Abstract
The Ess1/Pin1 peptidyl-prolyl isomerase (PPIase) is thought to control mitosis by binding to cell cycle regulatory proteins and altering their activity. Here we isolate temperature-sensitive ess1 mutants and identify six multicopy suppressors that rescue their mitotic-lethal phenotype. None are cell cycle regulators. Instead, five encode proteins involved in transcription that bind DNA, modify chromatin structure or are regulatory subunits of RNA polymerase II. A sixth suppressor, cyclophilin A, is a member of a distinct family of PPIases that are targets of immuno suppressive drugs. We show that the expression of some but not all genes is decreased in ess1 mutants, and that Ess1 interacts with the C-terminal domain (CTD) of RNA polymerase II in vitro and in vivo. The results forge a strong link between PPIases and the transcription machinery and suggest a new model for how Ess1/Pin1 controls mitosis. In this model, Ess1 binds and isomerizes the CTD of RNA polymerase II, thus altering its interaction with proteins required for transcription of essential cell cycle genes.
Keywords: CTD/mitosis/PPIase/transcription/WW domain
Introduction
Peptidyl-prolyl cis/trans isomerases (PPIases) catalyze the rotation about the peptide bond preceding proline, a step that can be rate-limiting for the folding of newly synthesized proteins (Hemenway and Heitman, 1993; Schmid, 1995; Fischer et al., 1998). PPIases are also thought to regulate the activity of mature proteins, by promoting assembly or transport of subunits (Rutherford and Zuker, 1994; Hunter, 1998; Gothel and Marahiel, 1999). PPIases can act stoichiometrically rather than catalytically, as in the processing of rhodopsin by the NinaA protein in the Drosophila eye (Baker et al., 1994) and the assembly of infectious virions of human immunodeficiency virus (Luban et al., 1993).
The best-studied PPIases are the cyclophilins and FKBPs, which bind the immunosuppressive drugs cyclosporin A and FK506, respectively, and block T-cell activation (Walsh et al., 1992; Kunz and Hall, 1993; Marks, 1996). In yeast, none of the eight cyclophilins and four FK506-binding proteins (FKBPs) is essential for growth, as shown by individual gene deletion and by deleting all 12 (Dolinski et al., 1997). Members of a third family, termed parvulins after an Escherichia coli protein of the same name, are structurally distinct from the cyclophilins and FKBPs and do not bind either cyclosporin A or FK506 (Rahfeld et al., 1994; Rudd et al., 1995). The first eukaryotic parvulin to be discovered was Ess1 (Hanes, 1988; Hani et al., 1995), which is essential for growth in budding yeast (Hanes et al., 1989).
Ess1 is highly conserved. Homologs isolated from a variety of organisms, including Drosophila (Dodo) and humans (Pin1), substitute for Ess1 in yeast (Lu et al., 1996; Maleszka et al., 1996). Surprisingly, complete removal of the dodo gene from the Drosophila genome yields flies that are both viable and fertile (Maleszka et al., 1996), suggesting that Ess1 homologs are not essential in metazoans, perhaps due to the existence of ESS1/PIN1-related genes such as PIN1-like or hPAR14 (Campbell et al., 1997; Uchida et al., 1999). Indeed, Pin1 homozygous knockout mice are viable and show no overt phenotype (Fujimori et al., 1999).
Ess1 contains an N-terminal WW domain and a C-terminal PPIase domain. The WW domain is a protein–protein interaction module with a pair of invariant tryptophans found in a variety of signaling proteins that bind proline-rich peptides (Staub and Rotin, 1996; Sudol, 1996). The WW domain of Pin1 binds peptides in which phosphorylated serine or threonine precedes proline (Yaffe et al., 1997). The X-ray model of Pin1 shows a two-lobed structure in which the WW and PPIase domains are connected by an unstructured linker (Ranganathan et al., 1997).
Early studies revealed that ess1 mutants have cell cycle defects, possibly in cytokinesis or cell separation (Hanes et al., 1989). Using promoter shut-off experiments in yeast, depletion of Ess1 was shown to cause mitotic arrest and nuclear fragmentation, while in human cells, treatment with PIN1 antisense RNA results in chromatin condensation (Lu et al., 1996). These results, and the fact that PIN1 was originally isolated in a two-hybrid screen with the G2/M-specific kinase NIMA from Aspergillus nidulans, suggested a role in mitosis (Lu et al., 1996).
Biochemical interactions have been detected between Pin1 protein and mitotic phosphoproteins, including Cdc25 phosphatase and Polo-like kinase (Crenshaw et al., 1998; Shen et al., 1998). However, the functional relevance of these interactions has not been established, as in vivo and genetic data are lacking. The PPIase activity of Ess1 has been demonstrated in vitro (Hennig et al., 1998); however, whether Ess1/Pin1 controls the function of cell cycle regulators by direct interaction remains an open question. Pin1 was also found to activate transcription weakly when bound to DNA via GAL4 in mammalian cells (Komuro et al., 1999); however, DNA-bound Ess1 in yeast does not (V.Madelian and S.D.Hanes, unpublished data). Mutations in ESS1 (also called PTF1), are defective in mRNA 3′ end formation when tested with certain reporter constructs (Hani et al., 1999). Finally, Pin1 in Xenopus oocycte extracts was shown to be required for the DNA replication checkpoint (Winkler et al., 2000). In these experiments, Pin1 was important for regulating the G2/M transition, but not for exit from mitosis. Thus, in different systems, Ess1/Pin1 seems to function in distinct ways.
Here, we took a genetic approach to understanding Ess1/Pin1 function. We isolated a large number of ess1 temperature-sensitive (ts) mutants which indicate that both the WW and PPIase domains are critical for in vivo function. The mutants were used for a multicopy suppressor screen to identify proteins in the Ess1/Pin1 pathway. The suppressors identified were not cell cycle regulators, but rather proteins important for RNA polymerase II (RNA pol II) transcription and its regulation. These include YKL005C, a TFIIS-like protein; Fcp1, a C-terminal domain (CTD) phosphatase; Sap30, a member of the Sin3–Rpd3 repression complex; Cth1, a putative transcription factor; and Rpb7, a subunit of RNA pol II. We also show that Ess1 interacts genetically and physically with RNA pol II. The results establish a clear link between this essential prolyl isomerase and the general transcription machinery, and suggest that Ess1 and Pin1 act indirectly in mitosis by controlling the transcription of genes required for cell cycle progression. We also link cyclophilin A, a major target of immunosuppressive drugs, to this pathway, providing the first evidence of cross-talk among different PPIase families. A model for Ess1/Pin1 function is presented.
Results
Isolation of ts ess1 mutations
The ESS1 gene was mutagenized in vitro and transformed into a host strain bearing an ess1::URA3 disruption and an integrated GAL1–PIN1 fusion gene. Viability of the host strain was maintained by growth in galactose-containing medium to drive expression of human PIN1, which substitutes for ESS1 in yeast (Lu et al., 1996). To identify ts mutants, expression of PIN1 was repressed by growth in glucose-containing medium, and cells were replica-plated and incubated at permissive (23°C) and non-permissive (37°C) temperatures. Using this method, 15 ts and three null alleles (no growth at either temperature) of ESS1 were identified (Table I).
Table I. Ess mutations identified in this study.
| Ess1mutation | Residue in Pin1 | Phenotype | Location |
|---|---|---|---|
| S7P | D3 | null | precedes WW |
| W15R | W11 | ts | WW signature Trp |
| G43V | G39 | tsa | WW domain |
| K51E | K46 | tsb | linker |
| C62R | C57 | tsb | PPIase near active site |
| H64P | H59 | null | PPIase, catalyticc |
| L66P | L61 | tsa | PPIase |
| S77P | S72 | tsa | PPIase |
| L94P | I89 | tsa | PPIase, α1 helix |
| T96P | G91 | ts | PPIase, α1 helix |
| D102Y | K97 | null | PPIase, α1 helix |
| F110S | F103 | tsb | PPIase, h.p.d |
| S118L | S111 | tsa | PPIase |
| C120R | C113 | tsb | PPIase, catalyticc |
| S122P | S115 | ts | PPIase |
| G127D | G120 | ts | PPIase |
| A144T | A137 | ts | PPIase |
| H164R | H157 | ts | PPIase, catalyticc |
aSlow growth; bleaky; cputative catalytic residue; dhydrophobic patch on surface.
