Abstract
The Zap1 transcriptional activator of Saccharomyces cerevisiae controls zinc homeostasis. Zap1 induces target gene expression in zinc-limited cells and is repressed by high zinc. One such target gene is ZAP1 itself. In this report, we examine how zinc regulates Zap1 function. First, we show that transcriptional autoregulation of Zap1 is a minor component of zinc responsiveness; most regulation of Zap1 activity occurs post-translationally. Secondly, nuclear localization of Zap1 does not change in response to zinc, suggesting that zinc regulates DNA binding and/or activation domain function. To understand how Zap1 responds to zinc, we performed a functional dissection of the protein. Zap1 contains two activation domains. DNA-binding activity is conferred by five C-terminal C2H2 zinc fingers and each finger is required for high-affinity DNA binding. The zinc-responsive domain of Zap1 also maps to the C-terminal zinc fingers. Furthermore, mutations that disrupt some of these fingers cause constitutive activity of a bifunctional Gal4 DNA-binding domain–Zap1 fusion protein. These results demonstrate a novel function of Zap1 zinc fingers in zinc sensing as well as DNA binding.
Keywords: gene expression/regulation/Saccharomyces cerevisiae
Introduction
Metal ions such as iron, copper and zinc are essential nutrients but can also be cytotoxic if accumulated in excess amounts. Therefore, in the face of ever-changing extracellular or dietary levels, organisms must maintain adequate intracellular supplies of these metals for cellular metabolism while preventing their overaccumulation. In general, under metal-limiting conditions, pathways are upregulated to allow efficient scavenging of the metals from the extracellular environment or utilization of intracellular stores. Under conditions of metal excess, other systems are induced that facilitate metal ion efflux from the cell or promote intracellular sequestration in membrane-bound organelles or binding to macromol ecules such as metallothionein, phytochelatin and ferritin.
Regardless of the specific homeostatic regulatory mechanism in question, intracellular metallosensing proteins play key roles in controlling these processes. For example, MTF-1 is a zinc sensor protein in vertebrates that upregulates the transcription of metallothionein genes in response to excess zinc levels (Heuchel et al., 1994). The iron sensor proteins IRP1 and IRP2 control both the translation of ferritin mRNA as well as the stability of the transferrin receptor mRNA to regulate simultaneously iron storage and uptake (Eisenstein and Blemings, 1998). These are just two examples of a growing number of metallosensing regulatory proteins that have been identified in studies at all phylogenetic levels. A common theme emerging from these many studies is that metallosensors monitor intracellular metal ion status by direct binding of the metal (or metal-containing complexes, e.g. heme) to regulatory sites within the protein that then modulate the activity of the metallosensor (O’Halloran, 1993). Thus, an understanding of the precise mechanism that these regulatory proteins use to sense metal ion levels is clearly essential for understanding of metal ion homeostasis.
Our recent studies have focused on the mechanisms of zinc homeostasis in the yeast Saccharomyces cerevisiae. In this yeast, zinc uptake is mediated by the Zrt1 and Zrt2 zinc transporters found in the plasma membrane (Zhao and Eide, 1996a,b; Gitan et al., 1998). The activity of these transporters is controlled by two separate zinc-responsive mechanisms. First, their activity is regulated at a post-translational level by controlling the rate of their removal from the cell surface in response to zinc status (Gitan et al., 1998; Gitan and Eide, 2000). Treating cells with high concentrations of zinc triggers endocytosis of the zinc transporters and this regulation prevents overaccumulation of the metal. The zinc-responsive metallosensor(s) controlling this process has not been identified.
The second zinc-responsive regulatory mechanism in yeast occurs at the transcriptional level. Both the ZRT1 and ZRT2 genes are expressed at high levels in zinc-limited cells and their expression is shut off at high zinc levels (Zhao and Eide, 1996a,b). The Zap1 transcriptional activator is directly responsible for this regulation (Zhao and Eide, 1997). In addition to the zinc transporter genes, Zap1 upregulates expression of its own promoter via a positive autoregulatory mechanism. This autoregulation was proposed to increase the magnitude of the transcriptional response to zinc deficiency (Zhao and Eide, 1997). Zap1 also controls the export of stored zinc from the vacuole by regulating expression of Zrt3, a putative vacuolar zinc efflux transporter (MacDiarmid et al., 2000). In addition to these genes, DNA microarray analysis suggested that Zap1 controls the expression of as many as 42 other genes in response to zinc status (Lyons et al., 2000). Clearly, this protein is a major component in the regulation of cellular zinc homeostasis.
Zap1 is a 93 kDa protein with seven potential zinc finger domains. Five of these domains are clustered at the C-terminus of the protein and constitute the intact DNA-binding domain (Zhao et al., 1998; Bird et al., 2000). These five fingers are required for high-affinity and sequence-specific DNA binding to sites, called zinc-responsive elements (ZREs), which are found in the promoters of Zap1 target genes. The current consensus ZRE sequence, derived from mutational studies as well as comparison of many such elements from potential Zap1 target gene promoters, is 5′-ACCTTNAAGGT-3′ (Zhao et al., 1998; Lyons et al., 2000). Zap1 also contains two regions of high acidic residue content that were previously proposed to be activation domains (Zhao and Eide, 1997).
Two key questions regarding Zap1’s role as a zinc metallosensory protein are (i) what is the mechanism of zinc sensing used by this regulatory system and (ii) how does the zinc signal control Zap1 function? We show here that Zap1 activity is regulated by zinc largely at a post-translational level. To address the mechanism of how Zap1 senses zinc, we have mapped the zinc-responsive domain (ZRD) of the protein to the five zinc fingers also required for DNA binding. Furthermore, our data demonstrate that at least some of these fingers, in addition to their role in DNA binding, are required for the zinc-responsive regulation of Zap1 activity. These results suggest a novel role for zinc fingers in both metal sensing and DNA binding.
