Abstract
To humanize the glycosylation pathway in the yeast Pichia pastoris we developed several combinatorial genetic libraries and used them to properly localize active eukaryotic mannosidases and sugar transferases. Here we report the details of the fusion of up to 66 N-terminal targeting sequences of fungal type II membrane proteins to 33 catalytic domains of heterologous glycosylation enzymes. We show that while it is difficult to predict which leader/catalytic domain will result in the desired activity, analysis of the fusion protein libraries allows for the selection of the leader/catalytic domain combinations that function properly. This combinatorial approach together with a high throughput screening protocol has allowed us to humanize the yeast glycosylation pathway to secrete human glycoprotein with complex N-glycosylation.
Keywords: Pichia pastoris, glycosylation, combinatorial genetic libraries, Golgi targeting
Introduction
Recent advances in genomics and proteomics have fueled the increasing demand for large quantities of therapeutic proteins. Because the majority of these therapeutically relevant proteins require certain posttranslational modifications, like N-glycosylation, for proper function [Helenius and Aebi, 2001], most glycoproteins of commercial importance are currently expressed in mammalian cell culture. Fungal protein-expression systems are viewed as a potential alternative because of high volumetric productivity [Werten et al., 1999; Durand and Clanet, 1988], low media cost, lack of retroviral contamination and ease of generation of stable cell lines. However the presence of high-mannose type glycans renders glycoproteins derived from fungal expression systems unfit for human applications [Ballou, 1990].
Proteins produced in fungal and mammalian systems share the same (mannose)8-(N-acetylglucosamine)2 ((Man)8-(GlcNAc)2) glycan structures leaving the ER, but upon entering the Golgi the glycan processing pathways differ significantly. While the mammalian Golgi contains mannosidases and GlcNAc transferases that form the (GlcNAc)2-(Man)3-(GlcNAc)2 precursor for complex mammalian glycans, the yeast Golgi, where this process has been studied extensively [Dean, 1999], contains several mannosyl- and phosphomannosyl transferases that can extend the glycosylation chain to 100 mannose units or more.
Early attempts to reengineer the fungal glycosylation pathway have been described with varying success. Maras and coworkers found that about one third of the N-glycans from cellobiohydrolase I obtained from Trichoderma reesei can be trimmed to (Man)5-(GlcNAc)2 by Aspergillus saitoi α-1,2-mannosidase in vitro [Maras et al., 1999]. Less than 1% of those glycans, however, could serve as a productive substrate for GlcNAc-transferase I. Chiba et al. localized an Aspergillus saitoi α-1,2-mannosidase to the ER of Saccharomyces cerevisiae with modest intracellular mannosidase activity, but reported no activity at all when they fused the mannosidase to the transmembrane domain of Och1p [Chiba et al., 1998].
Although much is known about the signals that target endogenous proteins in the secretory pathway of S. cerevisiae and other yeasts [Gleeson, 1998], prior to GlycoFi's use of the combinatorial genetic library disclosed here and in prior publications there was no reliable way of predicting whether a particular heterologously expressed glycosyltransferase or mannosidase in a lower eukaryote will be (1) sufficiently translated, (2) catalytically active, or (3) located to the proper organelle within the secretory pathway.
To overcome these problems we developed a combinatorial genetic library consisting of an array of different fusion-protein constructs, each containing a fungal cellular targeting sequence fused in frame to a catalytic domain of a heterologous glycosylation enzyme. We also developed a 96 well high throughput protein expression protocol that allows us to analyze large numbers of yeast strains in parallel for their ability to modify N-glycans of recombinant reporter proteins.
Whereas small parts of this project have already been described [Choi et al., 2003; Hamilton et al., 2003], here we provide a previously unpublished comprehensive report of the construction and analysis of libraries of several hundred MnsI, GnTI, MnsII, GnTII and GnTIII fusion protein constructs.
Materials and Methods
Strains Plasmids and Reagents
E. coli strain DH5α was used for recombinant DNA work. All yeast strains used in this study were derived from JC308 [Lin Cereghino et al., 2001], which has the phenotype ade1arg4his4ura3 (a gift from James M. Cregg, Keck Graduate Institute, Claremont, CA). Strains PBP1 [Choi et al., 2003], YJN168 [Choi et al., 2003], YSH1 [Hamilton et al., 2003] and RDP27 [Bobrowicz et al., 2004] have been described previously. Strains YJN169 and YJN191 were generated by transformation of PBP1 with plasmids pBC8 and pBC18 that had been linearized using SalI, and selection on complete synthetic medium lacking histidine. The fusion vectors for library construction were pJN347 (MnsI), pJN346 (GnTI), pJN348 (MnsII and GnTIII) and pPB124 (GnTII) [Choi et al., 2003; Hamilton et al., 2003]. Restriction and modification enzymes were from New England Biolabs. Oligonucleotides were obtained from the Dartmouth College Core Facility (Hanover, NH) or Integrated DNA Technologies (Coralville, IA). The enzymes N-glycosidase F, mannosidases, and oligosaccharides were obtained from Glyko (San Rafael, CA). DEAE ToyoPearl resin was from TosoHaas. Metal chelating “HisBind” resin was from Novagen (Madison, WI). 96-well lysate-clearing plates were from Promega (Madison, WI). Protein-binding 96-well plates were from Millipore (Bedford, MA). 96 well plates for protein expression were from Qiagen (Valencia, Ca). Shakers for 96 well protein expression were model DMS-2500 from VWR (West Chester, PA) or the High Speed Micro Plate Shaker form Troemner (Thorofare, NJ). Salts and buffering agents were from Sigma (St. Louis, MO). MALDI matrices were from Aldrich (Milwaukee, WI).
Construction of targeting domain and catalytic domain libraries
All targeting domains were amplified by PCR using ExTaq (Takara) and the respective fungal genomic DNA as template, cloned into plasmids pCR2.1 (Invitrogen) or pVM1 (pUC19 with additional NotI and AscI sites in the MCS) and sequenced. The 5′-oligo introduced a NotI site and the CACC Kozak consensus sequence just upstream of the native ATG. In the case of the leaders derived from PpSEC12 or ScSEC12 the codon for the first amino acid of the targeting domain was changed to ATG. The 3′-oligo introduced an AscI site and an additional C, which resulted in a fusion linker encoding GlyArgAla. The oligonucleotides used to amplify targeting domains from PpKTR1, PpKTR3 and PpKRE2 are given in Table 3. The sequences of any of the other oligos are available upon request.
Table 3.
| Name | Sequence | Use |
|---|---|---|
| PpKTR1-5 | ATATGTGAGCGGCCGCCACCATGGAATTA GTGCGCCTGGCCAATCTTGTCAACGTC |
5′-oligo for amplification of PpKTR1 leaders |
| PpKTR1-1 | GGCGCGCCCCCCTCTTGATAGGATGAATC CTAACAGTAC |
3′-oligo for amplification of PpKTR1-s leader |
| PpKTR1-2 | GGCGCGCCCCAGTTCAGTTGCACCACTGC TGCCTTGGGAA |
3′-oligo for amplification of PpKTR1-m leader |
| PpKTR1-3 | GGCGCGCCCCCACTCCGGGAACGACCAA TGTTCCTTTGG |
3′-oligo for amplification of PpKTR1-l leader |
| PpKTR3-5 | CTGAATGTGCGGCCGCCACCATGATGCGA GCAAGATTAAGCCTTGAACGAGTTAACTTG |
5′-oligo for amplification of PpKTR3 leaders |
| PpKTR3-1 | GGCGCGCCCCAAAGAGATGAAAAGAACTG CAACTGAAGCC |
3′-oligo for amplification of PpKTR3-s leader |
| PpKTR3-2 | GGCGCGCCCGCCTATGGTGCCTATGTGTT GAGTTGTCTGT |
3′-oligo for amplification of PpKTR3-m leader |
| PpKTR3-3 | GGCGCGCCCCGGAAATCTCCAATGAGAG GCCGGTACTTGA |
3′-oligo for amplification of PpKTR3-l leader |
| PpKRE2-5 | TTTCTTAGGCGGCCGCCACCATGGTACAC ATAGGGTTCAGAAGCTTGAAAGCGGTG |
5′-oligo for amplification of PpKRE2 leaders |
| PpKRE2-1 | GGCGCGCCCCCCGTCAAAGGTCGTGACA ATCCCGTACAGA |
3′-oligo for amplification of PpKRE2-s leader |
| PpKRE2-2 | GGCGCGCCCGTCCTCTACTTTCTTTATTGA TTGGATGATT |
3′-oligo for amplification of PpKRE2-m leader |
| PpKRE2-3 | GGCGCGCCCGTGCTGGTCTTCTAACTGCC TTCTTGACTCT |
3′-oligo for amplification of PpKRE2-l leader |
After amplification of the targeting domain containing plasmids 36 μg of each was digested with 50 units AscI for 4 hr. The linearized DNA was then ethanol precipitated, digested with 60 units NotI for 15 hr and the resulting inserts were purified by two consecutive rounds of separation on agarose gels. For fragments ranging from 87 to 219 nucleotides we used 2 % agarose, for fragments from 240 to 426 nucleotides 1.5 % agarose, and for fragments from 435 to 1098 nucleotides 1.0 % agarose. While the bulk of the material was then stored undiluted at −80° C, 1.2 pmole of each was diluted to 100 μl, arranged in a 96 well plate and also stored at −80° C.