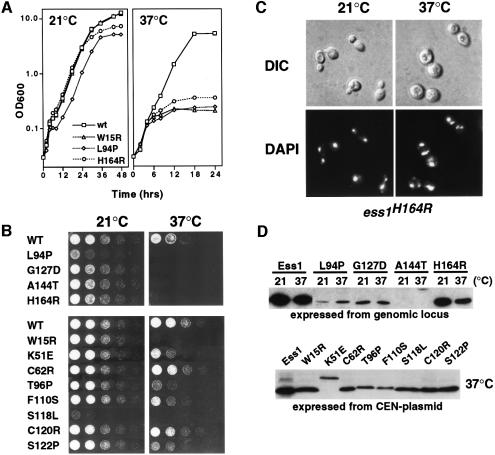
To confirm that the ESS1 mutations caused a ts growth phenotype when expressed from the genomic ESS1 locus, several alleles (L94P, G127D, A144T and H164R) were used to replace the chromosomal ESS1 gene. All were ts for growth (Figure 1A and B). An additional eight ts alleles were tested from centromeric plasmids in an ess1Δ background, and these exhibit various ts growth phenotypes (Figure 1B, lower panel). The arrest kinetics for all mutants are relatively slow; cells do not exhibit first cycle arrest, but instead arrest after several doublings (Figure 1A and data not shown), suggesting either that Ess1 might not be required during each cell cycle or that Ess1 target proteins, once isomerized, are stable. Cells accumulate in mitosis after 2–3 generations (4 h) at the restrictive temperature, and mitotic arrest is maximal by 8–12 h (data not shown).
Fig. 1. Growth properties of ess1ts mutants. (A) Growth of wild-type (W303-1A) and selected mutants in rich medium (YEPD) at permissive and non-permissive temperatures. The ess1W15R allele was on a centromeric plasmid in an ess1Δ background. The ess1L94P and ess1H164R alleles were integrated into the genome. (B) Upper panel: growth of strains containing integrated ess1ts alleles. Cells were grown to mid-log phase in YEPD medium, and serial 1:5 dilutions were spotted onto plates and incubated for 1–2 days at the indicated temperatures. Lower panel: growth of strains carrying plasmid-borne (CEN) copies of ess1ts alleles in an ess1Δ host. Cells were grown and spotted as in the upper panel. (C) Mitotic arrest and nuclear fragmentation of ess1ts mutants. Cells (ess1H164R) were grown to mid-log phase and shifted to 37°C for 8 h. Cells were fixed, stained with DAPI, and visualized under Nomarski optics (DIC) or UV light (DAPI). (D) Western analysis of mutant proteins. Cells were grown at permissive temperature and shifted to 37°C for 4 h prior to harvesting. Equal amounts of total cell lysates were analyzed. Rabbit anti-Ess1 antiserum was used at a 1:4000 dilution.
The terminal phenotype was examined in strains in which the ess1ts alleles were chromosomally integrated (L94P, G127D, A144T and H164R) or expressed from a centromeric plasmid (W15R). At the restrictive temperature, all the mutants undergo mitotic arrest and nuclear fragmentation within 4–8 h (Figure 1C and data not shown). These results were critical, since in earlier experiments these phenotypes appeared only after relatively long incubation times (12–24 h) after promoter shut-off (Lu et al., 1996), and thus might have been an indirect consequence of prolonged arrest. These results establish that ESS1 is required for mitotic progression and that mutations in either the WW or the PPIase domain cause a similar loss-of-function phenotype.
All of the mutant proteins with the exception of Ess1(S7P) were detected in cells grown at the restrictive temperature, 37°C (Figure 1D and data not shown). Several mutant proteins (e.g. W15R, C120R and S122P) were present at near wild-type levels, while others (K51E, F110S and H164R) were reduced 2- to 10-fold. A few were reduced >50-fold at both permissive and non-permissive temperatures (A144T and L94P). Despite the large reduction, the A144T mutant shows robust growth at the permissive temperature, indicating that very low concentrations of Ess1 are sufficient for viability. These results and studies with ESS1 promoter fusions (not shown) suggest that the reduced levels of the mutant proteins per se were not causing the ts phenotype, but that the proteins lack either binding or catalytic activity, or fail to localize correctly in the cell. The mobility of four proteins, K51E, A144T (Figure 1D), S77P and D102Y (not shown), was retarded for unknown reasons.
Structure–function analysis of Ess1
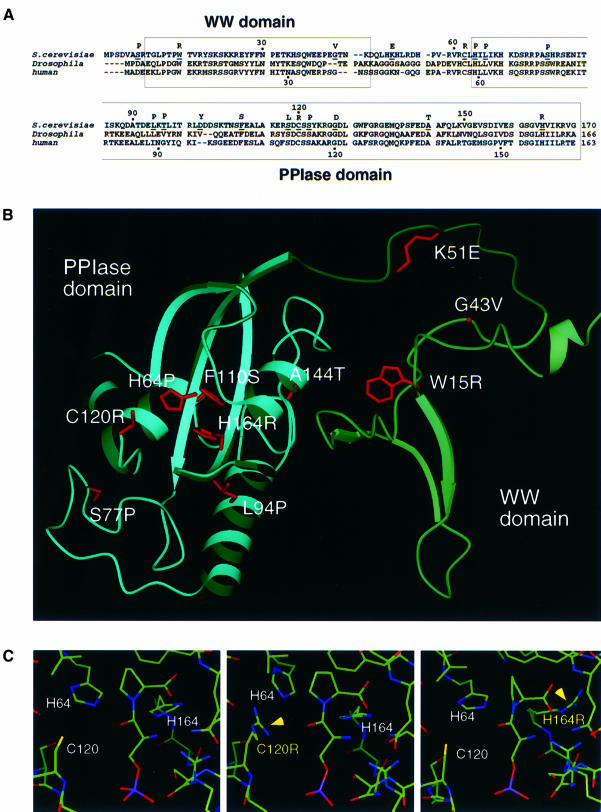
Given that Ess1 and Pin1 are 46% identical in sequence and are functionally interchangeable in yeast, their structures are likely to be similar. We therefore generated a model of Ess1 based on the Pin1 structure (Ranganathan et al., 1997; Figure 2B). The r.m.s. deviation between the Ess1 model and Pin1 structure was <0.66 Å over the WW and PPIase domains.
Fig. 2. Identity of ts and lethal substitutions in Ess1. (A) Alignment of yeast Ess1 and homologs from Drosophila (Dodo) and human (Pin1), showing the position (underlined) and identity of the substitutions. (B) Model of the Ess1 structure based on X-ray crystallographic data of Pin1 (Ranganathan et al., 1997) showing the location of selected mutations. (C) Model of the active site of wild-type Ess1, C120R and H164R mutants, each containing a bound phosphoserine–proline dipeptide in the cis configuration. Although there are minor rearrangements, none appears to preclude binding of the substrate peptide.
The ess1 mutations are distributed throughout the protein (Figure 2A), and selected mutations are highlighted (Figure 2B). One mutation, K51E, maps to the linker that joins the two modules, in a region that is disordered in the X-ray structure of Pin1, suggesting that the linker might become ordered upon binding substrate. Within the WW domain, mutation of the first signature tryptophan (W15R) results in a ts phenotype. This mutation, and G43V, demonstrate the importance of the WW domain for Ess1 mitotic function. In the A144T mutation, substitution of alanine by a polar residue probably disrupts the hydrophobic interface formed between the α4 helix of the PPIase domain and the β2/β3 loop of the WW domain, destabilizing the pocket formed between the two domains where peptide is thought to bind.
Three mutations, L94P (Figure 2B), T96P and D102Y (not shown), map to the long α1 helix in the PPIase domain that is unique to Ess1-type parvulin-class PPIases. This helix projects a hydrophobic face into the pocket between the WW and PPIase domains, and these mutations might interfere with substrate binding, causing the observed growth defect. Ranganathan et al. (1997) proposed that a protein substrate might bind in the WW domain pocket, wrap around Pin1 contacting a solvent-exposed hydrophobic patch consisting of residues I96, F103, M146 and L160, and loop into the PPIase active site. Consistent with this model, the F110S mutation (analogous to F103 in Pin1) would disrupt these interactions by introducing a polar residue into this hydrophobic patch.
Mutations in three of the four residues seen to interact most directly with an Ala–Pro peptide in the active site of Pin1 (H59, C113, S154 and H157; Ranganathan et al., 1997) were identified in our screen (H64P, C120R and H164R; Table I). As expected, mutation of H64, whose counterpart in Pin1 is thought to catalyze isomerization, results in a null phenotype. In contrast, the C120R mutation in Ess1 is not lethal, but confers a ts phenotype. Arginine modeled at this position does not preclude binding of a phosphorylated Ser–Pro dipeptide (Figure 2C); however, it would not allow formation of the covalent intermediate proposed for C113 in Pin1 (Ranganathan et al., 1997). Thus, Ess1 might catalyze isomerization by a mechanism independent of this cysteine intermediate.