Results
Regulation of Zap1 activity by zinc occurs at a post-translational level
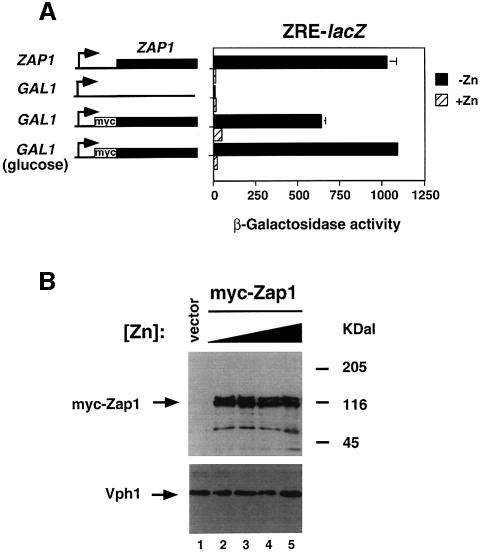
In wild-type cells where Zap1 is expressed from its own promoter, expression of a ZRE-lacZ fusion is induced ∼90-fold in zinc-limited cells relative to cells grown under zinc-replete conditions (Figure 1A). In previous studies, we demonstrated that Zap1 activates its own expression during zinc deficiency, which presumably amplifies the transcriptional response. To determine what contribution this autoregulation makes to the overall zinc responsiveness of Zap1p-regulated gene expression, we fused the ZAP1 protein coding region to the GAL1 promoter and examined regulation in a zap1 mutant strain (Figure 1A). Expression of the GAL1 promoter was previously found to be unaffected by zinc availability (Lyons et al., 2000). Six myc epitope tags were also introduced at the N-terminus of GAL1-expressed Zap1 to facilitate the detection and localization of the protein (see below). This epitope-tagged protein (myc-Zap1) fully complemented the growth defect of a zap1 mutant strain in low zinc, indicating that the fusion retained wild-type function (data not shown). The GAL1 promoter vector alone conferred no ZRE-lacZ expression in either low or high zinc. When the myc-Zap1 protein was expressed from this promoter at a high level in galactose-grown cells, they showed ∼8-fold induction of ZRE-lacZ expression in low zinc, i.e. only 10% of the wild-type regulation. This apparently lower level of regulation is largely due to increased basal lacZ expression in the cells expressing high levels of Zap1. Low level expression of myc-Zap1 from the GAL1 promoter in cells grown on glucose, a carbon source that represses GAL1 expression 1000-fold (St John and Davis, 1981; Johnston et al., 1994), was still sufficient to fully complement the zap1 mutation (data not shown) and confer nearly wild-type zinc-regulated expression (∼50-fold). Thus, while autoregulation may contribute slightly to overall zinc responsiveness, most of the regulation occurs through post-transcriptional control of Zap1p activity.
Fig. 1. Regulation of Zap1 activity by zinc occurs at a post-translational level. (A) Wild-type (DY1457) and zap1 mutant (ZHY6) cells containing either the pYef2 vector or pMyc-Zap11–880 were grown to exponential phase in LZM-galactose supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2. Zap1 activity in each strain was assessed using the pDg2 ZRE-lacZ reporter. ZHY6 pMyc-Zap11–880 transformants were also assayed after growth in glucose, a carbon source that represses most but not all expression from the GAL1 promoter. A representative experiment is shown and the error bars indicate 1 SD. (B) The stability of Zap1 is not affected by zinc status. Wild-type (DY1457) cells transformed with the pYef2 vector and zap1 mutant (ZHY6) cells bearing pMyc-Zap11–880 were grown in LZM-galactose to exponential phase. The concentrations of ZnCl2 added to the medium were 5 (lane 2), 250 (lane 3), 500 (lane 4) and 1000 µM (lanes 1 and 5). Crude protein extracts were prepared, fractionated by SDS–PAGE analysis, and assayed for Zap1 and Vph1 protein levels by immunoblotting.
Post-transcriptional control of Zap1 could occur through zinc-regulated changes in translation efficiency or protein stability. To test these hypotheses, we used immunoblotting to determine the effects of differing zinc availability on Zap1 level (Figure 1B). An immunoblot of the Vph1 vacuolar ATPase subunit showed equal protein loading in all lanes. In a vector-transformed zap1 mutant, no Zap1 protein was observed in protein extracts when probed with the anti-myc antibody. Several bands, ranging in size from ∼45 to 120 kDa, were detected in protein extracts of cells expressing myc-Zap1. The predicted molecular mass of myc-Zap1 is 105 kDa and two bands were observed that were of this approximate size. We noted that Zap1 was particularly sensitive to proteolysis during preparation of these samples (data not shown), so smaller forms may result from partial degradation of the protein. Alternatively, the highest molecular mass form may result from protein modification (e.g. phosphorylation) of full-length Zap1. Whatever their source(s), all forms of Zap1 detected were present in similar relative amounts across a broad range of zinc concentrations, indicating that none represented forms of the protein associated with its post-transcriptional control by zinc. Thus, we concluded that Zap1 zinc responsiveness occurs at a post-translational level.
Zap1 is present in the nucleus of zinc-deficient and zinc-replete cells
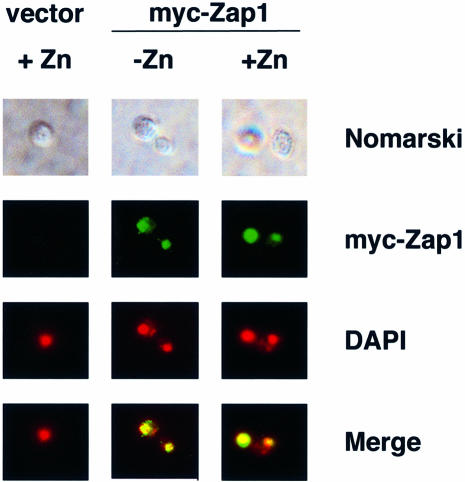
Many transcription factors are regulated post-translationally by controlling their nuclear localization. To determine whether zinc similarly affects Zap1 protein trafficking, we used indirect immunofluorescence to observe myc-Zap1’s subcellular location in zinc-deficient and -replete cells (Figure 2). Little fluorescence was observed in vector-transformed cells. Myc-Zap1 fluorescence was present in cells expressing the tagged allele and grown under both deficient and replete conditions. This fluorescence co-localized with the DNA-staining reagent 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI), demonstrating that Zap1 was localized to the nucleus in both low and high zinc. These results show that regulation of Zap1 by zinc does not occur through altered protein trafficking.
Fig. 2. The subcellular localization of Zap1 is not affected by zinc status. Wild-type (DY1457) cells bearing the vector pYef2 and zap1 mutant (ZHY6) cells bearing pMyc-Zap11–880 were grown to exponential phase in LZM-galactose supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2. Cells were viewed by Nomarski optics or epifluorescence. DAPI was used to stain the nucleus and the myc-Zap1 protein was detected by indirect immunofluorescence. The blue fluorescence of DAPI staining was converted to red and the DAPI and myc-Zap1 images were overlaid using Adobe Photoshop (merge). Yellow color in the merged images indicates colocalization of the markers.
Mapping the ZRD of Zap1 with Gal4 DNA-binding domain fusions
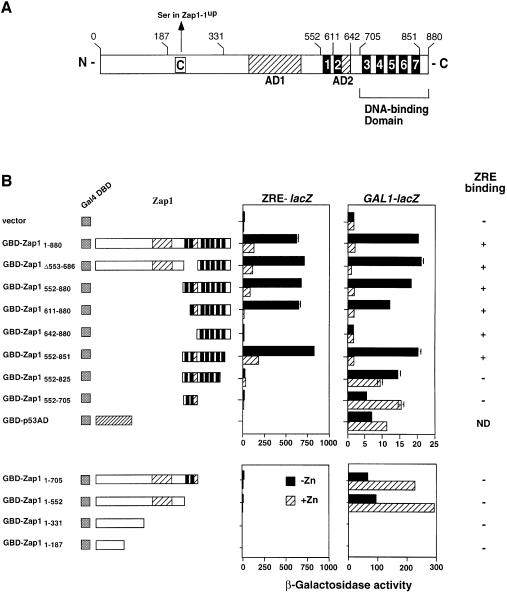
By a process of elimination, we concluded from the data shown in Figures 1 and 2 that the activity of Zap1 is regulated through the control of its DNA-binding and/or activation domain function. To gain insight into these possible mechanisms of regulation, we conducted a functional dissection of Zap1 using a ‘one-hybrid’ protein fusion approach. The main goal of this analysis was to identify the ZRD of Zap1. The primary amino acid sequence of Zap1 suggested a number of domains that are common to transcriptional activators (Figure 3A). These include a C-terminal region that contains seven putative zinc finger domains (Zhao and Eide, 1997) and two regions rich in acidic residues (glutamate and aspartate) that could potentially be transcriptional activation domains. We have recently demonstrated that the five C-terminal zinc fingers of Zap1 are both necessary and sufficient for ZRE binding activity in vivo and in vitro (Zhao et al., 1998; Bird et al., 2000).
