Abstract
We have shown that a hybrid motor consisting of proton-type Rhodobacter sphaeroides MotA and sodium-type Vibrio alginolyticus PomB, MotX and MotY, can work as a sodium-driven motor in Vibrio cells. In this study, we tried to substitute the B subunits, which contain a putative ion-binding site in the transmembrane region. Rhodobacter sphaeroides MotB did not work with either MotA or PomA in Vibrio cells. Therefore, we constructed chimeric proteins (MomB), which had N-terminal MotB and C-terminal PomB. MomB proteins, with the entire transmembrane region derived from the H+-type MotB, gave rise to an Na+ motor with MotA. The other two MomB proteins, in which the junction sites were within the transmembrane region, also formed Na+ motors with PomA, but were changed for Na+ or Li+ specificity. These results show that the channel part consisting of the transmembrane regions from the A and B subunits can interchange Na+- and H+-type subunits and this can affect the ion specificity. This is the first report to have changed the specificity of the coupling ions in a bacterial flagellar motor.
Keywords: coupling ion specificity/flagella/motor proteins/Vibrio
Introduction
Bacterial cells have two kinds of rotary machine: the F0F1-ATPase and the flagellar motor (Khan, 1997). Both motors use the electrochemical potential of a specific ion (a proton or a sodium ion, depending on the organism) for the energy source of rotation. The F0F1-ATPase exists in bacteria as well as mitochondria and chloroplasts, and produces ATP by the ion flux. The flagellar motor is only present in bacteria and converts the ion flux to the locomotive force of the cell. These are all the rotary motors that are known in nature.
In the case of the F0F1-ATPase, rotation of the water-soluble F1 part as well as of the membrane-embedded F0 part was observed (Noji et al., 1997; Sambongi et al., 1999). The F1 part is composed of three α and three β, γ, δ and ε subunits, with the β subunits having ATPase activity. On the other hand, the membrane-embedded F0 part consists of an a, two b and 10–12 c subunits (Dimroth, 1997; Fillingame, 1997). It has been found that a hybrid enzyme composed of the F0 part from the sodium type and the F1 part from the proton type shows a sodium-type phenotype (Laubinger et al., 1990; Kaim and Dimroth, 1993, 1994). Thus, the F0 part seems to determine the coupling ion. It has been suggested that the a and c subunits of F0 are involved in the ion translocation, because mutation of either subunit changes the coupling ion specificity (Kaim et al., 1997, 1998; Valiyaveetil and Fillingame, 1997). In the c subunits, some residues that are involved in ion recognition have been identified, and include the conserved dicyclohexylcarbodiimide (DCCD)-reactive acidic residue (Zhang and Fillingame, 1995; Kaim et al., 1997). Recently, the three-dimensional structure (Dmitriev et al., 1999) and the conformational changes between protonated and deprotonated forms (Rastogi and Girvin, 1999) of Escherichia coli c subunits have been resolved.
Other rotary machines, namely bacterial flagellar motors, rotate helical flagella and propel cells toward favorable stimuli. Among various bacteria, the proton-driven motor of E.coli is most studied in terms of structure, genetics, biochemistry and biophysics (Blair, 1995; DeRosier, 1998; Berry and Armitage, 1999). The inner part of the motor consists of FliG, FliM and FliN, and is called C-ring or switch complex. On the other hand, the stator part is composed of MotA and MotB in proton-driven motors (DeRosier, 1998), or PomA and PomB in sodium-driven motors (Asai et al., 1997; Jaques et al., 1999). In these sodium motors there are two additional stator subunits: MotX and MotY (McCarter, 1994a,b; Okunishi et al., 1996).
The A subunits, MotA and PomA, have four transmembrane regions and a large cytoplasmic loop. It was proposed that some charged residues in this loop in E.coli MotA interact with oppositely charged residues of FliG during torque generation (Zhou et al., 1998a). The B subunits, MotB and PomB, have a single transmembrane region in the N-terminus, including an important Asp residue thought to be involved in ion transfer (Sharp et al., 1995; Zhou et al., 1998b). A peptidoglycan-binding motif is conserved in the C-terminus of B subunits, which plays a role in anchoring the motor to the cell wall. The region between the transmembrane and the C-terminal region was thought to be a linker. This was also supported by analysis of a series of 10 amino acid deletions in Salmonella MotB (Muramoto and Macnab, 1998). MotA and MotB interact with each other via their transmembrane regions and are thought to form ion-conducting channels (Blair and Berg, 1990; Stolz and Berg, 1991; Blair, 1995). PomA and PomB of Vibrio alginolyticus have been shown to interact functionally with each other (Yorimitsu et al., 1999). Recently, it was reported that when PomA and PomB were purified and reconstituted into proteoliposomes, they catalyzed 22Na+ uptake activity (Sato and Homma, 2000). In addition, mutations conferring resistance to phenamil, a specific inhibitor of sodium-driven motors or sodium channels (Atsumi et al., 1990), were found in both pomA and pomB in V.alginolyticus (Kojima et al., 1999), and in their homologs in Vibrio parahaemolyticus (Jaques et al., 1999). These data strongly support the hypothesis that PomA and PomB form a sodium-conducting channel.
The proteins MotX and MotY have single transmembrane regions and are unique components of the sodium-driven motor. They do not have similarity to the proton motor components or any other proteins, although a peptidoglycan-binding motif is in the C-terminal region of MotY (McCarter, 1994a,b). MotX is inferred to be a sodium channel component (McCarter, 1994a). Their functions are rather unclear.
It should be possible to investigate the coupling mechanisms of proton or sodium ion flux for force generation in flagella. As reported previously, we were able to produce and study a hybrid motor made of the proton and sodium types (Asai et al., 1999). The components from Rhodobacter sphaeroides (for the proton type) and from V.alginolyticus (for the sodium type) were used, because both A and B subunits of R.sphaeroides were found to be most similar to those of V.alginolyticus (Shah and Sockett, 1995; Shah et al., 1995; Asai et al., 1997). The photosynthetic bacterium R.sphaeroides has a single flagellum extending from the center of the cell body. The flagellar motor of R.sphaeroides rotates unidirectionally in the clockwise direction, and stops and restarts periodically (Armitage and Macnab, 1987). Both RsMotA (MotA of R.sphaeroides) and VaPomA (PomA of V.alginolyticus) are 253-amino-acid proteins, and they have ∼40% identity over their entire length. A PomA-deficient Vibrio strain expressing RsMotA from plasmids can swim using sodium ion flux. This showed that A subunits do not determine the ion specificity. In this study, we tried to clarify whether the B subunits were involved in coupling ion recognition and whether the nature of the B subunit transmembrane region affected the ion specificity. We constructed and characterized bacterial strains with motors containing either RsMotA or VaPomA, each with one of a series of chimeric B subunits formed between RsMotB and VaPomB.