The glycolytic enzyme catalytic domains were amplified using either Ex Taq (Takara) or Pfu DNA polymerase (Stratagene) from sources given in Table 2. Generally the full length enzymes were amplified first, cloned into pCR2.1 (Invitrogen) and sequenced. Then the truncated fragments were amplified using those plasmids as template and the 5′ oligonucleotides to introduce an AscI site and the 3′ oligos to introduce a PacI site directly after the stop codon. The sequences of any of the oligonucleotides are available upon request. The fragments were then again cloned into vector pCR2.1 and sequenced. From there they were excised as AscI/PacI fragments and ligated into the respective fusion vectors. The MNSI derived fragments were cloned into pJN347, the GNTI derived fragments into pJN346, the MNSII derived fragments into pJN348, the GNTII derived fragments into pPB124, and the GNTIII derived fragments into pJN348.
Table 2.
Cat. Domain
| Cat. Domain | Gene | Accession # | Source | Trunc. [Nuc.] | Plasmid |
|---|---|---|---|---|---|
| AB | CeMNS1A | CAB01415 | C.e. λACT-RBI cDNA library, Robert Barstead, Oklahoma research foundation |
Delta 90 | pRCD123 |
| AC | CeMNS1A | Delta 150 | pRCD124 | ||
| AD | CeMNS1A | Delta 387 | pRCD125 | ||
| BB | CeMNS1B | CAA98114 | Delta 93 | pRCD127 | |
| BC | CeMNS1B | Delta 240 | pRCD128 | ||
| CB | DmMNS1A | AAF46570 | D.m. λZAP ovary cDNA library | Delta117 | pRCD209 |
| CC | DmMNS1A | Delta 312 | pRCD210 | ||
| CD | DmMNS1A | Delta 561 | pRCD211 | ||
| DB | HsMNS1B | AF027156 | Human placenta, cDNA lib.Clontech | Delta 177 | pJN383 |
| DC | HsMNS1B | Delta 240 | pJN384 | ||
| DD | HsMNS1B | Delta 297 | pJN385 | ||
| EB | PcMNS1 | D45839 | Plasmid pEPM14, T.Yoshida, Tohoku University, Japan |
Delta 63 | pSH168 |
| FB | MmMNS1A | NM_008548 | Mouse liver cDNA library, Clontech | Delta 561 | pSH146 |
| GB | MmMNS1B | NM_010763 | Mouse brain marathon ready cDNA library, Clontech |
Delta 174 | pSH147 |
| GC | MmMNS1B | Delta 297 | pSH148 | ||
| GD | MmMNS1B | Delta 510 | pSH149 | ||
| HC | AnMNS1A | AF129497 | Delta 294 | pSH162 | |
| HD | AnMNS1A | Delta 495 | pSH163 | ||
| JB | AnMNS1C | AF233287 | Delta 168 | pSH166 | |
| KB | DmMNSII | X77652 | D.m. λZAP ovary cDNA library | Delta 144 | pSH199 |
| KC | DmMNSII | Delta 297 | pSH200 | ||
| KD | DmMNSII | Delta 222 | pSH220 | ||
| LB | CeMNSII | NM_0735941 | C.e. λACT-RBI cDNA library, Robert Barstead, Oklahoma research foundation |
Delta 93 * | pSH236 |
| LC | CeMNSII | Delta 324 * | pSH237 | ||
| LD | CeMNSII | Delta 93 ** | pSH238 | ||
| LE | CeMNSII | Delta 324 ** | pSH239 | ||
| MA | HsMNSII | U31520 | Human kidney cDNA lib. Clontech | Delta 162 | pSH206 |
| NA | HsGNT1 | NM_002406 | Plasmid pHG4.5, ATCC | Delta 114 | pPB-NA |
| TC | RnGNTII | Q09326 | Rat liver cDNA, Clontech | Delta 261 | pPB-TC |
| UA | HsGNTII | Q10469 | Human liver cDNA, Clontech | Delta 252 | pPB-UA |
| VA | MmGNTIII | Q10470 | Pamela Stanley, Albert Einstein College of Medicine, Yeshiva University, New York, NY |
Delta 96 | pPB-VA |
| VB | MmGNTIII | Delta 198 | pPB-VB | ||
| VC | MmGNTIII | Delta 630 | pPB-VC |
Amino acid 133 is K
Amino acid 133 is E
High throughput fusion library construction and expression
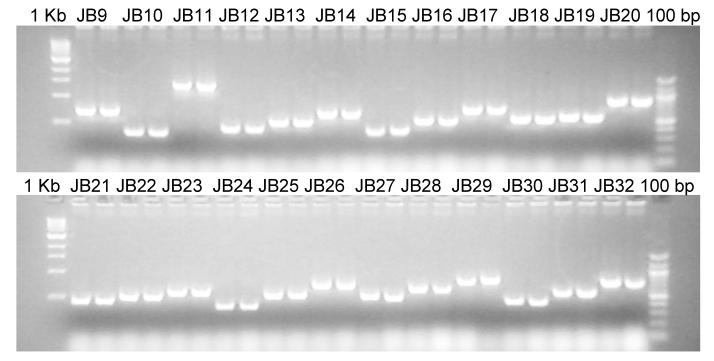
For construction of the leader/catalytic domain chimeras, 30 μg of each glycolytic domain containing fusion vector (see Table 2) was digested with 10 units AscI overnight. In the morning another 10 units were added and the incubation was continued for another 1 hr. The linearized DNA was Ethanol precipitated and incubated with 50 units NotI and 100 units SwaI at RT. After 1 hr the temperature was increased to 37° C and the incubation was continued for another 4 hr. Then 20 units calf intestinal alkaline phosphatase were added and the incubation was continued for another 1 hr. After purification by agarose gel extraction the DNA was diluted to 3 nM and 2.5 μl was loaded into each of the first 69 wells of a 96 well PCR plate. Then 2.5 μl of a 12 nM solution of the isolated targeting domain NotI/AscI fragments (see above) was added. After addition of 5 μl 2x ligation buffer and 0.5 μl Quick ligase (NEB) the ligation was allowed to proceed for 5 min at RT, and 2 μl of the ligation mix was added to 50 μl E. coli strain DH5α made competent according to the method of Hanahan [Hanahan et al., 1991] that were arranged in a 96 well plate. The mixture was incubated for 20 min on ice, heat shocked at 42° C for 1 min, 200 μl SOC were added to each well of the 96 well plate and the cells were allowed to recover at 37° C for 1 hr. Of each transformation mix 200 μl were then plated on a single LB Amp plate and incubated overnight. We routinely obtained 10 to 1000 colonies per plate as compared to 0 to 10 colonies on the no insert controls. For each construct two colonies were checked by colony PCR using GAPScr. (5′-GTCCCTATTTCAATCAATTGAACAAC-3′) as forward primer and a reverse primer that was specific to the individual catalytic domain. A sample gel of the colony PCR for constructs JB9 – JB32 is given in Fig. 8. Plasmid DNA was isolated from positive clones and 1 μg was digested with 2.5 units AscI for 2 hr to determine whether the subcloning had resulted in an in frame fusion of both fragments (the case in more than 99 % of all tested clones). In preparation for their transformation into yeast strains the fusion constructs were then linearized using a unique restriction site in the auxotrophic marker region of the plasmids (or in the PMA1 promoter in the case of the GNTII containing constructs). Restriction enzymes used for linearization were SalI for fusions containing catalytic domains A, D, F, G, M, U and V, EcoNI for fusions containing catalytic domains C, E, H and J, ApaI for fusion constructs containing K and L, AatII for fusion constructs containing catalytic domain N, EcoRI for fusion constructs containing catalytic domain T and XbaI for fusion constructs MA63, MA64 VA63, VA64, VB63, VB64, VC63 and VC64. Fusion constructs UA20 to UA67 were transformed without prior linearization. All leader/catalytic domain chimeras were then introduced into yeast cells via electroporation. MNSI derived fusions were transformed into strain PBP1 and selected on synthetic defined medium lacking histidine (synthetic defined medium is 1.34% yeast nitrogen base, 2% dextrose, 1.5% agar and 4×10−5% biotin with amino acids supplemented as appropriate). GNTI containing chimeras were transformed into strains YJN168, YJN169 and YJN191 and selected on synthetic defined medium lacking arginine. MNSII derived fusions were transformed into strain YSH1 and selected on synthetic defined medium lacking uracil. GNTII containing chimeras were transformed into yeast strain RDP27 and selected on YPD supplemented with 100 μg of the drug Nourseothricin (clonNAT from Hans Knoell Institute fuer Naturstofforschung, Jena, Germany). GNTIII containing fusions were transformed into YSH1 and selected on synthetic defined medium lacking uracil. Six individual clones of each transformation were then analyzed in liquid culture in a high throughput format. To this end the repatched transformants were grown for 3 days in 0.6 ml buffered glycerol complex medium (BMGY) consisting of 1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer, pH 6.0, 1.34% yeast nitrogen base, 4 × 10 −5% biotin, and 1% glycerol in deep well 96 well plates (Flat Bottom Blocks from Qiagen) on a heavy duty orbital shaker. Each starter plate was then used to inoculate 0.6 ml BMGY in 6 identical 96 deep well plates to an optical density of 1 – 2 at 600 nm and grown overnight. After the cultures had reached an optical density of 20 – 30 at 600 nm three plates each were combined and washed once in buffered methanol-complex medium (BMMY) consisting of 1.5% methanol instead of glycerol in BMGY. Reporter protein expression in the resulting two identical plates was then induced by growth in 0.6 ml BMMY for 24 h. The two plates were then combined, the yeast cells removed by centrifugation and 0.8 ml of each supernatant was used for purification of K3 and glycan analysis.