Fig. 3. The functional domains of Zap1. (A) Schematic representation of Zap1. Shown are the functional activation domains (AD1 and AD2, hatched boxes) and the DNA-binding domain. The seven putative zinc finger domains are represented by black boxes and are numbered 1–7. Amino acid positions relevant to the constructed plasmid fusions are also numbered. The position of the Cys→Ser substitution in the Zap1-1up protein is shown. (B) Mapping the ZRD of Zap1. The activity of GBD–Zap1 fusion proteins expressed from the ADH1 promoter in the strain DEY1538 (zap1 gal4 gal80) was measured using either a ZRE-lacZ (pDg2) or a GAL1-lacZ (pRY171) reporter. Cells were grown in LZM supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2 prior to the β-galactosidase activity assays. A representative experiment is shown and the error bars indicate 1 SD. Accumulation of all fusion proteins was confirmed by immunoblot analysis using an anti-GBD antibody. EMSA with a ZRE oligonucleotide probe (Zhao et al., 1998) was used to assess DNA-binding activity. ND, not determined.
To identify the ZRD of Zap1, we first fused the Gal4 DNA-binding domain (GBD) to the N-terminus of either full-length Zap1 or various Zap1 truncates to generate a series of fusion proteins designated GBD–Zap1x–y (where x and y denote the endpoints of the Zap1 region included; deletion endpoints are denoted by Δx–y) (Figure 3B). Because the GBD can bind to its own upstream activation sequence, the activity of these potentially bifunctional fusion proteins could be assayed using either a GAL1-lacZ reporter or a ZRE-lacZ reporter. As controls, immunoblot analysis using an anti-GBD antibody was used to ensure that the fusion proteins were produced and electrophoretic mobility shift assays (EMSA) were used to determine which fusion proteins retained ZRE binding activity. Nuclear localization of inactive fusions was also confirmed by indirect immunofluorescence.
The GBD fusion proteins were expressed from the ADH1 promoter in a zap1 gal4 mutant strain (DEY1538). Transformation of this strain with the GBD vector alone failed to confer expression on either ZRE-lacZ or GAL1-lacZ reporters (Figure 3B). The full-length GBD–Zap1 fusion (GBD–Zap11–880) fully complemented the growth defect of the zap1 mutation in low zinc (data not shown) and resulted in significant zinc-responsive gene expression on the ZRE-lacZ reporter. Given that this fusion is expressed from the ADH1 promoter, whose expression is also not upregulated in low zinc (Lyons et al., 2000), these results support our other experiments, indicating that autoregulation of the ZAP1 promoter plays a minor role in the overall regulation. The results also demonstrated that the GBD fusion does not interfere with the repression of Zap1 activity by this high zinc condition (see below).
When activity of GBD–Zap11–880 was tested on the GAL1-lacZ reporter, a similarly high degree of zinc regulation was also observed. Thus, Zap1 can confer regulation on a heterologous DNA-binding domain fused to its N-terminus. This ability indicated that we could use this approach to map the Zap1 ZRD independently of its DNA-binding activity. Given that our previous studies indicated that the first two zinc fingers of Zap1, ZnF1 and ZnF2, were not required for DNA binding, an attractive hypothesis was that these two fingers function in zinc-responsive regulation (Bird et al., 2000). However, this was clearly not the case; deletion of both ZnF1 and ZnF2 (GBD–Zap1Δ553–686) retained wild-type zinc-responsive gene expression on both ZRE-lacZ and GAL1-lacZ reporters. The N-terminal deletions GBD–Zap1342–880 (data not shown), GBD–Zap1552–880 and GBD–Zap1611–880 also showed wild-type levels of zinc-responsive expression on both reporters, indicating that each retained fully functional ZRD activity. GBD–Zap1642–880 had no activity on either promoter despite accumulating to high levels in the nucleus and having ZRE-binding activity. This suggested that residues 611–642 contained activation domain function (see below). Truncations were also generated from the C-terminal end; GBD–Zap1552–850, in which the last 29 amino acids of Zap1 were removed, showed wild-type zinc regulation. Therefore, this region, which contains an eighth potential zinc finger with the non-canonical sequence of C-X2-C-X12-Q-X3-C, is not required for either DNA binding or zinc responsiveness. Deletion of ZnF7 (GBD–Zap1552–825) resulted in loss of ZRE-lacZ expression, consistent with this finger being required for DNA binding (Bird et al., 2000). Remarkably, this fusion had nearly constitutive expression on the GAL1-lacZ reporter. Taken together, these results mapped the ZRD of Zap1 to amino acids 642–850. Furthermore, they demonstrated that residues 825–851, which contain ZnF7, are required for both DNA binding and zinc-responsive regulation of Zap1.
GBD fusions were also useful in mapping the activation domains of Zap1. GBD–Zap1552–705 conferred expression on the GAL1-lacZ reporter. These data, together with other results described in Figure 3B, indicated that one Zap1 activation domain, designated as AD2, is located between amino acids 611 and 642. Expression of the GAL1-lacZ reporter by GBD–Zap1552–705 was actually higher in zinc-replete cells than in zinc-limited cells, probably due to the negative effects of zinc deficiency on overall gene expression that we have observed previously (Zhao and Eide, 1996a). This conclusion was further supported by the observation that a GBD fusion to the p53 activation domain showed similarly increased activity in high zinc (GBD–p53AD).
That additional activation domain function is found elsewhere in Zap1 was suggested by the activity of GBD–Zap1Δ553–686, which lacks AD2. Consistent with this hypothesis, GBD–Zap11–552, which also lacks AD2, conferred expression on the GAL1-lacZ reporter (Figure 3C). Neither GBD–Zap11–331 nor GBD–Zap11–187 showed GAL1-lacZ expression despite being stable, nuclear (data not shown) proteins. These results demonstrated that another activation domain, designated AD1, is located between amino acids 331 and 552. As expected, none of the fusions lacking the Zap1 DNA-binding domain showed ZRE binding in vitro or expression from the ZRE-lacZ reporter in vivo.
