INTRODUCTION
The ERCC1-XPF complex is a structure-specific endonuclease involved in the repair of damaged DNA. ERCC1-XPF performs a critical incision step in nucleotide excision repair (NER), and is also involved in the repair of DNA interstrand crosslinks (ICLs) and some double-strand breaks (DSBs) [1–7]. A fraction of ERCC1-XPF is localized at telomeres, where it is implicated in recombination of telomeric sequences and loss of telomeric overhangs at deprotected chromosome ends [8, 9]. Deficiency of either ERCC1 or XPF in humans results in a variety of conditions, which include the skin cancer-prone disease xeroderma pigmentosum (XP), a progeroid syndrome of accelerated aging, or cerebro-oculo-facio-skeletal syndrome (COFS) [10, 11]. These diseases are extremely rare in the general population and therefore mice with low levels of either ERCC1 or XPF have been generated and studied extensively. These murine models clearly illustrate the importance of DNA repair in preventing aging-related tissue degeneration (See also review by Diderich et al. in this issue of DNA Repair). The purpose of this review is to provide an overview on the phenotypes of patients with mutations in ERCC1 or XPF, and the mouse models used to study the diseases that result from decreased levels of ERCC1-XPF.
1. FUNCTIONS OF ERCC1-XPF NUCLEASE
1.1 NUCLEOTIDE EXCISION REPAIR
Ultraviolet light damages DNA, resulting in a myriad of lesions, most predominantly cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts [12]. NER is the only mechanism by which these photodimers can be removed from DNA in human cells, and ERCC1-XPF functions as the nuclease that incises the damaged strand 5’ to the adduct [13–16]. This incision creates a 3’ end that is used as a primer by the replication machinery to replace the excised nucleotides. XPF contains the catalytic activity with its conserved nuclease domain, and ERCC1 is required for binding to DNA [17–21] (see also article by Fagbemi et al. in this issue of DNA Repair). Defects in the proteins required for NER can result in xeroderma pigmentosum (XP), trichothiodystrophy (TTD), and Cockayne Syndrome (CS), highlighting the importance of DNA repair in preventing UV-induced skin cancer and developmental abnormalities. XP is a disease characterized by extreme photosensitivity and a 10,000-fold increased risk of cutaneous and ocular neoplasms [22]. Cells from all of the XP complementation groups (XP-A to XP-G, and XP-V) are hypersensitive to UV radiation [10, 13, 23–25]. ERCC1-XPF deficient cells are distinct from other XP patient-derived cells because of their extreme sensitivity to chemicals that induce DNA ICLs [7, 26–28]. Another critical piece of evidence indicating that ERCC1-XPF has functions distinct from NER is that ERCC1 and XPF knockout mice exhibit a much more severe phenotype than XPA null mice, which are completely deficient in NER [7, 29–32].
1.2 INTERSTRAND CROSSLINK REPAIR
The mechanism of DNA ICL repair in mammalian cells is not as well defined as NER. In replicating cells, crosslinking agents lead to DSBs created by endonuclease(s) near the site of stalled replication machinery [33]. In the absence of ERCC1-XPF, replication-dependent crosslink-induced DSBs occur, indicating that ERCC1-XPF cannot be solely responsible for creating these DSBs [28]. There is clear evidence that ERCC1-XPF participates in the same mechanism of ICL repair as the Fanconi anemia proteins. In the absence of ERCC1-XPF, FANCD2 is still monoubiquitylated by FANCL, but translocation of FANCD2 to chromatin is impaired [34]. In addition, when FANCD2 is depleted, replication-dependent incisions of ICLs are dramatically reduced [35].
Recently it was demonstrated that XPF binds SLX4, a related endonuclease, and this interaction is critical for ICL repair [36–39]. Fanconi anemia patients, mice deficient in ERCC1-XPF, and Slx4(Btbd12)−/− mice share many spontaneous developmental and degenerative phenotypes, supporting roles for all of these proteins in a common pathway and illustrating the dramatic consequences of failure to repair endogenous ICLs [40, 41]. Recent reports describe the discovery of biallelic mutations in SLX4 in two patients who exhibited clinical features of Fanconi anemia [42, 43]. Based on evidence that reintroduction of wild-type SLX4 into the patients’ cells rescued sensitivity to crosslinking agents, SLX4 is considered as a new complementation group of Fanconi anemia: FANCP.
1.3 DOUBLE-STRAND BREAK REPAIR
Orthologs of ERCC1-XPF in lower eukaryotes such as Arabidopsis thaliana, Drosophila melanogaster, and Saccharomyces cerevisiae play a vital role in the repair of DSBs and meiosis [44–47]. The two primary mechanisms of DSB repair are non-homologous end-joining (NHEJ) and homologous recombination (HR). Work in budding yeast has contributed tremendously to defining the role of ERCC1-XPF in DSB repair in mammalian cells. Mutation of rad10 or rad1, the orthologs of ERCC1 and XPF in S. cerevisiae, suppresses HR between sequence repeats [48–50]. The function of the RAD10-Rad1 nuclease in HR is to remove non-homologous 3’ termini of single-stranded overhangs of broken ends to facilitate single-strand annealing, an error-prone sub-pathway of HR [46, 47, 50, 51]. Like, single-strand annealing, there is an error prone sub-pathway of NHEJ that utilizes short stretches of homology to join two broken DNA ends termed micro-homology mediated end-joining. Rad10-Rad1 also participates in this end-joining pathway in yeast [52]. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences [52]. Mammalian cells deficient in ERCC1-XPF are modestly sensitive to ionizing radiation (IR), a source of DSBs [3]. Like in yeast, HR and end-joining of DSBs is attenuated in ERCC1-XPF-deficient mammalian cells [53–57]
The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells [3, 57]. Therefore, it is proposed that ERCC1-XPF nuclease facilitates both HR and NHEJ pathways (single-strand annealing and microhomology-mediated end-joining) but only if the broken DNA ends contain 3’-overhanging unmatched sequences or that cannot be used to prime DNA synthesis [3].
1.4 AT TELOMERES
ERCC1-XPF deficiency is linked with accelerated aging, and telomere shortening is associated with aging, therefore it was important to understand if the nuclease impacts telomere length or function [58]. Telomeres in humans with mutations in XPF, or Ercc1 knockout mice are not shorter than controls [8]. Furthermore, there is no difference in sister chromatid exchange at telomeres in the absence of ERCC1-XPF [59]. However, ERCC1 colocalizes with TRF2 at telomeres [8]. In a TRF2 dominant negative background, ERCC1-XPF deficient cells accumulate telomeric double-minutes. This led to the conclusion that ERCC1-XPF cleaves the G-rich, 3’- overhang, rendering chromosomes vulnerable to end-to-end fusions [8]. Hence the absence of ERCC1-XPF apparently does not have a deleterious impact on telomere length or function. Consistent with that, correction of XP-F cells or overexpression of XPF in normal human cells leads to telomere shortening [60]. Therefore, accelerated aging associated with ERCC1-XPF deficiency is presumed to arise from cellular senescence and cell death and not as a consequence of telomere-dependent replicative senescence.
2. XPF-DEFICIENCY IN HUMANS
Humans with mutations in XPF can be classified into two groups based on the clinical manifestations of their disease (see Table 1). The majority of XP-F patients present with mild symptoms of XP, which include sun sensitivity, freckling of the skin, and basal or squamous cell carcinomas typically occurring after the second decade of life. This is in contrast to many XP-A and XP-C patients, in which skin cancer occurs even before 2 yrs of age [61]. The second group of XP-F patients exhibit neurological deterioration in addition to their XP-like symptoms. There has been one published case of a patient with mutations in XPF with dramatically accelerated aging [10]. The mutation in XPF, its impact on protein expression, function and subcellular localization are all critical determinants in the clinical manifestations [62]. Of note, all XP-F patients carry a missense mutation in at least one allele, and none of these affect the catalytic domain of the protein (Table 1). This has led to speculation that ERCC1-XPF is essential for human life [11]. This is supported by the observation that mice homozygous for null alleles of these genes are not viable except in select genetic backgrounds (see the end of Section 3).
Table 1.
