Abstract
A polarized chromosomal arrangement with clustered telomeres in a meiotic prophase nucleus is often called bouquet and is thought to be important for the pairing of homologous chromosomes. Fluorescence in situ hybridization in fission yeast indicated that chromosomal loci are positioned in an ordered manner as anticipated from the bouquet arrangement. Blocking the formation of the telomere cluster with the kms1 mutation created a disorganized chromosomal arrangement, not only for the regions proximal to the telomere but also for interstitial regions. The kms1 mutation also affected the positioning of a linear minichromosome. Consistent with this cytological observation, the frequency of ectopic homologous recombination between a linear minichromosome and a normal chromosome increased in the kms1 background. Intragenic recombination between allelic loci is reduced in the kms1 mutant, but those between non-allelic loci are unaffected or slightly increased. Thus, telomere-led chromosome organization facilitates homologous pairing and also restricts irregular chromosome pairing during meiosis.
Keywords: bouquet/chromosome pairing/fission yeast/meiosis/telomere
Introduction
Faithful segregation of chromosomes generally relies upon the preceding formation of a physical link between the pair of chromosomes to be separated. In mitosis, this link is formed between sister chromatids that were produced by a mechanism coupled with semiconservative DNA replication (Skibbens et al., 1999; Toth et al., 1999). In meiosis, however, the link is typically formed between homologous chromosomes, each of which was brought into the same cell from previously separated gametic cells. How the search for meiotic partner chromosomes is attained in the cell has long been a basic biological problem (Loidl, 1990; Roeder, 1997). Although it is evident that the homology in the nucleotide sequences of chromosomal DNA is crucial for the partner search, it is speculated that other chromosomal features are also used to ensure the fidelity and efficiency of homologous pairing. Non-random distribution of chromosomes and/or the loose pairing of homologous chromosomes found in the premeiotic cells of several organisms might be involved in meiotic synapsis (Cremer et al., 1993; Scherthan et al., 1994; Weiner and Kleckner, 1994). In Drosophila, heterochromatin has been implicated in the achiasmate chromosome segregation (Dernburg et al., 1996). Another and more important example is the characteristically polarized chromosome arrangement with telomeres grouped within a small area near the nuclear membrane, a configuration often referred to as bouquet. This structure has long been inferred to be important to chromosome synapsis, mainly because the configuration as well as the timing of its formation by itself implies so and also because of its occurrence in a wide range of species (Dernburg et al., 1995; Scherthan et al., 1996; Bass et al., 1997; Roeder, 1997; Hiraoka, 1998). Trelles-Sticken et al. (1999) recently reported that in Saccharomyces cerevisiae, telomeres also form a single cluster near the spindle pole body (SPB) in the meiotic prophase and that this event occurs prior to and independently of recombination and synaptonemal complex (SC) formation. This suggests that telomere clustering is an early and distinct step towards recombination, and reinforces the generality and importance of the bouquet arrangement. Telomeres facilitate homologous pairing, probably through bringing homologs into close proximity (Rockmill and Roeder, 1998). Until recently, however, the biological function of the bouquet structure could not be addressed experimentally because of the lack of appropriate agents to disrupt the bouquet structure specifically.
Fission yeast Schizosaccharomyces pombe provides one of the best experimental systems to address this issue. The bouquet formation upon induction of meiosis is well documented in this yeast (Chikashige et al., 1994, 1997), where telomeres of all chromosomes form a tight cluster beneath the SPB, the microtubule organization center (MTOC) equivalent in the yeast, during the meiotic prophase. Furthermore, several mutants that are defective in telomere clustering but still capable of traversing through meiosis/sporulation have been isolated. In one class of these mutants, genes named taz1 (Cooper et al., 1998) and lot2 (Nimmo et al., 1998) are mutated, both of which are implicated in the regulation of the telomere structure. The other class includes the kms1 mutant (Shimanuki et al., 1997), in which the telomere cluster tends to split into multiple subclusters, probably due to the disintegrated structure of the SPB. Kms1p is a component of the SPB, and its loss specifically affects the integrity of the SPB structure during the sexual process (Tange et al., 1998 and this study). In all of these mutants, there was a several-fold reduction in recombination and increased missegregation, suggesting the importance of telomere clustering and the bouquet arrangement in meiotic recombination and chromosome segregation (de Lange, 1998). It has not been formally ruled out, however, that the defect in telomere clustering brings about a general reduction of recombination activity; therefore the observed reduction in recombination might not be a result of the problem in chromosome pairing.
Chikashige et al. (1997) reported that a fission yeast linear minichromosome (Ch16) was always located near the tip of the horse tail nucleus. This observation was interesting because it was shown for years that Ch16 and its shorter or longer derivatives were unable to interact with chromosome III during meiosis, despite the fact that the chromosomes have homologous segments of several hundred kilobases (Niwa et al., 1986, 1989). As it is unlikely that the lack of meiotic interaction between the minichromosome and chromosome III is due to rearrangement in the primary sequence of the relevant chromosome segments (Niwa et al., 1989; T.Matsumoto and O.Niwa, unpublished data), the positioning of the minichromosome in the nucleus provides an explanation for the absence of recombination (Chikashige et al., 1997). In the present study, we used this minichromosome to determine whether the observed reduction of the recombination rate in the kms1 mutant is related specifically to the spatial arrangement of chromosomes formed in the meiotic prophase nucleus.