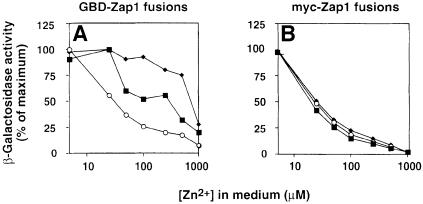
The experiments described in Figure 3 are consistent with a single ZRD in Zap1 localizing to residues 642–851. An alternative hypothesis is that there are multiple independent ZRDs in the protein that, because of their differences in sensitivity to zinc, might allow for regulation of gene expression over a broader range of concentrations than a single domain could provide. Subtle differences in the zinc responsiveness of different fusion proteins that this model predicts could be missed when only examining their activities at very high and very low zinc concentrations. To determine whether the different GBD fusions respond differently to zinc, we assayed ZRE-lacZ expression over a range of zinc concentrations. On the ZRE-lacZ reporter, these fusions were similarly repressed at the highest concentration of zinc used (Figures 3B and 4A). However, marked differences were observed at intermediate zinc concentrations (Figure 4A). Activity of GBD–Zap1552–880 was repressed at lower zinc concentrations than GBD–Zap11–880. Furthermore, even higher levels of zinc were required to repress the activity of GBD–Zap1Δ553–686. The differential regulation of these various fusions in response to zinc initially suggested that more than one ZRD might be present in Zap1. Further analysis demonstrated that this was not the case and that the differential effects were an artefact of the GBD fusions. When these same Zap1 proteins were analyzed as myc fusions, all were found to have identical zinc dose– response curves (Figure 4B). Thus, a single zinc-responsive mechanism is at work in these various proteins. These data also independently support the GBD mapping of the ZRD to the C-terminus of Zap1; i.e. the only region common to all of these similarly regulated fusions is residues 687–880.
Fig. 4. Comparison of GBD–Zap1 and myc-Zap1 activities in response to a range of zinc concentrations. (A) A zap1 mutant strain (ZHY6) bearing the pDg2 ZRE-lacZ reporter and pGBD–Zap11–880 (filled squares), pGBD–Zap1Δ553–686 (filled diamonds) or pGBD–Zap1552–880 (open circles) was grown to exponential phase in LZM supplemented with the indicated concentrations of ZnCl2 prior to assay for β-galactosidase activity. (B) The same zap1 mutant strain bearing the pDg2 ZRE-lacZ reporter and pMyc-Zap11–880 (filled squares), pMyc-Zap1Δ553–686 (filled diamonds) or pMyc-Zap1552–880 (open circles) was grown to exponential phase in LZM supplemented with the indicated concentrations of ZnCl2 prior to assay for β-galactosidase activity. Shown are representative experiments in which the standard deviations were <10% of the corresponding mean.
Mapping the ZRD of Zap1 with Gal4 activation domain fusions
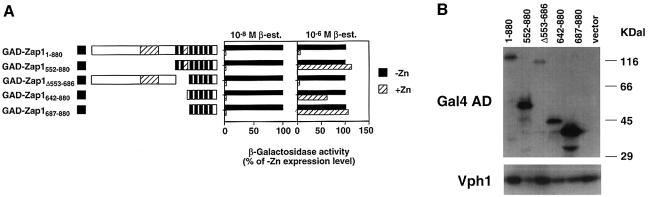
Mapping of the Zap1 ZRD to its DNA-binding domain suggested the usefulness of a complementary approach to examine this regulation, i.e. via activation domain fusions. This method potentially allows mapping of the ZRD independently of Zap1’s activation domain function. The Gal4 activation domain (GAD) was fused to the N-terminus of full-length Zap1 and Zap1 truncates, and examined for regulation of a ZRE-lacZ reporter in response to zinc. Because the Gal80 protein inhibits GAD function in glucose-grown cells, these fusions were analyzed in a zap1 gal4 gal80 mutant. Furthermore, the proteins were expressed from the GAL1 promoter in this strain through action of the β-estradiol-responsive GEV activator protein, a fusion of the GBD, the human estrogen receptor hormone response domain and the herpes virus VP16 activation domain (C.Y.Gao and J.L.Pinkham, manuscript submitted). This expression system has the additional advantage of controlling the levels of protein expression by increasing doses of the inducer, β-estradiol.
At low levels of expression (β-estradiol = 10–8 M), the GAD–Zap1–880 fusion complemented a zap1 mutation (data not shown) and retained highly zinc-responsive regulation (Figure 5A). This indicated that the GAD did not interfere with the normal regulation of Zap1 when expressed at a low level. Furthermore, the results demonstrated that the ZRD of Zap1 can confer this regulation not only on a heterologous DBD but also on a foreign activation domain. Fusions of various Zap1 truncates to the GAD confirmed mapping of the ZRD to the same region of the protein required for DNA binding, i.e. amino acids 687–880. All fusions conferred strong zinc-responsive regulation of expression including the smallest fusion, GAD–Zap1687–880. These results confirm the mapping of the ZRD to this region of the protein.
Fig. 5. Mapping the ZRD with GAD fusions. The indicated fusions were expressed in a zap1 gal4 gal80 mutant strain (DEY1538) bearing the pDg2 ZRE-lacZ reporter and pGEV-HIS3. pGEV-HIS3 encodes a hybrid transcriptional activator that is induced by β-estradiol. (A) Cells were grown to exponential phase in LZM supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2 and either 10–8 or 10–6 M β-estradiol. β-galactosidase activities are represented as a percentage of the corresponding –Zn expression level and the standard deviations were <10% of the corresponding mean. (B) The same transformants as in (A) were grown to exponential phase in LZM supplemented with 1000 µM ZnCl2; protein extracts were prepared and analyzed by immunoblotting using anti-GAD or anti-Vph1 antibodies. Apparent proteolytic products were observed in some samples; i.e. the largest band in each lane corresponds to the expected molecular mass of each fusion.
At higher levels of expression (i.e. β-estradiol = 10–6 M), some of the fusions (e.g. GAD–Zap1552–880, GAD–Zap1642–880 and GAD–Zap1687–880) no longer showed zinc responsiveness. While the precise cause of this effect is not known (see Discussion), immunoblots demonstrated that these particular fusion proteins accumulated to higher levels than those forms that retained zinc regulation when their mRNAs were overexpressed (Figure 5B). Thus, as is true for other transcription factors, studies of Zap1 require careful control of expression levels to obtain meaningful results.
DNA-binding zinc finger domains are structural determinants of the ZRD
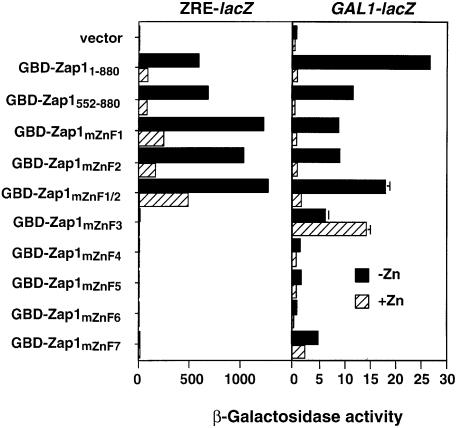
The ZRD of Zap1 contains the five zinc fingers ZnF3–ZnF7 (Figure 3A), each of which we have shown previously to be required for DNA binding. Colocal ization of the zinc-responsive and DNA-binding domains suggests that these zinc fingers may also play a role in zinc-responsive regulation of Zap1 activity. To test this hypothesis, we again took advantage of the ability of Zap1 to confer zinc regulation on the GBD. Mutations were generated in each Zap1 zinc finger such that their ability to bind zinc would be significantly impaired; i.e. the two histidine ligands in each motif were converted to glutamines (Bird et al., 2000). These mutant fusion proteins were then analyzed for regulation of the GAL1-lacZ reporter (Figure 6). As shown previously, GBD–Zap11–880 and GBD–Zap1552–880 were both highly regulated by zinc. Mutation of either ZnF1 or ZnF2 in the context of the GBD–Zap1552–880 fusion had little effect on zinc regulation. These results confirmed our earlier conclusion that these two fingers play little if any role in zinc regulation of Zap1. Interestingly, mutation of both ZnF1 and ZnF2 resulted in increased basal level expression. The reason for this effect is not known (see Discussion) but our deletion analysis results indicated that this is not due to loss of zinc responsiveness.