Results
Introduction of R.sphaeroides motB into the pom mutants of V.alginolyticus
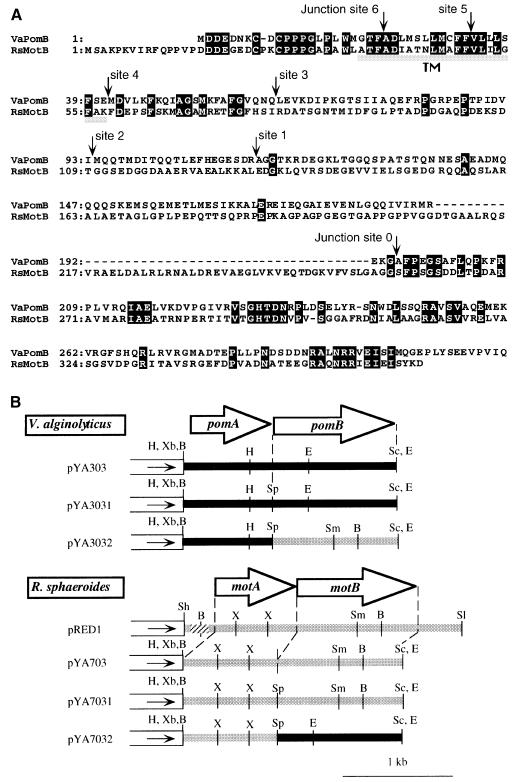
While this study was in progress we found that the published sequence of R.sphaeroides motB gene contained some errors in the 3′ region. We determined the nucleotide sequence of the whole motB gene again. Figure 1A shows the corrected amino acid sequence. It was found that RsMotB has a peptidoglycan-binding motif in the C-terminal region, like all members of the MotB/PomB family. The linker region of RsMotB between the transmembrane region and the peptidoglycan-binding motif is longer than those of the B subunits from other species. The transmembrane region of the proton-type R.sphaeroides MotB (RsMotB) is very similar to the transmembrane region of the sodium-type PomB of V.alginolyticus.
Fig. 1. (A) Amino acid alignments of V.alginolyticus PomB and R.sphaeroides MotB. White letters in black boxes and arrowheads show identical residues and the junction site of chimeric protein, respectively. Gray bars indicate putative transmembrane regions. (B) Restriction maps of plasmids. White boxes and the direction of solid arrowheads in the boxes indicate the vector part of pSU41 and the direction of transcription from the lac promoter, respectively. Inserted fragments from V.alginolyticus and R.sphaeroides, respectively, are indicated by a black or gray bold line. The hatched part of pRED1 is the region of the native R.sphaeroides promoter. The open arrows show the coding regions of pomA, pomB, motA and motB. VaPomB, V.alginolyticus PomB; RsMotB, R.sphaeroides MotB; H, HindIII; Xb, XbaI; B, BamHI; Sc, SacI; E, EcoRI; Sp, SpeI; Sh, SphI; X, XhoI; Sm, SmaI; Sl, SalI.
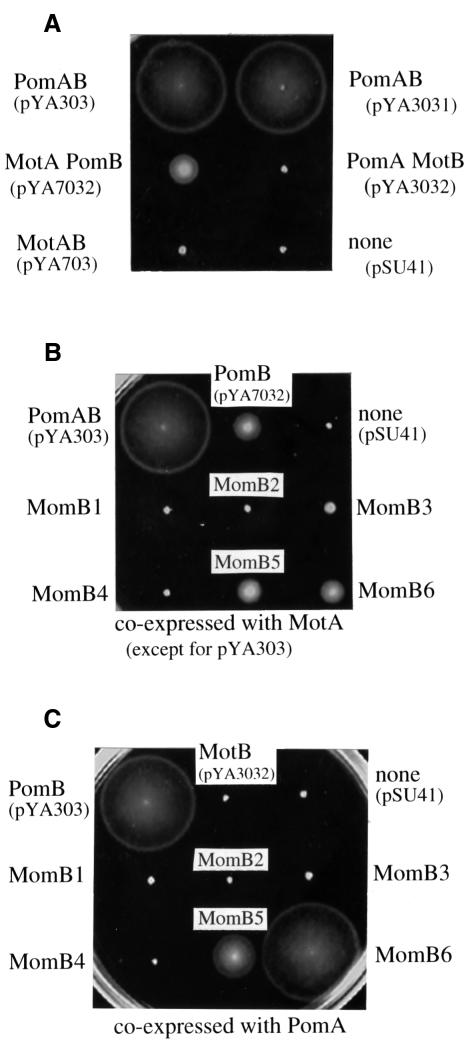
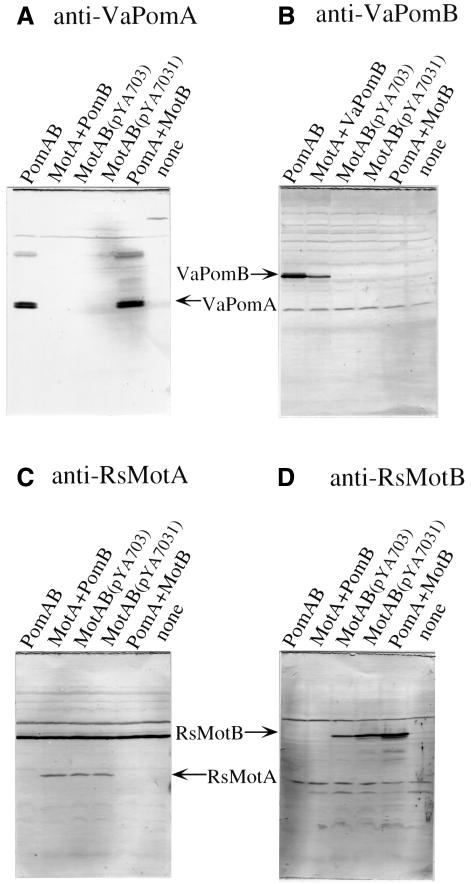
As shown in Figure 1B, the pomAB and RsmotAB genes could be controlled from the lac promoter in pYA303, pYA3031, pYA703 and pYA7031. We put a SpeI site between the stop codon of the A subunit gene and the Shine–Dalgarno sequence for the B subunit gene of pYA303 and pYA703, to give pYA3031 and pYA7031, respectively. This allowed us to switch the SpeI–SacI fragments of pYA3031 and pYA7031, to give pYA3032 (pomA and motB) and pYA7032 (motA and pomB). These plasmids were introduced into the pomAB mutant NMB191. Figure 2A shows the motility of the transformants. The pomAB mutant was restored to motility by pYA303, pYA3031 and pYA7032. All of these plasmids contain the pomB gene. On the other hand, pomAB mutant cells carrying pYA703, pYA7031 (data not shown) or pYA3032 were not motile. Thus, the whole motB gene could not complement the pomB defect of Vibrio polar flagella even when PomA or MotA was co-expressed with it. We tested whether the proteins were expressed in non-swarming cells by immunoblotting (Figure 3). In all of the swarming strains as well as non-swarming strains, both the A and B subunits were detected. For example, in pomAB mutant cells containing pYA3032, bands of PomA and MotB were detected (Figure 3A and D). Thus, we concluded that non-swarming cells also expressed both subunits, and that MotB can not function in Vibrio cells.