Finally, the Ess1 H164R mutant is ts for growth. The equivalent histidine (H159) in Pin1 stabilizes the covalent intermediate formed with substrate peptide. Indeed, arginine modeled at this position does not disrupt the overall architecture of the active site (Figure 2C) and the guanido group of arginine might partially substitute for the imido nitrogen of histidine, allowing sufficient isomerase activity at permissive temperature.
Multicopy suppressors of ess1ts mutants suggest a role in transcription
To identify proteins in the Ess1 pathway, we searched for yeast genes that, when present on multicopy plasmids, suppress the lethal phenotype of ess1ts mutants at 37°C. Five suppressors were identified (Figure 3) plus an additional one identified in unrelated experiments (see below). All six suppress at least three different ess1ts mutants: H164R, A144T and L94P. Four of them, YKL005C, CTH1, CPR1 and CaRPB7, suppressed a complete deletion of ESS1 (ess1Δ) (Table II), suggesting that they act downstream of Ess1 or in a parallel pathway.
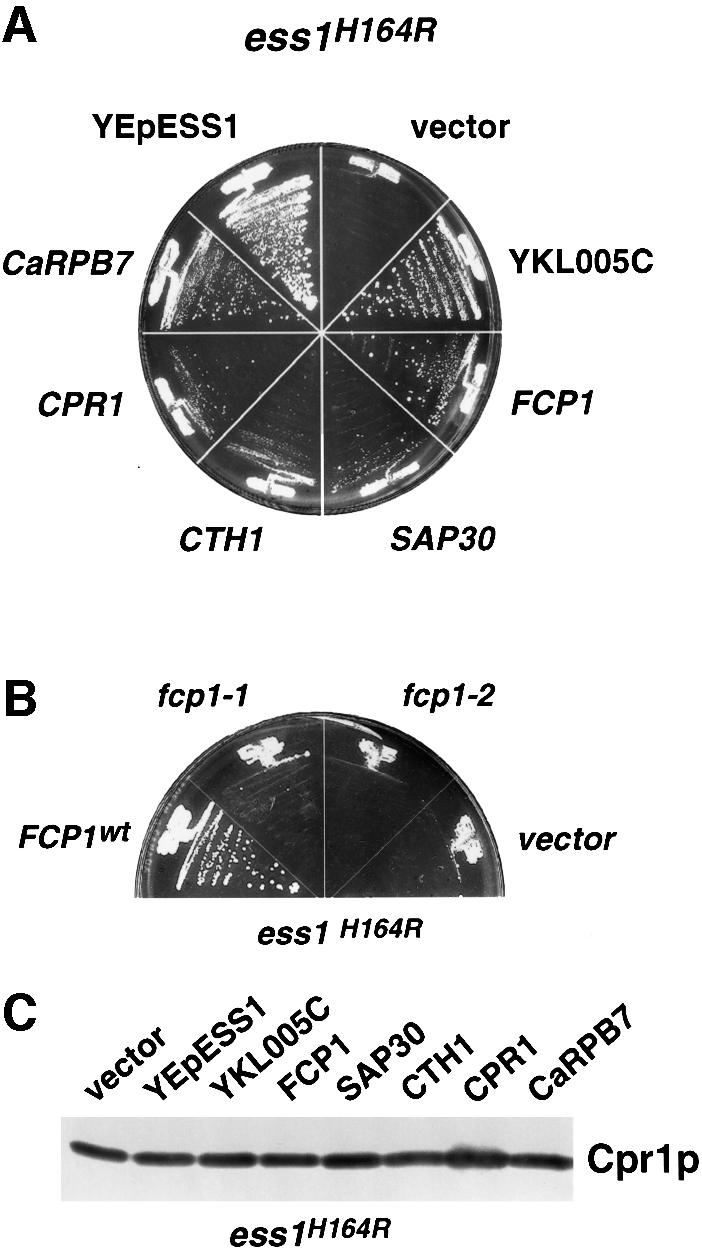
Fig. 3. High-copy suppressors of the lethal ess1 mutant phenotype. (A) Suppression of ess1H164R host cells at 37°C by six different high-copy plasmids carrying the indicated genes. Control plasmids were YEpESS1 or an empty vector (pRS426). CaRPB7 is from Candida albicans. Cells were streaked onto medium that lacked uracil and incubated at 37°C for 5 days. (B) Fcp1 phosphatase activity is required for suppression of ess1ts mutants. Suppression of ess1H164R mutant cells at 37°C was tested using high-copy plasmids (2µ; TRP1) encoding wild-type (FCP1) or mutants alleles (fcp1-1 and fcp1-2) that lack CTD phosphatase activity (Kobor et al., 1999). (C) Western analysis to detect cyclophilin A (Cpr1) in whole-cell lysates from ess1H164R mutant cells carrying the indicated suppressors, or control plasmids, YEpESS1 or pRS426. Cells were grown at permissive temperature and shifted to 37°C for 4 h. Rabbit anti-cyclophilin A serum was used at a 1:3000 dilution.
Table II. Multicopy suppressors of ess1ts mutants.
| Suppressor | Protein/similarities | Suppress ess1Δ? | Knockout phenotype |
|---|---|---|---|
| YKL005C | TFIIS-like | yes | viable |
| FCP1 | CTD phosphatase | no | lethal |
| YMR263W | SAP30, HDAC complex | no | viable |
| CTH1 | human Tis11, txn factor | yes | viable |
| CPR1 | cyclophilin A | yes | viable |
| CaRPB7 | Candida albicans Rpb7 | yes | n.d. |
The references for the knockouts are as follows: YKL005C (this study), FCP1 (Archambault et al., 1997), YMR263W (Zhang et al., 1998), CTH1 (Thompson et al., 1996) and CPR1 (Davis et al., 1992). n.d., not determined.
The suppressor most commonly isolated (11 times) was the yeast open reading frame YKL005C, which predicts a 68 kDa protein with 40% similarity to the Drosophila and human transcription elongation factor TFIIS over about one-third of the protein (amino acids 177–354). The YKL005C protein also contains a PHD finger (amino acids 72–133), a cysteine-rich motif (Aasland et al., 1995) found in Polycomb and Trithorax group proteins, which, in Drosophila participates in global chromatin-mediated regulation of homeotic genes (Paro, 1990; Tamkun, 1995). Based on these similarities, the YKL005C protein might be important for transcription activation (or elongation) by chromatin remodeling.
Another suppressor, YMR263W, which encodes a homolog of human Sap30, a component of the Sin3A–Rpd3 histone deacetylase complex (HDAC), is recruited to promoters by sequence-specific DNA-binding proteins where it represses transcription (Zhang et al., 1998). In the accompanying paper (Arévalo-Rodríguez et al., 2000), Ess1 was shown to bind Sap30 indirectly via other members of this complex (Sin3–Rpd3) and to antagonize HDAC activity. Both Sin3 and Rpd3 are Ser/Thr–Pro rich, suggesting that they might be targets of Ess1-dependent isomerization. Suppression of ess1 mutants by Sap30 is likely to result from a reduction in Sin3–Rpd3 HDAC activity by a titration effect.
Two suppressors are essential components of the RNA pol II complex itself. The first, FCP1, encodes a CTD phosphatase (Archambault et al., 1998; Kobor et al., 1999). Dephosphorylation of the CTD is required for recycling of RNA pol II for re-initiation, and is associated with paused polymerase complexes, which require re-phosphorylation to resume elongation (Dahmus, 1996). Suppression of ess1ts mutants by FCP1, and the observed interaction between Ess1 and the CTD (see below) suggest that isomerization of the CTD by Ess1 might promote CTD dephosphorylation by Fcp1. In ess1 mutants, high concentrations of Fcp1 would overcome the need for isomerization, allowing dephosphorylation of the CTD. According to this model, the phosphatase activity of Fcp1 should be required for suppression. Indeed, two fcp1 mutants that fail to dephosphorylate the RNA pol II CTD in vivo (Kobor et al., 1999) do not suppress ess1ts mutants (Figure 3B).