List of patients with verified mutations in XPF or ERCC1
| Patient Code |
Mutation | Amino Acid Changes |
Age1 (M or F) |
% UDS2 | UV-S3 | Neuro4 | Abnormal pigmentation5 |
Skin Carcinoma6 |
Ref |
|---|---|---|---|---|---|---|---|---|---|
| XP26BR | Base sub 2377 (C➔T) Homozygous |
Arg799➔Trp Arg799➔Trp |
15 | − | + | + | [62] | ||
| XP32BR | Arg589➔Trp Pro379➔Ser |
12 | 10 | + (2X) | − | + | + | [62] | |
| XP230S | lbp insert 1330 Heterozygous |
Lys455➔Stop4 82 |
45(F) | 10 | + (4X) | − | + | − | [63] |
| XP126LO | Base sub 2377 (C➔T) | Arg799➔Trp | 22(F) | 45 | + | − | − | [13, 67] | |
| XP7NE | Pro379➔Ser Silent |
28 | 25 | + (2X) | − | + | [73] | ||
| XP25KO | 8(F) | 12 | + (3X) | − | + | − | [66] | ||
| XP27KO | 11(F) | 12 | + (2.5X) | − | + | − | [68] | ||
| XP28KO | 8(F) | 12 | + (3X) | − | + | − | [68] | ||
| XP38KO | 44(F) | 20–25 | + (2.3X) | − | + | − | [69] | ||
| XP46KO | 61(F) | 12 | + (2.8X) | − | + | − | [66] | ||
| XP90TO | 42 (M) | 12 | + (3X) | − | + | + (42) | [70] | ||
| XP92TO | 40(F) | 12 | + (3X) | − | + | + (41) | [70] | ||
| XP29MA | 24 (M) | >5 | + (6X) | − | + | − | [71] | ||
| XP30MA | 29 (M) | >5 | + (6X) | − | + | − | [71] | ||
| XP42RO | Base sub 2377 (C➔T) Homozygous |
Arg799➔Trp | 62 (M) | 22 | + (2X) | + (47) | + | + (27) | [76] |
| XP24BR | Base sub 2377 (C➔T) Homozygous |
Arg799➔Trp Arg799➔Trp |
29 | 5 | + (3X) | + | + | + | [73] |
| XP24KY | 10bp deletion (1575) Base sub 2377 (C➔T) |
Val536➔Stop5 33 Arg799➔Trp |
48 (M) | 7 | + (3X) | + | + | − | [74] |
| XP62RO | Base sub 2377 (C➔T) Homozygous |
Arg799➔Trp Arg799➔Trp |
20 | + (2X) | + (late onset) | [62] | |||
| AS871 | Arg589➔Trp Deletion of Exon 3 |
15 | + (2X) | + | [62] | ||||
| XP51RO | Base sub 458 (G➔C) Homozygous |
Argl53➔Pro | 15 (M) | >5 | + (10X) | + (6) | − | [10] | |
| XP48DC | Base sub 2377 (C➔T) Heterozygous |
Arg799➔Trp | + | [129] | |||||
| C014TA | Base sub 2377 (C➔T) Heterozygous |
Arg799➔Trp | + | [129] | |||||
| CO107TA | Base sub 2377 (C➔T) Heterozygous |
Arg799➔Trp | + | [129] | |||||
| XP202DC | Nonsense Splice mutation in ERCC1 |
Lys226➔STOP IVS6-26G➔A |
15 | + | − | [129] | |||
| 165TOR | Glul58➔STOP Phe231➔Leu |
congenital | 15 | + (5X) | + | − | − | [11] |
Age of patient at time of diagnosis and patient’s gender.
Unscheduled DNA synthesis as a percentage compared to control cells
Fold increase in UV sensitivity of patient fibroblasts compared to control cells
Presence (+) or absence (−) of neurodegenerative symptoms.
Presence (+) or absence (−) of abnormal pigmentation including freckling, blistering, epidermal atrophy and keratoses.
Presence (+) or absence (−) of skin carcinomas (basal cell and squamous cell carinomas). Blue region indicates XP-F patients without symptoms of neurodegeneration; pink region indicates XP-F patients with symptoms of neurodegeneration; green region indicates patients with mutations in ERCC1.
The first XPF-deficient human patient was reported in 1979, several years before the XPF gene was identified and cloned [63]. The patient, referred to as XP23OS, was confirmed as XP-F by genetic complementation analysis, and exhibited mild XP symptoms including freckling and photosensitivity. Primary cells from patient XP23OS have only 10% of the normal level of NER as measured by UV-induced unscheduled DNA synthesis (UDS), but only modest sensitivity to UV as measured by clonogenic survival. The seeming discrepancy can be explained by the fact that UDS measures NER that occurs in the first 3 hours following UV irradiation, whereas in a clonogenic survival assay cell growth is measured in the 7–10 days following UV irradiation. Thus XP23OS cells must have low levels of NER, but that is adequate to prevent cell death and replicative senescence given adequate time to repair the genome [64]. Furthermore, host cell reactivation of reporter expression following UV damage was only modestly impaired. These results suggest that although the efficiency of NER was impaired in this patient, the pathway must be intact to explain the relatively mild symptoms in this 45 year-old patient. In the years that followed, several patients with XP group F were described, most of them from Japan, having mild to moderate symptoms, similar to patient XP23OS [65–71]. The majority of XP-F patients had UV sensitivity and freckling of the skin, but severe ocular and neurological symptoms were rare in the XP-F complementation group (see Table 1) [72, 73]. It is important to note that there have been reports of additional XP-F patients that are not included in Table 1 because the mutations have yet to be verified in genomic DNA [74].
The human XPF gene was cloned in 1996 with the identification of a human gene homologous to yeast Rad1 [13, 75]. This cDNA corrected the defect in cells from XP group F patients. Additionally, causative mutations were identified in XP-F patients that corresponded to this gene. Following the cloning of XPF, an unusual XP-F patient was described displaying progressive late-onset neurologic decline [76]. This patient, referred to as XP42RO, had mild ocular photophobia with severe erythema on sun exposure. Basal and squamous cell carcinomas were detected in the second decade of life and by the fourth decade the patient exhibited profound neurodegenerative symptoms including ataxia, cerebral and cerebellar atrophy. The mutation found in XP42RO cells is a C➔T transition at nucleotide 2377, which results in a change of the conserved arginine residue at 799 to a tryptophan. The R799W mutation was identified in at least eight other XP-F patients, in six of whom neurodegeneration was reported (see Table 1). Patient XP126LO harbors the R799W mutation but without neurologic symptoms to date. One possible explanation is that neurologic symptoms may not appear until the 4th or 5th decade of life, and this patient was assessed and diagnosed at 22 years of age.
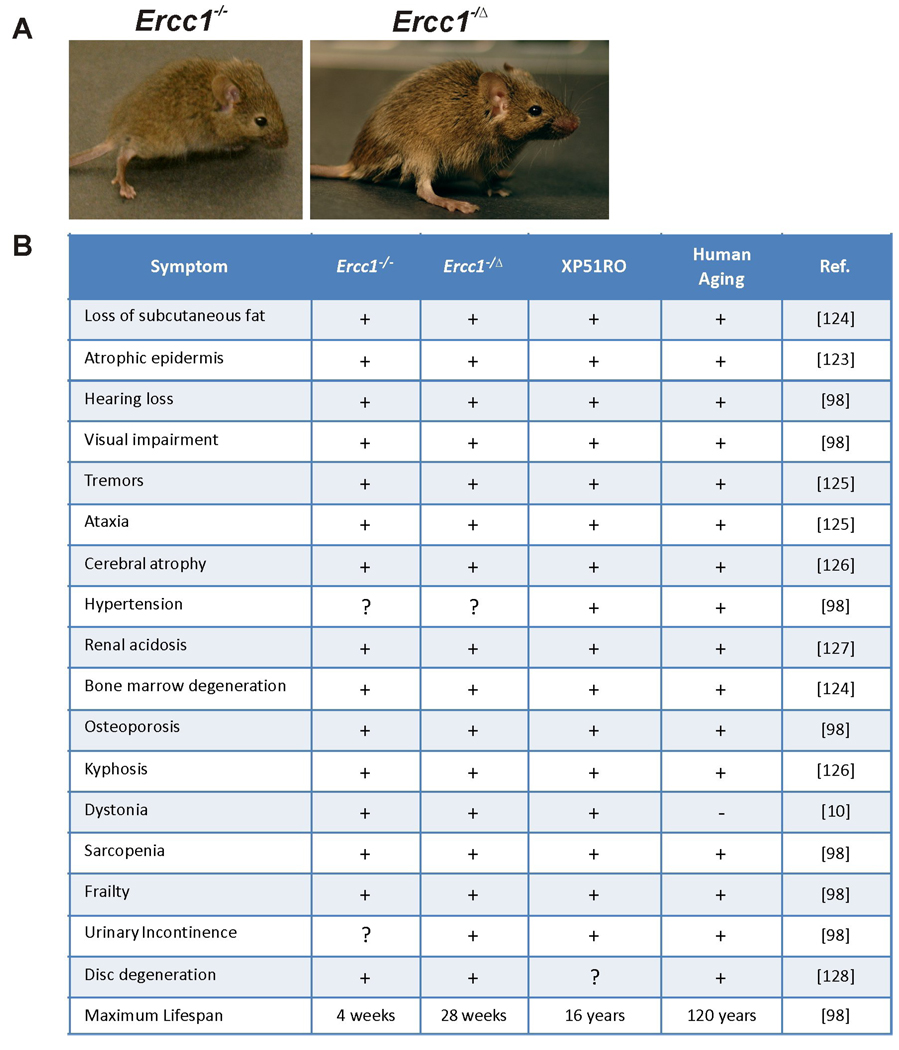
More recently, mutations in XPF were linked to a novel progeroid syndrome or disease of accelerated aging (called XFE progeroid syndrome). The patient, referred to XP51RO, had severe photosensitivity, which led investigators to hypothesize that NER was defective, and genetic complementation with XP patient cells revealed that XPF was affected [10]. In addition to sun sensitivity and the classical symptoms of XP, patient XFE had severe symptoms of accelerated aging that affected the neurologic, hepatobiliary, musculoskeletal, and hematopoietic systems. Mutation analysis revealed a G➔C transversion at position 458 which resulted in a non-conservative substitution of arginine at residue 153 to proline. The mutation in XPF was unexpected because patient XFE had severe symptoms of accelerated aging unlike most other XP-F patients who had mild XP. R153 is located within a conserved helicase motif of XPF which is also a leucine-rich region postulated to be important for protein-protein interactions. Primary fibroblasts from patient XP51RO are highly sensitive to UV and the crosslinking agent mitomycin C. Patient XP51RO had normal early postnatal development, but progeroid symptoms began to appear in early prepubescence. The symptoms included an old, wizened appearance, loss of subcutaneous fat, liver dysfunction, vision and hearing loss, renal insufficiency, muscle wasting, osteopenia, kyphosis and cerebral atrophy. These symptoms of accelerated aging are strikingly similar to those seen in Ercc1−/− and Xpfm/m mice (see Figure 1).