Fig. 2. Swarming abilities of transformants. Fresh colonies were inoculated in 0.3% agar VPG plates containing 100 µg/ml kanamycin and incubated at 30°C for 5 h. The pomAB mutant (NMB191) was used as the host strain. Proteins expressed in each strain, various combinations of A and B subunits of the proton-driven or sodium-driven motors (A), each MomB protein with proton-type MotA (B) and each MomB protein with sodium-type PomA (C), are noted around swarms.
Fig. 3. Detection of each protein by the anti-peptide antibody. NMB191 cells expressing each protein were cultured to exponential phase, harvested and suspended in distilled deionized water. Immunoblottings were carried out using anti-VaPomA (A), anti-VaPomB (B), anti-RsMotA (C) and anti-RsMotB antibody (D).
Construction of chimeric proteins between MotB and PomB
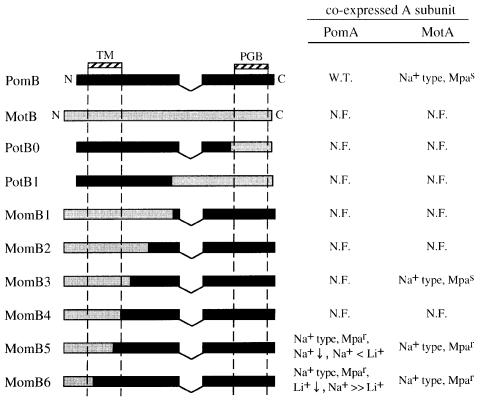
As we had found that the intact MotB protein can not work in the polar flagellar motors of V.alginolyticus, we wished to determine whether differences between N- or C-terminal regions of MotB and PomB were responsible for this result. To do this we constructed two chimeric proteins, named PotB0 and PotB1, consisting of N-terminal regions of PomB and C-terminal regions of MotB. The chimeric junctions are shown in Figure 1A (junction site 0 for PotB0 and site 1 for PotB1). Although most of PotB0 and PotB1 is derived from PomB, neither of the chimeras was functional in the presence of either MotA or PomA (data not shown). Accordingly, we constructed six other chimeric proteins, named MomB, consisting of N-terminal regions of MotB and C-terminal regions of PomB. The swarming abilities of pomAB mutant cells co-expressing MotA with each different MomB are shown in Figure 2B. The three chimeric proteins, MomB3, MomB5 and MomB6, expressed together with MotA, restored the motility of the pomAB mutant. The swarm sizes of the strains containing MomB5 and MomB6 were comparable to that of control cells expressing MotA with PomB, although the MomB3-expressing strain made a smaller swarm than the others. When expressed with PomA, MomB5 and MomB6 were also functional in pomAB mutant cells. Cells expressing MomB6 swarmed as fast as cells expressing wild-type PomB. Unlike with MotA, MomB3 did not restore the motility with PomA. The other chimeric proteins, MomB1, MomB2 and MomB4, did not support swarming regardless of whether they were co-expressed with PomA or MotA. We checked the expression of the MomB proteins by immunoblotting using antibodies raised against RsMotB and VaPomB peptides (data not shown). All of the MomB proteins were recognized by both anti-RsMotB and anti-PomB antibodies. Most of the chimeric proteins were detected as bands with apparent molecular masses similar to wild-type PomB and MotB.
Characterization of the MomB proteins co-expressed with PomA or MotA
We examined which ion is coupled to generate torque in the motile cells expressing the MomB proteins. As shown in Table I, we tested whether cells could swim in Na+-free buffer (TMK300 containing 300 mM K+) and whether their motility was affected by a protonophore, carbonylcyanide m-chlorophenylhydrazone (CCCP). The results showed that none of the functional motors rotated in the Na+-free buffer nor did the motor consisting of PomA and PomB (pYA303/NMB191), which uses Na+ but not H+ as a coupling ion. Moreover, the former and the latter motors did not stop in TMN300 (pH 8.5) containing 20 µM CCCP, but stopped completely with the further addition of 20 µM 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO), a specific inhibitor of the primary Na+ pump (Tokuda and Unemoto, 1982). In alkaline Tris motility buffer, YM19 (Pof– Laf+) cells swam in the absence of CCCP, but stopped completely in the presence of 20 µM CCCP, because lateral flagella have proton-driven motors. We concluded from these results that all of the MomB-based motors are driven by Na+-motive force. We found a few motile cells among cultures expressing MomB1 or MomB2 with MotA in TMN300. However, motile fractions of these strains were too low to measure the swimming speeds and to determine their coupling ions.
Table I. Characterization of chimeric proteins.
| Salt (300 mM) | pH | CCCP (µM) | Swimming speed (µm/s) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PomA+ |
MotA+ |
MotA +PomB (pYA7032) | YM19 | |||||||
| PomAB (pYA303) | MomB5 | MomB6 | MomB3 | MomB5 | MomB6 | |||||
| Na+ | 8.5 | 0 | 80 (3.8)a | 11 (0.9) | 70 (4.7) | 20 (2.3) | 26 (2.5) | 53 (3.9) | 28 (2.0) | 20 (1.8) |
| 20 | 64 (6.9) | 8 (1.3) | 51 (4.8) | 14 (2.0) | 20 (1.9) | 42 (3.6) | 27 (1.7) | 0 | ||
| 7.5 | 0 | 80 (3.7) | 10 (1.0) | 67 (7.9) | 23 (2.2) | 31 (5.5) | 54 (1.7) | 28 (1.7) | 27 (1.4) | |
| 20 | 10 (1.7) | 3 (1.5) | 6 (0.9) | 3 (1.7) | 5 (1.0) | 5 (2.2) | 9.9 (1.5) | 0 | ||
| K+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 (2.4) | |
| Li+ | 0 | 35 (4.5) | 24 (2.9) | 8.5 (1.2) | L.F.b | 5.5 (0.7) | L.F. | 4.4 (0.8) | 24 (2.2) | |
aSD is shown in parentheses.
bL.F., low motile fraction.