The second suppressor that is a component of the RNA pol II complex is Rpb7, a σ factor-like protein that associates with the holoenzyme during stress, such as starvation and heat shock, and is required for initiation (McKune et al., 1993; Khazak et al., 1995). The version of Rpb7 we identified is encoded by a gene from the pathogenic fungus Candida albicans (G.Devasahayam, V.Chaturvedi and S.D.Hanes, unpublished). Curiously, neither the Saccharomyces cerevisiae nor the human RPB7 genes suppressed ess1ts mutants in our assays (not shown). Perhaps this is because yeast Rpb7 requires Rpb4 for function (McKune et al., 1993), or because Rpb7 from C.albicans functions more efficiently at 37°C, which is favorable for C.albicans.
The last transcription-related suppressor is CTH1, which encodes a CCCH-type Zn2+ finger protein that is a homolog of the mammalian serum-induced protein Tis11 (Varnum et al., 1991). Tis11 activates transcription (Ma and Herschman, 1995), but was also shown to translocate to the cytoplasm, bind AU-rich sequences in the 3′ ends of mRNA and promote mRNA degradation (Carballo et al., 1998). When overexpressed, however, Tis11 stabilizes tumor necrosis factor-α (TNF-α) mRNA (Lai et al., 1999). Overexpression of CTH1 in yeast might stabilize mRNAs important for cell cycle progression, thereby obviating the need for Ess1 to promote transcription, or compensating for defects in 3′ end formation (Hani et al., 1999).
Finally, we identified CPR1, which encodes the prolyl isomerase cyclophilin A, as a suppressor of ess1ts mutants. This was a surprise given that the substrate specificities of parvulin-class PPIases (Ess1) and cyclophilins are distinct (Hennig et al., 1998), and that cyclophilin A is cytoplasmic and does not contain a WW domain, which is thought to be critical for Ess1/Pin1 binding to target proteins (Lu et al., 1999; Morris et al., 1999). The PPIase activity of cyclophilin A is required for this suppression (Arévalo-Rodríguez et al., 2000).
It is possible that the five suppressors implicated in transcription suppress ess1ts mutations by increasing expression of cyclophilin A. To test this idea, northern and western analyses were used to measure steady-state levels of CPR1 mRNA and protein in ess1ts mutants carrying each of the multicopy suppressors. CPR1 mRNA and protein levels were not significantly altered (Figures 6B and 3C). More importantly, three of the five genes (YKL005C, FCP1 and CaRPB7) suppressed ess1ts mutants in strains in which CPR1 is disrupted (Arévalo-Rodríguez et al., 2000). These results indicate that suppression is not due to increased transcription of CPR1.
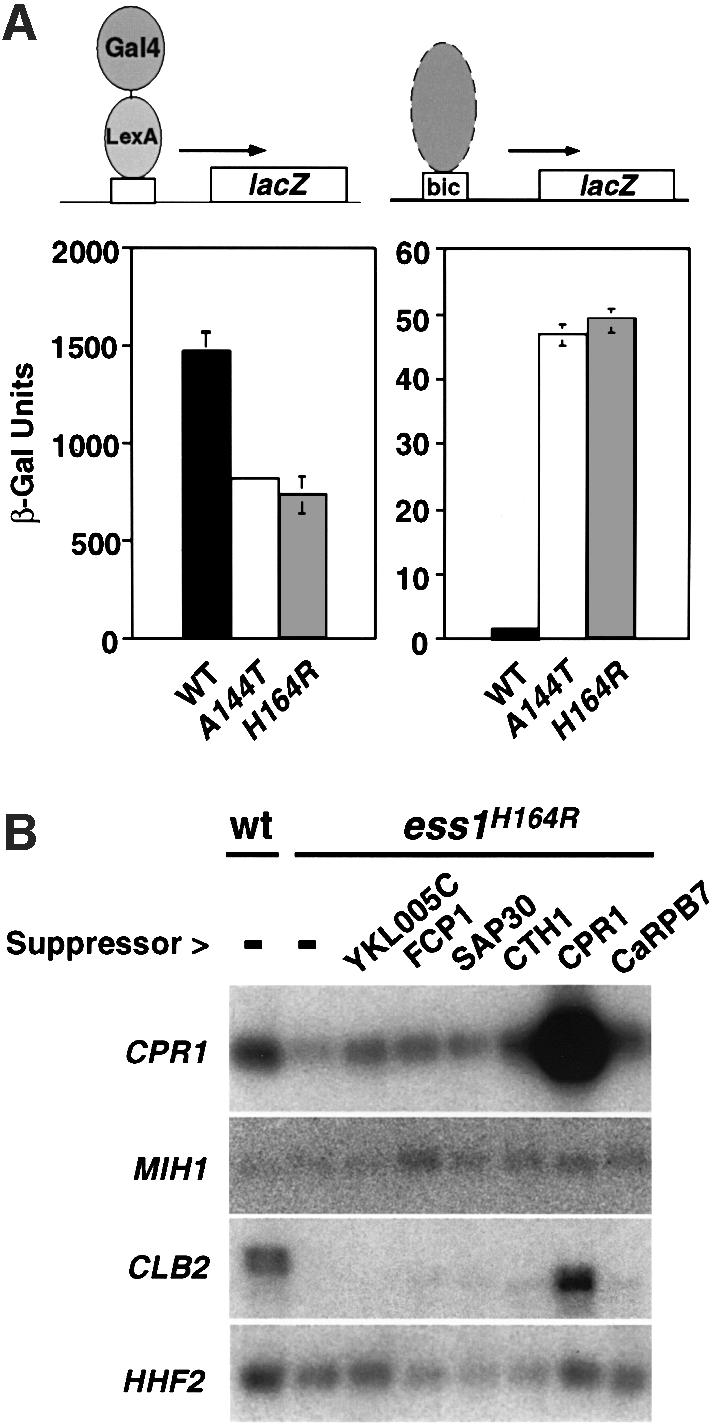
Fig. 6. Selective effects on transcription in ess1 mutant cells. (A) LacZ reporter gene expression in yeast. Left: wild-type or ess1ts mutant yeast were transformed with a plasmid expressing LexA–Gal4 (pSH17-4; Hanes and Brent, 1989), and a 4×-LexA operator-lacZ reporter construct (pSH18-8; S.D.Hanes and R.Brent, unpublished) at 23°C. β-galactosidase activity was measured in liquid cultures of five independent isolates. Similar results were obtained at 37°C (not shown). Right: cells were transformed with a Bicoid-site lacZ reporter (pWZ11-1; Zhu and Hanes, 2000) and treated as above, except that cells were incubated at 35°C. The differences between wild-type and the ess1 mutants were less pronounced at 23°C (not shown). (B) Northern analysis of genes encoding mitotic regulators in ess1H164R mutant cells. Wild-type cells or ess1H164R mutant cells with control vector pRS426 (–) or the indicated multicopy suppressors were grown in selective medium and shifted to 37°C for 4 h prior to harvesting. A 6 µg aliquot of total RNA was loaded per lane. Duplicate blots were hybridized with random primed 32P-labeled probes for CPR1 (cyclophilin A) and MIH1 (Cdc25 phosphatase), or CLB2 (cyclin B) and HHF2 (histone H4).
What do the suppressors of ess1 have in common? With the exception of cyclophilin A, all are likely to function during the various stages of transcription. Work from many laboratories has shown that all these stages, including initiation, elongation, termination and mRNA processing, require the CTD of RNA pol II large subunit (Rpb1) (Carlson, 1997; Corden and Patturajan, 1997). Results presented below suggest that the suppressors work by overcoming defects in CTD function resulting from lack of Ess1.
Ess1 interacts physically with the CTD of RNA polymerase II
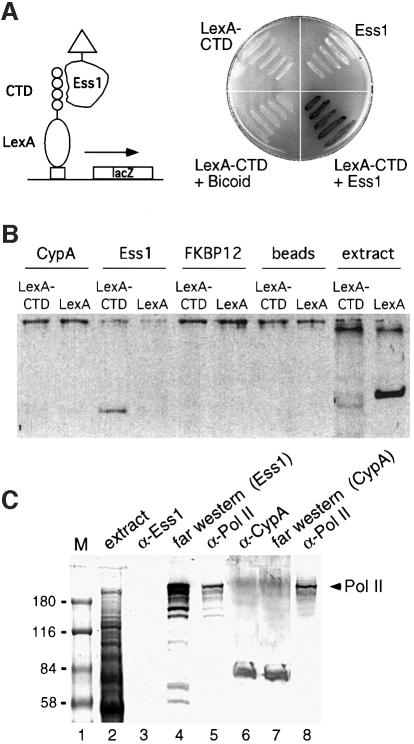
The CTD of RNA pol II carries multiple copies (26 in yeast; 52 in humans) of the heptapeptide YSPTSPS, or close variants. Previous work showed that Pin1 binds to peptides that contain phosphorylated Ser–Pro motifs (Yaffe et al., 1997). Both Ser2 and Ser5 within the YSPTSPS motif are known to be phosphorylated in vivo (Dahmus, 1996; Yuryev and Corden, 1996; Ho and Shuman, 1999), resulting in two Pin1-binding sites per repeat. We tested whether Ess1 interacts with the CTD using a two-hybrid assay. Activation-tagged Ess1 interacts strongly with a LexA fusion protein carrying nine repeats of the CTD heptapeptide motif, whereas control proteins did not (Figure 4A and data not shown).