Figure 1.
(A) Representative images of Ercc1−/− (left; 3 wks of age) and Ercc1−/Δ (right; 18 wks of age) mice. (B) Table listing the symptoms of aging observed in Ercc1−/− mice, Ercc1−/Δ mice, and patient XP51RO who had a progeroid syndrome (or disease of accelerated aging) due to a homozygous mutation in XPF. Symptoms are compared to normal human aging. (+) indicates presence and (−) indicates absence of the symptom. References are listed in the righthand column. Also indicated are whether or not the same symptoms are associated with old age in humans and (+) or (−) indicates presence or absence in the Ercc1−/− mice, Ercc1−/Δ mice, or progeroid patient XP51RO.
Very recently a patient with features similar to XP51RO was identified in the UK. The patient has the facial appearance of CS, sun-sensitivity, microcephaly, neurological problems and developmental delay, together with pancytopenia and renal failure. Cellular studies revealed low UDS and assignment to the XP-F group (D. Pilz, D. McGibbon, R. Sarkany, M. Stefanini and A Lehmann, personal communication). XP-F patients with early onset and more severe symptoms appear to have mutations that lead to mislocalization of ERCC1-XPF to the cytoplasm, thereby decreasing the cellular capacity for efficient repair of nuclear DNA [62].
3. ERCC1 MUTATIONS IN HUMANS
ERCC1 was the first human DNA repair gene cloned [77]. For decades, however, no patients were identified with ERCC1 mutations; hence the gene does not have the standard XP nomenclature (XP-x) like other NER factors associated with xeroderma pigmentosum. Recently, a single patient was discovered who had mutations in ERCC1 resulting in severe pre- and postnatal developmental defects [11]. The patient, referred to as 165TOR, had severe skeletal defects at birth, including microcephaly, arthrogryposis and rocker-bottom feet. These abnormalities were seen in conjunction with neurological alterations including cerebellar hypoplasia and blunted cortical gyri. The clinical diagnosis was cerebro-oculo-facio-skeletal syndrome, or COFS syndrome. COFS syndrome was first reported by Lowry in 1971, and further characterized in the Manitoba aboriginal population by Pena and Shokeir in 1974 [78, 79]. COFS syndrome is a rare autosomal recessive disorder in which patients undergo rapid neurologic decline. Symptoms include but are not limited to brain microcephaly and atrophy with calcifications, cataracts, optic atrophy, progressive joint contractures, and severe postnatal growth failure [80]. Patients with COFS syndrome are reported to have mutations in genes encoding DNA repair proteins ERCC6/CSB, ERCC5/XPG and ERCC2/XPD [81, 82].
Two mutations were found in the coding region of ERCC1 in patient 165TOR. The maternal allele harbors a C➔T transition that converts Gln158 into an amber translational stop codon. The result is a truncated polypeptide that lacks the entire C-terminal domain, essential for binding XPF [83]. The paternal allele has a C➔G transversion, resulting in the conversion of Phe231 to leucine. This amino acid falls within the C-terminal tandem helix-hairpin-helix domain of ERCC1, critical for binding XPF, and is conserved in invertebrates and mammals [83]. ERCC1 mRNA levels were normal in this patient, although the protein levels of ERCC1 and XPF in the nucleus were reduced 4–5-fold. The truncated protein was not detectable by immunoblot. Accordingly, fibroblasts from patient 165TOR had 15% of the normal level of NER, representing a modest defect, suggesting that the missense mutation affects stability of ERCC1-XPF and/or its nuclear localization, but not enzymatic activity.
Unexpectedly, the expression of ERCC1-XPF in 165TOR cells is reduced but comparable to a patient with mild XP-F. Several hypotheses have been proposed to explain this incongruity, the simplest being that ERCC1 and XPF play distinct roles in vivo. Evidence against this hypothesis includes the fact that Ercc1−/− and Xpfm/m mice have apparently identical phenotypes [29, 32]. Additionally, ERCC1 and XPF are required to stabilize one another in vivo [83, 84]. It is important to note that COFS is also caused by mutations in the TFIIH subunit XPD and the NER endonuclease XPG that stabilizes TFIIH [80, 85–87]. ERCC1-XPF and the other members of the NER machinery have been demonstrated to play a role in regulating transcription that has been suggested to be independent of their role in DNA repair [88]. Therefore, the clinical severity of patient 165TOR may be due to a transcriptional transcription-coupled nucleotide excision repair defect during development. It is also possible that 165TOR was genetically replete of functional ERCC1-XPF during development, but selective pressure led to partial reversion of the cellular phenotype postnatally. Reversion with mosaicism has been reported in many genome instability disorders including Fanconi anemia, Werner and Bloom syndromes, and observed for ERCC1-XPF deficient cells chronically exposed to crosslinking agents [40, 89–91]. In addition, there are undoubtedly modifier genes that affect the severity of symptoms caused by mutations in ERCC1 or XPF as illustrated by the fact that Ercc1−/− mice are not viable in inbred C57Bl/6 or FVB/n inbred backgrounds but are born with Mendelian frequency in an f1 mixed background [10]. Finally, little is known about regulation of ERCC1-XPF expression, which could be tissue-specific and therefore contribute to heterogeneous phenotypes. Identifying modifier genes, identifying regulators of nuclease expression and modeling additional patient mutations in mice will be essential for deciphering genotype:phenotype correlations. A second patient with mutations in ERCC1 was briefly described recently [129]. The patient had a nonsense mutation affect amino acid 226, which lies early in the helix-hairpin-helix domain necessary for binding XPF. The second allele contains a splicing mutation (IVS6-G➔A). The patient displayed neurologic symptoms beginning at age 15 years and died by the age of 37. Neurodegeneration was progressive and severe resulting in dementia and cortical atrophy. The symptoms are very similar to XPF patients with neurologic involvement (Table 1) supporting the conclusion that ERCC1 and XPF function exclusively as a complex in vivo.
4. MOUSE MODELS OF ERCC1-XPF DEFICIENCY
4.1 ERCC1 KNOCKOUT MICE
To understand the biological significance of ERCC1, the gene was knocked-out in the mouse by two independent laboratories [29, 92]. The two knockout alleles were created by interrupting different exons. McWhir et al created the first knockout mouse model by disrupting exon 5 of Ercc1 leading to a truncated transcript missing the last four exons, which contain the XPF interaction domain [17, 29, 84]. The second knockout strain was generated by inserting a neomycin resistance cassette into exon 7 of Ercc1 [92]. The result was a truncation in the helix-hairpin-helix motif required for interaction with XPF [83]. Ercc1 mRNA was not detected in these mice [92]. The former strain is born with Mendelian frequency; the latter is sub-Mendelian, likely due to differences in the genetic background [29, 92]. Deletion of ERCC1 is lethal in a fully inbred genetic background, indicating that there are modifier genes that influence the severity of the phenotype [10]. In both knockout strains, postnatal growth is severely retarded and the mice die at approximately 3 weeks of age when they weigh only about 20% compared to their normal littermates [29, 92]. The median lifespan of Ercc1−/− mice in an f1 mixed genetic background of 50:50 C57BL/6:FVB/n is 21 days and the maximum lifespan 28 days [10]. The Ercc1−/− mice spontaneously develop symptoms characteristic of progressive neurodegeneration, including dystonia, trembling and ataxia [10].