Li+ coupling abilities of functional MomBs
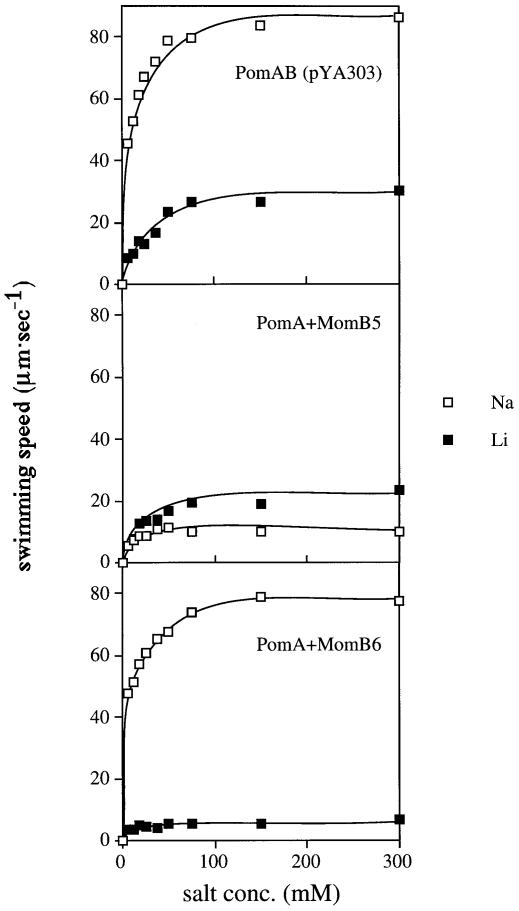
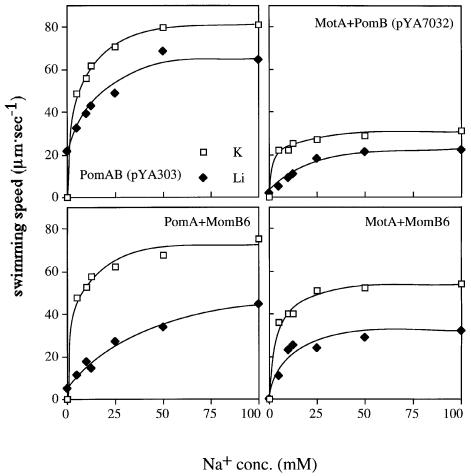
The wild-type polar flagellar motor of V.alginolyticus can use Li+ as a coupling ion less efficiently than Na+. The maximal swimming speed produced by Li+ is only ∼30% of that of Na+ ions (Liu et al., 1990; Imae, 1991). To test the ion specificity of the MomB-based motors, we measured swimming speeds of cells expressing MomB proteins in 300 mM Li+ (Table I). In 300 mM Li+, the motile fractions and the swimming speeds of the pomAB strain expressing MomB3 or MomB6 with MotA were too low for quantitative measurement. The motors consisting of PomA and MomB5 or MomB6 showed an interesting property. Unlike cells expressing PomA and PomB, cells expressing MomB5 with PomA swam faster in 300 mM Li+ than in 300 mM Na+. Cells expressing MomB6 with PomA swam much slower in 300 mM Li+ than those expressing PomA and PomB, although in 300 mM Na+ the former cells swam nearly as fast as the latter cells. We examined the effect of salt concentration on swimming speed (Figure 4). Cells of any strain did not swim in the absence of Na+ or Li+, and their swimming speeds increased with an increase in either salt concentration. Cells expressing MomB5 with PomA swam faster with Li+ than with Na+ at any fixed concentration. The maximal swimming speed of cells expressing PomA and PomB in Li+ was approximately one-third of that in Na+, as reported before. On the other hand, the maximal speed of cells expressing MomB6 with PomA in Li+ is one-tenth of that in Na+. Next, we examined the competitive effect of Li+ against Na+, because it is thought that Li+ shares the same binding site(s) in the motor as Na+. We measured the swimming speed in Tris motility buffer, which contained various concentrations (from 0 to 100 mM) of Na+ with or without 200 mM Li+. The total salt concentration of every condition was constantly adjusted to 300 mM with K+ (Figure 5). The addition of Li+ reduced the maximum speed of cells expressing PomA and PomB from 80 to 65 µm/s, suggesting that Na+ and Li+ compete for the same binding site(s) in the motor. In the absence of Li+, cells expressing a motor consisting of MomB6 with PomA showed a similar swimming speed to that of cells expressing PomA and PomB. However, Li+ had a larger effect on the PomA and MomB6 motor than the PomA and PomB motor: the maximum speeds in the presence of Li+ decreased to 45 and 65 µm/s, respectively. Cells expressing MomB6 with MotA gave similar results to those with PomA, although the speed of the former motor was always lower than the latter. These results may suggest that Li+ binds to motors containing MomB6, but is not conducted, or that Li+ flux is not coupled to rotation in these motors.
Fig. 4. Swimming speed of NMB191 cells expressing PomAB (pYA303), MomB5 with PomA or MomB6 with PomA. The cells were suspended in Tris-motility buffer (TMN50) and diluted 100-fold into buffers containing various concentration of Na+ (open squares) or Li+ (closed squares) as coupling ions, and the swimming speeds were measured.
Fig. 5. Competition of Na+ with Li+ for swimming speed of each strain. NMB191 cells expressing PomAB(pYA303), PomA+MomB6, RsMotA+PomB or RsMotA+MomB6 were suspended in buffers at various concentrations of Na+ with 200 mM K+ (open squares) or 200 mM Li+ (closed diamonds) and their swimming speeds were measured. The total salt concentration was adjusted to 300 mM with K+.
Resistance to inhibitors of hybrid motors containing MomB
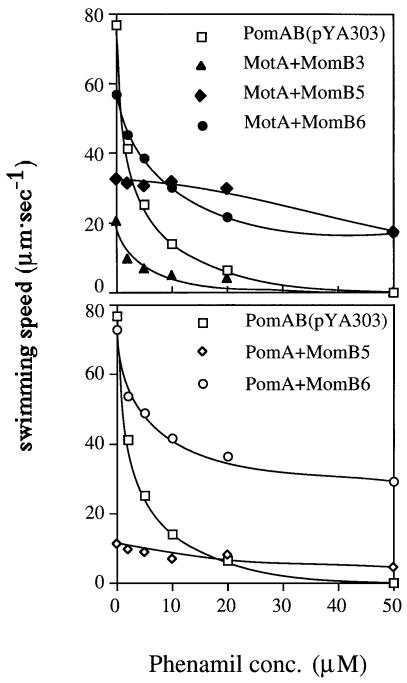
If the ion specificity was changed in motors consisting of MomB and PomA, the effects of phenamil and amiloride, specific inhibitors of the sodium-driven motor, may also be changed. As shown in Figure 6, cells expressing MomB5 or MomB6 with either MotA or PomA showed phenamil-resistant phenotypes. For MomB6 motors, the swimming speed slowed initially on phenamil addition, but then reached a phenamil-resistant plateau of 20–40 µm/s. It is noteworthy that cells expressing MomB5 with either MotA or PomA were hardly inhibited even by 50 µM phenamil, the highest concentration tested, which completely inhibits the wild-type motor. The IC50 value, the concentration of inhibitor required for 50% inhibition of motility, for phenamil in cells expressing MomB3 with MotA was comparable to that for cells expressing PomA and PomB. This suggests that changing motor conformation alters phenamil-binding sites, yielding MomB strains with resistance to phenamil. On the other hand, all the strains tested showed the same sensitivity phenotypes to amiloride as cells expressing PomA and PomB (data not shown). The noteworthy features of all the chimeric B proteins are summarized in Figure 7.