Fig. 4. Ess1 interacts with the CTD of RNA pol II in vivo and in vitro. (A) A schematic of the two-hybrid experiment is shown on the left. Yeast cells expressing the indicated proteins were streaked on an X-gal-containing plate and incubated overnight at 30°C (right). The LexA1–87–CTD bait protein contains nine repeats of the YSPTSPS motif and interacts with activation domain-tagged Ess1 but not with the control protein, Bicoid. Neither LexA1–87–CTD nor Ess1 alone activates transcription. (B) Ess1, but not cyclophilin A, interacts with LexA1–87–CTD in vitro. Cyclophilin A- (CypA), Ess1- or FKBP12-conjugated affinity beads or control beads were reacted with total protein extracts from cells expressing either a LexA1–87–CTD fusion protein or intact LexA1–202. Bound proteins were eluted and analyzed by western blotting with LexA antisera. Extract denotes whole-cell lysates not reacted with affinity beads. (C) Far-western analysis shows that Ess1 reacts with a protein that co-migrates with hyperphosphorylated RNA pol II. Lanes 1 and 2 are marker proteins and Coomassie-stained total yeast extracts, respectively. Lanes 4 and 7 are far-western samples using His-tagged Ess1 or CypA proteins as probes, and detected using the cognate antibodies. Lanes 3, 5, 6 and 8 are standard western blots reacted with the indicated antisera.
Ess1 also interacted with the CTD in vitro as shown by affinity pull-down assays. Ess1-conjugated beads pull down LexA–CTD (but not LexA alone) from yeast cell extracts, whereas beads conjugated with cyclophilin A or FKBP12 did not (Figure 4B). These results show that Ess1 interacts with the CTD, but do not distinguish whether this interaction is direct or indirect. Far-western analysis indicates that this interaction is direct. Proteins from whole-cell extracts were fractionated on SDS–polyacrylamide gels, transferred to nitrocellulose and incubated with purified His-tagged Ess1. Filters were washed and reacted with antibodies to Ess1. The major Ess1-reactive band co-migrates with RNA pol II (Figure 4C). Control blots using antibodies to Ess1 or RNA pol II show that anti-Ess1 serum does not cross-react with RNA pol II. Since the antibodies used against pol II are specific for the phospho-CTD form of the enzyme, these results suggest that Ess1 interacts with the phosphorylated form (II0) of RNA pol II. Biochemical interaction between Ess1 and the phospho-CTD of RNA pol II has also been reported by Morris et al. (1999). Analogous experiments with cyclophilin A failed to detect an interaction with RNA pol II (Figure 4C).
ESS1 interacts genetically with RNA polymerase II and SRB2
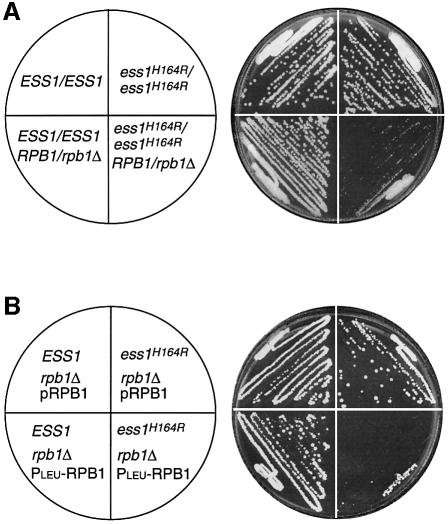
If the interaction between Ess1 and the CTD of RNA pol II is biologically meaningful, then we should detect genetic interactions between ESS1 and RPB1, the gene encoding the large subunit of RNA pol II. To test this, three approaches were used. First, we deleted one copy of the RPB1 gene in isogenic wild-type and ess1ts mutant diploid strains. These strains were tested for growth at permissive temperature. As shown in Figure 5A, homozygous ess1 mutant cells bearing one copy of RPB1 (ess1H164R/ess1H164R RPB1/rpb1Δ) were severely growth inhibited compared with control strains. This growth defect is not seen in cells with either one or two copies of wild-type ESS1 (Figure 5A and data not shown). Thus, ess1ts mutations are synthetically lethal with RPB1 heterozygous mutations.
Fig. 5. ESS1 shows a genetic interaction with RPB1. (A) The ess1H164R mutant is synthetically lethal with a reduced dosage of the RPB1 gene. Cells of the indicated genotype (W303-1A background) were streaked to rich medium and grown for 3 days at permissive temperature (30°C). (B) The ess1H164R mutant is synthetically lethal with reduced levels of RPB1 expression. Cells of the indicated genotype (W303-1A background) were streaked to selective medium and grown for 3 days at permissive temperature (25°C). pRPB1 is a CEN-ARS (URA3) plasmid that expresses the large subunit of RNA pol II from its natural promoter. PLEU-RPB1 is a CEN-ARS (TRP1) plasmid (also known as pLEU-RPO21; Archambault et al., 1996) that expresses the large subunit of RNA pol II from the repressible LEU2 promoter. The plate shown contains 2 mM leucine, threonine and isoleucine, which represses the PLEU promoter, driving low-level expression of RPB1.
Secondly, we tested the sensitivity of ess1ts haploid cells to lowered levels of RPB1 expression using a repressible promoter strategy (Kobor et al., 1999). Strains were generated in which RPB1 was deleted from the chromosome in wild-type and ess1ts cells. Since RPB1 is an essential gene, viability was maintained by a plasmid-borne copy of RPB1. However, when ess1ts mutant cells carrying the plasmid PLEURPB1 were shifted to repressing conditions (2 mM leucine) where RPB1 expression is low, a severe growth defect was observed (Figure 5B). Wild-type cells were only modestly affected, and ess1ts cells bearing a control plasmid that expresses RPB1 from its own promoter (pRPB1) showed no growth defects (Figure 5B).
Thirdly, we found that ess1ts mutants are hypersensitive to the dominant-negative effects of CTD truncation mutants. This was discovered in efforts to test whether ess1ts mutations enhance (or suppress) the phenotypes reported for CTD truncations and substitutions (West and Corden, 1995). Attempts to generate the appropriate strains were thwarted because introduction of CTD mutants proved toxic in ess1ts cells at permissive temperature, even though they have a normal chromosomal RPB1 gene (Table III, upper). This toxicity was reversed by co-transformation with wild-type ESS1 (Table III, lower).IV The simplest explanation is that RNA pol II function is already compromised in ess1 mutants, and therefore cells are hypersensitive to the dominant-negative effects of CTD mutations. Note that a similar dominant-negative effect was reported for some srb mutants (Yuryev and Corden, 1996), which were originally isolated as suppressors of CTD mutants, and are known to play important roles in RNA pol II function (Thompson et al., 1993).
Table III. Ess1 mutant strains are hypersensitive to CTD truncation alleles.
| Vector | pRPB1 | pWT0 | pWT9 | pA5(15) | |
|---|---|---|---|---|---|
| Wild type | +++ | +++ | +++ | +++ | +++ |
| H164R | ++ | ++ | – | – | – |
| A144T | ++ | ++ | – | – | – |
| Wild type + pESS1 | +++ | +++ | +++ | +++ | +++ |
| H164R + pESS1 | ++ | ++ | ++ | ++ | ++ |
| A144T + pESS1 | ++ | + | ++ | ++ | ++ |
Wild-type or ess1ts mutants (H164R, A144T) were transformed with equal amounts (∼0.2–0.5 µg) of plasmids encoding wild-type or mutant forms of RNA pol II large subunit and grown for 3 days at 30°C. Strains in the bottom half of the table were co-transformed with pESS1 (CEN, HIS3), which encodes wild-type ESS1. Transformation efficiency is summarized as follows: +++ (>10 000); ++ (1001–10 000); + (100–1000); – (<100). Plasmid pRPB1 expresses wild-type Rpb1. A total of eight different CTD mutant plasmids was tested and the results were identical. Representative data for three mutants are shown. Plasmids pWT0, pWT7, pWT8 and pWT9 express truncated RNA pol II derivatives bearing zero, seven, eight or nine copies of the wild-type heptapeptide repeat (YSPTSPS), respectively. Plasmids pA2(18), pE2(15), pA5(15) and pE5(18) express RNA pol II bearing the indicated number of repeats in which Ser2 or Ser5 of the heptad repeat is mutated to alanine (A) or glutamic acid (E). The RPB1 plasmids have been described (West et al., 1995; Yuryev et al., 1996).
