Abstract
The nearly one million Alu repeats in human chromosomes are a potential threat to genome integrity. Alus form dense clusters where they frequently appear as inverted repeats, a sequence motif known to cause DNA rearrangements in model organisms. Using a yeast recombination system, we found that inverted Alu pairs can be strong initiators of genetic instability. The highly recombinagenic potential of inverted Alu pairs was dependent on the distance between the repeats and the level of sequence divergence. Even inverted Alus that were 86% homologous could efficiently stimulate recombination when separated by <20 bp. This stimulation was independent of mismatch repair. Mutations in the DNA metabolic genes RAD27 (FEN1), POL3 (polymerase δ) and MMS19 destabilized widely separated and diverged inverted Alus. Having defined factors affecting inverted Alu repeat stability in yeast, we analyzed the distribution of Alu pairs in the human genome. Closely spaced, highly homologous inverted Alus are rare, suggesting that they are unstable in humans. Alu pairs were identified that are potential sites of genetic change.
Keywords: Alu repeats/human genome stability/inverted repeats/recombination/yeast
Introduction
The approximately one million Alu elements in the human genome constitute the major class of repetitive DNA (reviewed in Schmid, 1996). Although the average Alu homology is 85% (Shen et al., 1991), recombination between Alus is often cited as a source of human genome instability. Alus form dense clusters where they frequently appear as inverted repeats (IRs). Since IRs can induce DNA rearrangements in model organisms, it is important to know whether inverted Alu repeats are contributors to genomic change in humans.
DNA sequence motifs comprised of long identical IRs can be unstable in prokaryotes and eukaryotes, inducing both homologous and illegitimate recombination (reviewed in Ehrlich, 1989; Leach, 1994). It is generally assumed that IR-stimulated genomic instability results from intrastrand complementary interactions between IRs that lead to hairpin or cruciform secondary structures. While there are several explanations for the instability, two common models have emerged. Extruded hairpins can be recognized and processed by structure-specific nucleases resulting in double-strand break (DSB) formation and subsequent end-joining or recombinational repair (Ehrlich, 1989; Leach, 1994; Connelly and Leach, 1996; Akgun et al., 1997; Leach et al., 1997; Lewis, 1999). Alternatively, a hairpin structure that might arise prior to or during DNA synthesis can block replication fork progression either leading to DSBs or causing intra- or intermolecular template switching of DNA polymerase between short direct repeats (Egner and Berg, 1981; Foster et al., 1981; Gordenin et al., 1993; Lobachev et al., 1998).
The frequency with which IRs stimulate DNA rearrangements depends on various factors that might affect secondary structure formation or stability of a hairpin stem. They include the length of an IR and the spacer (distance between the repeats); base composition of IRs and/or spacer; type and position of replication origin; IR location in the genome; and genetic background (reviewed in Ehrlich, 1989; Leach, 1994). The greatest level of IR-induced instability in prokaryotes and eukaryotes is observed with long palindromes (perfect head-to-head IRs), which are expected to be the most efficient at forming stable hairpins or cruciforms, as opposed to IRs separated by a spacer. In wild-type bacteria, palindromes longer than 150–200 bp introduced on a plasmid are highly prone to deletion and cannot be propagated (Collins et al., 1982; Hagan and Warren, 1983; Yoshimura et al., 1986). Although long palindromes can be maintained in lower and higher eukaryotes, they are extremely unstable during mitotic growth and meiosis (Henderson and Petes, 1993; Ruskin and Fink, 1993; Collick et al., 1996; Akgun et al., 1997; Nag and Kurst, 1997; Lobachev et al., 1998; Lewis, 1999; Lewis et al., 1999). For example, a perfect palindrome formed by two 1.0 kb IRs in yeast can stimulate intra- and interchromosomal recombination in the adjacent region nearly 10 000-fold (Lobachev et al., 1998). Hyper instability of artificially created palindromic sequences suggests that naturally occurring long IRs in the human genome that are capable of efficient formation of secondary structures may represent a threat to genomic integrity.