Fig. 6. Effects of zinc finger mutations on zinc responsiveness. A zap1 gal4 gal80 mutant strain (DEY1538) containing GBD–Zap11–880, GBD–Zap1552–880 or GBD–Zap1mZnFX (where X indicates the mutated zinc finger domain) was grown in LZM supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2. β-galactosidase activity was measured in each strain using either a ZRE-lacZ (pDg2) or a GAL1-lacZ (pRY171) reporter. A representative experiment is shown and the error bars indicate 1 SD.
Mutation of ZnF3 resulted in a loss of ZRE-lacZ expression consistent with the importance of this finger in DNA binding. A striking observation was that this allele resulted in constitutive expression from the GAL1 promoter. A similar effect was observed when ZnF7 was disrupted. The effect of this mutation was similar to that observed when the region was deleted altogether (Figure 3B). Thus, the regulatory function of this region of the protein correlates with the ability of ZnF3 and ZnF7 to form folded zinc finger structures. These results indicate that these two fingers, both of which are required for high-affinity ZRE binding, are also structural determinants of the ZRD.
When ZnF4, 5 and 6 were mutated, no expression was observed on either ZRE-lacZ or GAL1-lacZ reporters. We have shown previously (Bird et al., 2000) that these mutant proteins are not localized to the nucleus but rather accumulate as cytoplasmic aggregates. Because of this localization defect, these mutants were not informative in determining the role of these particular zinc fingers in zinc responsiveness in vivo.
What’s ‘up’
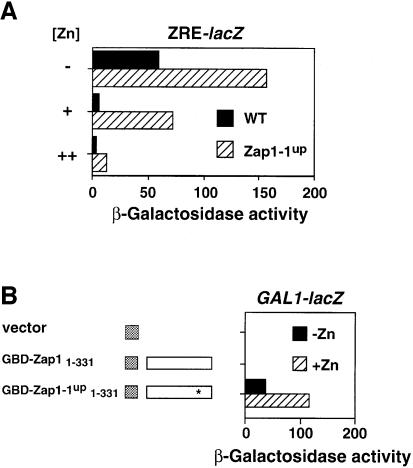
Mapping of the ZRD to the C-terminus of Zap1 created an interesting paradox. The ZAP1 gene was initially identified because a mutant allele was isolated, ZAP1-1up, which caused high level expression of ZRT1 and ZRT2 in both zinc-replete and zinc-limited cells (Zhao and Eide, 1997). This result suggested that the ZRD of Zap1 was directly affected by this mutation. However, the ZAP1-1up allele is a single nucleotide mutation causing a cysteine to serine substitution at amino acid 203, i.e. far removed from the C-terminal ZRD identified in this study (Figure 3A). Thus, it seemed unlikely that this mutation directly altered the zinc responsiveness of Zap1. An alternative hypothesis was that the ‘up’ allele increased that activity of a cryptic activation domain in Zap1. Consistent with this hypothesis, Zap1-1up showed greater expression of a ZRE-lacZ reporter in zinc-limited cells (Figure 7A). Furthermore, the protein was still zinc regulated, albeit it required higher levels of zinc to shut off expression. To test directly for new activation domain function in this allele, we compared expression of GBD fusions (Figure 7B). As was shown previously, GBD–Zap11–331 did not activate expression of GAL1-lacZ. The corresponding GBD–Zap1-1up1–331 fusion constructed with the C203S mutation activated high level expression. The GBD–Zap1-1up1–331 fusions showed a similar response to zinc to the GDB–p53AD fusion (Figure 3A), indicating that this allele is not zinc regulated. The wild-type fusion protein accumulated to even higher levels than the Zap1-1up fusion, and both were similarly localized to the nucleus (data not shown), arguing that the observed effects are not due to differences in protein level or localization. Thus, the C203S mutation in the ZAP1-1up allele activates a normally quiescent activation domain.
Fig. 7. The ZAP1-1up mutation turns on a quiescent activation domain. (A) ZAP1-1up retains zinc-responsive regulation. Wild-type (DY1457) and ZAP1-1up (ZHY7) strains bearing pDg2 were grown to exponential phase in SD media (+) or in SD media that had been supplemented with 1 mM EDTA, 100 µM ZnCl2 (–) or 100 µM ZnCl2 alone (++) prior to β-galactosidase activity assays. (B) The ZAP1-1up allele contains an activation domain not present in the wild-type protein. Strain DEY1538 (zap1 gal4 gal80) containing the GAL1-lacZ reporter (pRY171) and either the vector (pMA424), pGBD–Zap11–331 or pGBD Zap1-1up1–331 was grown to exponential phase in LZM supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2 prior to β-galactosidase assay. The asterisk denotes the mutation in the Zap1-1up allele. Representative experiments are shown.
Repression of Zap1 function by zinc is not mediated by the recruitment of a repressor protein
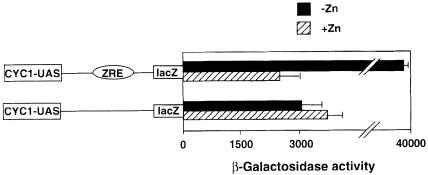
The ability of Zap1 to confer zinc responsiveness on both fused heterologous DNA-binding and activation domains suggested a specific mechanism by which zinc controls Zap1 function, i.e. zinc- and Zap1-dependent recruitment of a repressor protein to its target promoters. Similar regulatory mechanisms are well known. For example, activity of the Ume6 transcription factor is repressed by recruitment of a complex containing the corepressor Sin3 and the histone deacetylase Rpd3 proteins (Kadosh and Struhl, 1997), while the α2-Mcm1 activator is repressed by recruiting the Ssn6–Tup1 complex to their target promoters (Keleher et al., 1992). One unique feature of these mechanisms is the ability of the recruited repressor complex to repress epistatically the activity of transcriptional activators bound nearby in the promoter by altering the chromatin structure of the chromosomal region. Therefore, to test whether a similar repression model was at work for Zap1, we inserted a ZRE element into the intact CYC1 promoter and determined the ability of Zap1 to confer zinc-responsive regulation on the activity of the nearby CYC1 UAS elements (Figure 8). β-galactosidase activity generated from the CYC1 reporter in which a single ZRE element was inserted was high in zinc-limiting media and low in zinc-replete media. Thus, the ZRE-conferred Zap1 regulation is functional in the context of this promoter. The expression of the CYC1 promoter in high zinc is presumably mediated by the Hap1 transcription factor. There was no difference in reporter expression in high zinc with or without the ZRE. These results indicate that the ZRE is not capable of repressing expression driven by neighboring transcriptional activators. Furthermore, zinc regulation occurred normally in a tup1 mutant strain (data not shown). These results suggest that zinc does not control Zap1 activity by triggering the recruitment of a repressor protein or complex that alters the chromatin structure of Zap1 target promoters.