Table IV. Ess1 mutants are synthetically lethal with srb2 mutants.
| Relevant genotype | 5-FOARcolonies/total | CaESS1 plasmid lossa |
|---|---|---|
| ESS1 SRB2 | 742/10 992 | 68 |
| ESS1 srb2Δ | 620/7480 | 83 |
| ess1H164R SRB2 | 286/25 050 | 11 |
| ess1H164R srb2Δ | 0/39 690 | 0 |
Strains of the indicated genotype carrying a functional ESS1 gene on a CEN-based URA3 plasmid were grown overnight in the absence of selection (in YEPD) and plated to 5-FOA-containing medium. All growth was at permissive temperature (30°C). 5-FOAR colonies were scored versus the total number of colony-forming units (c.f.u.) as measured by duplicate platings to YEPD. The fraction of 5-FOAR colonies reveals the frequency of plasmid loss (a), which is given per 1000 c.f.u. The C.albicans ESS1 gene (CaESS1), which is functionally homologous to S.cerevisiae ESS1 (G.Devasahayam, V.Chaturvedi and S.D.Hanes, unpublished), was used to prevent unwanted recombination of the plasmid-borne copy of ESS1 and the chromosomal ess1H164R allele.
The above experiments suggest that Ess1 positively regulates RNA pol II function. If so, then ESS1 mutations should be synthetically lethal with mutations in other genes that positively regulate RNA pol II, such as SRB2, which encodes a component of the mediator complex that interacts with the CTD (Thompson et al., 1993). Indeed, this is true, as shown using a plasmid loss assay in which double mutant cells (ess1ts srb2Δ) were unable to lose an ESS1-containing plasmid even at permissive temperature (Table I). This result provides additional genetic linkage between ESS1 and the transcription machinery.
Ess1 mutants show selective effects on gene transcription
Genetic and physical evidence indicate that Ess1 functions in transcription. If true, then the expression of some or all genes might be affected in ess1 mutants. We therefore measured expression levels of lacZ reporter genes in wild-type and ess1 mutant cells. The results indicate that one reporter, whose expression was driven by the LexA–GAL4 activator, is dependent on Ess1. Mutant cells show an ∼2-fold reduction in β-galactosidase activity (Figure 6A, left). This effect was not due to a change in the relative amounts of intracellular LexA–GAL4 protein as detected by immunoblotting (not shown). In contrast, another reporter, which carried upstream Bicoid-binding sites, was stimu lated nearly 40-fold in ess1 mutants (Figure 6A, right), suggesting that Ess1 helps keep this gene silent. Activation of this reporter was by an endogenous yeast activator, since Bicoid was not expressed in these experiments. The results show that different genes have different requirements for Ess1.
We also examined the expression of two G2/M-specific genes in wild-type and ess1ts mutant cells grown at the restrictive temperature (Figure 6B). Relative to the control gene HHF2 (histone H4), the expression of mitotic regulator MIH1 (cdc25 phosphatase) was not changed significantly in ess1 mutant cells, whereas CLB2 (cyclin B) levels were dramatically reduced. With the exception of CPR1, the multicopy suppressors did not restore CLB2 expression to wild-type levels. These results suggest that Ess1 is important for the expression of a subset of genes, but not all genes. Note that in budding yeast CLB2 is not essential (Fitch et al., 1992), so it is unlikely that this reduction alone is responsible for the mitotic arrest observed in ess1 mutants.
Discussion
The results presented here provide genetic and biochemical evidence that Ess1, a parvulin-class PPIase, interacts with the transcription machinery and that this interaction is essential for cell viability. Isomerization of the CTD provides a mechanism by which Ess1 might regulate the interaction of RNA pol II with accessory proteins required for initiation, elongation, termination and splicing. Genetic suppressors of ess1 mutants encode proteins involved in several of these processes, suggesting that Ess1 function might be required at multiple steps in transcription.
This work is consistent with the observations by Hani et al. (1995, 1999) that ess1 (ptf1) mutants are defective in mRNA processing. However, this work does not support the current model that Ess1/Pin1 proteins control mitosis via direct interaction with cell cycle regulatory proteins, and suggests either that Ess1/Pin1 proteins are multifunctional, or that biochemical interactions with mitotic regulatory proteins might not be physiologically relevant. Instead, we propose that Ess1 and the mammalian homolog Pin1 are important for transcription of genes whose products can be rate-limiting for mitosis in eukaryotic cells. Using whole-genome approaches available in S.cerevisiae, it should be possible to determine the identity of those target genes.
Both the WW and PPIase domains are necessary for mitotic function
Our mutational analyses provide insight into the function of eukaryotic parvulin-class prolyl isomerases. Several of the mutants support predictions made based on the structural model of Pin1. However, some of the results were unexpected. For example, two of the three predicted PPIase catalytic residues could be replaced (C120R and H164R) without abolishing the mitotic activity of Ess1 at permissive temperature. This was not expected given their absolute conservation among Ess1/Pin1 homologs, and their observed interaction with the substrate peptide in the co-crystal. These results suggest that isomerization occurs by a mechanism different from that previously proposed (Ranganathan et al., 1997), or that efficient isomerization is not required for mitotic function under some conditions.
Ess1 suppressors and interactions with RNA pol II suggest a role in transcription
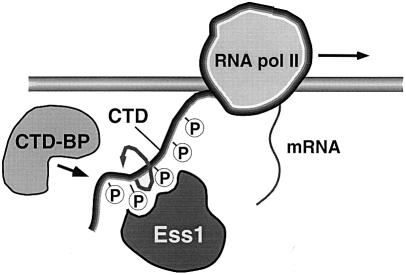
All of the multicopy suppressors of ess1ts mutants except one are known or suspected components of the large complex known as the transcriptosome. This finding and our demonstration that Ess1 interacts genetically and physically with RNA pol II suggest a model depicted in Figure 7.
Fig. 7. Model for the action of Ess1 in transcription. Ess1 binds the phosphorylated CTD of RNA pol II, catalyzing its isomerization, thus acting as a regulatory switch for loading of proteins required for initiation, elongation, termination and 3′ end formation. In this model, Ess1 coordinates the sequential steps of transcription by changing the three-dimensional structure of the CTD, altering the affinity of protein–CTD interactions. Binding of Ess1 to the CTD would be regulated by phosphorylation–dephosphorylation by CTD kinases and CTD phosphatases (e.g. Fcp1). Ess1 might instead work stoichiometrically; Ess1 would sterically block (or promote) binding of RNA pol II-associated proteins to the phosphorylated CTD.
In this model, Ess1 would act catalytically to isomerize the CTD of RNA pol II, altering its structure to promote the binding or dissociation of CTD-interacting proteins that function in the sequential steps of transcription and mRNA processing. The signal for Ess1–CTD interaction would be phosphorylation at serines 2 and/or 5 of the heptad repeat, thereby increasing its affinity for the WW domain of Ess1, which would effectively tether the PPIase domain to the CTD. Isomerization of the CTD would then trigger dephosphorylation by Fcp1, allowing other proteins to bind to (or dissociate from) the CTD. Interaction of Ess1 with the CTD might explain why Hani et al. (1999) observed mRNA 3′-processing defects in ess1 mutants; proteins involved in 3′-processing are likely to require interaction with the CTD.
In our model, Fcp1 would act after Ess1 on the CTD. Consistent with this idea, we find that whereas high-copy expression of FCP1 suppresses ess1 mutants (Figure 3), the reverse is not true. High-copy expression of ESS1 did not suppress fcp1 mutants (data not shown), indicating that FCP1 is genetically downstream of ESS1.
Ess1 might not be the only prolyl isomerase that associates with RNA pol II via the CTD. A set of conserved proteins called CASPs (CTD-associated SR-like proteins), which are thought to bind mRNA and to be important for mRNA processing, has been shown to interact with the CTD. One of these proteins, mammalian CASP10, contains an N-terminal cyclophilin-like domain, shown to have PPIase activity in vitro (Bourquin et al., 1997). As yet, no regulatory activity on RNA pol II by CASP10 has been described. A recent report found an interaction between human Pin1 and the CTD, although no functional significance was established (Albert et al., 1999).