Fig. 8. Colony PCR sample.
Colony PCR screen for formation of fusion constructs JB9 through JB32. Bacterial colonies were analyzed using oligonucleotides annealing to the GAPDH promoter as forward primer and Aspergillus nidulans MNS1C (construct JB) as reverse primer.
Reporter protein expression and purification and release of N-linked glycans
The K3 domain reporter protein, under the control of the alcohol oxidase 1 (AOX1) promoter, was used as a model protein and was purified using the 6xHistidine tag as reported previously [Choi et al., 2003]. The glycans were released and separated from the glycoproteins by a modification of a previously reported method [Papac et al., 1998]. After the proteins were reduced and carboxy methylated, and the membranes blocked, the wells were washed three times with water. The protein was deglycosylated by the addition of 30 μl of 10 mM NH4HCO3 pH 8.3 containing one milliunit of N-glycanase (Glyko). After 16 hours at 37° C, the solution containing the glycans was removed by centrifugation and evaporated to dryness.
MALDI/time-of-flight (TOF) mass spectrometry
Molecular weights of the glycans were determined by using a Voyager DE PRO linear MALDI-TOF (Applied Biosciences) mass spectrometer with delayed extraction. The dried glycans from each well were dissolved in 15 μl of water, and 0.5 μl was spotted on stainless steel sample plates and mixed with 0.5 μl of S-DHB matrix (9 mg/ml of dihydroxybenzoic acid, 1 mg/ml of 5-methoxysalicylic acid in 1:1 water/acetonitrile 0.1% TFA) and allowed to dry. Ions were generated by irradiation with a pulsed nitrogen laser (337 nm) with a 4-ns pulse time. The instrument was operated in the delayed extraction mode with a 125-ns delay and an accelerating voltage of 20 kV. The grid voltage was 93%, guide wire voltage was 0.1%, the internal pressure was less than 5 × 10−7 torr, and the low mass gate was 875 daltons. Spectra were generated from the sum of 100 to 200 laser pulses and acquired with a 500 MHz digitizer. (Man)5-(GlcNAc)2 oligosaccharide was used as an external molecular weight standard. All spectra were generated with the instrument in the positive ion mode.
Mannosidase Assays
Fluorescence-labeled (Man)8(GlcNAc)2 (0.5 μg) was added to 20 μl of supernatant and incubated for 30 hr at room temperature. Following incubation the sample was analyzed by HPLC using an Econosil NH2 4.6 × 250 mm, 5 micron bead, amino-bound silica column (Altech, Avondale, PA). The flow rate was 1.0 ml/min for 40 min and the column was maintained to 30°C. After eluting isocratically (68% A:32% B) for 3 min a linear solvent gradient (68% A:32% B to 40% A:60% B) was employed over 27 min to elute the glycans (18). Solvent A (acetonitrile) and solvent B (ammonium formate, 50 mM, pH 4.5. The column was equilibrated with solvent (68% A:32% B) for 20 min between runs.
Results
Cloning of ER and Golgi targeting sequences
Most of the glycosyltransferases in the ER and Golgi of mammals and yeast are type II membrane proteins [Gleeson, 1998]. Although there is very little sequence similarity between these enzymes they all share the same domain structure. A short amino terminal cytoplasmic tail is followed by a transmembrane domain [TMD], a short stem region and a large carboxy terminal catalytic domain in the lumen of the ER or the Golgi (see Fig. 1). It is generally believed that the targeting information resides largely in the TMD and the cytoplasmic tail [Chapman and Munro, 1994; Lussier et al., 1995; Tang et al., 1997; Graham and Krasnov, 1995], however there have been reports that also the lumenal domains can contain Golgi localization signals [Graham and Krasnov, 1995].
Fig. 1. Leader and catalytic domain nomenclature.
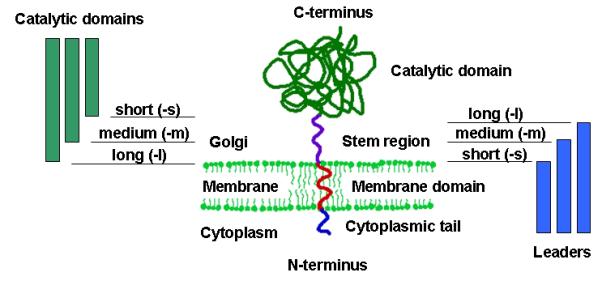
Cartoon depicting the structure of type II membrane proteins and the compositional nature of leader/catalytic domain fusion constructs. For details on the sizes of individual leaders and catalytic domains please refer to Tables 1 and 2.
Because we wanted to test a large variety of targeting sequences, we amplified DNA fragments encoding amino terminal regions of varying length of ER and Golgi residing type II membrane proteins of S. cerevisiae, Pichia pastoris and Kluyveromyces lactis. The shortest fragments (designated ‘-s’) contained only the cytoplasmic tail and the TMD. The medium length fragments (designated ‘-m’) also contained parts of, or the entire stem region. The longest fragments (designated ‘-l’) contained the entire stem region and some, also parts of the catalytic domain (see Fig. 1). We also included fragments of the S. cerevisiae and P. pastoris SEC12 gene, whose gene product Sec12p is a type II transmembrane protein that acts as a nucleotide exchange factor for the GTP-binding protein Sar1 [Barlowe and Schekman, 1993]. In yeast Sec12p is mainly localized in the ER, but it has also been shown to cycle between the early Golgi and the ER [Nakano et al., 1988; Boehm, et al., 1994]. Its cytosolic amino terminal catalytic domain is followed by a single transmembrane domain and the carboxy terminus in the lumen of the ER. In the case of the S. cerevisiae SEC12 gene, our shortest targeting fragment contained a short stretch of cytoplasmic domain and the TMD. The medium length fragment contained also a part of the ER lumenal domain to serve as a stem region, and the longest fragment contained the TMD and the complete carboxy terminal region. In the case of the P. pastoris SEC12 gene, we only included the shortest fragment in our studies because the carboxy terminal domain proved to be toxic to bacterial hosts. The other ER targeting domains utilized were amino terminal fragments of the S. cerevisiae α-glucosidase I [Tang et al., 1997] encoded by GLS1 and the S. cerevisiae α-1,2-mannosidase encoded by MNS1.