Fig. 8. Zap1 activity in zinc-replete cells is not repressed by a repressor capable of acting on nearby UASs. Wild-type (DY1457) cells transformed with pLGΔ312 or pZRE-LGΔ312 were grown to exponential phase in LZM supplemented with either 5 µM (–Zn) or 1000 µM (+Zn) ZnCl2 prior to β-galactosidase assay. A representative experiment is shown and the error bars indicate 1 SD.
Discussion
The Zap1 transcriptional activator is responsible for controlling the expression of a large number of genes in response to zinc status. At this time, there are eleven confirmed Zap1 targets in the yeast genome, including ZRT1, ZRT2, ZRT3 and ZAP1 itself, and many other potential targets (Zhao and Eide, 1997; Lyons et al., 2000; MacDiarmid et al., 2000). It is our current hypothesis that Zap1 is the primary zinc sensor in the cell, directly sensing the level of a loosely bound or ‘labile’ regulatory pool of zinc and translating that signal into altered levels of target gene expression. A primary goal of our studies is to understand how Zap1’s activity is regulated by zinc. In this report we show that, in addition to the transcriptional autoregulation that controls Zap1 synthesis, the activity of the protein is regulated by zinc at a post-translational level. In fact, the transcriptional control of Zap1 plays a minor role in the overall zinc responsiveness of target gene expression; the degree of zinc regulation was similar when Zap1 was expressed from either its own zinc-responsive promoter or from the zinc-insensitive (Lyons et al., 2000) GAL1 or ADH1 promoters. However, it should be noted that our results do not exclude the possibility that the autoregulation of Zap1 transcription may be important under more severely zinc-limiting conditions than those examined in this study.
Many transcription factors, e.g. glucocorticoid receptor, NF-κB, etc. (Picard and Yamamoto, 1987; Baeuerle and Baltimore, 1988), are regulated by controlling their nuclear localization, but this is not the case for Zap1. Zap1’s nuclear localization was unaffected by zinc treatment. This observation indicates that zinc controls the DNA-binding and/or activation domain function of Zap1. Furthermore, if Zap1 and not some other protein (see below) is the primary zinc sensor, the constitutively nuclear localization of Zap1 indicates that the regulatory pool being monitored is labile zinc in the nucleus rather than the cytoplasm. This nuclear zinc is likely to be in equilibrium with the cytoplasmic pool because the metal ion can probably diffuse unimpeded through large channel structures that are found in the nuclear pore complexes (Paine et al., 1975; Hinshaw et al., 1992).
To gain insight into how zinc regulates Zap1 activity, we mapped the functional domains of the protein. Domains of yeast transcription factors that are responsible for activating transcription are typified by an abundance of acidic residues (Hahn, 1993). The primary amino acid sequence of Zap1 suggested the presence of two such activation domains, which were designated AD1 and AD2 (Zhao and Eide, 1997). AD2 was predicted to lie between residues 603 and 703, and this location was confirmed and mapped more precisely to residues 611–642. In contrast, AD1 was predicted to lie between residues 190 and 331 but was functionally mapped to residues 331–562. While not as rich in acidic amino acids as the 190–331 interval, the latter region still contains an abundance of acidic residues that may confer activation domain function. AD1 and AD2 appear to be fully redundant given that mutant proteins lacking either single domain still fully complement a zap1 mutant for growth under zinc-limiting conditions. Furthermore, these deletion mutants have similar abilities to activate expression of a ZRE-lacZ reporter. One intriguing observation is that deleting the Zap1 DNA-binding domain resulted in a 10-fold increase in AD1 activity when assayed in the context of a GBD fusion on the GAL1-lacZ reporter (Figure 3B). One explanation for this result is that binding of the Zap1 DNA-binding domain to low-affinity sites in the GAL1 promoter could sterically hinder activity of AD1. This is apparently not the case because mutations in the Zap1 zinc fingers that also eliminated DNA binding did not have the same effect. Thus, there may be protein–protein interactions between AD1 and the Zap1 DNA-binding domain that somehow attenuate the activity of AD1. A similar effect on AD2 activity was not observed.
In our previous studies, we mapped the DNA-binding domain to amino acids 687–880, i.e. the C-terminal region of the protein that contains ZnF3–ZnF7 (Bird et al., 2000). Fusing this region to the GAD complemented a zap1 mutation for low zinc growth and also conferred high level expression on a ZRE-lacZ reporter. In vitro, the purified 194 residue fragment bound to DNA with a high affinity (Kd ∼1 nM) similar to that of longer fragments of Zap1. Furthermore, by deletion and site-directed mutagenesis, we demonstrated that each one of the five C-terminal zinc fingers is required for high-affinity DNA binding. Given that they are required for DNA binding in zinc-limited cells, ZnF3–ZnF7 presumably bind Zn2+ with high affinity such that these fingers are properly folded to confer ZRE binding. Consistent with this hypothesis, purified Zap1 proteins capable of DNA binding have five molar equivalents of zinc (Bird et al., 2000).
ZnF1 and ZnF2 were previously shown to be unnecessary for high-affinity ZRE DNA binding (Bird et al., 2000). Moreover, determinations of the zinc stoichiometries of purified Zap1 proteins indicated that these two fingers bind zinc with low affinity. These results suggested that ZnF1 and ZnF2 might play a role in regulating Zap1 activity in response to zinc. The results of this study indicate that these fingers are probably not involved in this response; deletion of both fingers had no effect on Zap1 zinc responsiveness. However, it seems unlikely that these domains have no function whatsoever and this conclusion is supported by the recent sequencing of a ZAP1-related gene (DDBJ/EMBL/GenBank accession No. AF140504) from Candida albicans. While the function of this protein has not been established, the C-terminal half of the protein (amino acids 448–876) is 31% identical to the C-terminus of Zap1, suggesting that the C.albicans protein is a true Zap1 ortholog. The Candida Zap1 amino acid sequence contains all seven of the S.cerevisiae Zap1 zinc fingers including ZnF1 and ZnF2. Given this conservation, it appears that these domains of Zap1 do play important roles under some as yet unrecognized conditions. The increased activity of the GBD–Zap1mZnf1/2 mutant (Figure 6) suggests one model in which metal binding by these fingers specifically attenuates the activity of AD2. The contribution of this effect to overall zinc regulation is unclear.
Mapping of the ZRD of Zap1 to the C-terminal DNA-binding domain region also raised a paradox regarding the effects of the ZAP1-1up mutation (Zhao and Eide, 1997). ZAP1-1up was originally isolated because it resulted in what was first viewed as constitutive expression of Zap1 target genes. This mutation, a substitution of a cysteine at position 203 with serine, is located in a region of Zap1 that we originally postulated was within AD1. These observations led to the hypothesis that the C203S mutation alters a zinc regulatory site within a functional activation domain. This is clearly not the case given that we have mapped both activation domain and ZRD functions to elsewhere in the protein. This paradox is explained in Figure 7; the ZAP1-1up mutation creates a new activation domain in Zap1. First, the ZAP1-1up mutation increased expression of a ZRE-lacZ reporter ∼2-fold in low zinc, indicating that the protein is a more potent activator of transcription when maximally active. Secondly, the ZAP1-1up mutant is still zinc regulated, albeit more zinc is required to repress Zap1-dependent expression completely. Finally, and most importantly, fusion of amino acids 1–331 of the wild-type and Zap1-1up proteins to the GBD shows that while the wild-type truncate cannot activate transcription, the same region of Zap1-1up can induce high levels of activity.