Results
Ordered arrangement of chromosomes in a meiotic prophase nucleus of fission yeast
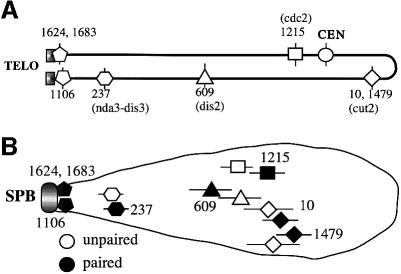
We examined the arrangement of homologous chromosomes in meiotic prophase nuclei of fission yeast using fluorescence in situ hybridization (FISH) with fixed nuclei and several cosmid DNAs as probes for unique sequences on chromosome II (see Figure 1). Three other probes were also used: YIp10.4 for rRNA gene clusters (rDNA) located at both termini of chromosome III, cos212 for telomeres of chromosomes I and II, and pRS140 for the centromeres of all chromosomes. Meiosis was induced in diploid cells by nitrogen starvation, and cells with a characteristically elongated nucleus (often referred to as a ‘horse tail’ nucleus, formed during meiotic prophase) were observed 5–6 h later with the aid of computerized optical sectioning and noise removal. The horse tail nuclei continuously change their shape due to the oscillatory movement mediated by the SPB (Chikashige et al., 1994). In the present study, only those nuclei with a stretched morphology were chosen. In accordance with previous reports (Chikashige et al., 1994, 1997; Scherthan et al., 1994), probes from telomeric regions (cos1624, cos1683 and cos1106) always made single or closely juxtaposed doublet signals near the tip of the nuclei where rDNA was also closely located, while probes from centromere-proximal regions or interstitial regions were located away from the tip and were not always paired.
Fig. 1. (A) A physical map of cosmids used in this study. The cosmid number is as described in Mizukami et al. (1993). Marker genes carried by a respective cosmid are shown in parentheses. (B) Positioning of each cosmid sequence in the meiotic prophase nucleus as revealed using FISH. Horizontal bars indicate the standard deviation.
With respect to the bouquet arrangement of the chromosomes, we examined whether each of these chromosomal regions was positioned at a fixed distance from the nuclear tip. The distance to each signal (paired or unpaired) from the center of the rDNA signal was determined and expressed as the ratio to the length of the respective nucleus. In this measurement, the depth of the signal was not taken into account, because this would only slightly affect the conclusion due to the elongated morphology of the nuclei. The results indicate that the relative position of each sequence coincides well with its physical map or distance from the nearest telomere (Figure 1). It should be emphasized that the positions of these chromosomal loci in the meiotic prophase nucleus did not differ significantly between paired and unpaired loci (Figure 1B). Furthermore, both of the unpaired signals were positioned at approximately the same distance from the tip in each individual nucleus (data not shown). This suggests that chromosome segments only need to traverse the nucleus perpendicularly to the long axis to search for their homologous partners. Therefore, the chromosome arrangement provides a possible advantageous condition toward efficient homologous pairing. Centromeres were not located at the distal end of the horse tail nuclei; in preference, cos10 was located more distal than all centromeres in 80% of nuclei. cos10 was mapped around the middle point of chromosome II, so that it was ∼2.3 Mb from the telomere. None of the centromeres were mapped >2 Mb from their nearest respective telomere (Mizukami et al., 1993). Thus, the positions of the centromeres appear to be determined by the distance from their respective telomere.
The kms1 mutation disrupts the integrity of the SPB
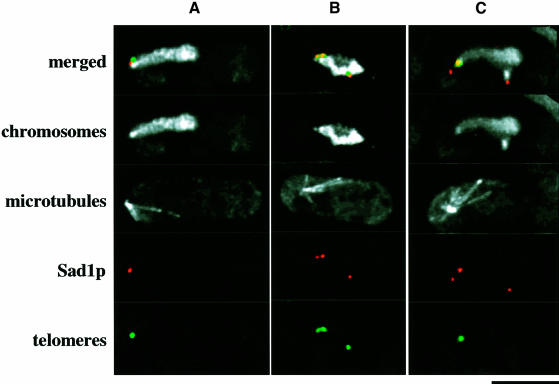
The clustering of telomeres is impaired in the kms1 mutant, probably due to the disintegration of the SPB (Shimanuki et al., 1997). To examine the defective phenotype more closely, the telomeres and the SPB were stained simultaneously using the cos212 probe and the anti-Sad1 antibody. In wild-type nuclei, the SPB was associated with tightly clustered telomeres (Figure 2A). Figure 2B shows a mutant nucleus in which the anti-Sad1 antibody stained multiple sites, each of which was associated with a telomere signal. In another example of the mutant nuclei (Figure 2C), there were also multiple Sad1 sites, but only one was associated with a single telomere cluster. In 20 nuclei observed, >80% of the telomere signals were associated with the Sad1 stain. These results indicate that in the kms1 mutant, telomeres fail to cluster because they are drawn to multiple sites provided by the disintegration of the SPB. In the mutant nuclei, the MTOC was either associated (Figure 2C) or not associated (Figure 2B) with one of the multiple Sad1-containing bodies. This defect is probably related to the staggered movement of the horse tail nucleus in the kms1 mutant (Shimanuki et al., 1997; M.Shimanuki, F.Miki and O.Niwa, unpublished result).
Fig. 2. Multiple Sad1 stains observed in meiotic prophase nuclei in the kms1 mutant. Chromosomes, microtubules, Sad1p and telomeres were stained with DAPI, TAT1, anti-Sad1 and cos212 DNA probe, respectively. Merged images, chromosomes and mictrotubules: white; Sad1p: red; telomeres: green. (A) Wild type, (B and C) kms1-1 mutant. Scale bar, 5 µm.