The hematopoietic system of Ercc1−/− mice develops normally [93]. However, by the end of life, Ercc1−/− mice are leukopenic and thrombocytopenic, and there is extensive adipose transformation of the bone marrow, hallmark features of normal aging in mice [93]. Proliferation of multi-potent and lineage-committed progenitors from Ercc1−/− mice is profoundly impaired [93]. Collectively, these data suggest that ERCC1-deficient mice undergo rapid turnover of hematopoietic cells leading to premature exhaustion of stem cell reserves. In addition, bone marrow progenitors from Ercc1−/− mice are exquisitely sensitive to crosslinking agents, similar to murine models of Fanconi anemia [93, 94].
The liver of Ercc1−/− mice is prominently affected, with hepatocellular polyploidy, aneuploidy and G2 arrest [29, 95, 96]. The structural changes correlate with impaired liver function as demonstrated by significantly increased liver enzymes in the serum [29]. Ercc1−/− mice develop kyphosis, sarcopenia, dystonia and ataxia, indicative of musculoskeletal and nervous system defects [10]. There is a suppression of the somatotroph, lactotroph and thyrotroph hormonal axes in the Ercc1−/− mice, which explains their growth delay and diminutive size [10]. Many of the degenerative and endocrine abnormalities are similar to what occurs with old age in mice [97, 98]. To further investigate the relationship of DNA repair deficiency to normal aging, genome-wide expression changes in Ercc1−/− mice relative to wild-type littermates was compared to changes that occur with natural aging (differences in gene expression between old wild-type and young wild-type mice). There is a highly significant overlap between these two profiles whether comparing gene-by-gene or by comparing over-represented biological pathways [10]. This provided some of the early support for the notion that DNA damage may contribute to aging.
4.2 XPF MUTANT MICE
Tian et al recreated the XPF mutation in patient XP23OS in the mouse [32]. The patient had a single base insertion after nucleotide 1330 leading to a frameshift mutation after Lysine 455 and a stop codon 38 residues later [74]. This leads to truncation of XPF upstream of the catalytic domain and its ERCC1-interaction domain [17]. The clinical phenotype of the patient was mild, having reached her 4th decade without neurodegeneration or skin cancer [74]. In keeping with this, the level and length of the XPF transcript in XP23OS cells are comparable to normal cells, illustrating that the patient must have a 2nd XPF allele. To further emphasize this, a mouse homozygous for the frameshift mutation has undetectable levels of Xpf mRNA and a severe phenotype identical to that of ERCC1 null mice [32]. The Xpfm/m mice develop normally and are born with Mendelian frequency. However, postnatal growth is delayed such that by 2 wks of age the Xpfm/m mice are approximately 25% the size of littermates, and die by 3 wks of age. Hepatocellular polyploidy was prominent, as in the Ercc1−/− mice [32]. The phenotypic parallels between ERCC1 and XPF null mice strongly suggest that the proteins function exclusively as a complex.
Importantly, ERCC1 and XPF-deficient mice appear to have a normal complement of mature and immature B cells [32, 99, 100]. This provides definitive evidence that ERCC1-XPF is not essential for NHEJ, the DSB repair pathway required for class switch recombination (CSR) [101]. However, ex vivo CSR is mildly attenuated in splenocytes isolated from Ercc1−/− mice and the mutation pattern in the switch region of the immunoglobulin locus is significantly different from that of normal littermate mice, suggesting that ERCC1-XPF may contribute to DNA end-processing of DSBs at the Ig locus [100]. Consistent with this, ERCC1-deficient mice are hypersensitive to IR, which induces DNA DSBs [3].
4.3 LIVER CORRECTED Ercc1−/− MICE
To investigate the cause of death in Ercc1−/− mice, Selfridge et al crossed the mice with a transgenic strain expressing ERCC1 specifically in the liver, using transthyretin (TTR) regulatory sequences to control ERCC1 expression [102]. The resulting mice (Ercc1−/− + TG) have dramatically improved growth, reaching 58% of normal body weight for their age. Furthermore, their lifespan is significantly increased, with a median survival of ∼75 days. Hepatocellular polyploidy and abnormal liver functions are largely corrected by expression of ERCC1 in the liver. Interestingly, by 7 wks of age, the transgenic mice begin to display evidence of renal dysfunction (significantly elevated serum creatinine and proteinuria) and renal histopathology (glomerulosclerosis, hyaline casts, renal tubular epithelial anisokaryosis, karyomegaly, hyperchormasia, pyknosis and keryorrhexis) [102, 103]. These data suggest that Ercc1−/− mice die of liver failure and that hepatocytes and renal cells are the most vulnerable to loss of ERCC1-XPF-dependent DNA repair.
Since the Ercc1−/− + TG mice live longer than Ercc1−/− mice, they are a practical system for identifying other tissues affected by ERCC1-deficiency. Performance of Ercc1−/− + TG mice on an opto-kinetic response test to measure visual acuity is impaired by 4 wks and worsens with age [103]. Structural abnormalities of the eye were not detected, indicating that loss of vision is not due to a developmental defect [103]. Also, there is no evidence for retinal degeneration typical of CSB mice, another NER-defective mutant strain [104].
Like Ercc1−/−, symptoms associated with neurodegeneration were observed in Ercc1−/− + TG mice. The Ercc1−/− + TG mice displayed dystonia and ataxia indicative of a cerebellar defect. While mild atrophy of the neocortex and cerebellum were observed, there were no abnormalities or loss of Purkinje cells to explain these phenotypes. Also, there were no signs of degeneration at the neuromuscular junctions. Therefore these symptoms were attributed to uraemic encephalopathy, caused by kidney failure [103].
Male and female Ercc1−/− + TG mice were found to be infertile [105]. Testes of these mice were approximately 50% the normal size at puberty and contained significantly less spermatocytes. Spermatogenesis is not arrested at a particular stage, as expected for a meiotic defect, but instead there is abundant apoptosis in these rapidly dividing cells [105, 106]. This leads to a resulting 97% reduction in the sperm count [106]. Ovaries from adult Ercc1−/− + TG mice showed a reduced number of oocytes and an absence of primary follicles [105].
4.4 ERCC1 MUTANT MICE
To further probe the DNA repair function of ERCC1 in vivo, a premature stop codon was engineered at position 292 of mErcc1 [92]. This results in a C-terminal deletion of 7 amino acids of the murine protein, including a phenylalanine residue at position 293, thought to be essential for binding to XPF [83]. Thus the prediction was that this mutation would ablate DNA repair function without compromising the protein stability [84]. Unlike either of the null alleles, normal levels of the mutant Ercc1 transcript are detected in the tissues from the mutant mice [92]. Homozygous Ercc1*292 (also referred to as Ercc1Δ/Δ) mice live up to 6 months, which is 6X longer than ERCC1 null mice. Similar to the Ercc1−/− mice, the Ercc1*292 mice are infertile and their skin is atrophic and lacks subcutaneous fat [92]. The spleen of Ercc1*292 mice contains increased ferritin and hemosiderin deposits, indicative of a high turnover of erythrocytes. The kidney exhibits dilated renal tubules with hyaline casts. Nuclear polyploidy is common in the liver and kidney. Thus virtually all of the phenotypes of Ercc1−/− mice are recapitulated in this longer-lived mutant strain. Primary mouse embryonic fibroblasts (MEFs) from the Ercc1−/− mice are modestly more sensitive to the crosslinking agent mitomycin C than MEFs from Ercc1*292 mice, suggesting that their increased longevity is due to increased DNA repair capacity [92]. Despite this, topical application of the tumor initiator DMBA to Ercc1*292 mice, leads to acute toxicity rather than carcinogenesis, illustrating a dramatic difference from other NER-deficient mice [92].
4.5 ERCC1 HYPOMORPHIC MICE
Combination of one null and one mutant Ercc1 allele yields mice (Ercc1−/*292; Ercc1−/Δ or Ercc1d/−) that are born with Mendelian frequency and have an even greater maximal lifespan of 7 months in an f1 background of 50:50 C57Bl/6J:FVB/n [107]. Ercc1−/Δ mice are runted compared to their wild-type littermates [108]. However, they develop normally until sexual maturity, at which point they began to exhibit signs of rapid aging [109]. They live 24–30 weeks while progressively developing dystonia, tremors, kyphosis and ataxia [108].