A role for Ess1 in chromatin remodeling and cell cycle progression
It is well established that chromatin remodeling by histone deacetylation is important for control of cell cycle genes (Brehm et al., 1998). The results presented here and in Arévalo-Rodríguez et al. (2000) also connect Ess1 to the Sin3–Rpd3 HDAC, and suggest that Ess1 antagonizes HDAC activity. Loss of Ess1 function would increase deacetylase activity and repression of cell cycle genes, leading to mitotic arrest. Interestingly, yeast mutants lacking an essential protein called Esa1 show a mitotic arrest phenotype very similar to that seen in ess1 mutants (Clarke et al., 1999), and Esa1 was shown to be a histone acetyl-transferase (HAT). Thus, in both ess1 and esa1 mutants, histone deacetylation would be favored over acetylation, perhaps leading to similar misregulation of genes required for mitosis.
Parvulin- and cyclophilin-class PPIases act in concert to promote mitosis
In yeast, as in other eukaryotes, the natural targets of the cyclophilins have been difficult to identify, because mutations of their genes, alone or in combination, have little or no phenotype (Dolinski et al., 1997). Here, we identify the major cytoplasmic cyclophilin, cyclophilin A, as a suppressor of ess1 mutants. No cyclophilins have previously been implicated in essential cell cycle activities. The simplest interpretation of our results is that overexpression of cyclophilin A suppresses ess1 mutants, because at higher concentrations it binds certain Ess1 substrates non-specifically. Since overexpression of cyclophilin A suppresses ess1 deletion mutants, it suggests that cyclophilin A and Ess1 act in parallel pathways and bind to common targets, such as Sin3–Rpd3 HDAC, as shown in the accompanying paper (Arévalo-Rodríguez et al., 2000). The idea is supported by the fact that ess1ts cpr1Δ double mutants are synthetic lethal at permissive temperature and that ess1ts mutants are cyclosporin A sensitive (Arévalo-Rodríguez et al., 2000; X.Wu and S.D.Hanes, unpublished data). Thus, in addition to linking the parvulin-class Ess1/Pin1 prolyl isomerase to transcriptional regulatory mechanisms, these studies provide the first evidence for cross-talk among different families of prolyl isomerases.
Materials and methods
Plasmids and strains
DNA manipulations were performed according to standard procedures (Ausubel et al., 1987). Details of plasmid construction and gene knockouts of YKL005C, RPB1 and SRB2 are available in the Supplementary data or upon request. Chromosomal integration of ess1ts mutations into strain W303-1A (MATa ura3-1 leu2-3,112 trp1-1 can1-100 ade2-1 his3-11,15 [psi+]; R.Rothstein) was carried out by integration/excision using YIpEss1ts plasmids. The presence of the ess1ts alleles in the genome was confirmed by phenotype and by PCR amplification and DNA sequencing.
Isolation of ts ess1 mutants
ESS1 was mutagenized by error-prone PCR and transformed into yeast using gapped-plasmid methodology (Muhlrad et al., 1992) (see Supplementary data). From a total of 6600 transformants, 15 ts alleles bearing single substitutions were identified.
Molecular modeling
Sequence alignment and initial homology modeling of Ess1, from the X-ray crystal structure of Pin1 (Ranganathan et al., 1997) with a bound Ser–Pro dipeptide, were carried out using Look (Molecular Applications Group), with additional modeling using Insight II and Discover 3 (MSI). Energy minimizations were performed as described in the Supplementary data.
Identification of multicopy suppressors
Yeast strain YGD-ts22W, a W303-1A derivative bearing the ess1H164R allele, was transformed with a yeast genomic library and suppressors identified by selecting for Ura+ transformants at 37°C. From an estimated 2 × 105 Ura+ transformants, 156 clones grew at 37°C. To confirm that the suppression was plasmid linked, cells were replica-plated to 0.1% 5-fluoro-orotic acid (5-FOA) at 37°C. From the 113 5-FOAS clones, plasmids were rescued and restriction analysis and PCR showed that 80 carried inserts, 25 of which were ESS1. Eighteen of the remaining clones suppressed when retested, and by DNA sequencing they fell into five classes. Subcloning and retransformation identified the gene responsible for the suppression. Suppression of an ess1 deletion mutant was tested by tetrad dissection and plasmid curing, and the results concurred.
Interaction of Ess1 with the CTD
The two-hybrid assays were performed as described (Gyuris et al., 1993) using pSH18-34, which contains eight LexA operators driving GAL1-lacZ (S.D.Hanes and R.Brent, unpublished). Expression of the LexA1–87–CTD was verified by western blotting using anti-LexA antisera (R.Brent). Details of the affinity pull-down experiments and far-western analysis are available in the Supplementary data. For far-western analysis, anti-phospho-CTD antibodies were a gift of Arno Greenleaf.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal online.
Acknowledgments
Acknowledgements
We thank Roger Brent, Howard Bussey, Barak Cohen, Russ Finley, James Friesen, Erica Golemis, Jack Greenblatt, Jenó Gyuris and Rod Rothstein for plasmids and yeast strains, Clara Alarcon for the yeast genomic library, Arno Greenleaf for reagents and advice, and David Porter for cyclosporin A. We are grateful to Jeff Bell, Chris Waddling, Ivan Auger and Patrick VanRoey for molecular modeling, Mark Verdecia, Joe Noel and Arno Greenleaf for communicating unpublished results, and Dave Burz, Randy Morse, Vergine Madelian, Dilip Nag and Wencheng Zhu for helpful discussions and/or comments on the manuscript. We thank the Wadsworth Center Core Facilities for oligonucleotide synthesis, DNA sequencing, media preparation, antibody production and photography. This work was supported by Health Research Inc. (S.D.H.), the Howard Hughes Medical Institute (J.H.), and by grants from the National Institutes of Health to M.E.C. and S.D.H.
References
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Albert A., Lavoie,S. and Vincent,M. (1999) A hyperphosphorylated form of RNA polymerase II is the major interphase antigen of the phosphoprotein antibody MPM-2 and interacts with the peptidyl-prolyl isomerase Pin1. J. Cell Sci., 112, 2493–2500. [DOI] [PubMed] [Google Scholar]
- Archambault J., Jansma,D.B. and Friesen,J.D. (1996) Underproduction of the largest subunit of RNA polymerase causes temperature sensitivity, slow growth, and inositol auxotrophy in Saccharomyces cerevisiae. Genetics, 142, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Chambers,R.S., Kobor,M.S., Ho,Y., Cartier,M., Bolotin, D., Andrews,B., Kane,C.M. and Greenblatt,J. (1997) An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 14300–14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Pan,G., Dahmus,G.K., Cartier,M., Marshall,N., Zhang,S., Dahmus,M.E. and Greenblatt,J. (1998) FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem., 273, 27593–27601. [DOI] [PubMed] [Google Scholar]
- Arévalo-Rodríguez M., Cardenas,M.E., Wu,X., Hanes,S.D. and Heitman,J. (2000) Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3–Rpd3 histone deacetylase. EMBO J., 19, 3739–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R., Moore,D., Seidman,J., Smith,J. and Struhl,K. (1987) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY. [Google Scholar]
- Baker E.K., Colley,N.J. and Zuker,C.S. (1994) The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J., 13, 4886–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin J.P., Stagljar,I., Meier,P., Moosmann,P., Silke,J., Baechi,T., Georgiev,O. and Schaffner,W. (1997) A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res., 25, 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Campbell H.D., Webb,G.C., Fountain,S. and Young,I.G. (1997) The human PIN1 peptidyl-prolyl cis/trans isomerase gene maps to human chromosome 19p13 and the closely related PIN1L gene to 1p31. Genomics, 44, 157–162. [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai,W.S. and Blackshear,P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science, 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Carlson M. (1997) Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol., 13, 1–23. [DOI] [PubMed] [Google Scholar]
- Clarke A.S., Lowell,J.E., Jacobson,S.J. and Pillus,L. (1999) Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol., 19, 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J.L. and Patturajan,M.A. (1997) CTD function linking transcription to splicing. Trends Biochem. Sci., 22, 413–416. [DOI] [PubMed] [Google Scholar]