As targeting domains for the early or cis-Golgi we chose amino terminal fragments of the P. pastoris Och1p, and of the proteins that make up the S. cerevisiae mannan polymerases M-Pol I [Jungman et al., 1999] and M-Pol II, namely Mnn9p, Van1p, Anp1p, Hoc1p, Mnn10p and Mnn11p. Targeting signals for the medial Golgi [Brigance et al., 2000] were derived from S. cerevisiae Kre2p, Ktr1p, Mnn2p, Mnn5p, and Yur1p, from K. lactis Gnt1p and from P. pastoris proteins with homology to Ktr1p, Ktr3p and Kre2p (unpublished results). As targeting domains for the late Golgi we included amino terminal regions of S. cerevisiae Mnn1p and Mnn6p. The complete list of targeting domains is given in Table1.
Table 1.
| Leader# | Gene | Nucleotides | Plasmid | Leader# | Gene | Nucleotides | Plasmid | |
|---|---|---|---|---|---|---|---|---|
| 1 | ScGLS1-s | 1-102 | pJN355 | 35 | ScKRE2-l | 1-306 | pJN279 | |
| 2 | ScGLS1-m | 1-255 | pJN356 | 36 | PpKTR1-s | 1-216 | pJN351 | |
| 3 | ScGLS1-l | 1-411 | pJN373 | 37 | PpKTR1-m | 1-336 | pJN372 | |
| 4 | ScMNS1-s | 1-90 | pJN278 | 38 | PpKTR1-l | 1-588 | pJN352 | |
| 5 | ScMNS1-m | 1-246 | pJN280 | 39 | PpKTR3-s | 1-90 | pJN353 | |
| 6 | ScMNS1-l | 1-429 | pJN282 | 40 | PpKTR3-m | 1-255 | pJN354 | |
| 7 | ScSEC12-s | 988-1140 | pJN297 | 41 | PpKTR3-l | 1-489 | pJN387 | |
| 8 | ScSEC12-m | 988-1296 | pJN305 | 42 | PpKRE2-s | 1-93 | pJN366 | |
| 9 | ScSEC12-l | 988-1413 | pJN291 | 43 | PpKRE2-m | 1-252 | pJN350 | |
| 10 | PpSEC12-s | 1003-1089 | pJN328 | 44 | PpKRE2-l | 1-450 | pJN367 | |
| 12 | PpOCH1-s | 1-150 | pJN274 | 45 | ScKTR1-s | 1-117 | pJN301 | |
| 13 | PpOCH1-m | 1-240 | pJN275 | 46 | ScKTR1-l | 1-465 | pJN344 | |
| 14 | PpOCH1-l | 1-402 | pJN276 | 47 | ScKTR2-s | 1-120 | pJN295 | |
| 15 | ScMNN9-s | 1-120 | pJN271 | 48 | ScKTR2-m | 1-300 | pJN288 | |
| 16 | ScMNN9-m | 1-273 | pJN272 | 49 | ScKTR2-l | 1-510 | pJN345 | |
| 17 | ScMNN9-l | 1-462 | pJN342 | 50 | KlGNT1-s | 1-93 | pJN377 | |
| 18 | ScVAN1-s | 1-279 | pJN294 | 51 | KlGNT1-m | 1-243 | pJN378 | |
| 19 | ScVAN1-m | 1-294 | pJN368 | 52 | KlGNT1-l | 1-516 | pJN379 | |
| 20 | ScVAN1-l | 1-609 | pJN357 | 53 | ScMNN2-s | 1-108 | pJN281 | |
| 21 | ScANP1-s | 1-180 | pJN302 | 54 | ScMNN2-m | 1-291 | pJN273 | |
| 22 | ScANP1-m | 1-246 | pJN303 | 55 | ScMNN2-l | 1-450 | pJN277 | |
| 23 | ScANP1-l | 1-315 | pJN304 | 56 | ScMNN5-s | 1-105 | pJN361 | |
| 24 | ScHOC1-s | 1-102 | pJN370 | 57 | ScMNN5-m | 1-255 | pJN362 | |
| 25 | ScHOC1-m | 1-252 | pJN359 | 58 | ScMNN5-l | 1-450 | pJN363 | |
| 26 | ScHOC1-l | 1-435 | pJN360 | 59 | ScYUR1-s | 1-108 | pJN289 | |
| 27 | ScMNN10-s | 1-219 | pJN382 | 60 | ScYUR1-m | 1-270 | pJN290 | |
| 28 | ScMNN10-m | 1-363 | pJN358 | 61 | ScYUR1-l | 1-450 | pJN296 | |
| 29 | ScMNN10-l | 1-516 | pJN369 | 62 | ScMNN1-s | 1-126 | pJN292 | |
| 30 | ScMNN11-s | 1-156 | pJN375 | 63 | ScMNN1-m | 1-279 | pJN343 | |
| 31 | ScMNN11-m | 1-303 | pJN376 | 64 | ScMNN1-l | 1-459 | pJN293 | |
| 32 | ScMNN11-l | 1-495 | pJN381 | 65 | ScMNN6-s | 1-90 | pJN364 | |
| 33 | ScKRE2-s | 1-174 | pJN340 | 66 | ScMNN6-m | 1-255 | pJN371 | |
| 34 | ScKRE2-m | 1-240 | pJN380 | 67 | ScMNN6-l | 1-480 | pJN357 |
Cloning of heterologous glycosylation enzymes
To be able to identify catalytic domains that would function properly in the yeast ER and Golgi we decided to test enzymes with a wide range of pH and temperature optima. Similar to the targeting domains, the glycolytic enzymes were amplified as DNA fragments of varying length. The longest fragments contained the enzyme without the TMD. The medium length fragments were lacking part of the stem region, and the shortest fragments only contained the catalytic domain (see Fig. 1). Some of the mannosidases were selected on the basis of sequence homology to other known mannosidases and had not been characterized previously. This library of heterologous glycosylation enzymes contained α-1,2-mannosidases (MnsI)) from Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Penicillium citrinum, Mus musculus and Aspergillus nidulans, β-1,2-N-acetylglucosaminyltransferase I (GnTI) from H. sapiens, mannosidases II (MnsII) from D. melanogaster, C. elegans and H. sapiens, β-1,2-N-acetylglucosaminyltransferase II (GnTII) from H. sapiens and Rattus norvegicus and β-1,4-N-acetylglucosaminyltransferase III (GnTIII) from M. musculus. The complete list of catalytic domains is given in Table 2.
High throughput fusion and expression of leader/catalytic domain chimeras
To facilitate the fusion of targeting sequences to catalytic domains, the targeting domains were amplified by PCR and cloned into plasmids pCR2.1 (Invitrogen) or pVM1 (a pUC19 based plasmid containing a NotI and an AscI site) with NotI sites at the 5′-terminus followed by the Kozak consensus sequence CACC and the start codon. At the 3′ terminus of the targeting domains the sequence GGGCGCGCC had been added. This introduced an AscI restriction site and resulted in a fusion linker encoding GlyArgAla. The catalytic domains were amplified introducing an AscI site at the 5′ end and a PacI site at the 3′ end and cloned into integration plasmids under the control of the strong PpGAP or PpPMA1 promoters. The agarose gel purified targeting domain fragments were then, in a high throughput format, ligated in frame into these plasmids to create the fusion libraries. For details see Materials and Methods. After transformation into bacteria, two colonies per fusion construct were examined by colony PCR, which confirmed that more than 95% of all constructs contained the correct fusions (see Fig. 8). To assess whether the ligation reaction had resulted in in-frame fusions, plasmid DNA was isolated and an AscI restriction digest was performed. This was based on the fact that only if the AscI site had been recreated a genuine in frame fusion had occurred. This proved to be true in over 99% of all cases.
Before transformation into yeast strains, the fusion constructs were linearized using an appropriate restriction enzyme that was absent in the leader/catalytic domain chimera but present in the integration locus. For details see Materials and Methods. The linearized DNA was then transformed into yeast strains expressing the Kringle 3 domain of human plasminogen (K3) by electroporation and six individual clones of each transformation were analyzed in liquid culture as described in Materials and Methods.