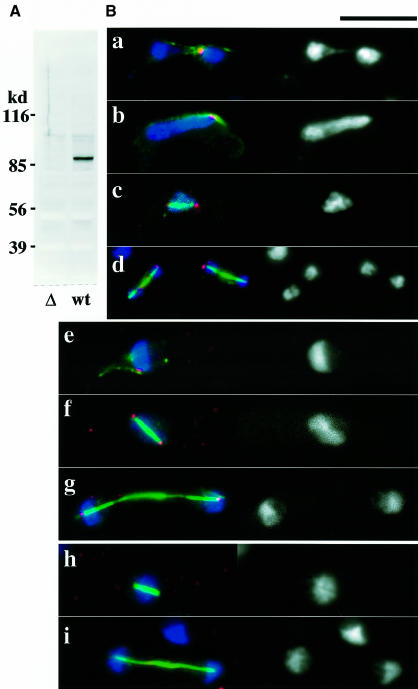
To understand the nature of the kms1 phenotype, the localization of the kms1+ product was examined. A polyclonal antibody was raised against Kms1p (Figure 3A) and indirect fluorescent staining was performed. During sexual development of wild-type cells, this antibody stained the leading edge of the two haploid nuclei to be fused (Figure 3B, a), the tip of horse tail nuclei (b), and the spindle poles of meiosis I and II (c and d). Although the kms1+ gene is not required for vegetative growth, Kms1p was also present in vegetative cells and localized to the SPB, as shown in Figure 3B (e–g). As anticipated, in cells with a null allele of the kms1 gene, no particular sites were stained with this antibody (Figure 3B, h and i). These data indicate that the kms1+ product is localized at the SPB. This is consistent with the finding that green fluorescent protein (GFP)-tagged Kms1p localized to the SPB (Tange et al., 1998; M.Shimanuki, unpublished result). Kms1p also interacts with Sad1p in a two-hybrid assay (data not shown; see Materials and methods). Furthermore, both Sad1p and Kms1p were recovered only in the insoluble fraction of a cell extract (data not shown). Taken together, these findings suggest that the kms1+ gene encodes an SPB component and its loss induces disintegration of the SPB in meiotic prophase nuclei. The disintegrated SPB, however, probably retains the function of providing a clustering site for telomeres.
Fig. 3. (A) Specificity of the anti-Kms1 antibody. Immunoblot analysis of a kms1-disrupted (left lane) and a wild-type (right) cell extract with the antibody. (B) Localization of the kms1+ product. (a–g) Wild type; (h and i) cells with the disrupted kms1. See the text for details. In each panel, the left side is a merged image of chromosomes (blue), microtubules (green) and Kms1p (red). The right hand side shows chromosomes stained with DAPI. Scale bar, 10 µm.
Disturbance of chromosome arrangement in the kms1 mutant
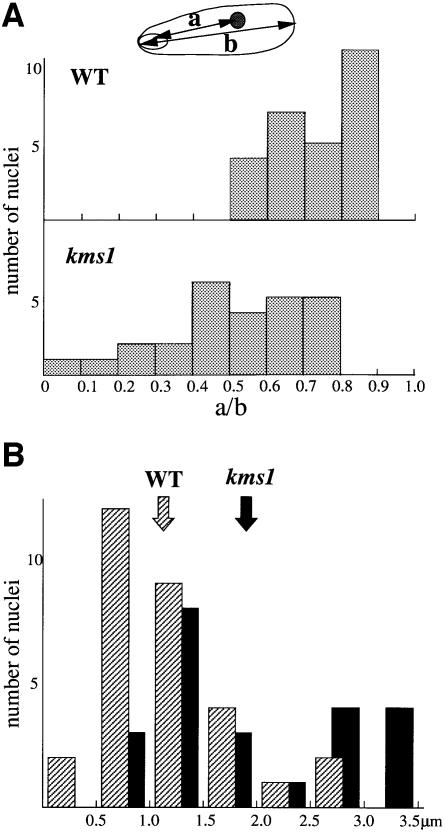
The kms1 mutant nuclei were then analyzed using FISH. As reported previously (Shimanuki et al., 1997), horse tail nuclei in the kms1 mutant tend to be deformed, i.e. the frequency of the smoothly elongated nuclei is low, which is probably due to the disintegrated structure of the SPB and the lack of smooth horse tail movement. We, however, recognize a fraction of nuclei as horse tail nuclei with little ambiguity due to their elongated morphology. In fact, these elongated nuclei always coexisted with cytoplasmic microtubules that were morphologically unique to the meiotic prophase (Svoboda et al., 1995; Ding et al., 1998; Figure 2). Thus, in the present study only nuclei with lengths similar to the wild type were used. The actual length of the observed nuclei was 4.4 ± 0.54 µm, 6% shorter than the wild type (4.7 ± 0.42 µm) on average. The chromosome arrangement in these mutant nuclei is disordered in several aspects, as described below.
Positioning and pairing of telomeric regions
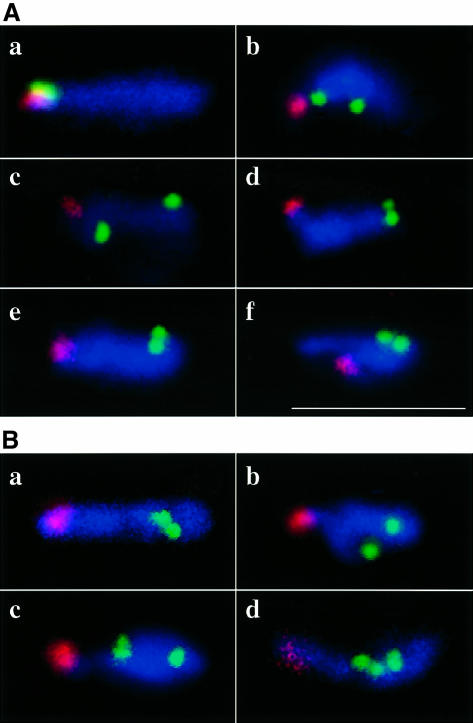
In the kms1 mutant, the formation of the telomere cluster in the meiotic prophase nucleus was impaired. Because this observation was made using a cosmid probe that hybridizes to telomeres of both chromosomes I and II, it was not certain whether the pairing between homologous telomeric segments was actually impaired. To clarify this point, unique probes were used for the terminal regions of chromosome II. In contrast to the configuration in the wild type, the telomeric sequences were detached from rDNA in >80% of the mutant nuclei (Figure 4A, b–f). Furthermore, in ∼30% of the mutant nuclei, these telomeric probes produced well separated doublet signals (Figure 4A, b, c and f). This was also in clear contrast to the wild type, in which the telomere probes always made tightly associated signals near the pointed end of the nucleus (Figure 4A, a). By staining both ends simultaneously, we found that the pairing at each terminus was affected independently from the other end (data not shown). The rDNA probe, the telomeric marker for chromosome III, only rarely made split signals even in the mutant, suggesting that the bundling of telomeric regions in this chromosome is controlled differently from the other two chromosomes.
