular genetic basis of ovarian cancer and its roadmap towards a better treatment. Gynecol Oncology. 2010 doi: 10.1016/j.ygyno.2010.02.012. on line. [DOI] [PubMed] [Google Scholar]
- Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- Dubeau L. The cell origin of ovarian epithelial tumours. Lancet Oncol. 2008;9:1191–1197. doi: 10.1016/S1470-2045(08)70308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Shimada M, Liu Z, Cahill N, Noma N, Wu Y, et al. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicular development and ovulation. Development. 2008;135:2127–2137. doi: 10.1242/dev.020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Liu Z, Paquet M, Wang J, Lydon JP, DeMayo FJ, et al. Cell type specific targeted mutation of Kras and Pten document proliferation arrest in granulosa cells versus oncogenic insult in ovarian surface epithelial cells. Cancer Res. 2009;69:6463–6472. doi: 10.1158/0008-5472.CAN-08-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, et al. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324:938–941. doi: 10.1126/science.1171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesken-Nikitin A, Choi K-C, Eng JP, Shmidt N, Nikitin AY. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- Gava N, Clarke CL, Bye C, Byth K, deFazio A. Global gene expression profiles of ovarian surface epithelial cells in vivo. J Mol Endocrinol. 40:281–296. doi: 10.1677/JME-07-0149. 281. [DOI] [PubMed] [Google Scholar]
- GlaxoSmithKline GSK. Cancer Cell Line Genomic Profiling Data. 2008. https//:cabig.nci.nih.gov/tools/caArray_GSK database.
- Gorringe KL, C. IG. Large-scale genomic analysis of ovarian carcinomas. Mol Oncology. 2009;3:157–164. doi: 10.1016/j.molonc.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. p53 Enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat Genet. 2002;32:408–410. doi: 10.1038/ng1003. [DOI] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Meng Y, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLos One. 2009;4:1–13. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation by canonical Wnt signaling. J Cell Biochem. 2009;108:216–224. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst AM, Drapkin R. Ovarian cancer pathogenesis: a model in evolution. J Oncology. 2010 doi: 10.1155/2010/932371. doi:10.1155/2010/932371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindelberger DW, Lee Y, Miron A, Hirsch MS, F. C, Medeiros F, et al. Intraepithelial carcinoma of the fimbria and pelvic serous adenocarcinoma: evidence for a causal relationship. Am J Surg Pathol. 2007;31:161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- Ko SY, Lengyel E, Naora H. The Mullerian HOXA10 gene promotes growth of ovarian surface epithelial cells by stimulating epithelial-stroma interactions. Mol Cell Endocrinol. 2010;317:112–119. doi: 10.1016/j.mce.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Medicine. 2008;5:1749–1760. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Sppentzos D, Cannistra SA. Gene-expression profiling in epithelial ovarian cancer. Nature Clin Practice. 2008;5:577–587. doi: 10.1038/ncponc1178. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriety G. miRNA-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Shih I-M. The origin and pathogeneis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lague MN, Paquet M, fan HY, Kaartinene MJ, Chu S, Jamin SP, et al. Synergistic effects of Pten loss and WNT/CTNNB1 signaling pathway activation in granulosa cell tumor development and progression. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn186. On line. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Miron A, N. M. Drapkin r, Medeiros F, Saleemuddin A, Garber J, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- Liang S, Yang N, Pan Y, Deng S, Lin X, Yang X, et al. Expression of activated PIK3CA in ovarian surface epithelium results in hyperplasia but not tumor formation. PLoS One. 2009;4:e4295. doi: 10.1371/journal.pone.0004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica A, Deavers MT, Lu K, Bodurka DC, Atkinson EN, Gershenson DM, et al. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004;28:496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- Miller KA, Yeager N, Baker K, Liao X-H, Referoff S, De Cristofano A. Oncogenic Kras requires simultaneous PI3K signaling to induce ERK activation and transform thyroid epithelial cells in vivo. Cancer Res. 2009;69:3689–3694. doi: 10.1158/0008-5472.CAN-09-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page CL, Ouellet V, Madore J, Ren F, Hudson TJ, Tonin PN, et al. Gene expression profiling of primary cultures of ovarian epithelial cell identifies novel molecular classifiers of ovarian cancer. Br J Cancer. 2006;94:436–445. doi: 10.1038/sj.bjc.6602933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee JH, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Raimondi AR, Molinolo A, Gutkind JS. Rapamycin prevents early onset of tumorigenesis inan oral-specific K-ras and p53 two-hit carcinogenesis model. Cancer Res. 2009;69:4159–4166. doi: 10.1158/0008-5472.CAN-08-4645. [DOI] [PubMed] [Google Scholar]
- Roh MH, Yassin Y, Miron A, Mehra KK, Mehrad M, Monte NM, et al. High-grade fimbrial-ovarian carcinomas are unified by altered p53, PTEN and PAX2 expression. Modern Pathology. 2010 doi: 10.1038/modpathol.2010.119. On line; doi: 10.1038/modpathhol.2010.119. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends in Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, B. C. Smirnova L, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Janva Treeview --extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, et al. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- Stafani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Wang D, DuBois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/b-Catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Jagani Z, Xiang XX, Loo A, Dorsch M, Yao Y-M, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- Wong KK, Lu KH, Malpica A, Bodurka DC, Shvartsman HS, Schmandt RE, et al. Significantly greater expression of ER, PR, and ECAD in advanced-stage low-grade ovarian serous carcinoma as revealed by immunohistochemical analysis. Int J Gynecol Pathol. 2007;26:404–409. doi: 10.1097/pgp.0b013e31803025cd. [DOI] [PubMed] [Google Scholar]
- Wu R, Hendrix-Lucas N, Kiuck R, zhai Y, Schwartz DR, Akyol A, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/b-catenin and PI3K/Ptten pathways. Cancer Cell. 2007;11:321–333. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Parkin RK, Mitchell PS, O. B. K. Fritz BR, Godwin AK, Urban N, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Scangas G, Nitta M, He L, Xu X, Ioffe YJM, et al. A role for BRCA1 in uterine leiomyosarcoma. Cancer Res. 2009;69:8231–8235. doi: 10.1158/0008-5472.CAN-09-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao J-J, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Ge G, Tong W, Hug C, Lodish HF. Angiopoietin-like proteins stimulate ex vivo expansion of hemapoitetic stem cells. Nat med. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Deve. 2010 doi: 10.1101/gad.1940710. on line. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.