Analysis of leader/catalytic domain libraries
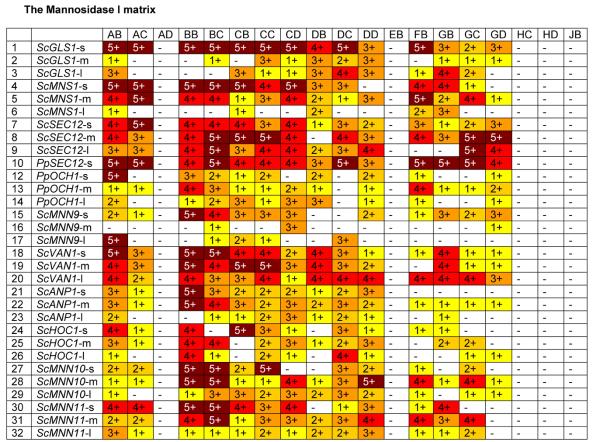
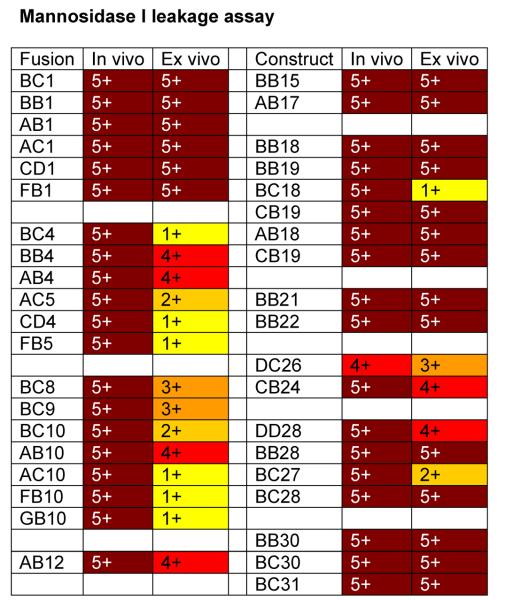
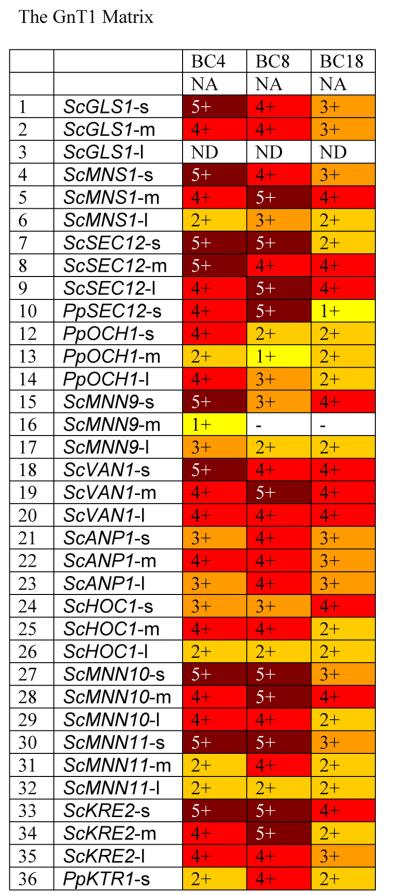
In the human, Golgi α-1,2-mannosidases (IA-IC) remove mannose from the predominantly (Man)8-(GlcNAc)2 containing glycoproteins that are secreted from the ER to yield the (Man)5(GlcNAc)2 structure, which forms the precursor for complex N-glycans. We therefore constructed a library of 19 different α-1,2-mannosidase fragments fused to fungal targeting domains from the ER and early Golgi (leaders 1 to 32) and transformed them into the och1 knockout strain PBP1 [Choi et al., 2003] to determine whether this would result in trimming of the fungal glycoforms. Fig. 2 shows that catalytic domains obtained from fungal sources (EB, HC, HD, JB) did not show any activity, whereas the catalytic domains from putative C. elegans mannosidases showed the highest activity, followed by mannosidases from Drosophila, Mouse and Human. The truncation that removes the N-terminal 129 amino acids (delta 387) of the putative C.elegans mannosidase IA also showed no activity, suggesting that an essential part of the catalytic domain had been removed. Significant differences were also observed in the ability of leader sequences to target fusion proteins with high mannosidase activity. Generally the longest leader fragments resulted in fusion proteins with lower activity, whereas the fusion proteins containing only the fungal TMDs had the highest activity. Exceptions were the medium sized leaders from ScMNN10 and ScMNN11 and the longest leader from ScVAN1, which also showed consistently high activity. Also all the leaders derived from SEC12 resulted in fusions with very high activity, whereas the leaders derived from PpOCH1 and ScMNN9 typically resulted in fusion proteins with poor apparent mannosidase activity. We also considered the possibility that some of the (Man)5(GlcNAc)2 observed was formed outside the cell by mannosidase fusion protein that had been secreted into the medium. To determine to which extent this was the case, we examined the supernatants of 39 strains using 2-aminobenzamide-labeled (Man)8(GlcNAc)2 as substrate (see Materials and Methods). Fig. 3 shows that for some of the (Man)5(GlcNAc)2- producing strains a high degree of mannosidase activity was observed in the culture medium, suggesting that some of the (Man)5(GlcNAc)2 structures produced by the strains might have been produced ex vivo. The amount of mannosidase activity was independent of the optical density at which protein expression in the culture was induced and also independent of the amount of methanol that was used for induction (results not shown). The leaders derived from ScMNS1 (#s 4-6) and SEC12 (#s 7-10) typically showed relatively low mannosidase activity in the medium, whereas other leaders tested in the mannosidase library displayed a significant level of leakage into the medium. It is noteworthy that apparently also the length of the mannosidase domain influences the amount of mannosidase that is secreted into the medium. For example whereas constructs BB4 and BB18 display the maximum amount of activity, constructs BC4 and BC18 that contain a shorter mannosidase domain show only background levels of mannosidase activity in the medium.
Fig. 2. The Mannosidase I Matrix.
Glycans released from protein expressed in strains transformed with mannosidase fusions were analyzed by MALDI TOF as described in Materials and Methods. For each fusion a number and color was assigned based on the level of (Man)5-(GlcNAc)2 detected (as a % of total glycans): - / white = none detected; 1 / yellow = 0 to 10%; 2 / gold = 10 to 20%; 3 / orange = 20 to 50%; 4 / red = 50 to 75%; 5 / dark red = >75%. Codes for glycosylation enzymes are given in Table 2. Details on leaders are given in Table 1.
Fig. 3. Mannosidase leakage assay results.
Fluorescence-labeled (Man)8-(GlcNAc)2 was added to culture supernatant, incubated and then analyzed by HPLC as described in Materials and Methods. For each sample a number and color was assigned based on the level of (Man)5-(GlcNAc)2 detected (as a % of total glycans): - / white = none detected; 1 / yellow = 0 to10%; 2 / gold = 10 to 20%; 3 / orange = 20 to 50%; 4 / red = 50 to 75%; 5 / dark red = >75%.
The second step in the formation of complex N-glycans in the mammalian Golgi is the addition of N-acetylglucosamine to the 1,3-arm of (Man)5-(GlcNAc)2 to generate GlcNAc-(Man)5-(GlcNAc)2. To replicate this reaction in P. pastoris we fused the soluble domain of human GnTI to leader sequences 1 through 36 of our leader library and transformed the resulting constructs into three different α-1,2-mannosidase containing yeast strains. YJN168, YJN169 and YJN191 were created from PBP1 by transformation with mannosidase fusion constructs BC4, BC8 and BC18 respectively. We chose strains that contained mannosidases that were targeted to different locations in the secretory pathway, BC4 to the ER, BC8 to the ER/early Golgi and BC18 to the early Golgi, because we also wanted to determine the effect of localization of the mannosidase on the formation of GlcNAc-(Man)5-(GlcNAc)2. As expected we observed lower amounts of GlcNAc-(Man)5-(GlcNAc)2 when the GnTI fusion library was transformed into strain YJN191 which contained the mannosidase in a later secretory compartment (see column BC18 in Fig. 4). However the fact that a GnTI chimera that is targeted to the ER by fusion with the TMD of ScGls1p or ScMns1p still shows some residual activity in a strain that contains an early Golgi targeted mannosidase is also consistent with the view, that early Golgi enzymes are not stationary in their compartments, but rather undergo continuous recycling through the ER [Todorow et al., 2000]. In our case this would mean that the early Golgi targeted mannosidase BC18 is recycled to the ER where it could form (Man)5-(GlcNAc)2, which in turn would be the substrate for the ER targeted GnTI fusions NA1 or NA4. An alternative explanation would be that the ER targeted GnTI is leaking into the early Golgi and is acting on the (Man)5-(GlcNAc)2 that is formed there.
Fig. 4. The GnTI Matrix.
Glycans released from protein expressed in 3 different mannosidase I containing strains transformed with human GnTI fusions were analyzed by MALDI TOF as described in Materials and Methods. For each fusion a number and color was assigned based on the level of GlcNAc-(Man)5-(GlcNAc)2 detected (as a % of total glycans): - / white = none detected; 1 / yellow = 0 to10%; 2 / gold = 10 to 20%; 3 / orange = 20 to 50%; 4 / red = 50 to 75%; 5 / dark red = >75%. Codes for glycosylation enzymes are given in Table 2. Details on leaders are given in Table 1.