The Ercc1−/Δ mice were crossed with a transgenic lacZ reporter strain to measure the mutation frequency in vivo [107]. The mutation frequency is modestly elevated in the liver of 5–6 month old Ercc1−/Δ mice compared to normal littermates. Interestingly, the mutations are primarily chromosomal rearrangements characteristic of old wild-type mice rather than point mutations characteristic of NER-deficient mice [107]. This observation extends the parallels between the progeroid Ercc1−/Δ mice and aged normal mice.
Ercc1−/Δ mice are hypersensitive to IR [3]. Even though these mice are hypomorphic for ERCC1-XPF, they were equally as sensitive to IR as DNA-PKcs knockout mice [110]. IR causes persistent γH2AX foci in ERCC1-deficient cells and mice, supporting a role for ERCC1-XPF in the repair of DSBs. The involvement of ERCC1-XPF in DSB repair is presumably a Ku-independent pathway since Ercc1−/− Ku86−/− mice are not viable [3]. Ercc1−/− MEFs have normal levels of spontaneous and mitomycin C-induced sister chromatid exchanges, illustrating that ERCC1-XPF is not essential for homologous recombination [56, 111]. In contrast, DSBs with 3’ overhangs cause large deletions in Ercc1−/− cells [3, 56]. These genetic and in vitro data support a role for ERCC1-XPF in processing a subset of DSBs (those with 3’ overhangs) and a role in the alternative end-joining pathway of DSB repair [112].
Ercc1−/−, Ercc1−/− + TG, and Ercc1−/Δ mice all develop progressive dystonia, tremors and ataxia, highly suggestive of neurodegeneration [10, 103, 108]. The former strains display cerebellar hypoplasia, similar to the ERCC1 patient 165TOR, but this could not be completely dissected from developmental abnormalities, due to their young age [10, 11, 103]. Evidence for degenerative processes in the central nervous system is quite clear in Ercc1−/Δ mice [108]. De Waard et al found profound astrocytosis and microgliosis in the spinal cord of 4 month old Ercc1−/Δ mice, compared to normal littermates and 1 month old mutant animals. This is accompanied by a significant reduction in the number of motor neurons in the ventral horn of the spinal cord and a concomitant denervation of the skeletal muscle. There is an approximately 50% reduction in neurons from 4–8 wks of life, and then again from 8–16 wks of life in the Ercc1−/Δ mice. It is also evident that the pre-synaptic motor nerve terminals have degenerated in the aged Ercc1−/Δ mice with characteristics similar to those seen in amyotrophic lateral sclerosis and aging motor neurons [108].
Genome-wide expression profiling of Ercc1−/Δ mice revealed a highly significant correlation with the transcriptome of numerous long-lived models, including Ames and Snell dwarf mice and/or calorically restricted mice [97]. In addition, there is a significant correlation with the transcriptome of old wild-type mice. This indicates that Ercc1−/Δ mice look biologically “old”, but also that the failure to repair DNA damage in these mice triggers activation of a stress response that is transcriptionally regulated and promotes longevity. Schumacher et al showed that this same stress response is also triggered by nutritional deprivation and is mediated through suppression of GH-IGF1 signaling as evidenced by the significant correlation with Ames and Snell dwarf mice. All of these mice displayed suppression of the somatotroph axis, oxidative metabolism and peroxisomal biogenesis coupled with an upregulation of antioxidants, DNA damage and apoptosis [97].
This was further supported by analysis of metabolites in the serum and urine of Ercc1−/Δ mice [109]. There is no difference between Ercc1−/Δ mice and normal littermates at 8 and 12 weeks of age. However by 16 weeks, there are significant differences, which become further amplified by 20 weeks of age. Collectively, these data support the conclusion that Ercc1−/Δ mice develop normally into adulthood, but then undergo degenerative changes. Several of the metabolic changes mimic those that occur with caloric restriction, including increased HDL, decreased LDL and VLDL, and ketosis. However, a number of metabolic changes in the Ercc1−/Δ mice are also consistent with degeneration, such as increased glucose, citrate and succinate in the urine indicative of kidney dysfunction, and metabolic alkalosis indicative of liver dysfunction [109]. These observations are consistent with the model that DNA damage accumulates in Ercc1−/Δ mice leading to metabolic reprogramming in response to stress [97]. This may be beneficial, but in the long-run is insufficient to sustain the organism in the face of continued DNA damage [10].
Remarkably, Ercc1−/Δ mice spontaneously develop numerous diseases associated with old age in humans. This includes osteoporosis and intervertebral disc degeneration [113]. There is progressive attrition of disc extracellular proteoglycans with age, leading to loss of disc height and its cushioning function [114, 115]. Similar changes were observed in discs of 5 month old Ercc1−/Δ mice and exacerbated in mice treated with genotoxic chemotherapeutic agents [113]. These observations support the conclusion that DNA damage, if not repaired, can promote common aging-related degenerative diseases, even in post-mitotic tissues.
4.6 TISSUE-SPECIFIC DELETION OF ERCC1
Tissue-specific deletion of a gene is a powerful tool for dissecting a complex phenotype, such as that of the ERCC1-deficient mice, allowing for dissection of whether a particular symptom or pathology is a direct consequence of a deletion of a gene or merely a secondary consequence (e.g., is neurodegeneration due to loss of ERCC1 in neurons or uraemic encephalopathy due to loss of ERCC1 in the kidneys?). Tissue specific knockout of protein expression occurs if the gene of interest (or an exon vital to its function) is flanked (floxed) by recombination signals (loxP or FRT) and mice expressing the two copies of the floxed allele are crossed with transgenic mice expressing recombinase (CRE or FLP, respectively) under a tissue specific promoter [116]. Another important attribute of this approach is that it is possible to distinguish developmental from degenerative changes by selecting promoters that are active only postnatally.
A floxed allele of Ercc1 was generated by inserting loxP sites in intron 2 and 5, such that Cre recombinase excises exons 3–5 of the Ercc1 locus [117]. Mice harboring two floxed alleles of Ercc1 were crossed with transgenic mice expressing Cre-recombinase under a tyrosinase promoter [118]. The goal was to knockout expression of ERCC1 in melanocytes to generate a murine melanoma model. Unexpectedly, the mice die by 6 months of age due to severe colonic obstruction. Tyrosinase is expressed in all neural crest cell-derived lineages, including parasympathetic neurons that innervate the gastrointestinal tract and are required for colonic peristalsis. Knocking-out ERCC1 expression in neural crest cells causes p53 activation and apoptosis of ganglion cells in the mesenteric plexus. Denervation of the bowel explains the colonic obstruction and demonstrates that ERCC1-XPF dependent DNA repair is critical for protecting neurons from degeneration [118]. This provides strong evidence that the neurological symptoms observed in ERCC1-deficient mice are due, at least in part, to loss of functional neurons rather than a defect in supportive glial cells.
ERCC1 was also knocked-out specifically in the skin to create a model of UV-induced skin cancer. Ercc1flox/− mice were crossed with transgenic mice expressing Cre recombinase under the keratin 5 (K5) promoter, which is only expressed in the basal layer of the epidermis [117]. To facilitate UV carcinogenesis studies, these mice were produced in an albino, hairless background. Deletion of Ercc1 in the skin leads to a 20-fold reduction in the minimal erythemal dose in response to UV-B irradiation, leading to dramatic, but transient hyperplasia [117]. Furthermore the mice develop significantly more skin tumors, early, and at a lower dose of UVB than normal controls. The cumulative dose of UV-B required to induce tumors in half of the ERCC1-deficient mice was 37X lower than normal controls in a chronic exposure study [117]. The mice also developed actinic keratosis and squamous cell carcinomas. Thus this tissue-specific knockout strain offers an accurate and rapid (tumors within 8 weeks) model of UV-induced skin cancer, which may be useful in the study of XP. Subsequently, these mice have been used to test topical treatments that protect against UV-induced skin cancer [119].
4.7 DOUBLE MUTANT MICE
TRF1 and TRF2 are sheltrin proteins required to protect telomeres at chromosomal ends [120]. Overexpression of either protein leads to a dominant negative effect exemplified by telomere shortening, loss of telomeric 3’ G-rich overhangs and end fusions [9, 121]. Overexpression of TRF2 in the skin of mice by putting the cDNA under control of the keratin 5 promoter (K5TRF2), leads to skin atrophy, hyperpigmentation and increased skin cancer in sun-exposed areas. These results demonstrate that dysregulation of telomeres promotes UV-induced skin cancer [9]. Skin and keratinocytes isolated from either K5TRF1 or K5TRF2 mice contain telomeric defects, which were rescued by knocking-out Xpf [9, 121]. This strongly supports the previous observation that the absence of ERCC1-XPF does not negatively impact telomere length or function, but instead has an unexpected beneficial effect [8].