The ZRD mapped precisely to the same region of the protein as the DNA-binding domain. This colocalization suggests that zinc regulates Zap1 DNA-binding activity. However, the ability of Zap1 to confer zinc-responsive gene expression on the GBD also suggests that zinc impairs activation domain function. While others are possible, we propose the following models of how Zap1 activity is regulated by zinc. First, we hypothesize that Zap1 is the direct zinc sensor and contains one or more additional and low-affinity Zn2+-binding sites in the DNA-binding domain in addition to the five high-affinity C2H2 zinc finger sites. Binding of Zn2+ to the regulatory site may stabilize a conformer that precludes DNA binding. This zinc-induced conformer (e.g. a multimeric complex) not only sterically impairs the DNA-binding interface but also interferes with the accessibility of the activation domain(s) to general transcription factors.
Alternatively, Zap1 may be inhibited through the action of another protein that specifically represses Zap1 through protein–protein interactions or through post-translational modifications. In this model, either Zap1 or the accessory protein could contain the regulatory Zn2+-binding site that controls the interaction. One prediction of this model is that it should be possible to isolate loss-of-function mutations in the gene encoding the accessory protein that would result in constitutive expression of Zap1 target genes. Despite substantial effort (A.J.Bird and D.J.Eide, unpublished observations), such mutations have not yet been isolated.
The observation that the particular Zap1 fusion proteins that accumulate to high levels cause constitutive expression of a ZRE-lacZ reporter (Figure 5) is consistent with either hypothesis. If zinc binds to Zap1 to repress its function directly, one explanation is that Zap1 overexpression leads to an increase in the normally small pool of active Zap1 that persists in zinc-replete cells. For example, if the labile zinc in zinc-replete cells is sufficient to inactivate 99% of Zap1, overexpression by 100-fold could result in maximal target gene expression mediated by the remaining 1% of active molecules. Alternatively, overexpression of Zap1 may result in constitutive activation through titration of a repressor molecule such that the excess Zap1 protein is not inhibited. If the latter is true, the constitutive expression of these Zap1 fusion proteins may provide a strategy to clone the gene that encodes the repressor through the isolation of overexpression suppressors.
Regardless of the precise mechanism of regulation, an exciting lesson arising from this study is that ZnF3 and ZnF7 are not only required for DNA binding to the ZRE but these fingers are also structural elements of the ZRD. Glutamine substitution mutations of the histidine ligands in these fingers, or complete deletion of ZnF7, resulted in constitutive expression of a GBD fusion analyzed on the GAL1-lacZ reporter. In the context of the models described above, formation of these fingers by classical high-affinity Zn2+ binding may be required to establish the appropriate coordination geometry for a low-affinity, regulatory Zn2+-binding site. Alternatively, the folded fingers may form essential parts of a protein–protein interaction domain to facilitate intermolecular repression. Whatever the case, our results demonstrate a novel dual role of zinc fingers in Zap1 as components of both a DNA-binding domain and a zinc-sensing domain.
Materials and methods
Strains and culture conditions
The strains used in this study were DY1457 (MATα ade6 can1 his3 leu2 trp1 ura3), ZHY6 (DY1457 zap1::TRP1) (Zhao and Eide, 1997), ZHY7 (DY1457 ZAP1-1up) (Zhao and Eide, 1997) and DEY1538 (MATa ura3 his3 ade2 lys2 leu2 trp1 tyr1 gal4 gal80 ade5::hisG zap1::TRP1). DEY1538 is a zap1 mutant derivative of YM4271 (Liu et al., 1993). Yeast were grown in YP medium containing 2% glucose or in synthetic defined medium supplemented with 2% glucose and any necessary auxotrophic supplements. Low zinc media (LZM) was prepared as described in Gitan et al. (1998) with either 2% glucose (LZM) or 2% galactose (LZM galactose) as carbon source, and supplemented with the indicated concentration of ZnCl2.
Reporter plasmids
ZRE-lacZ and GAL1-lacZ reporters were pDg2 (Zhao et al., 1998) and pRY171 (Yocum et al., 1984), respectively. The ZRE element was inserted into the XhoI site of pLGΔ312 (Guarente and Mason, 1983) to generate pZRE- LGΔ312.
Plasmid construction
All plasmid constructs were confirmed by DNA sequencing. To create a plasmid containing epitope-tagged ZAP1 under the control of the GAL1 promoter, the ZAP1 open reading frame was PCR amplified using a 5′ primer that contained added StuI, SalI and NdeI sites, and a 3′ primer that contained added XhoI and NotI sites. A fragment containing six copies of the c-myc epitope (Kolodziej and Young, 1991) was excised from the vector CS2 + MT (Rupp et al., 1994) as a BamHI–StuI fragment and subcloned into pHolly (R.Palmiter, University of Washington) to create Myc-pHolly. The ZAP1 PCR product was then inserted into StuI–XhoI-digested Myc-pHolly generating an N-terminal myc-tagged ZAP1. The myc-Zap1 fusion was then excised as a ClaI–NotI fragment and subcloned into the vector pYef2 (Cullin and Minvielle-Sebastia, 1994) to generate pMyc-Zap11–880. Plasmid pMyc-Zap1Δ553–686 was made by two-step overlapping PCR (Ho et al., 1989) using pMyc-Zap1 as template to generate a 2.2 kb ClaI–NotI fragment containing the region encoding amino acids 1–552 fused to the region encoding residues 687–880. The resulting fragment was inserted into ClaI–NotI-digested pYef2. To create pMyc Zap1552–880, two-step overlapping PCR was used with pMyc-Zap11–880 as template to generate a 1 kb ClaI–NotI fragment containing the region encoding the myc epitope tags and Zap1 amino acids 1–5 fused to residues 552–880. This fragment was inserted into ClaI–NotI-digested pYef2. LEU2 derivatives of these plasmids were generated by in vivo homologous recombination with a 4.7 kb SmaI ura3::LEU2 fragment isolated from the vector pUL9 (Cross, 1997).
GBD–Zap1 fusions were constructed as follows. The ZAP1 open reading frame was PCR amplified using primers with added 5′ and 3′ SalI sites. The resulting fragment was inserted into SalI-digested pMA424 (Ma and Ptashne, 1987) to create pGBD–Zap11–880. Related deletion plasmids (pGBD–Zap1552–880, pGBD–Zap1611–880, pGBD–Zap1642–880, pGBD–Zap1552–851 and pGBD–Zap1552–825) were created with PCR amplicons containing the indicated regions of the ZAP1 open reading frame. To assist cloning of these fragments, all 5′ primers contained an added EcoRI site, all 3′ primers contained an added BamHI site, and the fragments were inserted into a BamHI–EcoRI-digested pMA424. pGBD–Zap11–705 and pGBD–Zap1552–705 were generated in a similar fashion using 3′ primers containing an added SalI site and 5′ primers containing an added SalI or EcoRI site. All other GBD fusions were created in pMA424 by in vivo homologous recombination (Kunes et al., 1987) using PCR fragments flanked on either side by 40 bp of homology to the vector site of insertion. Plasmids containing the ZAP1-1up mutation used a mutant allele plasmid as the PCR template. The zinc finger mutants were constructed in pGBD–Zap1552–880 (Bird et al., 2000).