Fig. 4 also shows that, as seen in the mannosidase I fusion library, the short and medium length leaders resulted in fusion proteins with higher GnTI activity whereas the long leaders generally yielded lower GnTI activity. Again almost all of the SEC12 and VAN1 derived leaders produced fusions with high activity, whereas only few of the PpOCH1 and ScMNN9 derived leaders did.
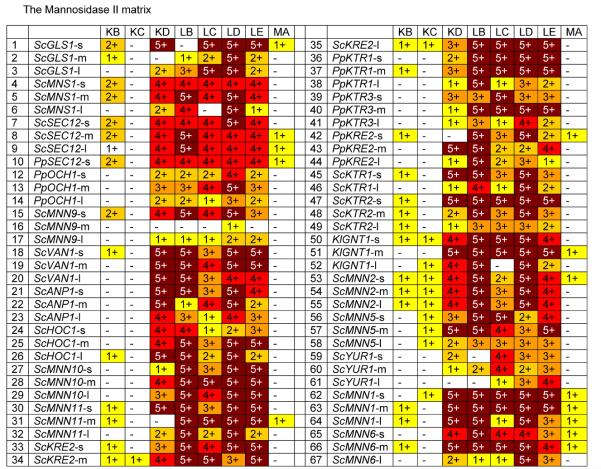
The third step in the formation of complex N-glycans in the mammalian Golgi is the removal of the two terminal α-1,3- and α-1,6-linked mannose residues from GlcNAc-(Man)5-(GlcNAc)2 by mannosidase II to form GlcNAc-(Man)3-(GlcNAc)2. To replicate this reaction in P. pastoris, we transformed a mannosidase II / leader fusion library into the GlcNAc-(Man)5-(GlcNAc)2 producing strain YSH1 [Hamilton et al., 2003], which is an och1 knockout containing the gene for the Kluyveromyces lactis uridine 5′-diphosphate (UDP)-GlcNAc transporter, the mannosidase fusion FB8 and the GnTI fusion NA15. As evident from Fig. 5, the human mannosidase II (MA) showed only low levels of activity, as did D. melanogaster MnsII having N-terminal 48 or 99 amino acid truncations (KB and KC, respectively). However the fusion constructs containing the D. melanogaster mannosidase II with the N-terminal 74 amino acids removed (KD) showed ample activity when combined with a variety of leader constructs. In the case of the third tested enzyme, the C. elegans mannosidase II, there were only minor differences, but overall the longer catalytic domains (LB, LD) scored slightly higher than the further truncated ones (LC, LE). When combined with a highly active mannosidase II catalytic domain the large majority of all leader constructs resulted in active fusion constructs, with notable laggards again being leaders derived from PpOCH1, the ScMNN9-m and −l leaders, and leaders derived from ScYUR1. Again it should be noted that despite the GnTI being targeted to the early Golgi, some ER-targeted mannosidase II fusions showed very high activity. This corroborates the results obtained with the GnTI fusion library that our fusion proteins apparently are not targeted to exclusively one compartment in the ER or the Golgi, but rather are dispersed in a wide area in the secretory pathway.
Fig. 5. The Mannosidase II Matrix.
Glycans released from protein expressed in strains transformed with mannosidase II fusions were analyzed by MALDI TOF as described in Materials and Methods. For each fusion a number and color was assigned based on the level of GlcNAc-(Man)3-(GlcNAc)2 detected (as a % of total glycans): - / white = none detected; 1 / yellow = 0 to10%; 2 / gold = 10 to 20%; 3 / orange = 20 to 50%; 4 / red = 50 to 75%; 5 / dark red = >75%. Codes for glycosylation enzymes are given in Table 2. Details on leaders are given in Table 1.
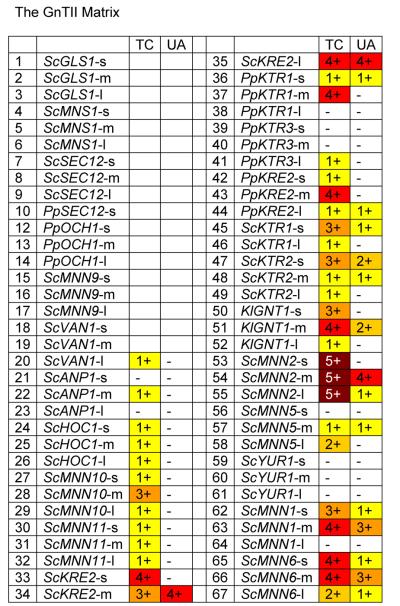
The next step in the formation of complex human N-glycans is the addition of another GlcNAc to GlcNAc-(Man)3-(GlcNAc)2, a step that is catalyzed by GnTII. We constructed a library of fusion constructs of human and rat GnTII with our 66 targeting domains and tested them in RDP27 [Bobrowicz et al., 2004]. This yeast strain is an och1 knockout containing the K. lactis (UDP)-GlcNAc transporter, the mannosidase I fusion FB8 and the GnTI fusion NA15. It also contains a deletion of the P. pastoris homologue of ALG3 [Davidson et al., 2004], an α-1,3-mannosyl-tranferase involved in the dolichol-linked oligosaccharide biosynthesis. Instead of (Man)8-(GlcNAc)2, knockouts of alg3 transfer the truncated (Man)5-(GlcNAc)2 glycan from dolichol to newly synthesized proteins in the ER. Action of MnsI and GnTI then leads to the formation of GlcNAc-(Man)3-(GlcNAc)2, which is the substrate for GnTII. As shown in Fig. 6 the activity of GnTII / leader fusions appears to be more dependent on the leader than what had been seen in the cases of MnsI, GnTI or MnsII. The rat GNTII / S.c.MNN2 chimeras TC53, TC54 and TC55 resulted in fusion proteins with full activity, whereas the human GnTII / leader fusions typically exhibited slightly lower activity. Other leaders that resulted in fusion proteins with high activity were derived from ScKRE2, PpKTR1, PpKRE2, KlGNTI, ScMNN1 and ScMNN6, however, as opposed to what had been seen in the cases of the MnsI and GnTI libraries, here the medium length leaders typically had the highest activity.
Fig. 6. The GnTII Matrix.
Glycans released from protein expressed in strains transformed with GnTII fusions were analyzed by MALDI TOF as described in Materials and Methods. For each fusion a number and color was assigned based on the level of (GlcNAc)2-(Man)3-(GlcNAc)2 detected (as a % of total glycans): - / white = none detected; 1 / yellow = 0 to10%; 2 / gold = 10 to 20%; 3 / orange = 20 to 50%; 4 / red = 50 to 75%; 5 / dark red = >75%. Codes for glycosylation enzymes are given in Table 2. Details on leaders are given in Table 1.
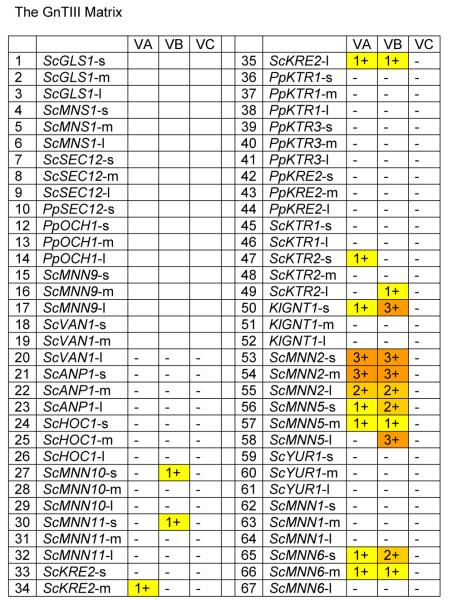
A modification of glycosylation that is commonly found on human antibodies is the addition of a β-1,4-GlcNAc residue to the (GlcNAc)2-(Man)3-(GlcNAc)2 complex glycan, the so called bisecting GlcNAc, a reaction catalyzed by N-acetylglucosaminyltransferase III (GnTIII) [Narasimhan, 1982; Bhaumik et al., 1995]. The bisecting GlcNAc has been implicated in the modulation of antibody-dependent cellular cytotoxicity (ADCC) [Umana et al., 1999; Davies et al., 2001; Shinkawa et al., 2003], and might be a potential way to generate antibodies with optimized effector functions. We fused three different truncations of mouse GNTIII [Bhaumik et al., 1995] to our 66 targeting domains and transformed the fusion constructs into P. pastoris in an attempt to generate a yeast strain with glycans containing bisecting GlcNAc. The recipient yeast strain that was chosen was YSH1 (see above), a P. pastoris strain expressing the reporter protein K3 with the glycan structure GlcNAc-(Man)5-(GlcNAc)2. Fig. 7 shows a picture very similar to what was seen in the GnTII fusion library. While some of the leader constructs resulted in fusions with no or only trace amounts of activity, some fusion constructs yielded glycans containing a significant amount of bisecting GlcNAc. As seen in the case of GnTII the leaders resulting in the highest activity were derived from ScMNN2, KlGNT1, ScMNN5 and ScMNN6. The fact that we didn't observe complete conversion to the bisecting GlcNAc containing glycan in any of the cases might be due to the fact that GlcNAc-(Man)5-(GlcNAc)2 is not the natural substrate for GnTIII, therefore a strain producing a biantennary sugar chain like (GlcNAc)2-(Man)3-(GlcNAc)2 might prove to be a better host for GnTIII.