Fig. 4. (A) FISH analysis of wild type (a) and kms1 mutant (b–f) meiotic prophase nuclei. Red, rDNA; green, cos1683. Nuclei are stained with DAPI (blue). (B) Wild type (a) and kms1 mutant (b–d). Red, rDNA; green, cos1479; blue, DAPI. In (c and d), prematurely separated sister chromatids are shown. Scale bar, 10 µm.
Positioning and pairing of centromere-proximal regions
In the wild type, the centromere-proximal cos1479 probe always produced signals away from the rDNA signal and near the distal end of the horse tail nucleus (Figure 4B, a). In the kms1 mutant, however, this probe was observed in the rDNA-proximal half of the nucleus in ∼50% of cases (Figure 5A). Consistent with this observation and in contrast to the wild type, where the centromere probe pRS140 produced more or less grouped signals, the signals in the mutant nuclei were more scattered (Figure 5B). These data indicated that in the kms1 mutant, the chromosome arrangement was disordered not only in the terminal regions but also in the centromere-proximal regions. With respect to chromosome pairing, we use the term ‘paired’ as defined by Scherthan et al. (1994), i.e. indicating two FISH signals that are either overlapping or touching. The cos1479 probe produced a paired signal at a frequency of 56% (n = 50) in the wild type 5 h after the induction of meiosis, whereas in the mutant, the frequency decreased slightly to 42% (n = 77) (Table I; Figure 4B). Another centromeric probe, cos1215, produced similar results, suggesting slightly reduced pairing in the mutant (Table I). Due to the small number of samples, however, it remains to be determined whether these differences are significant. In contrast, signals from detached sister chromatids were observed with a frequency of 17.5% (27 of 154 chromosomes) in the mutant, while it was only 4% (four of 100 chromosomes) in the wild type (Table I; see also Figure 4B, c and d). All of these detached sister chromatids were observed for unpaired chromosomes; therefore, these results indicate that some forms of unpaired chromosomes increased in the kms1 mutant.
Fig. 5. (A) Positioning of the centromere-proximal sequence in the meiotic nuclei. (a) The distance from the center of the rDNA signal to the center of the cos1479 signal; (b) the length of the respective nucleus. (B) Scattering of centromeres in the meiotic nuclei. Distribution of the longest distance between pairs of centromeres in individual zygote. Shaded bars, wild type; solid bars, kms1. The arrows indicate average distances.
Table I. Frequencies of homologous pairing observed using FISH.
| Probe | Strain | Paired (%) | Unpaired (%) | Sister chromatids separated in one or both homologs (%) | No. of cells | |
|---|---|---|---|---|---|---|
| 1215 | WT | 80 | 20 | 0 | 0 | 20 |
| kms1 | 52 | 16 | 8 | 24 | 25 | |
| 1479 | WT | 56 | 36 | 8 | 0 | 50 |
| kms1 | 42 | 34 | 14 | 13 | 77 | |
Positioning of a linear minichromosome in the kms1 mutant
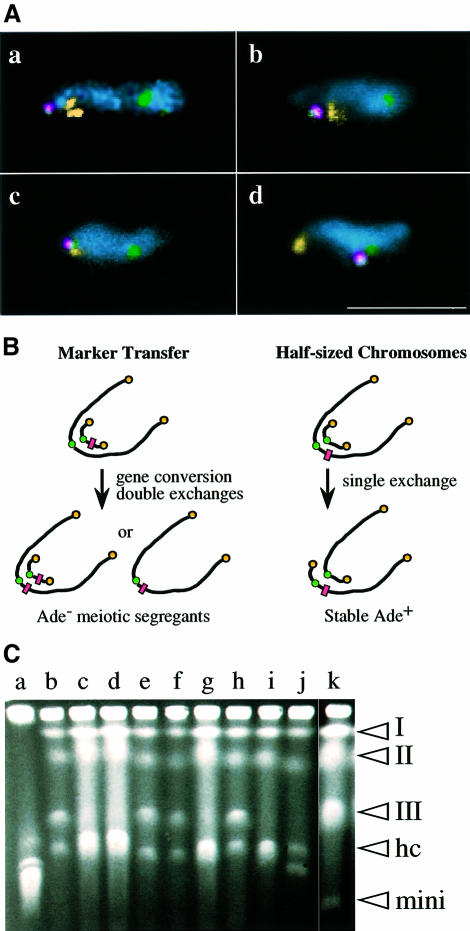
We examined the positioning of a minichromosome in the meiotic prophase nucleus using FISH. In wild-type cells, consistent with a previous report (Chikashige et al., 1997), a minichromosome signal was found close to the rDNA signal in all observed cells (20 out of 20; Figure 6A, a). In the kms1 mutant, however, the minichromosome was associated with rDNA in only three of 44 observed nuclei (Figure 6A, b). In many cases (16 out of 22), the minichromosome was located close to one of the cos212 telomere signals (Figure 6A, c). Furthermore, the minichromosome was positioned near centromeres rather than telomeres in 16% of the nuclei (nine of 56; Figure 6A, d). In the mutant background, it appears that the minichromosome is more accessible to centromeres or to the homologous chromosomal segment. In almost all cases, the minichromosome was associated either with telomeres or centromeres.