GAD–Zap1 fusions were constructed as follows. The ZAP1 open reading frame was amplified by PCR with primers containing added 5′ and 3′ SalI sites. This fragment was inserted into SalI-digested pGAD424 to create pGAD–Zap11–880. All other GAD fusion constructs were created by amplifying from the relevant region of the ZAP1 ORF and inserting them into pGAD424. The GAD fusions were introduced into BamHI and BstXI sites of pRS316-GAL1 by homologous recombination (Kunes et al., 1987).
Immunoblot analysis and immunofluorescence microscopy
Crude protein extracts were prepared as described by Gitan et al. (1998) and protein quantified by the method of Bradford (1976). Immunoblots were performed essentially as described (Harlow and Lane, 1988). Proteins were fractionated by SDS–PAGE (7.5% acrylamide) and then transferred to nitrocellulose. Blots were incubated with anti-c-myc (Boehringer-Mannheim), anti-Vph1 (Molecular Probes), anti-Gal4 DBD (Clontech) or anti-Gal4AD (Clontech) antibodies, washed and then incubated with goat anti-mouse IgG antibody coupled to horseradish peroxidase (Pierce). Detection was by enhanced chemiluminescence (ECL; Amersham). Indirect immunofluorescence microscopy was performed as described by Gitan et al. (1998).
β-galactosidase assays
Cells were grown for 15–20 h to mid-exponential phase (A600 0.3–0.7) in LZM supplemented with the indicated amount of Zn2+. β-galactosidase assays were performed by either of two methods. For most assays, β-galactosidase activity was measured in permeabilized cells as described by Guarente (1983) and is expressed in Miller units that are calculated as follows (ΔA420 × 1000)/(min × ml of culture used × absorbance of the culture at 600 nm). For the experiment described in Figure 8, yeast were grown to the desired density, harvested by centrifugation, and washed with 1 ml of buffer (85 mM Na2HPO4, 45 mM NaH2PO4, 10 mM KCl and 1 mM MgSO4) before freezing at –70°C. After thawing, the cells were resuspended in 200 µl of the same buffer and lysed by vortexing with glass beads. β-galactosidase activities were then assayed in these crude homogenates and the units reported are (ΔA420 × 1000)/(min × ml of extract assayed × mg/ml protein).
Acknowledgments
Acknowledgements
We thank Mark Johnston and Stan Fields for plasmids used in this study. This work was supported by NIH grant GM58265.
References
- Baeuerle P.A. and Baltimore,D. (1988) IκB: a specific inhibitor of the NF-κB transcription factor. Science, 242, 540–545. [DOI] [PubMed] [Google Scholar]
- Bird A.J., Evans-Galea,M., Blankman,E., Zhao,H., Luo,H., Winge,D.R. and Eide,D.J. (2000) Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem., 275, 16160–16166. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Cross F.R. (1997) ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast, 13, 647–663. [DOI] [PubMed] [Google Scholar]
- Cullin C. and Minvielle-Sebastia,L. (1994) Multipurpose vectors designed for the fast generation of N- or C-terminal epitope-tagged proteins. Yeast, 10, 105–112. [DOI] [PubMed] [Google Scholar]
- Eisenstein R.S. and Blemings,K.P. (1998) Iron regulatory proteins, iron responsive elements and iron homeostasis. J. Nutr., 128, 2295–2298. [DOI] [PubMed] [Google Scholar]
- Gitan R.S. and Eide,D.J. (2000) Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J., 346, 329–336. [PMC free article] [PubMed] [Google Scholar]
- Gitan R.S., Lou,H., Rodgers,J., Broderius,M. and Eide,D. (1998) Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem., 273, 28617–28624. [DOI] [PubMed] [Google Scholar]
- Guarente L. (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol., 101, 181–191. [DOI] [PubMed] [Google Scholar]
- Guarente L. and Mason,T. (1983) Heme regulates transcription of the CYC1 gene of S.cerevisiae via an upstream activation site. Cell, 32, 1279–1286. [DOI] [PubMed] [Google Scholar]
- Hahn S. (1993) Structure (?) and function of acidic transcription activators. Cell, 72, 481–483. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Heuchel R., Radtke,F., Georgiev,O., Stark,G., Aguet,M. and Schaffner,W. (1994) The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J., 13, 2870–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J.E., Carragher,B.O. and Milligan,R.A. (1992) Architecture and design of the nuclear pore complex. Cell, 69, 1133–1141. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Johnston M., Flick,J.S. and Pexton,T. (1994) Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 3834–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell, 89, 365–371. [DOI] [PubMed] [Google Scholar]
- Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6–Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- Kolodziej P.A. and Young,R.A. (1991) Epitope tagging and protein surveillance. Methods Enzymol., 194, 508–519. [DOI] [PubMed] [Google Scholar]
- Kunes S., Ma,H., Overbye,K., Fox,M.S. and Botstein,D. (1987) Fine structure recombinational analysis of cloned genes using yeast transformation. Genetics, 115, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wilson,T.E., Milbrandt,L. and Johnston,M. (1993) Identifying DNA-binding sites and analyzing DNA binding domains using a yeast selection system. Methods, 5, 125–137. [Google Scholar]
- Lyons T.J., Gasch,A.P., Gaither,L.A., Botstein,D., Brown,P.O. and Eide,D. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. and Ptashne,M. (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell, 48, 847–853. [DOI] [PubMed] [Google Scholar]
- MacDiarmid C.W., Gaither,L.A. and Eide,D.J. (2000) Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J., 19, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Halloran T.V. (1993) Transition metals in control of gene expression. Science, 261, 715–725. [DOI] [PubMed] [Google Scholar]
- Paine P.L., Moore,L.C. and Horowitz,S.B. (1975) Nuclear envelope permeability. Nature, 254, 109–114. [DOI] [PubMed] [Google Scholar]
- Picard D. and Yamamoto,K.R. (1987) Two signals mediate hormone dependent nuclear localization of the glucocorticoid receptor. EMBO J., 6, 3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp R.A.W., Snider,L. and Weintraub,H. (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev., 8, 1311–1323. [DOI] [PubMed] [Google Scholar]
- St John T.P. and Davis,R.W. (1981) The organization and transcription of the galactose gene cluster of Saccharomyces. J. Mol. Biol., 152, 285–315. [DOI] [PubMed] [Google Scholar]
- Yocum R.R.S., Hanley,S., West,J.,R. and Ptashne,M. (1984) Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. and Eide,D. (1996a) The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl Acad. Sci. USA, 93, 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. and Eide,D. (1996b) The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem., 271, 23203–23210. [DOI] [PubMed] [Google Scholar]
- Zhao H. and Eide,D.J. (1997) Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Butler,E., Rodgers,J., Spizzo,T., Duesterhoeft,S. and Eide,D. (1998) Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem., 273, 28713–28720. [DOI] [PubMed] [Google Scholar]