Fig. 7. The GnTIII Matrix.
Glycans released from protein expressed in strains transformed with GnTIII fusions were analyzed by MALDI TOF as described in Materials and Methods. For each fusion a number and color was assigned based on the level of (GlcNAc)2-(Man)5-(GlcNAc)2 detected (as a % of total glycans): - / white = none detected; 1 / yellow = 0 to10%; 2 / gold = 10 to 20%; 3 / orange = 20 to 50%. Codes for glycosylation enzymes are given in Table 2. Details on leaders are given in Table 1.
Discussion
The lack of a suitable mammalian expression system is a significant obstacle to the low-cost and safe production of recombinant human glycoproteins for therapeutic applications. Whereas lower eukaryotes such as fungi and yeasts generally offer higher production yields and ease of fermentation they also suffer from potential problems associated with the non-human glycosylation pattern that is found on proteins expressed in these organisms. To overcome these problems we have focused our efforts on engineering commercially relevant yeast and filamentous fungi to produce N-glycans with human-like glycosylation structures. In eukaryotes sugar transferases and glycosidases (e.g. mannosidases) line the inner (luminal) surface of the ER and Golgi to provide a “catalytic” surface that allows for the sequential processing of glycoproteins as they proceed through the secretory pathway. Much work has been dedicated to revealing the mechanism by which these glycosylation enzymes are retained and anchored to their respective organelle. Evidence suggests that stem region, transmembrane domain and cytoplasmic tail, individually or in concert, direct enzymes to the membrane of a specific compartment and thereby localize the associated catalytic domain to that locus [Gleeson, 1998]. Before our work on optimization of targeting sequences, attempts to engineer the fungal secretory pathway to express functional heterologous glycosylation enzymes have been described with limited success. Chiba et al. localized an α-1,2-mannosidase of Aspergillus saitoi to the ER of an och1mnn1mnn4 mutant S. cerevisiae strain using the C-terminal motif HDEL as a targeting signal [Chiba et al., 1998]. They were able to show that only 27 % of the N-glycans of the endogenous marker protein caboxypeptidase Y were trimmed from (Man)8-(GlcNAc)2 to (Man)5-(GlcNAc)2. Kalsner et al. transformed Aspergillus nidulans with the full-length gene for rabbit GNTI [Kalsner et al., 1995]. While they could detect intracellular GnTI activity, they did not observe any formation of GlcNAc-(Man)5-(GlcNAc)2 despite the fact that Aspergillus nidulans expresses several α1,2-mannosidases [Eades and Hintz, 2000].
In light of these problems, and because prior to our work there was no reliable way of predicting whether a particular heterologously expressed glycosyltransferase or mannosidase in a lower eukaryote will be targeted to a specific location in the secretory pathway, we developed a systematic fusion library approach to select for the proper combination of targeting signal and catalytic domain. Our targeting domain library consisted of transmembrane domains and adjacent regions of known ER and Golgi residing type II membrane proteins selected from S. cerevisiae, P. pastoris and K. lactis. For some of these proteins the regions responsible for proper localization had been characterized previously [Chapman and Munro, 1994; Lussier et al., 1995; Tang et al., 1997; Graham and Krasnov, 1995; Sato et al., 1996], but for the majority of the proteins we employed, the targeting had not been examined in detail before. To replicate the mammalian glycosylation machinery in P. pastoris we fused these targeting sequences to mannosidases and GlcNAc transferases from different species and tested for their functional expression in P. pastoris strains producing the necessary substrates. Because the removal of 1,2-mannose is the first step in the formation of complex N-glycans we decided to target the α-1,2-mannosidases to an early compartment of the secretory pathway, fusing them to leaders 1 through 32. While some mannosidases showed no activity at all, others were able to almost quantitatively remove the α-1,2-mannoses from (Man)8-(GlcNAc)2 to generate (Man)5-(GlcNAc)2 when fused to a large variety of leader sequences. When we examined the amount of mannosidase that was secreted into the medium, a phenomenon that had been described previously [Callewaert et al., 2001], it became obvious, that only a small subset of fusion constructs displayed no extracellular mannosidase activity. While the fact that some leader/mannosidase chimeras leak into the medium does not necessarily mean that some or all of their observed activity happens ex vivo, it is currently not possible to determine where the (Man)5-(GlcNAc)2 is being generated in these strains.
The next step in the in the formation of hybrid- and complex-type glycans involves the correct localization of GnTI, the enzyme responsible for the formation of GlcNAc-(Man)5-(GlcNAc)2 from (Man)5-(GlcNAc)2. We transformed the GnTI fusion library into three different yeast strains each expressing a different highly active, non-leaking mannosidase construct that was targeted to either the ER (MNS1), the ER/early Golgi (SEC12) or the early Golgi (VAN1). Whereas a large number of GnTI fusion constructs showed very high activity when expressed in the first two strains, the same constructs typically showed a lower level of activity in the third. This is consistent with the view that the mannosidase is optimally targeted to a very early compartment of the secretory pathway to allow GnTI the maximum amount of time to act on the substrate generated by it. On the other hand the fact that a GnTI that is targeted to the ER has any activity at all in a strain with an early Golgi targeted mannosidase also confirms the view that the glycosylation enzymes apparently are able to move within the secretory pathway and are continuously retrieved to their original compartments [Todorow et al., 2000; Sato et al., 1996; Harris and Waters, 1996].
The third enzyme in the formation of mammalian complex N-glycans is MnsII, responsible for removal of 1,3- and 1,6-mannose from GlcNAc-(Man)5-(GlcNAc)2. Analysis of our MnsII fusion protein library shows the importance of testing glycosylation enzymes with varying lengths of stem regions. In the case of MnsII from D. melanogaster the truncation missing the N-terminal 74 amino acids showed full activity with a number of leaders, whereas the truncations missing the N-terminal 48 or 99 amino acids only had trace amounts of activity.
The next step in complex N-glycan formation is the addition of a second GlcNAc residue to GlcNAc-(Man)3-(GlcNAc)2 by GnTII. To avoid any potential complications arising from leaking MnsII, we tested the GnTII fusion library in an och1 knockout strain that in addition to expressing MnsI and GnTI leader/fusions was deleted for alg3, an α-1,3-mannosyl-tranferase that is responsible for the first Dol-P-Man-dependent mannosylation step at the luminal side of the ER [Sharma et al., 2001]. In this strain GlcNAc-(Man)3-(GlcNAc)2, the substrate for GnTII, is generated without the activity of MnsII. Contrary to what was seen with the earlier enzymes MnsI, GnTI and MnsII, the activity of the GnTII fusions was largely dependent on the targeting domain. Leaders derived from ScMNN2 gave rise to fusion proteins with maximum GnTII activity. A similar picture was seen in the fusion library of GnTIII, which is the enzyme that adds the bisecting GlcNAc to (GlcNAc)2-(Man)3-(GlcNAc)2. Here too the leaders derived from ScMNN2 lead to the fusions with the highest GnTIII activity.
Taken together our results demonstrate that it is almost impossible to predict beforehand precisely which leader/catalytic domain fusion will exhibit optimal activity and whether the observed activity actually occurs in vivo. On the other hand we also show that our combinatorial genetic library approach and high throughput screening protocol are extremely powerful tools that can be employed to find the exact combinations of leader/catalytic domain fusion proteins that allow for the reengineering of the glycosylation machinery of commercially important yeast such as P. pastoris to produce human-like glycan structures. Here we have used these tools to generate yeast strains that attach the complex human-like N-glycan (GlcNAc)2-(Man)3-(GlcNAc)2 to an expressed protein, and have laid the foundation for the complete humanization of the glycosylation machinery of Pichia pastoris.
Acknowledgements
The expert technical assistance of Sebastian Rausch, Harry Wischnewski, and Eduard Renfer is gratefully acknowledged.