5. CONCLUDING REMARKS
This review provides a comprehensive overview of the physiological impact of reduced expression or activity of ERCC1-XPF DNA repair endonuclease. There is tremendous variability between patients with mutations in ERCC1 or XPF, ranging from mild cutaneous symptoms to severe neurodegeneration. ERCC1 patient 165TOR had severe developmental abnormalities that are not normally seen in XP-F patients. These differences cannot be fully predicted by the patients’ mutations or the level of residual NER (UDS) in patient fibroblasts. Clearly, further work is needed to decipher how expression and activity of ERCC1-XPF is regulated. This in turn is likely to yield better methods for predicting the physiological consequences of a particular mutation [122]. The studies in mice have been crucial for a number of reasons. First, the identical phenotypes of Ercc1−/− and Xpfm/m mice, which are null for XPF, provide the strongest possible evidence that ERCC1 and XPF must function exclusively as a heterodimer. Second, the unexpected premature aging phenotypes of the ERCC1 mutant mice led to the discovery of a new rare genetic disease (XFE progeroid syndrome) and contributed to the body of evidence that DNA damage is one type of cellular damage that promotes aging-related degenerative changes (e.g., neurodegeneration). Third, it is clear that the severity of symptoms, associated with ERCC1-XPF deficiency, are somehow linked to variable levels of expression or activity. This implies that functional single nucleotide polymorphisms in either gene may be important for predicting risk of cancer or degenerative diseases. Fourth, Ercc1 mutant mice are unique amongst the NER-deficient mutant strains because they spontaneously develop neurodegeneration, which may be used to screen therapies for treating XP, CS and TTD patients. Finally, tissue-specific ERCC1 knockout strains will be crucial for identifying which tissues and cell types are most vulnerable to DNA damage and are responsible for triggering systemic stress responses.
ACKNOWLEDGEMENTS
The authors are supported by the National Institutes of Health grants ES016114 and -032. L.J.N. is also supported but the University of Pittsburgh Claude D. Pepper Center (P30AG024827). The authors are extremely grateful to Dr. Lehmann and colleagues for contributing unpublished information and to the reviewers for critical additions to this manuscript.
List of Abbreviations
- CS
Cockayne syndrome
- COFS
cerebro-oculo-facio-skeletal syndrome
- CPD
cyclopyrimidine dimer
- CSR
class switch recombination
- DSB
double-strand break
- ERCC1
excision repair cross complementation group 1
- HR
homologous recombination
- ICL
interstrand crosslink
- Ig
immunoglobulin
- IR
ionizing radiation
- K5
keratin 5
- MEF
mouse embryonic fibroblast
- NER
nucleotide excision repair
- NHEJ
non-homologous end-joining
- SSA
single-strand annealing
- TG
transgenic
- TTD
trichothiodystrophy
- TTR
transthyretin
- UDS
unscheduled DNA synthesis- a measure of NER
- UV
ultraviolet light
- XPF
xeroderma pigmentosum complementation group F
- XP
xeroderma pigmentosum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 2.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlake C, Ostermann K, Schmidt H, Gutz H. Analysis of DNA repair pathways of Schizosaccharomyces pombe by means of swi-rad double mutants. Mutat Res. 1993;294:59–67. doi: 10.1016/0921-8777(93)90058-o. [DOI] [PubMed] [Google Scholar]
- 6.Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–627. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busch DB, van Vuuren H, de Wit J, Collins A, Zdzienicka MZ, Mitchell DL, Brookman KW, Stefanini M, Riboni R, Thompson LH, Albert RB, van Gool AJ, Hoeijmakers J. Phenotypic heterogeneity in nucleotide excision repair mutants of rodent complementation groups 1 and 4. Mutat Res. 1997;383:91–106. doi: 10.1016/s0921-8777(96)00048-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 9.Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 10.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 11.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JH, Vermeulen W. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 13.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, Rademakers S, de Rooij J, Jaspers NG, Hoeijmakers JH, Wood RD. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 14.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardwell AJ, Bardwell L, Tomkinson AE, Friedberg EC. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 16.Park CH, Bessho T, Matsunaga T, Sancar A. Purification and characterization of the XPF-ERCC1 complex of human DNA repair excision nuclease. J Biol Chem. 1995;270:22657–22660. doi: 10.1074/jbc.270.39.22657. [DOI] [PubMed] [Google Scholar]
- 17.Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, Oberman R, Scharer OD, Wagner G, Ellenberger T. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 2007;26:4768–4776. doi: 10.1038/sj.emboj.7601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci U S A. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc Natl Acad Sci U S A. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripsianes K, Folkers G, Ab E, Das D, Odijk H, Jaspers NG, Hoeijmakers JH, Kaptein R, Boelens R. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–1858. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Kraemer KH, Levy DD, Parris CN, Gozukara EM, Moriwaki S, Adelberg S, Seidman MM. Xeroderma pigmentosum and related disorders: examining the linkage between defective DNA repair and cancer. J Invest Dermatol. 1994;103:96S–101S. doi: 10.1111/1523-1747.ep12399329. [DOI] [PubMed] [Google Scholar]
- 23.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 24.Weeda G, Eveno E, Donker I, Vermeulen W, Chevallier-Lagente O, Taieb A, Stary A, Hoeijmakers JH, Mezzina M, Sarasin A. A mutation in the XPB/ERCC3 DNA repair transcription gene, associated with trichothiodystrophy. Am J Hum Genet. 1997;60:320–329. [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh T, Linn S, Ono T, Yamaizumi M. Reinvestigation of the classification of five cell strains of xeroderma pigmentosum group E with reclassification of three of them. J Invest Dermatol. 2000;114:1022–1029. doi: 10.1046/j.1523-1747.2000.00952.x. [DOI] [PubMed] [Google Scholar]
- 26.Hoy CA, Thompson LH, Mooney CL, Salazar EP. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 1985;45:1737–1743. [PubMed] [Google Scholar]
- 27.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet. 1993;5:217–224. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- 30.de Vries A, van Oostrom CT, Hofhuis FM, Dortant PM, Berg RJ, de Gruijl FR, Wester PW, van Kreijl CF, Capel PJ, van Steeg H, et al. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature. 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 31.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, et al. High incidence of ultraviolet-B-or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature. 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 32.Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D'Andrea A, Niedernhofer LJ, McHugh PJ. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DM, Rouse J. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard PH, McIntyre RE, Gallagher F, Kettunen MI, Lewis DY, Brindle K, Arends MJ, Adams DJ, Patel KJ. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AW, Jaspers NG, Bettecken T, Joenje H, Schindler D, Rouse J, de Winter JP. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 44.Hefner E, Preuss SB, Britt AB. Arabidopsis mutants sensitive to gamma radiation include the homologue of the human repair gene ERCC1. J Exp Bot. 2003;54:669–680. doi: 10.1093/jxb/erg069. [DOI] [PubMed] [Google Scholar]
- 45.Baker BS, Carpenter AT, Ripoll P. The Utilization during Mitotic Cell Division of Loci Controlling Meiotic Recombination and Disjunction in DROSOPHILA MELANOGASTER. Genetics. 1978;90:531–578. doi: 10.1093/genetics/90.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 47.Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein HL. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiestl RH, Prakash S. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol Cell Biol. 1990;10:2485–2491. doi: 10.1128/mcb.10.6.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sargent RG, Rolig RL, Kilburn AE, Adair GM, Wilson JH, Nairn RS. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc Natl Acad Sci U S A. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000;28:3771–3778. doi: 10.1093/nar/28.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 2000;19:5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satoh H, Hiyama K, Takeda M, Awaya Y, Watanabe K, Ihara Y, Maeda H, Ishioka S, Yamakido M. Telomere shortening in peripheral blood cells was related with aging but not with white blood cell count. Jpn J Hum Genet. 1996;41:413–417. doi: 10.1007/BF01876332. [DOI] [PubMed] [Google Scholar]