Fig. 6. The swimming speeds of each strain were measured at various concentrations of the specific inhibitor of sodium motor, phenamil, in TMN50. The results of MomB chimeras with RsMotA (upper panel) or with PomA (lower panel) are shown.
Fig. 7. Summary of the chimeric protein features. Black and gray boxes show regions that are derived from VaPomB and RsMotB, respectively. TM, transmembrane region; PGB, peptidoglycan-binding motif; W.T., wild type; MpaS, motility that is phenamil sensitive; N.F., non-functional; Mpar, motility that is phenamil resistant; Na+, motility coupling to sodium ion; Li+, motility coupling to lithium ion; ↓, impaired motility.
Discussion
Recently, we reported that a hybrid motor consisting of R.sphaeroides MotA, V.alginolyticus PomB, MotX and MotY, can work as a sodium-driven motor in Vibrio cells (Asai et al., 1999). This showed that the A subunits do not determine the ion specificity and raises the question: which components decide the ion selectivity? The most likely candidate seemed to be the B subunits. The PomB proteins are known to form a sodium channel together with the PomA proteins (Sato and Homma, 2000), and contain a conserved Asp residue, which is located in the transmembrane region and seems to be essential for the ion transfer. In this study, we examined whether specific regions of the B subunits are involved in the ion specificity of the motor.
Initially, we constructed plasmids encoding various combinations of A and B subunits of the proton-type (R. sphaeroides) or the sodium-type motors (V.alginolyticus), i.e. MotA and PomB, or PomA and MotB. These plasmids were introduced into the pomAB mutant of Vibrio cells. As shown in Figure 2A, MotB could not work with either MotA or PomA in the pomAB mutant. In spite of the higher GC content of the motB gene compared with the Vibrio pomB gene, the MotB protein seemed to be expressed at sufficient levels to give function. A possible reason for the lack of complementation is that the peptidoglycan-binding domain of MotB can not associate with the Vibrio peptidoglycan layer. This speculation was supported by the evidence that the chimeric proteins, PotB, between the N-terminal region of PomB and the C-terminal (peptidoglycan-binding) region of MotB, did not function at all (data not shown) and that the mutation of the highly conserved Ser residue (but not conserved in RsMotB) in the peptidoglycan-binding domain caused a defect of the PomB function in Vibrio NMB161 (Asai et al., 1997; our unpublished data).
Next, we constructed the chimeric protein series, named MomB, consisting of N-terminal regions of MotB and C-terminal regions of PomB. The features of all chimeras are shown in Figure 7. MomB1, MomB2, MomB3 and MomB4 have junction sites in a non-conserved ‘linker’ region, which connects the transmembrane region and the peptidoglycan-binding motif. The other chimeric proteins, MomB5 and MomB6, have junction points within the transmembrane region that are highly conserved. These six MomB proteins were expressed with either MotA or PomA in the pomAB mutant. Out of all the MomB chimeras, only MomB5 and MomB6 functioned as sodium-driven motor components. However, the specificities for Na+ and Li+ were changed in the motors with MomB5 and MomB6. The MomB6-containing motor was driven by Na+ as effectively as the wild-type motor, but motility could barely be driven by Li+. The MomB5-containing motor was driven slowly by Na+, but rotation was coupled to Li+ more effectively than to Na+. Structural changes to the channel in the transmembrane regions of the A and B subunits may affect the efficiency of the ion transfer and/or the energy coupling for Li+ or Na+. We speculate that the differing ion specificities for rotation shown by MomB5- and MomB6-containing motors may correlate with ion size. This may be because the MomB5 protein, whose transmembrane region derived mostly from the proton-type protein MotB, works better with the smaller ion Li+, and because MomB6, whose transmembrane region derived mostly from the sodium-type protein PomB, works better with the larger Na+ ion. There are few non-conserved amino acids between the MotB and PomB proteins in the region between the MomB6 and MomB5 junction points. While these small differences may account for ion-specificity changes, it is more likely that the periplasmic region of PomB proximal to the surface of the membrane (which is present in MomB6 but not in MomB5) plays a role in ion specificity or interaction with other sodium-motor proteins. Thus, the ion size discrimination of channels containing MomB6 is different to that of channels containing MomB5. This idea is further supported by the MomB3 data.
MomB3 retains the sequence of the sodium-type B subunit from the linker region to the C-terminus, and MomB3 was also functional in a pomAB strain with MotA, although the swimming ability was poor. However, MomB3 did not function with PomA. The region around the junction site 3 may need to interact with A subunits precisely. Moreover, from the evidence that the motor consisting of MotA and MomB3 is sodium type, it seems that the transmembrane regions of the B subunit as well as the A subunit do not determine the ion specificity directly. MomB1, MomB2 and MomB4 expressed with either PomA or MotA were non-functional. The MomB4 junction site may be more important for maintaining the structure of an ion-specific channel composed of the transmembrane regions of the A and B subunits. Garza et al. (1996) also suggested that in E.coli the MotA periplasmic region interacts with the linker region of MotB, or that this region helps the transmembrane domain to position properly to interact with the transmembrane regions of MotA. We have investigated the PomA periplasmic region (loop1–2 and loop3–4) by cysteine scanning mutagenesis, and it is inferred that loop1–2 may be covered with other proteins or may be embedded in the pore region of the channel (Asai et al., 2000). We speculate that loop1–2 interacts with the linker region of the B subunit.