Fig. 6. (A) FISH analysis of wild-type (a) and kms1 mutant (b–d) meiotic prophase nuclei containing the minichromosome ChR33-Tr29. Yellow, rDNA (a, b and d) or telomeres (cos212 in c); green, centromeres (pRS140); red, minichromosome (YIp32). Note that the minichromosome also contains the pRS140 sequence. Nuclei are stained with DAPI (cyan). Scale bar, 10 µm. (B) Experimental designs for the detection of meiotic recombination between a minichromosome and chromosome III (see the text). The rectangles represent the Ade– marker. (C) Pulsed-field gel electrophoresis of stable Ade+ segregants (see the text for details). (Lane a) Chromosomes of S.cerevisiae; (lanes b–j) Ade+ segregants from the mutant cross; (lane k) S.pombe strain containing Ch16 minichromosome. The positions of chromosomes I, II and III, half-sized chromosomes (HC), and minichromosome Ch16 (mini) are indicated on the right hand side.
Minichromosomes interact with the normal chromosome more frequently in the kms1 mutant
The minichromosome is more accessible to its homologous counterpart in the mutant. Therefore, we examined whether meiotic recombination between linear minichromosomes and chromosome III occurs at a higher rate in the kms1 mutant using two types of genetic analysis.
The frequency of a marker transfer from the minichromosome Ch16 to chromosome III was first determined; more specifically, the ade6-216 mutation on the minichromosome can be transferred to chromosome III, which originally carried the ade6+ allele, through double exchange or gene conversion (Figure 6B). If meiotic segregants had the ade6-216 allele on chromosome III but did not have the ade6+ allele together, Ade– red colonies that can be readily discerned from Ade+ white colonies would be produced. The results indicated that in the kms1 mutant, viable Ade– segregants that were produced through meiotic recombination appeared 10-fold more frequently than in the wild type (Table II; crosses 1–4). A comparable result was obtained using another minichromosome, ChR33, which is a deletion derivative of Ch16 (Niwa et al., 1989; Table II, crosses 5 and 6). We also found that the ectopic recombination between Ch16 and chromosome III was enhanced in the taz1 mutant (data not shown). We looked only at viable spores, therefore some bias towards selecting for Ade– segregants in the mutant cross was possible. For example, mutant Ade+ segregants might be less viable, but this is highly unlikely. Therefore, we concluded that the meiotic recombination frequency between the minichromosome and chromo some III was actually increased in the mutant. Note that the Ade– phenotype could only be exhibited in the absence of the minichromosome carrying the ade6+ allele, which could be produced from the meiotic recombination. Meiotic transmission frequency and mitotic stability of the minichromosome, therefore, could somehow influence the frequency of Ade– colonies. There was no significant difference, however, in either of these parameters (data not shown). Furthermore, disomy of chromosome III in the meiotic segregants could affect the Ade– frequency. In fact, we reported previously that the kms1 mutant produced such segregants at an elevated frequency (Shimanuki et al., 1997). This factor should contribute to reduce the frequency of Ade– segregants, and therefore the frequency of the marker transfer in the mutant would be underestimated.
Table II. Frequency of a meiotic marker transfer from minichromosome to chromosome III.
| Crossa | Spore viability (%) | Frequency of Ade– segregants per viable sporesb (relative frequency) |
|---|---|---|
| 1 | 73 | 9.8 × 10–5 (1.0) |
| 2 | 77 | 7.1 × 10–5 (0.72) |
| 3 | 82 | 8.5 × 10–5 (0.87) |
| 4 | 45 | 1.0 × 10–3 (10.2) |
| 5 | 75 | 8.3 × 10–5 (1.0) |
| 6 | 47 | 4.5 × 10–4 (5.4) |
aCross 1, HM365 (h+ leu1 fur1) × HM672 (h– his2 fur1 Ch16); cross 2, HM670 (h+ leu1 fur1 kms1) × HM672; cross 3, HM365 × HM671 (h– his2 fur1 kms1 Ch16); cross 4, HM670 × HM671; cross 5, HM480 (h– leu1 kms1) × h+ leu1 ChR33; cross 6, HM480 × h+ leu1 kms1 ChR33.
bValues are the average of four independent experiments.
If increased frequency of the Ade– segregants was due to increased meiotic recombination between Ch16 and chromosome III, a higher rate of production of ‘half-sized chromosomes’ from a single crossover would be expected (Figure 6B). To test this, a second experiment was performed. Briefly, we made use of the fact that Ch16 is readily eliminated from a cell when the cell is exposed to the microtubule agent thiabendazole (TBZ), producing an ‘unstable’ Ade+ property. If, however, Ch16 recombines with chromosome III to produce two half-sized chromosomes and if any meiotic segregants contain only these two chromosome fragments, such segregants would become stable for the Ade+ phenotype, because the loss of either of the chromosome fragments is lethal for the cell. There were very few such stable segregants (one of 1800) from a control cross, whereas in an experimental cross there was a significantly increased number of stable Ade+ segregants (12 of 2200).
Pulsed-field gel electrophoresis (PFGE) was performed on these Ade+ segregants, and all of these segregants contained rearranged chromosomes. There were three types of segregants with respect to the structure of chromosome III (Figure 6C). Type I (six of 12; Figure 6C, lanes c, d, g and i) lost chromosome III and contained two half chromosomes that had instead a length similar to the longest chromosome in S.cerevisiae, as expected. Type II (five of 12; Figure 6C, lanes b, e, f and h) contains an apparently intact chromosome III and probably one but not both of the half chromosomes, judging from the intensity of ethidium bromide staining. Type III (one of 12; Figure 6C, lane j) contained a half chromosome and one additional chromosome fragment that was shorter than the half chromosome but much longer than the minichromosome. The stable Ade+ segregant recovered from the wild-type cross was type III. Southern hybridization analysis using a chromosome III-specific probe (rRNA gene sequence) revealed that these half-sized chromosomes carried rRNA gene cluster(s) (data not shown). The shorter chromosome fragment in the type III strain appeared to have a reduced number of rRNA genes. Why type II segregants were recovered is not clear. Although these type II segregants were isolated as being stable for the Ade+ phenotype in the first screening, they were no longer stable in the subsequent test (in fact, these were unstable, even in the absence of TBZ). Similar types of chromosomal aberrations were recovered following transformation of mitotic cells with minichromosomal DNA (Allshire, 1992).