- Crenshaw D.G., Yang,J., Means,A.R. and Kornbluth,S. (1998) The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J., 17, 1315–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M.E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem., 271, 19009–19012. [DOI] [PubMed] [Google Scholar]
- Davis E.S., Becker,A., Heitman,J., Hall,M.N. and Brennan,M.B. (1992) A yeast cyclophilin gene essential for lactate metabolism at high temperature. Proc. Natl Acad. Sci. USA, 89, 11169–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K., Muir,S., Cardenas,M. and Heitman,J. (1997) All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G., Tradler,T. and Zarnt,T. (1998) The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett., 426, 17–20. [DOI] [PubMed] [Google Scholar]
- Fitch I., Dahmann,C., Surana,U., Amon,A., Nasmyth,K., Goetch,L., Byers,B. and Futcher,B. (1992) Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell, 3, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori F., Takahashi,K., Uchida,C. and Uchida,T. (1999) Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G0 arrest. Biochem. Biophys. Res. Commun., 265, 658–663. [DOI] [PubMed] [Google Scholar]
- Gothel S.F. and Marahiel,M.A. (1999) Peptidyl-prolyl cis–trans isomerases, a superfamily of ubiquitous folding catalysts. Cell. Mol. Life Sci., 55, 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hanes S.D. (1988) Isolation, sequence and mutational analysis of ESSI, a gene essential for growth in Saccharomyces cerevisiae. PhD thesis, Brown University, Providence, RI. [DOI] [PubMed] [Google Scholar]
- Hanes S.D. and Brent,R. (1989) DNA specificity of the Bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell, 57, 1275–1283. [DOI] [PubMed] [Google Scholar]
- Hanes S.D., Shank,P.R. and Bostian,K.A. (1989) Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast, 5, 55–72. [DOI] [PubMed] [Google Scholar]
- Hani J., Stumpf,G. and Domdey,H. (1995) PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett., 365, 198–202. [DOI] [PubMed] [Google Scholar]
- Hani J., Schelbert,B., Bernhardt,A., Domdey,H., Fischer,G., Wiebauer,K. and Rahfeld,J.U. (1999) Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J. Biol. Chem., 274, 108–116. [DOI] [PubMed] [Google Scholar]
- Hemenway C. and Heitman,J. (1993) Proline isomerases in microorganisms and small eukaryotes. In Allison,A.C., Lafferty,K.J. and Fliri,H. (eds), Immunosuppressive and Antiinflammatory Drugs. The New York Academy of Sciences, New York, NY, pp. 38–46. [DOI] [PubMed] [Google Scholar]
- Hennig L., Christner,C., Kipping,M., Schelbert,B., Rucknagel,K.P., Grabley,S., Kullertz,G. and Fischer,G. (1998) Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry, 37, 5953–5960. [DOI] [PubMed] [Google Scholar]
- Ho C.K. and Shuman,S. (1999) Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell, 3, 405–411. [DOI] [PubMed] [Google Scholar]
- Hunter T. (1998) Prolyl isomerases and nuclear function. Cell, 92, 141–143. [DOI] [PubMed] [Google Scholar]
- Khazak V., Sadhale,P.P., Woychik,N.A., Brent,R. and Golemis,E.A. (1995) Human RNA polymerase II subunit hsRPB7 functions in yeast and influences stress survival and cell morphology. Mol. Biol. Cell, 6, 759–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M.S. et al. (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S.cerevisiae. Mol. Cell, 4, 55–62. [DOI] [PubMed] [Google Scholar]
- Komuro A., Saki,M. and Kato,S. (1999) Npw38, a novel nuclear protein posessing a WW domain capable of activating basal transcription. Nucleic Acids Res., 27, 1957–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J. and Hall,M.N. (1993) Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem. Sci., 18, 334–338. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.P., Hanes,S.D. and Hunter,T. (1996) A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature, 380, 544–547. [DOI] [PubMed] [Google Scholar]
- Lu P.J., Zhou,X.Z., Shen,M. and Lu,K.P. (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science, 283, 1325–1328. [DOI] [PubMed] [Google Scholar]
- Luban J., Bossolt,K.L., Franke,E.K., Kalpana,G.V. and Goff,S.P. (1993) Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell, 73, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Ma Q. and Herschman,H.R. (1995) The yeast homologue YTIS11, of the mammalian TIS11 gene family is a non-essential, glucose repressible gene. Oncogene, 10, 487–494. [PubMed] [Google Scholar]
- Maleszka R., Hanes,S.D., Hackett,R.L., DeCouet,H.G. and Miklos,G.L.G. (1996) The Drosophila melanogaster dodo (dod) gene, conserved in humans, is functionally interchangeable with the ESS1 cell division gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 93, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks A.R. (1996) Cellular functions of immunophilins. Physiol. Rev., 76, 631–649. [DOI] [PubMed] [Google Scholar]
- McKune K., Richards,K.L., Edwards,A.M., Young,R.A. and Woychik,N.A. (1993) RPB7, one of two dissociable subunits of yeast RNA polymerase II, is essential for cell viability. Yeast, 9, 295–299. [DOI] [PubMed] [Google Scholar]
- Morris D.P., Phatnanai,H.P. and Greenleaf,A.L. (1999) Phospho-CTD binding and the role of a prolyl isomerase in pre-mRNA 3′ end formation. J. Biol. Chem., 274, 31583–31587. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Paro R. (1990) Imprinting a determined state into the chromatin of Drosophila. Trends Genet., 6, 416–421. [DOI] [PubMed] [Google Scholar]
- Rahfeld J.U., Rucknagel,K.P., Schelbert,B., Ludwig,B., Hacker,J., Mann,K. and Fischer,G. (1994) Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases: amino acid sequence and recombinant production of parvulin. FEBS Lett., 352, 180–184. [DOI] [PubMed] [Google Scholar]
- Ranganathan R., Lu,K.P., Hunter,T. and Noel,J.P. (1997) Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell, 89, 875–886. [DOI] [PubMed] [Google Scholar]
- Rudd K.E., Sofia,H.J. and Koonin,E.V. (1995) A new family of peptidyl-prolyl isomerases. Trends Biochem. Sci., 20, 12–14. [DOI] [PubMed] [Google Scholar]
- Rutherford S.L. and Zuker,C.S. (1994) Protein folding and the regulation of signaling pathways. Cell, 79, 1129–1132. [DOI] [PubMed] [Google Scholar]
- Schmid F.X. (1995) Protein folding. Prolyl isomerases join the fold. Curr. Biol., 5, 993–994. [DOI] [PubMed] [Google Scholar]
- Shen M., Stukenberg,P.T., Kirschner,M.W. and Lu,K.P. (1998) The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev., 12, 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O. and Rotin,D. (1996) WW domains. Structure, 4, 495–499. [DOI] [PubMed] [Google Scholar]
- Sudol M. (1996) Structure and function of the WW domain. Prog. Biophys. Mol. Biol., 65, 113–132. [DOI] [PubMed] [Google Scholar]
- Tamkun J.W. (1995) The role of brahma and related proteins in transcription and development. Curr. Opin. Genet. Dev., 5, 473–477. [DOI] [PubMed] [Google Scholar]
- Thompson C.M., Koleske,A.J., Chao,D.M. and Young,R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell, 73, 1361–1375. [DOI] [PubMed] [Google Scholar]
- Thompson M.J., Lai,W.S., Taylor,G.A. and Blackshear,P.J. (1996) Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene, 174, 225–233. [DOI] [PubMed] [Google Scholar]
- Uchida T., Fujimori,F., Tradler,T., Fischer,G. and Rahfeld,J.U. (1999) Identification and characterization of a 14 kDa human protein as a novel parvulin-like peptidyl prolyl cis/trans isomerase. FEBS Lett., 446, 278–282. [DOI] [PubMed] [Google Scholar]
- Varnum B.C., Ma,Q.F., Chi,T.H., Fletcher,B. and Herschman,H.R. (1991) The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys–His repeat. Mol. Cell. Biol., 11, 1754–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.T., Zydowsky,L.D. and McKeon,F.D. (1992) Cyclosporin A, the cyclophilin class of peptidylprolyl isomerases, and blockade of T cell signal transduction. J. Biol. Chem., 267, 13115–13118. [PubMed] [Google Scholar]
- West M.L. and Corden,J.L. (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics, 140, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler K.E., Swenson,K.I., Kornbluth,S. and Means,A.R. (2000) Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science, 287, 1644–1647. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. et al. (1997) Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science, 278, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Yuryev A. and Corden,J.L. (1996) Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics, 143, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Zhu W. and Hanes,S.D. (2000) Identification of Drosophila Bicoid-interacting proteins using a custom two-hybrid selection. Gene, 245, 329–339. [DOI] [PubMed] [Google Scholar]