‘Where’ and ‘how’ do the motor proteins recognize the coupling ion? Many mutations that alter the ion specificity are known in various ion channels and transporters (Hama and Wilson, 1993; Heginbotham et al., 1994; Kellenberger et al., 1999). However, it is unclear how these residues recognize the different ions. In the other rotary motor of F0F1-ATPase, a hybrid enzyme composed of the F0 part from the sodium type and the F1 part from the proton type functioned as a sodium-type enzyme (Laubinger et al., 1990; Kaim and Dimroth, 1993, 1994). The residues involved in ion recognition were identified in the a and c subunit and the ions should interact with the subunits. The ion-binding residue, which is the DCCD-reactive residue, was postulated as Glu65 for Propionigenium modestum c subunit and as Asp61 for E.coli c subunit. In the case of sodium-type F0F1-ATPase c subunits, five residues that are involved in the ion recognition have been identified, and include the DCCD-reactive Glu65 residue, Ser66 and Gln32 (Kaim et al., 1997). The corresponding residues in proton-type c subunits are Asp61, Ala62 and Ile28, respectively. The substitution of Gln32 of sodium type to Ile abolished only the property of Na+ coupling. The substitution of Ser66 to Ala abolished the property of Na+ and Li+ coupling, and retained H+ coupling ability. From these results, it was proposed that the side chains of these residues coordinated an ion. Those side chains of corresponding residues in the proton-type c subunit were shown to face each other from the three-dimensional structure (Dmitriev et al., 1999). Another important example is the KcsA potassium channel, for which the crystal structure has been solved and the mechanism of ion recognition has been revealed (Doyle et al., 1998). Mutations at some residues in the pore region change the ability of the channel to discriminate K+ from Na+ (Heginbotham et al., 1994). The side chains of these residues face the outside of the pore and K+ ions interact with the oxygen atoms of these peptide backbones. The selective filter formed by four oxygen atoms fits and stabilizes a dehydrated K+ electrostatically. However, smaller Na+ ions can not fit into the filter and therefore fail to overcome electrostatic destabilization in the pore.
In the flagellar motor, Asp32 of E.coli MotB is speculated to function proximal to the surface of the membrane in conveying protons (Sharp et al., 1995; Zhou et al., 1998b) and the acidic residue is conserved in V.alginolyticus PomB and R.sphaeroides MotB. If the acidic residue has a similar function to the DCCD-reactive residue of F0, a few residues necessary for Na+ or Li+ binding might exist in the transmembrane region of the A and B subunits and the other components. It would be interesting to know whether the mechanism of ion recognition is common to the two kinds of ion-driven rotary motor. Otherwise, by the KscA analogy, the transmembrane regions of the A and B subunits may form a selective pore for ion recognition. The selective pore may be modulated by a specific part of the linker region of the B subunits or the other motor proteins, MotX and/or MotY. From our results with the motor consisting of MotA and MomB3 showing that the transmembrane region of the B subunit does not decide the ion specificity, the linker region might be required for the precise arrangement of the transmembrane regions. Accordingly, the linker regions of the proton- and sodium-type subunits might force the transmembrane regions to form small and large pores, which accommodate protons and sodium ions, respectively. Alternatively, MotX and/or MotY might recognize the ions and pass the ions to the PomA–PomB complex. FliG is also a candidate protein for determining the ion specificity, because it was implied that the charged residues at the MotA–FliG interface might form a pathway for the conduction of protons (Zhou et al., 1998a). It is possible that all of these proteins are essential for the precise ion specificity in the flagellar motor.
Materials and methods
Bacterial strains, plasmids, growth conditions and media
The strains and plasmids used are shown in Table II. Vibrio alginolyticus cells were cultured at 30°C in VC medium (0.5% Polypepton, 0.5% yeast extract, 0.4% K2HPO4, 3% NaCl, 0.2% glucose) or VPG medium (1% Polypepton, 0.4% K2HPO4, 3% NaCl, 0.5% glycerol). When necessary, kanamycin was added to a final concentration of 100 µg/ml. Escherichia coli cells were cultured at 37°C in LB medium (1% bactotryptone, 0.5% yeast extract, 0.5% NaCl). For E.coli, kanamycin was added to a final concentration of 50 µg/ml. The swimming speed was measured in Tris-motility buffers, TMN300 (300 mM NaCl, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 5 mM glucose), TMK300 (300 mM KCl, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 5 mM glucose) or mixtures of TMN300 and TMK300 to vary the sodium concentration.
Table II. Bacterial strains and plasmids.
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| V.alginolyticus strains | ||
| VIO5 | VIK4 laf (Rifr Pof+ Laf–) | Okunishi et al. (1996) |
| VIO586 | VIO5 pomA (Rifr Laf– Pom–) | Asai et al. (1997) |
| NMB191 | VIO5 ΔpomAB (0.94 bp DraI–HpaI fragment deletion) (Rifr Laf– Pom–) | Yorimitsu et al. (1999) |
| YM19 | 138-2 pof (Pof–) | Kawagishi et al. (1995) |
| E.coli strains | ||
| DH5α | F– λ–recA1 hsdR17 endA1 supE44 thi-1 relA1 gyrA96 Δ(argF-lacZYA)U169 φ80dlacZΔM15 | Grant et al. (1990) |
| Plasmids | ||
| pSU41 | kan (Kmr) PlaclacZ α | Bertolome et al. (1991) |
| pRED1 | pUC19 2.5 kb SphI–SalI fragment (motAB+) | Shah and Sockett (1995) |
| pYA303 | pSU41 1.9 kb BamHI–SacI fragment (pomAB+) | Kojima et al. (1999) |
| pYA3031 | pSU41 0.8 kb BamHI–SpeI fragment (pomA+) and 1.1 kb SpeI–SacI fragment (pomB+) | this work |
| pYA3032 | pSU41 0.8 kb BamHI–SpeI fragment (pomA+) and 1.1 kb SpeI–SacI fragment (motB+) | this work |
| pYA701 | pSU41 0.8 kb BamHI–EcoRI fragment (motA+) | Asai et al. (1999) |
| pYA703 | pSU41 1.9 kb BamHI–SacI fragment (motAB+) | this work |
| pYA7031 | pSU41 0.8 kb BamHI–SpeI fragment (motA+) and 1.1 kb SpeI–SacI fragment (motB+) | this work |
| pYA7032 | pSU41 0.8 kb BamHI–SpeI fragment (pomA+) and 1.1 kb SpeI–SacI fragment (pomB+) | this work |
Kmr, kanamycin resistant; Mpar, motility resistant to phenamil; Rifr, rifampicin resistant; Plac, lac promoter.
DNA manipulations and sequencing
Routine DNA manipulations were carried out according to standard procedures (Sambrook et al., 1989). Restriction endonucleases and other enzymes for DNA manipulations were purchased from Takara Shuzo (Kusatsu, Japan) and New England Biolabs (Beverly, MA). Nucleotide sequences were determined by using the dye terminator cycle sequencing kit (Perkin Elmer Co.) and an ABI PRISM sequencer Model 377 (PE Applied Biosystems).