Effect of the kms1 mutation on another type of ectopic interaction
The results with the minichromosome described above indicate the importance of the spatial arrangement of chromosomes in the meiotic prophase nucleus for efficient homologous pairing. To extend the generality of this finding, we used a genetic test system developed recently by Virgin and Bailey (1998). This system consists of two ade6 alleles (ade6-M26 and ade6-469), which are located either at the original ade6 locus (on chromosome III) or at artificially created loci (zzz15 and zzz7 on chromosomes I and II, respectively). Intragenic recombination between these alleles could produce the ade6+ allele, which can be readily detected. Consistent with previous results (Virgin and Bailey, 1998), the frequencies of non-allelic or ectopic recombination were markedly reduced compared with allelic recombination (Table III). The same intragenic recombination was then examined in the background of the kms1 mutant. Allelic recombination frequency was reduced to 30–60% of wild-type levels, while the frequency of ectopic recombination was either increased ∼2-fold (between zzz15 and ade6), unaffected (between zzz7 and ade6), or only slightly reduced (between zzz15 and zzz7) (Table III). In the mutant, therefore, the difference between allelic and ectopic recombination could be up to 9-fold less than in the wild type.
Table III. Effect of kms1 mutation on ectopic versus allelic recombination.
|
ade6-469 at |
||||
|---|---|---|---|---|
| ade6 | zzz15 | zzz7 | ||
| ade6-M26 at | ||||
| wild type | ade6 | 7900a | 1.6 | 4.9 |
| (3100) | (1.0) | (3.0) | ||
| zzz15 | 98 | 1500 | 64 | |
| (48) | (520) | (26) | ||
| zzz7 | 270 | 120 | 1400 | |
| (53) | (46) | (250) | ||
| kms1 | ade6 | 2200 | 3.2 | 4.3 |
| (1200) | (1.2) | (1.0) | ||
| zzz15 | 240 | 910 | 55 | |
| (53) | (350) | (14) | ||
| zzz7 | 360 | 64 | 810 | |
| (93) | (18) | (220) | ||
aNumber of Ade+ spores per 106 viable spores. Values are the average of four experiments. Standard deviation is shown in parentheses.
Discussion
The present study was designed to elucidate the functional significance of the polarized chromosome arrangement formed in meiotic prophase nuclei. The major findings were as follows: (i) ordered chromosome arrangement, usually formed during meiotic prophase, became disordered when the clustering of telomeres was blocked by the kms1 mutation; (ii) the positioning of a linear minichromosome was also changed so that the minichromosome became capable of locating near the centromeres where the homologous segments exist; and (iii) allelic recombination was reduced while ectopic recombination was enhanced or unaffected in mutants defective in telomere clustering. This differential effect on the recombination frequencies between different chromosomal segments cannot be readily explained by a general reduction in recombination activity, but is most plausibly explained in the context of a spatial chromosome arrangement. This is particularly clear for the interaction of the minichromosome with chromosome III.
Schizosaccharomyces pombe does not form chromosome synapsis mediated by the SC as do most of other eukaryotes (von Wettstein et al., 1984; Bahler et al., 1993). The frequency of paired loci as revealed by FISH analysis, however, increases for most of the genome as meiotic prophase proceeds (although it does not reach 100%) and this increase is accompanied by some changes in the arrangement of the linear elements, which make up the SC-related structure in fission yeast (Scherthan et al., 1994). It is therefore probable that some portion of the pairing defined by FISH represents meiotic interaction towards crossing over. In the present study, we could not determine whether the homologous pairing is significantly reduced in the kms1 mutant, except for the telomere-proximal regions where homologous pairing was impaired. Due to the low frequency of typically elongated nuclei in the mutant, it was difficult to observe a large number of samples. Also, because only those nuclei with an elongated morphology were observed, a biased observation for false-positive paired signals might have occurred. Nevertheless, the frequency of pairing appeared to be somewhat reduced, and prematurely separated sister chromatids, which were not engaged in the pairing, were observed at a significantly higher frequency in the mutant. It is not clear why the kms1 mutation tends to display such an abnormal separation of sister chromatids, but the result indicates that in the kms1 mutant an abnormal form of unpaired loci is present at an increased frequency. This result is consistent with the fact that univalent chromosomes tend to separate precociously in the first division (Flatters et al., 1995; Hunt et al., 1995). To what level the pairing frequency is reduced in the mutant, however, remains to be determined. It will also be interesting to examine whether the activity of meiosis-specific sister chromatid cohesion, e.g. that of Rec8 (Molnar et al., 1995; Watanabe and Nurse, 1999), is deregulated in the mutant.
The results of this study indicate that the bouquet arrangement of chromosomes functions not only to facilitate homologous allelic recombination but also to restrict irregular recombination in fission yeast. It is conceivable that the persistence of the bouquet structure throughout the prophase in fission yeast compensates for the absence of the SC and thereby ensures proper homologous pairing. It remains an important question then as to what extent the efficient synapsis relies on prior formation of the bouquet arrangement in most of the species that use the SC for the tight alignment of homologous chromosomes.
The kms1 mutant is unique among the mutants that are defective in the bouquet formation. In the taz1 and lot2 mutants, the SPB appears to be intact but telomeres are displaced from the SPB (Cooper et al., 1998; Nimmo et al., 1998). In the dhc1 mutant, which lacks the heavy chain of the cytoplasmic dynein motor, telomeres make a single cluster near the SPB but bouquet arrangement is somehow disturbed, probably due to impaired nuclear movement (Yamamoto et al., 1999). In the kms1 mutant, telomeres fail to make a single cluster, probably because they are focusing on multiple sites provided by the multiplication of a structure that contains the Sad1 protein. Thus, the kms1 mutant gives rise to an unusual situation where telomeres form subclusters yet retain the activity to associate with the Sad1 complex. Therefore, this mutant provides a useful experimental tool for investigating the structure and function of the telomere cluster.