We also would like to thank Jim Cregg for the gift of strain JC308.
This research was supported in part by a government grant from the National Institutes of Health (NIH Phase I Grant no. IR43GM66690-1) and a grant from the Department of Commerce, NIST-ATP Cooperative Agreement Number 70NANB2H3046.
References
- Ballou CE. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine nucleotide exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Bhaumik M, Seldin MF, Stanley P. Cloning and chromosomal mapping of the mouse Mgat3 gene encoding N-acetylglucosaminyltransferase III. Gene. 1995;164:295–300. doi: 10.1016/0378-1119(95)00260-d. [DOI] [PubMed] [Google Scholar]
- Bobrowicz P, Davidson RC, Li H, Potgieter TI, Nett JH, Hamilton SR, Stadheim TA, Miele RG, Bobrowicz B, Mitchell T, Rausch S, Renfer E, Wildt S. Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: production of complex humanized glycoproteins with terminal galactose. Glycobiology. 2004;14:757–766. doi: 10.1093/glycob/cwh104. [DOI] [PubMed] [Google Scholar]
- Boehm J, Ulrich HD, Ossig R, Schmitt HD. Kex2-dependent invertase secretion as a tool to study the targeting of transmembrane proteins which are involved in ER-Golgi transport in yeast. EMBO J. 1994;13:3696–3710. doi: 10.1002/j.1460-2075.1994.tb06679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigance WT, Barlowe C, Graham TR. Organization of the yeast Golgi complex into at least four functionally distinct compartments. Mol. Biol. Cell. 2000;11:171–182. doi: 10.1091/mbc.11.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert N, Laroy W, Cadirgi H, Geysens S, Saelens X, Min Jou W, Contreras R. Use of HDEL-tagged Trichoderma reesei mannosyl oligosaccharide 1,2-alpha-D-mannosidase for N-glycan engineering in Pichia pastoris. FEBS Lett. 2001;503:173–178. doi: 10.1016/s0014-5793(01)02676-x. [DOI] [PubMed] [Google Scholar]
- Chapman RE, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Suzuki M, Yoshida S, Yoshida A, Ikenaga H, Takeuchi M, Jigami, Y, Ichishima E. Production of human compatible high mannose type (Man5GlcNAc2) sugar chains in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:26298–26304. doi: 10.1074/jbc.273.41.26298. [DOI] [PubMed] [Google Scholar]
- Choi BK, Bobrowicz P, Davidson RC, Hamilton SR, Kung DH, Li H, Miele RG, Nett JH, Wildt S, Gerngross TU. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. 2003;100:5022–5027. doi: 10.1073/pnas.0931263100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N. Asparagine linked glycosylation in the yeast Golgi. Biochim. Biophys Acta. 1999;1426:309–322. doi: 10.1016/s0304-4165(98)00132-9. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Nett JH, Renfer E, Li H, Stadheim TA, Miller BJ, Miele RG, Hamilton SR, Choi BK, Mitchell TI, Wildt S. Functional analysis of the ALG3 gene encoding the Dol-P-Man: Man5GlcNAc2-PP-Dol mannosyltransferase enzyme of P. pastoris. Glycobiology. 2004;14:399–407. doi: 10.1093/glycob/cwh023. [DOI] [PubMed] [Google Scholar]
- Davies J, Jiang LY, Pan, LZ, LaBarre MJ, Anderson D, Reff M. Expression of GnTIII in a recombinant Anti-CD20 CHO production cell line: Expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FcγRIII. Biotechnol. Bioeng. 2001;74:288–294. [PubMed] [Google Scholar]
- Durand H, Clanet M. Genetic improvement of T. reesei for large scale cellulase production. Enzyme Microb. Technol. 1988;10:341–346. [Google Scholar]
- Eades CJ, Hintz WE. Characterization of the class I α-mannosidase gene family in the filamentous fungus Aspergillus nidulans. Gene. 2000;255:25–34. doi: 10.1016/s0378-1119(00)00298-5. [DOI] [PubMed] [Google Scholar]
- Gleeson PA. Targeting of proteins to the Golgi apparatus. Histochem. Cell. Biol. 1998;109:517–532. doi: 10.1007/s004180050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Krasnov VA. Sorting of yeast α-1,3-mannosyltransferase is mediated by a lumenal domain interaction, and a transmembrane domain signal that can confer clathrin-dependent Golgi localization to a secreted protein. Mol. Biol. Cell. 1995;6:809–824. doi: 10.1091/mbc.6.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SR, Bobrowicz P, Bobrowicz B, Davidson RC, Li H, Mitchell T, Nett JH, Rausch S, Stadheim TA, Wischnewski H, Wildt S, Gerngross TU. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Jessee J, Bloom FR. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J. Cell. Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- Kalsner I, Hintz W, Reid LS, Schachter H. Insertion into Aspergillus nidulans of functional UDP-GlcNAc: alpha 3-D-mannoside beta-1,2-N-acetyglucosaminyl-transferase I, the enzyme catalyzing the first commited step from oligomannose to hybrid and complex N-glycans. Glycoconj. 1995;12:360–370. doi: 10.1007/BF00731339. [DOI] [PubMed] [Google Scholar]
- Lin Cereghino GP, Lin Cereghino J, Jay Sunga A, Johnson MA, Lim M, Gleeson MAG, Cregg JM. New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene. 2001;263:159–169. doi: 10.1016/s0378-1119(00)00576-x. [DOI] [PubMed] [Google Scholar]
- Lussier M, Sdicu AM, Ketela T, Bussey H. Localization and targeting of the Saccharomyces cerevisiae Kre2p/Mnt1p α1,2-mannosyltransferase to a medial-Golgi compartment. J. Cell Biol. 1995;131:913–927. doi: 10.1083/jcb.131.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maras M, De Bruyn A, Vervecken W, Uusitalo J, Penttilä M, Busson R, Herdewijn P, Contreras R. In vivo synthesis of complex N-glycans by expression of human N-acetylglucosaminyltransferase I in the filamentous fungus Trichoderma reesei. FEBS Lett. 1999;452:365–370. doi: 10.1016/s0014-5793(99)00648-1. [DOI] [PubMed] [Google Scholar]
- Nakano A, Brada D, Schekman R. A membrane glycoprotein Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S. Control of glycoprotein synthesis. UDP-GlcNAc: glycopeptide β4-N-acetylglucosaminyltransferase III, an enzyme in hen oviduct which adds GalNAc in β 1-4 linkage to the β-linked mannose of the trimannosyl core of N-glycosyl oligosaccharides. J. Biol. Chem. 1982;257:10235–10242. [PubMed] [Google Scholar]
- Papac DI, Briggs JB, Chin ET, Jones AJ. A high-throughput microscale method to release N-linked oligosaccharides from glycoproteins for matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis. Glycobiology. 1998;8:445–454. doi: 10.1093/glycob/8.5.445. [DOI] [PubMed] [Google Scholar]
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, Hanai N, Shitara K. The absence of fucose but not the presence of galactose or bisecting N-Acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003;278:3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K, Nakano A. Endoplasmic reticulum localization of Sec12p is achieved by two mechanisms: Rer1p-dependent retrieval that requires the transmembrane domain and Rer1p-independent retention that involves the cytoplasmic domain. J. Cell. Biol. 1996;134:279–293. doi: 10.1083/jcb.134.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CB, Knauer R, Lehle L. Biosynthesis of lipid-linked oligosaccharides in yeast: the ALG3 gene encodes the Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol mannosyltransferase. Biol. Chem. 2001;382:321–328. doi: 10.1515/BC.2001.039. [DOI] [PubMed] [Google Scholar]
- Tang BL, Low SH, Hong W. Endoplasmatic reticulum retention mediated by the transmembrane domain of type II membrane proteins Sec12p and glucosidase 1. Eur. J. Cell Biol. 1997;73:98–104. [PubMed] [Google Scholar]
- Todorow Z, Spang A, Carmack E, Yates J, Schekman R. Active recycling of yeast Golgi mannosyltransferase complexes through the endoplasmic reticulum. Proc. Natl. Acad. Sci. 2000;97:13643–13648. doi: 10.1073/pnas.250472397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umana P, Jean-Mairet J, Moudry R, Amstutz H, Bailey JE. Engineered glycoforms of an anti-neuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat. Biotechnol. 1999;17:176–180. doi: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- Werten MW, van den Bosch TJ, Wind RD, Mooibroek H, de Wolf FA. High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast. 1999;15:1087–1096. doi: 10.1002/(SICI)1097-0061(199908)15:11<1087::AID-YEA436>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]