Plasmid construction and site-directed mutagenesis
We constructed a plasmid, pYA703, including the PCR-amplified motAB fragment from 40 bp upstream of the start codon to 8 bp downstream of the stop codon. The PCR amplification was performed as described previously (Kojima et al., 1999). In the reaction, we used the end primers, RsmotA.B1 and RsmotB.Z3, and pRED1 as templates. RsmotA.B1 has a BamHI site and is the sense primer anealing the upstream RsmotA gene, and was used when we constructed pYA701 (Asai et al., 1999). RsmotB.Z3, including a SacI and EcoRI site, is the antisense primer annealing the downstream of the gene, 5′-CGGAATTCGAGCTCGGA CAAGGTCAGTCCT-3′. This PCR-amplified fragment was inserted into pSU41 (Bartolome et al., 1991) between the BamHI site and the EcoRI site. The motAB gene of pYA703 is under the control of the lac promoter directly and does not have the native promoter.
Site-directed mutagenesis was performed by a two-step PCR reaction as described previously (Kojima et al., 1999). To introduce a SpeI site between the stop codon of the A subunit and Shine–Dalgarno sequence of the B subunit in pYA703 or pYA303, we used the pairs of mutant primers, RsmotA.spe1 and RsmotA.spe2 or VapomA.spe1 and VapomA.spe2. The sequences of the sense strand of RsmotA.speI and VapomA.speI were 5′-GGCGGCGTGACTAGTGAGGCCAGGCCATG-3′ and 5′-ACGAG TAACTAGTGGAGAGTCG-3′, respectively. We also constructed momB genes using the two-step PCR. The sense strand sequences of the mutant primers were as follows: MomB1, 5′-AGAAGGCGCTCGCTGGTGGC ACAAAACGT-3′; MomB2, 5′-AAGTCGGACACGATGCAGCAAAC CATG-3′; MomB3, 5′-TTCCATTCGATCCTTGAAGTGAAAGAC-3′; MomB4, 5′-GCTTCGCGAAGATGGATGTACTGAAGTTC-3′; Mom B5, 5′-ATGGCCTTCTTCGTTCTTCTGCTCTCG-3′; MomB6; 5′-CTT GCGACCTTTGCAGATTTGATGTCG-3′.
Transformation of Vibrio cells
Transformation of Vibrio cells was carried out by electroporation as described previously (Kawagishi et al., 1994). Cells were subjected to osmotic shock and washed thoroughly with 20 mM MgSO4. Electroporation was carried out with the Gene Pulser electroporation apparatus (Japan Bio-Rad Laboratories, Tokyo, Japan) at an electric field strength between 5.0 and 7.5 kV/cm.
Immunoblotting
Preparation of cells and immunoblotting were carried out as described previously (Yorimitsu et al., 1999). For the first antibody of immunoblotting, we used anti-peptide antibodies against V.alginolyticus PomA (VaPomA), VaPomB, R.sphaeroides MotA (RsMotA) or RsMotB. PomA91, PomB93 and RsMotA253 antibodies were used as anti-VaPomA, anti-VaPomB and anti-RsMotA antibodies, respectively (Asai et al., 1999; Yorimitsu et al., 1999). The production and affinity purification of anti-RsMotB antibodies (RsMotB2) were carried out by Sawady technology (Tokyo, Japan). The selected peptide fragment was synthesized artificially, with the sequence NH2- SAKPKVIRFQPPVPDDDEGEC-COOH, which is located at the N-terminus of RsMotB. A cysteine was added to the C-terminus and the synthesized fragment was conjugated to keyhole limpet hemocyanin (KLH). Rabbits were immunized with the conjugated KLH. For the second antibody, we used an alkaline phosphatase-conjugated goat anti-rabbit IgG antibody.
Measurement of swimming speed
An overnight culture in VC medium was inoculated into fresh VPG medium at a 100-fold dilution and grown at 30°C to exponential phase. Cells were harvested by centrifugation and suspended in TMN50 (50 mM NaCl, 250 mM KCl, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 5 mM glucose). The cell suspension was diluted ∼100-fold into Tris motility buffers containing various concentrations of NaCl, and 20 mM serine to suppress the directional change of swimming. Motility of the cells was observed under a dark-field microscope and recorded on videotape. Swimming speed was determined as described previously (Atsumi et al., 1996).
Acknowledgments
Acknowledgements
We thank Dr Deepan Shah for critically reading the manuscript. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (to I.K. and M.H.), the Japan Society for the Promotion of Science (to Y.A.), and the Royal Society and BBSRC (to R.E.S.).
References
- Armitage J.P. and Macnab,R.M. (1987) Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol., 169, 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai Y., Kojima,S., Kato,H., Nishioka,N., Kawagishi,I. and Homma,M. (1997) Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol., 179, 5104–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai Y., Kawagishi,I., Sockett,E. and Homma,M. (1999) Hybrid motor with the H+- and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J. Bacteriol., 181, 6332–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai Y., Shoji,T., Kawagishi,I. and Homma,M. (2000) Cysteine-scanning mutagenesis of the periplasmic loop regions of PomA, a putative channel component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol., 182, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T., Sugiyama,S., Cragoe,E.J.,Jr and Imae,Y. (1990) Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J. Bacteriol., 172, 1634–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T., Maekawa,Y., Yamada,T., Kawagishi,I., Imae,Y. and Homma,M. (1996) Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J. Bacteriol., 178, 5024–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome B. Jubete,Y., Martinez,E. and de la Cruz,F. (1991) Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene, 102, 75–78. [DOI] [PubMed] [Google Scholar]
- Berry R.M. and Armitage,J.P. (1999) The bacterial flagella motor. Adv. Microb. Physiol., 41, 291–337. [DOI] [PubMed] [Google Scholar]
- Blair D.F. (1995) How bacteria sense and swim. Annu. Rev. Microbiol., 49, 489–522. [DOI] [PubMed] [Google Scholar]
- Blair D.F. and Berg,H.C. (1990) The MotA protein of E.coli is a proton-conducting component of the flagellar motor. Cell, 60, 439–449. [DOI] [PubMed] [Google Scholar]
- DeRosier D.J. (1998) The turn of the screw: the bacterial flagellar motor. Cell, 93, 17–20. [DOI] [PubMed] [Google Scholar]
- Dimroth P. (1997) Primary sodium ion translocating enzymes. Biochim. Biophys. Acta, 1318, 11–51. [DOI] [PubMed] [Google Scholar]
- Dmitriev O.Y., Jones,P.C. and Fillingame,R.H. (1999) Structure of the subunit c oligomer in the F1F0 ATP synthase: model derived from solution structure of the monomer and cross-linking in the native enzyme. Proc. Natl Acad. Sci. USA, 96, 7785–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais,C.J., Pfuetzner,R.A., Kuo,A., Gulbis,J.M., Cohen,S.L., Chait,B.T. and MacKinnon,R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science, 280, 69–77. [DOI] [PubMed] [Google Scholar]
- Fillingame R.H. (1997) Coupling H+ transport and ATP synthesis in F1F0-ATP synthases: glimpses of interacting parts in a dynamic molecular machine. J. Exp. Biol., 200, 217–224. [DOI] [PubMed] [Google Scholar]