Materials and methods
Yeast strains
Yeast strains used were: h90 leu1, h90 leu1 kms1-1 (Shimanuki et al., 1997), HM365 (h+ leu1 fur1), HM480 (h– leu1 kms1-1), HM487 (h+ wild type), HM570 (h+ leu1 fur1 ade6-210 kms1-1 Ch16-23R), HM572S (h– leu1 fur1 ade6-210), HM573S (h– leu1 fur1 ade6-210 kms1-1), HM670 (h+ leu1 fur1 kms1-1), HM671 (h– his2 fur1 kms1-1 Ch16), HM672 (h– his2 fur1 Ch16), HM688 (h+ leu1 ChR33), HM689 (h+ leu1 kms1-1 ChR33), HM690 (h90 leu1 ade6-210 kms1-1), HM691 (h90 leu1 ade6-704 ChR33-Tr29), HM692 (h90 leu1 ade6-704 kms1-1 ChR33-Tr29), J27 (h+ ura4 taz1::ura4+; kindly donated by J.Cooper) and W206 (h– his2 fur1 ura4 taz1::ura4+ Ch16). Minichromosomes Ch16 and ChR33 have been described previously (Niwa et al., 1986, 1989). Minichromosomes Ch16 and ChR33 carry the ade6-216 allele, which is capable of intragenically complementing the ade6-210 mutation. ChR33-Tr29 was described by Chikashige et al. (1997). This minichromosome carries the ade6-216 allele together with multiply integrated YIp32-based plasmids containing the LEU2 gene. Strains GP1073, GP1123, GP1081, GP1135, GP1540 and GP1594 (Virgin and Bailey, 1998) were kindly provided by J.Virgin. These contain either ade6-M26 or ade6-469 alleles at the original ade6 locus or artificially located new loci. The mutation sites are positioned ∼1.8 kb apart in the ade6 gene. Intragenic recombination between these alleles produces the ade6+ allele. The kms1-1 mutation was introduced to these strains by appropriate genetic crossings. The disruption of the kms1 gene was described in Shimanuki et al. (1997).
Media and procedures
General genetic procedures have been described previously (Gutz et al., 1974; Moreno et al., 1991; Alfa et al., 1993). YEA is a complete solid medium containing 3% glucose, 0.5% yeast extract (Difco) and 1.6% agar. MEA contains 3% malt extract (OXOID, Basingstoke, Hampshire, UK) and 1.6% agar. Minimal medium EMM was made according to Moreno et al. (1991), with 0.5% sodium glutamate as a nitrogen source. When required, this nitrogen source was omitted to make EMM-N medium. Sporulation medium, SPA, was prepared according to Gutz et al. (1974). Genetic crosses were performed on an MEA or SPA plate at 30 or 26°C. Free spores were liberated by digestion with 0.5% β-glucuronidase (Sigma) at 30°C for 4 h. To count Ade– red colonies, ∼4000 viable spores per plate were spread on YEA plates, followed by 4–5 days incubation at 30°C. At least 1.2 × 105 colonies were surveyed for each cross. For the screening of stable Ade+ segregants, HM570 was mated with HM572S or HM573S and free spores were plated on EMM supplemented with leucine, followed by incubation at 30°C for 5 days. Ade+ colonies were replica-plated onto YEA plates containing 15 µg/ml of thiabendazole, followed by overnight incubation at 26°C, then further replica-plated or streaked on YEA plates, and incubated for another 4 days at 30°C. Those with no or very few Ade– mitotic segregants were subjected to PFGE analysis. To examine the ectopic interaction between two ade6 alleles (ade6-M26 and ade6-469), crosses were performed on SPA at 30°C for 2 days. Free spores were plated on EMM supplemented with leucine and uracil, and incubated at 30°C for 5 days. For FISH analysis of meiotic prophase nuclei of wild-type and kms1 mutants, diploid cells were first selected using the phloxin B-plate method (Alfa et al., 1993) and then transferred into EMM-N medium at 30°C to induce azygotic meiosis (Egel, 1989). For FISH analysis of minichromosome positioning, diploid cells were produced by prototroph selection and meiosis was induced on MEA plate at 26°C.
Two-hybrid screening of Kms1-interacting factors
The two-hybrid system used in the present study was purchased from Clontech Inc. (Palo Alto, CA). A segment of the kms1+ gene corresponding to amino acid residues 201–607 was inserted at the SmaI–BamHI site of the pAS2.1 vector. This construct was used as the bait for screening the S.pombe cDNA library. One of the positive clones contained a part of the sad1+ gene corresponding to amino acid residues 263–514. Further detailed analysis indicated that the C-terminal hydrophobic region in the Kms1 protein is involved in the two-hybrid interaction with the Sad1 protein (F.Miki, unpublished result).
Production of anti-Kms1 antibody
A segment of the kms1+-cDNA was cloned into a glutathione S-transferase (GST) gene fusion vector (pGEX4T-2; Pharmacia) and expressed in bacteria. The N-terminal segment of the Kms1 protein containing 209 amino acid residues was removed from the GST portion and used for immunizing rabbits as well as for affinity purification of the serum. Specificity of the purified serum was verified using western immunoblotting.
FISH and indirect immunostaining
DNA probes pRS140, cos212, YIp10.4 and YIp32 (Funabiki et al., 1993; Chikashige et al., 1994, 1997) were used as centromere-, telomere-, rRNA gene- and minichromosome-specific probes, respectively. Other cosmid DNAs, listed in Figure 1, contained unique DNA sequences mapped on chromosome II (Mizukami et al., 1993). Cy3-, Cy5- (Amersham) and FluorX- (Amersham or Biological Detection Systems) conjugated dCTPs were used for labeling probes as described in Shimanuki et al. (1997). The procedures for indirect immunostaining, hybridization of FISH probes, and data processing using a Deltavision System (Applied Precision) were also described in Shimanuki et al. (1997).