- 59.Hagelstrom RT, Blagoev KB, Niedernhofer LJ, Goodwin EH, Bailey SM. Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc Natl Acad Sci U S A. 107:15768–15773. doi: 10.1073/pnas.1006338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech Ageing Dev. 2008;129:602–610. doi: 10.1016/j.mad.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 62.Ahmad A, Enzlin JH, Bhagwat NR, Wijgers N, Raams A, Appledoorn E, Theil AF, JH JH, Vermeulen W, NG JJ, Scharer OD, Niedernhofer LJ. Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients. PLoS Genet. 6:e1000871. doi: 10.1371/journal.pgen.1000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arase S, Kozuka T, Tanaka K, Ikenaga M, Takebe H. A sixth complementation group in xeroderma pigmentosum. Mutat Res. 1979;59:143–146. doi: 10.1016/0027-5107(79)90202-1. [DOI] [PubMed] [Google Scholar]
- 64.Zelle B, Berends F, Lohman PH. Repair of damage by ultraviolet radiation in xeroderma pigmentosum cell strains of complementation groups E and F. Mutat Res. 1980;73:157–169. doi: 10.1016/0027-5107(80)90144-x. [DOI] [PubMed] [Google Scholar]
- 65.Nishigori C, Ishizaki K, Takebe H, Imamura S, Hayakawa M. A case of xeroderma pigmentosum group F with late onset of clinical symptoms. Arch Dermatol. 1986;122:510–511. [PubMed] [Google Scholar]
- 66.Yamamura K, Ichihashi M, Hiramoto T, Ogoshi M, Nishioka K, Fujiwara Y. Clinical and photobiological characteristics of xeroderma pigmentosum complementation group F: a review of cases from Japan. Br J Dermatol. 1989;121:471–480. doi: 10.1111/j.1365-2133.1989.tb15514.x. [DOI] [PubMed] [Google Scholar]
- 67.Norris PG, Hawk JL, Avery JA, Giannelli F. Xeroderma pigmentosum complementation group F in a non-Japanese patient. J Am Acad Dermatol. 1988;18:1185–1188. doi: 10.1016/s0190-9622(88)70121-8. [DOI] [PubMed] [Google Scholar]
- 68.Fujiwara Y, Ichihashi M, Uehara Y, Matsumoto A, Yamamoto Y, Kano Y, Tanakura Y. Xeroderma pigmentosum groups C and F: additional assignments and a review of the subjects in Japan. J Radiat Res (Tokyo) 1985;26:443–449. doi: 10.1269/jrr.26.443. [DOI] [PubMed] [Google Scholar]
- 69.Fujiwara Y, Uehara Y, Ichihashi M, Nishioka K. Xeroderma pigmentosum complementation group F: more assignments and repair characteristics. Photochem Photobiol. 1985;41:629–634. doi: 10.1111/j.1751-1097.1985.tb03538.x. [DOI] [PubMed] [Google Scholar]
- 70.Kondo S, Mamada A, Miyamoto C, Keong CH, Satoh Y, Fujiwara Y. Late onset of skin cancers in 2 xeroderma pigmentosum group F siblings and a review of 30 Japanese xeroderma pigmentosum patients in groups D, E and F. Photodermatol. 1989;6:89–95. [PubMed] [Google Scholar]
- 71.Thielmann HW, Popanda O, Edler L, Jung EG. Clinical symptoms and DNA repair characteristics of xeroderma pigmentosum patients from Germany. Cancer Res. 1991;51:3456–3470. [PubMed] [Google Scholar]
- 72.Moriwaki S, Nishigori C, Imamura S, Yagi T, Takahashi C, Fujimoto N, Takebe H. A case of xeroderma pigmentosum complementation group F with neurological abnormalities. Br J Dermatol. 1993;128:91–94. doi: 10.1111/j.1365-2133.1993.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 73.Berneburg M, Clingen PH, Harcourt SA, Lowe JE, Taylor EM, Green MH, Krutmann J, Arlett CF, Lehmann AR. The cancer-free phenotype in trichothiodystrophy is unrelated to its repair defect. Cancer Res. 2000;60:431–438. [PubMed] [Google Scholar]
- 74.Matsumura Y, Nishigori C, Yagi T, Imamura S, Takebe H. Characterization of molecular defects in xeroderma pigmentosum group F in relation to its clinically mild symptoms. Hum Mol Genet. 1998;7:969–974. doi: 10.1093/hmg/7.6.969. [DOI] [PubMed] [Google Scholar]
- 75.Brookman KW, Lamerdin JE, Thelen MP, Hwang M, Reardon JT, Sancar A, Zhou ZQ, Walter CA, Parris CN, Thompson LH. ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol Cell Biol. 1996;16:6553–6562. doi: 10.1128/mcb.16.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sijbers AM, van Voorst Vader PC, Snoek JW, Raams A, Jaspers NG, Kleijer WJ. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J Invest Dermatol. 1998;110:832–836. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- 77.Westerveld A, Hoeijmakers JH, van Duin M, de Wit J, Odijk H, Pastink A, Wood RD, Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- 78.Lowry RB, MacLean R, McLean DM, Tischler B. Cataracts, microcephaly, kyphosis, and limited joint movement in two siblings: a new syndrome. J Pediatr. 1971;79:282–284. doi: 10.1016/s0022-3476(71)80114-2. [DOI] [PubMed] [Google Scholar]
- 79.Pena SD, Shokeir MH. Autosomal recessive cerebro-oculo-facio-skeletal (COFS) syndrome. Clin Genet. 1974;5:285–293. doi: 10.1111/j.1399-0004.1974.tb01695.x. [DOI] [PubMed] [Google Scholar]
- 80.Graham JM, Jr, Anyane-Yeboa K, Raams A, Appeldoorn E, Kleijer WJ, Garritsen VH, Busch D, Edersheim TG, Jaspers NG. Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am J Hum Genet. 2001;69:291–300. doi: 10.1086/321295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nouspikel T, Lalle P, Leadon SA, Cooper PK, Clarkson SG. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci U S A. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Meira LB, Graham JM, Jr, Greenberg CR, Busch DB, Doughty AT, Ziffer DW, Coleman DM, Savre-Train I, Friedberg EC. Manitoba aboriginal kindred with original cerebro-oculo- facio-skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am J Hum Genet. 2000;66:1221–1228. doi: 10.1086/302867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Laat WL, Sijbers AM, Odijk H, Jaspers NG, Hoeijmakers JH. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 1998;26:4146–4152. doi: 10.1093/nar/26.18.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sijbers AM, van der Spek PJ, Odijk H, van den Berg J, van Duin M, Westerveld A, Jaspers NG, Bootsma D, Hoeijmakers JH. Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res. 1996;24:3370–3380. doi: 10.1093/nar/24.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamel BC, Raams A, Schuitema-Dijkstra AR, Simons P, van der Burgt I, Jaspers NG, Kleijer WJ. Xeroderma pigmentosum--Cockayne syndrome complex: a further case. J Med Genet. 1996;33:607–610. doi: 10.1136/jmg.33.7.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zafeiriou DI, Thorel F, Andreou A, Kleijer WJ, Raams A, Garritsen VH, Gombakis N, Jaspers NG, Clarkson SG. Xeroderma pigmentosum group G with severe neurological involvement and features of Cockayne syndrome in infancy. Pediatr Res. 2001;49:407–412. doi: 10.1203/00006450-200103000-00016. [DOI] [PubMed] [Google Scholar]
- 87.Thorel F, Constantinou A, Dunand-Sauthier I, Nouspikel T, Lalle P, Raams A, Jaspers NG, Vermeulen W, Shivji MK, Wood RD, Clarkson SG. Definition of a short region of XPG necessary for TFIIH interaction and stable recruitment to sites of UV damage. Mol Cell Biol. 2004;24:10670–10680. doi: 10.1128/MCB.24.24.10670-10680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 89.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40:721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bosma PT, van Eert SJ, Jaspers NG, Stoter G, Nooter K. Functional cloning of drug resistance genes from retroviral cDNA libraries. Biochem Biophys Res Commun. 2003;309:605–611. doi: 10.1016/j.bbrc.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 91.Ellis NA, Ciocci S, German J. Back mutation can produce phenotype reversion in Bloom syndrome somatic cells. Hum Genet. 2001;108:167–173. doi: 10.1007/s004390000447. [DOI] [PubMed] [Google Scholar]