- Garza A.G., Bronstein,P.A., Valdez,P.A., Harris,H.L. and Manson,M.D. (1996) Extragenic suppression of motA missense mutations of Escherichia coli. J. Bacteriol., 178, 6116–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.G., Jessee,J., Bloom,F.R. and Hanahan,D. (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl Acad. Sci. USA, 87, 4645–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H. and Wilson,T.H. (1993) Cation-coupling in chimeric melibiose carriers derived from Escherichia coli and Klebsiella pneumoniae. The amino-terminal portion is crucial for Na+ recognition in melibiose transport. J. Biol. Chem., 268, 10060–10065. [PubMed] [Google Scholar]
- Heginbotham L., Lu,Z., Abramson,T. and MacKinnon,R. (1994) Mutations in the K+ channel signature sequence. Biophys. J., 66, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae Y. (1991) Use of Na+ as an alternative to H+ in energy transduction. In Mukohata,Y. (ed.), New Era of Bioenergetics. Academic Press, Tokyo, Japan, pp. 197–221. [Google Scholar]
- Jaques S., Kim,Y.K. and McCarter,L.L. (1999) Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc. Natl Acad. Sci. USA, 96, 5740–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaim G. and Dimroth,P. (1993) Formation of a functionally active sodium-translocating hybrid F1F0 ATPase in Escherichia coli by homologous recombination. Eur. J. Biochem., 218, 937–944. [DOI] [PubMed] [Google Scholar]
- Kaim G. and Dimroth,P. (1994) Construction, expression and characterization of a plasmid-encoded Na+-specific ATPase hybrid consisting of Propionigenium modestum F0-ATPase and Escherichia coli F1-ATPase. Eur. J. Biochem., 222, 615–623. [DOI] [PubMed] [Google Scholar]
- Kaim G., Wehrle,F., Gerike,U. and Dimroth,P. (1997) Molecular basis for the coupling ion selectivity of F1F0 ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry, 36, 9185–9194. [DOI] [PubMed] [Google Scholar]
- Kaim G., Matthey,U. and Dimroth,P. (1998) Mode of interaction of the single a subunit with the multimeric c subunits during the translocation of the coupling ions by F1F0 ATPases. EMBO J., 17, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi I., Okunishi,I., Homma,M. and Imae,Y. (1994) Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology, 140, 2355–2361. [Google Scholar]
- Kawagishi I., Maekawa,Y., Atsumi,T., Homma,M. and Imae,Y. (1995) Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol., 177, 5158–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S., Gautschi,I. and Schild,L. (1999) A single point mutation in the pore region of the epithelial Na+ channel changes ion selectivity by modifying molecular sieving. Proc. Natl Acad. Sci. USA, 96, 4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. (1997) Rotary chemiosmotic machines. Biochim. Biophys. Acta, 1322, 86–105. [DOI] [PubMed] [Google Scholar]
- Kojima S., Asai,Y., Atsumi,T., Kawagishi,I. and Homma,M. (1999) Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel component. J. Mol. Biol., 285, 1537–1547. [DOI] [PubMed] [Google Scholar]
- Laubinger W., Deckers,H.G., Altendorf,K. and Dimroth,P. (1990) A hybrid adenosinetriphosphatase composed of F1 of Escherichia coli and F0 of Propionigenium modestum is a functional sodium ion pump. Biochemistry, 29, 5458–5463. [DOI] [PubMed] [Google Scholar]
- Liu J.Z., Dapice,M. and Khan,S. (1990) Ion selectivity of the Vibrio alginolyticus flagellar motor. J. Bacteriol., 172, 5236–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L.L. (1994a) MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol., 176, 5988–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L.L. (1994b) MotY, a component of the sodium-type flagellar motor. J. Bacteriol., 176, 4219–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto K. and Macnab,R.M. (1998) Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol., 29, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Noji H., Yasuda,R., Yoshida,M. and Kinosita,K.J. (1997) Direct observation of the rotation of F1-ATPase. Nature, 386, 299–302. [DOI] [PubMed] [Google Scholar]
- Okunishi I., Kawagishi,I. and Homma,M. (1996) Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol., 178, 2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi V.K. and Girvin,M.E. (1999) Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature, 402, 263–268. [DOI] [PubMed] [Google Scholar]
- Sambongi Y., Iko,Y., Tanabe,M., Omote,H., Iwamoto,K.A., Ueda,I., Yanagida,T., Wada,Y. and Futai,M. (1999) Mechanical rotation of the c subunit oligomer in ATP synthase (F0F1): direct observation. Science, 286, 1722–1724. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sato K. and Homma,M. (2000) Functional reconstituion of the Na+-driven polar flagellar motor components of Vibrio alginolyticus. J. Biol. Chem., 275, 5718–5722. [DOI] [PubMed] [Google Scholar]
- Shah D.S. and Sockett,R.E. (1995) Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop–start flagellum. Mol. Microbiol., 17, 961–969. [DOI] [PubMed] [Google Scholar]
- Shah D.S., Armitage,J.P. and Sockett,R.E. (1995) Rhodobacter sphaeroides WS8 expresses a polypeptide that is similar to MotB of Escherichia coli. J. Bacteriol., 177, 2929–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp L.L., Zhou,J. and Blair,D.F. (1995) Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry, 34, 9166–9171. [DOI] [PubMed] [Google Scholar]
- Stolz B. and Berg,H.C. (1991) Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol., 173, 7033–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda H. and Unemoto,T. (1982) Characterization of the respiration-dependent Na+ pump in the marine bacterium Vibrio alginolyticus. J. Biol. Chem., 257, 10007–10014. [PubMed] [Google Scholar]
- Valiyaveetil F.I. and Fillingame,R.H. (1997) On the role of Arg-210 and Glu-219 of subunit a in proton translocation by the Escherichia coli F0F1-ATP synthase. J. Biol. Chem., 272, 32635–32641. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Sato,K., Asai,Y., Kawagishi,I. and Homma,M. (1999) Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol., 181, 5103–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Fillingame,R.H. (1995) Changing the ion binding specificity of the Escherichia coli H+-transporting ATP synthase by directed mutagenesis of subunit c. J. Biol. Chem., 270, 87–93. [DOI] [PubMed] [Google Scholar]
- Zhou J., Lloyd,S.A. and Blair,D.F. (1998a) Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA, 95, 6436–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Sharp,L.L., Tang,H.L., Lloyd,S.A., Billings,S., Braun,T.F. and Blair,D.F. (1998b) Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp32 of MotB. J. Bacteriol., 180, 2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]