Acknowledgments
Acknowledgements
The authors wish to thank Satoko Kusaba for her technical assistance. Thanks are also due to J.Virgin and J.Cooper for strains, I.Hagan for the anti-Sad1 antibody, and Y.Tange for help with the artwork. This work was supported by the Kazusa DNA Research Institute Foundation and the Ministry of Education, Science and Culture of Japan.
References
- Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments With Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Allshire R.C. (1992) Manipulation of large minichromosomes in Schizosaccharomyces pombe with liposome-enhanced transform ation. Methods Enzymol., 216, 614–631. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wyler,T., Loidl J. and Kohli,J. (1993) Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol., 121, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H.W., Marshall,W.F., Sedat,J.W., Agard,D.A. and Cande,W.Z. (1997) Telomere cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol., 137, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Ding,D.-Q., Funabiki,H., Haraguchi,T., Mashiko,S., Yanagida,M. and Hiraoka,Y. (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science, 264, 270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding,D.-Q., Imai,Y., Yamamoto,M., Haraguchi,T. and Hiraoka,Y. (1997) Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J., 16, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.P., Watanabe,Y. and Nurse,P. (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature, 392, 828–831. [DOI] [PubMed] [Google Scholar]
- Cremer T. et al. (1993) Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb. Symp. Quant. Biol., 58, 777–792. [DOI] [PubMed] [Google Scholar]
- de Lange T. (1998) Ending up with the right partner. Nature, 392, 753–754. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., Sedat,J.W., Cande,W.Z. and Bass,H.W. (1995) Cytology of telomeres. In Blackburn,E.H. and Greider,C.W. (eds), Telomere. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 295–338. [Google Scholar]
- Dernburg A.F., Sedat,J.W. and Hawley,R.S. (1996) Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell, 86, 135–146. [DOI] [PubMed] [Google Scholar]
- Ding D.-Q., Chikashige,Y., Haraguchi,T. and Hiraoka,Y. (1998) Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci., 111, 701–712. [DOI] [PubMed] [Google Scholar]
- Egel R. (1989) Mating-type genes, meiosis, and sporulation. In Nasim,A., Young,P. and Johnson,B.F. (eds), Molecular Biology of the Fission Yeast. Academic Press, San Diego, CA, pp. 31–73. [Google Scholar]
- Flatters M., Maxfield,R. and Dawson,D. (1995) The effects of a ring chromosome on the meiotic segregation of other chromosomes in Saccharomyces cerevisiae. Mol. Gen. Genet., 249, 309–316. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Hagan,I., Uzawa,S. and Yanagida,M. (1993) Cell cycle-dependent positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol., 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H., Heslot,H., Leupold,U. and Loprieno,N. (1974) Schizo saccharomyces pombe. In King,R.C. (ed.), Handbook of Genetics. Plenum Press, New York, NY, pp. 395–446. [Google Scholar]
- Hiraoka Y. (1998) Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells, 3, 405–413. [DOI] [PubMed] [Google Scholar]
- Hunt P., LeMaire,R., Embury,P., Sheean,L. and Mroz,K. (1995) Analysis of chromosome behavior in intact mammalian oocytes: monitoring the segregation of a univalent chromosome during female meiosis. Hum. Mol. Genet., 4, 2007–2012. [DOI] [PubMed] [Google Scholar]
- Loidl J. (1990) The initiation of meiotic chromosome pairing: the cytological view. Genome, 33, 759–778. [DOI] [PubMed] [Google Scholar]
- Mizukami T. et al. (1993) A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell, 73, 121–132. [DOI] [PubMed] [Google Scholar]
- Molnar M., Bahler,J., Sipiczki,M. and Kohli,J. (1995) The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics, 141, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nimmo E.R., Pidoux,A.L., Perry,P.E. and Allshire,R.C. (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature, 392, 825–828. [DOI] [PubMed] [Google Scholar]
- Niwa O., Matsumoto,T. and Yanagida,M. (1986) Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Gen. Genet., 203, 397–405. [Google Scholar]
- Niwa O., Matsumoto,T., Chikashige,Y. and Yanagida,M. (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J., 8, 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B. and Roeder,G.S. (1998) Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev., 12, 2574–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Scherthan H., Bahler,J. and Kohli,J. (1994) Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J. Cell Biol., 127, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Weich,S., Schwegler,H., Heyting,C., Harle,M. and Cremer,T. (1996) Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol., 134, 1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M., Miki,F., Ding,D.-Q., Chikashige,Y., Hiraoka,Y., Horio,T. and Niwa,O. (1997) A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet., 254, 238–249. [DOI] [PubMed] [Google Scholar]
- Skibbens R.V., Corson,L.B., Koshland,D. and Hieter,P. (1999) Ctf7 is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda A., Bahler,J. and Kohli,J. (1995) Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma, 104, 203–214. [DOI] [PubMed] [Google Scholar]
- Tange Y., Horio,T., Shimanuki,M., Ding,D.-Q., Hiraoka,Y. and Niwa,O. (1998) A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J. Cell Biol., 140, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Ciosk,R., Uhlmann,F., Galova,M., Schleiffer,A. and Nasmyth,K. (1999) Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev., 13, 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E., Loidle,J. and Scherthan,H. (1999) Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci., 112, 651–658. [DOI] [PubMed] [Google Scholar]
- Virgin J.B. and Bailey,J.P. (1998) The M26 hotspot of Schizo saccharomyces pombe stimulates meiotic ectopic recombination and chromosomal rearrangements. Genetics, 149, 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen,S.W. and Holm,P.B. (1984) The synaptonemal complex in genetic segregation. Annu. Rev. Genet., 18, 331–413. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. and Nurse,P. (1999) Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature, 400, 461–464. [DOI] [PubMed] [Google Scholar]
- Weiner B.M. and Kleckner,N. (1994) Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell, 77, 977–991. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., West,R.R., McIntosh,J.R. and Hiraoka,Y. (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol., 145, 1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]