- 92.Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D, Hoeijmakers JH. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 93.Prasher JM, Lalai AS, Heijmans-Antonissen C, Ploemacher RE, Hoeijmakers JH, Touw IP, Niedernhofer LJ. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/−mice. EMBO J. 2005;24:861–871. doi: 10.1038/sj.emboj.7600542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parmar K, D'Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668:133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nunez F, Chipchase MD, Clarke AR, Melton DW. Nucleotide excision repair gene (ERCC1) deficiency causes G(2) arrest in hepatocytes and a reduction in liver binucleation: the role of p53 and p21. FASEB J. 2000;14:1073–1082. doi: 10.1096/fasebj.14.9.1073. [DOI] [PubMed] [Google Scholar]
- 96.Chipchase MD, O'Neill M, Melton DW. Characterization of premature liver polyploidy in DNA repair (Ercc1)-deficient mice. Hepatology. 2003;38:958–966. doi: 10.1053/jhep.2003.50421. [DOI] [PubMed] [Google Scholar]
- 97.Schumacher B, van der Pluijm I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van Steeg H, Niedernhofer LJ, van Ijcken W, Bartke A, Spindler SR, Hoeijmakers JH, van der Horst GT, Garinis GA. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Caruso LB, Silliman RA. Geriatric Medicine. In: Fauci AS, Braunwald E, Kasper DL, Hauser SL, Longo DL, Jameson JL, Loscalzo J, editors. Harrison's Principles of Internal Medicine. 17 ed. New York: McGraw Hill; 2006. [Google Scholar]
- 99.Winter AG, Samuel K, Hsia KT, Melton DW. The repair and recombination enzyme ERCC1 is not required for immunoglobulin class switching. DNA Repair (Amst) 2003;2:561–569. doi: 10.1016/s1568-7864(03)00021-1. [DOI] [PubMed] [Google Scholar]
- 100.Schrader CE, Vardo J, Linehan E, Twarog MZ, Niedernhofer LJ, Hoeijmakers JH, Stavnezer J. Deletion of the nucleotide excision repair gene Ercc1 reduces immunoglobulin class switching and alters mutations near switch recombination junctions. J Exp Med. 2004;200:321–330. doi: 10.1084/jem.20040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kotnis A, Du L, Liu C, Popov SW, Pan-Hammarstrom Q. Non-homologous end joining in class switch recombination: the beginning of the end. Philos Trans R Soc Lond B Biol Sci. 2009;364:653–665. doi: 10.1098/rstb.2008.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Selfridge J, Hsia KT, Redhead NJ, Melton DW. Correction of liver dysfunction in DNA repair-deficient mice with an ERCC1 transgene. Nucleic Acids Res. 2001;29:4541–4550. doi: 10.1093/nar/29.22.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lawrence NJ, Sacco JJ, Brownstein DG, Gillingwater TH, Melton DW. A neurological phenotype in mice with DNA repair gene Ercc1 deficiency. DNA Repair (Amst) 2008;7:281–291. doi: 10.1016/j.dnarep.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 104.Gorgels TG, van der Pluijm I, Brandt RM, Garinis GA, van Steeg H, van den Aardweg G, Jansen GH, Ruijter JM, Bergen AA, van Norren D, Hoeijmakers JH, van der Horst GT. Retinal degeneration and ionizing radiation hypersensitivity in a mouse model for Cockayne syndrome. Mol Cell Biol. 2007;27:1433–1441. doi: 10.1128/MCB.01037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsia KT, Millar MR, King S, Selfridge J, Redhead NJ, Melton DW, Saunders PT. DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development. 2003;130:369–378. doi: 10.1242/dev.00221. [DOI] [PubMed] [Google Scholar]
- 106.Paul C, Povey JE, Lawrence NJ, Selfridge J, Melton DW, Saunders PT. Deletion of genes implicated in protecting the integrity of male germ cells has differential effects on the incidence of DNA breaks and germ cell loss. PLoS One. 2007;2:e989. doi: 10.1371/journal.pone.0000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dolle ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, Niedernhofer LJ, van der Horst G, Hoeijmakers JH, van Steeg H, Vijg J. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 108.de Waard MC, van der Pluijm I, Zuiderveen Borgesius N, Comley LH, Haasdijk ED, Rijksen Y, Ridwan Y, Zondag G, Hoeijmakers JH, Elgersma Y, Gillingwater TH, Jaarsma D. Age-related motor neuron degeneration in DNA repair-deficient Ercc1 mice. Acta Neuropathol. 120:461–475. doi: 10.1007/s00401-010-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nevedomskaya E, Meissner A, Goraler S, de Waard M, Ridwan Y, Zondag G, van der Pluijm I, Deelder AM, Mayboroda OA. Metabolic profiling of accelerated aging ERCC1 d/- mice. J Proteome Res. 9:3680–3687. doi: 10.1021/pr100210k. [DOI] [PubMed] [Google Scholar]
- 110.Li XL, Shen SR, Wang S, Ouyang HH, Li GC. Restoration of T cell-specific V(D)J recombination in DNA-PKcs(−/−) mice by ionizing radiation: The effects on survival, development, and tumorigenesis. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2002;34:149–157. [PubMed] [Google Scholar]
- 111.Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vo N, Seo HY, Robinson A, Sowa G, Bentley D, Taylor L, Studer R, Usas A, Huard J, Alber S, Watkins SC, Lee J, Coehlo P, Wang D, Loppini M, Robbins PD, Niedernhofer LJ, Kang J. Accelerated aging of intervertebral discs in a mouse model of progeria. J Orthop Res. 28:1600–1607. doi: 10.1002/jor.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976) 2004;29:2691–2699. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 115.Roughley PJ, Alini M, Antoniou J. The role of proteoglycans in aging, degeneration and repair of the intervertebral disc. Biochem Soc Trans. 2002;30:869–874. doi: 10.1042/bst0300869. [DOI] [PubMed] [Google Scholar]
- 116.Tronche F, Casanova E, Turiault M, Sahly I, Kellendonk C. When reverse genetics meets physiology: the use of site-specific recombinases in mice. FEBS Lett. 2002;529:116–121. doi: 10.1016/s0014-5793(02)03266-0. [DOI] [PubMed] [Google Scholar]
- 117.Doig J, Anderson C, Lawrence NJ, Selfridge J, Brownstein DG, Melton DW. Mice with skin-specific DNA repair gene (Ercc1) inactivation are hypersensitive to ultraviolet irradiation-induced skin cancer and show more rapid actinic progression. Oncogene. 2006;25:6229–6238. doi: 10.1038/sj.onc.1209642. [DOI] [PubMed] [Google Scholar]
- 118.Selfridge J, Song L, Brownstein DG, Melton DW. Mice with DNA repair gene Ercc1 deficiency in a neural crest lineage are a model for late-onset Hirschsprung disease. DNA Repair (Amst) 9:653–660. doi: 10.1016/j.dnarep.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 119.Lawrence NJ, Song L, Doig J, Ritchie AM, Brownstein DG, Melton DW. Topical thymidine dinucleotide application protects against UVB-induced skin cancer in mice with DNA repair gene (Ercc1)-deficient skin. DNA Repair (Amst) 2009;8:664–671. doi: 10.1016/j.dnarep.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 120.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Munoz P, Blanco R, de Carcer G, Schoeftner S, Benetti R, Flores JM, Malumbres M, Blasco MA. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol Cell Biol. 2009;29:1608–1625. doi: 10.1128/MCB.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Callaghan TM, Wilhelm KP. A review of ageing and an examination of clinical methods in the assessment of ageing skin. Part I: Cellular and molecular perspectives of skin ageing. Int J Cosmet Sci. 2008;30:313–322. doi: 10.1111/j.1468-2494.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- 124.Kuk JL, Saunders TJ, Davidson LE, Ross R. Age-related changes in total and regional fat distribution. Ageing Res Rev. 2009;8:339–348. doi: 10.1016/j.arr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 125.Baron MS. Movement disorders in the older patient: differential diagnosis and general management. Cleve Clin J Med. 2005;72 Suppl 3:S38–S51. doi: 10.3949/ccjm.72.suppl_3.s38. [DOI] [PubMed] [Google Scholar]
- 126.Crimmins E, Vasunilashorn S, Kim JK, Alley D. Biomarkers related to aging in human populations. Adv Clin Chem. 2008;46:161–216. doi: 10.1016/s0065-2423(08)00405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Percy C, Pat B, Poronnik P, Gobe G. Role of oxidative stress in age-associated chronic kidney pathologies Adv Chronic Kidney Dis. 2005;12:78–83. doi: 10.1053/j.ackd.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 128.Hadjipavlou AG, Tzermiadianos MN, Bogduk N, Zindrick MR. The pathophysiology of disc degeneration: a critical review. J Bone Joint Surg Br. 2008;90:1261–1270. doi: 10.1302/0301-620X.90B10.20910. [DOI] [PubMed] [Google Scholar]
- 129.Imoto K, Boyle J, Oh K, Khan S, Ueda T, Nadem C, Slor H, Orgal S, Gadoth N, Busch D, Jaspers NG, Tamura D, JJ DiGiovanna, Kraemer KH. Patients with defects in the interacting nucleotide excision repair proteins ERCC1 or XPF show xeroderma pigmentosum with late onset severe neurological degeneration. J Invest Dermatol. 2007;127 Supp. 92 [Google Scholar]