In the human genome there are many long IRs, most of which are composed of Alu repeats (Deininger and Schmid, 1976). While inverted Alu repeats are only ∼300 bp and diverged, we proposed that they are likely to be at-risk motifs (ARMs) and a potential source of diseases (Gordenin and Resnick, 1998). It is therefore important to characterize the structural parameters that can cause inverted Alus to be unstable and the genetic factors that might contribute to their instability. We developed a yeast-based recombination system to address the role of sequence divergence, distance between repeats and genetic background in the ability of an inverted pair of Alu repeats to induce genetic instability. Guided by results in yeast we investigated the distribution of inverted Alu pairs in the human genome (the Alu pair database is available on line at http://dir.niehs.nih.gov/ALU). The inverted Alu motifs that are highly efficient at initiating genetic change in yeast were found to be excluded from the human genome. These results suggest that inverted Alus can have a strong impact on human genome stability and evolution.
Results
Experimental system
The destabilizing effect of inverted Alu repeats was assessed by their ability to stimulate mitotic ectopic recombination between two lys2 alleles in a haploid strain (Figure 1). The Alus were inserted into the LYS2 gene on chromosome II. A lys2-8 allele (Lobachev et al., 1998) was integrated at the LEU2 locus of chromosome III. Since reversion of the lys2-8 mutation or deletion of Alu insertions leading to restoration of the LYS2 function are extremely rare events (<10–9), all Lys+ prototrophs were considered to arise by recombination.
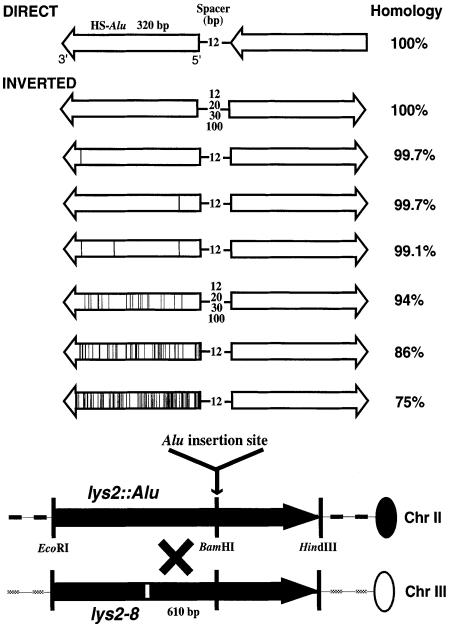
Fig. 1. Recombination system to study instability of Alu repeats in yeast. Alu repeats (open arrows) were inserted into the BamHI site of the LYS2 gene (black arrows). The Alu arrows describe the direction of transcription by RNA polymerase III; the arrowhead corresponds to the 3′ poly(A) tail (Schmid, 1996). The positions of nucleotide changes relative to the human-specific (HS) Alu consensus sequence (blank open arrow) are indicated as vertical lines within the arrows. The length of the spacer region separating the Alu repeats and the extent of homology in the Alu pairs are shown. The distances between the lys2-8 mutation (white rectangle) and the Alu insertion site are indicated (610 bp). The ‘X’ denotes a recombination event generating a wild-type LYS2 allele.
To examine the effect of sequence divergence on the ability of Alu IRs to stimulate homologous recombination, quasipalindrome inserts consisting of inverted Alu repeats with levels of identity from 75% to complete homology were developed (Figure 1). The 0.3 kb Alu repeats with different levels of homology relative to a human specific Alu consensus sequence (HS-Alu) (Batzer et al., 1994) were integrated into the LYS2 gene as inverted, as well as direct repeats. Alus that were 75, 86 and 94% diverged from HS-Alu were derived from the human HPRT gene. The single nucleotide changes (99.7% homology) were positioned at 49 and 43 bp from the 5′ or 3′ end of the Alu sequence, respectively, in order to test the consequences of a single mismatch at the base of a potential hairpin or closer to the loop. The Alu pair with three evenly distributed changes (99.1% homology) contained the two 3′ and 5′ single changes plus an additional nucleotide alteration 199 bp from the 5′ end of the Alu. Hairpins formed between the 99.7 and 99.1% identical inverted Alu would contain only base pair mismatches while the 94, 86 and 75% related pairs had small deletion/insertion (one or a few nucleotides) regions as well as base pair mismatches. The effect of spacer length, 12, 20, 30 and 100 bp, on the ability of inverted Alus to stimulate recombination was examined for 100 and 94% homologous pairs.
We measured stimulation of recombination by IRs as an increase over recombination in the presence of direct homologous or diverged Alu repeats; these would not be expected to form a secondary structure. Since direct repeats with different levels of identity and spacer distance had the same effect on recombination, only data for 100% homologous direct Alus separated by 12 bp are presented.
Effects of divergence and distance between Alu IRs on stimulation of recombination
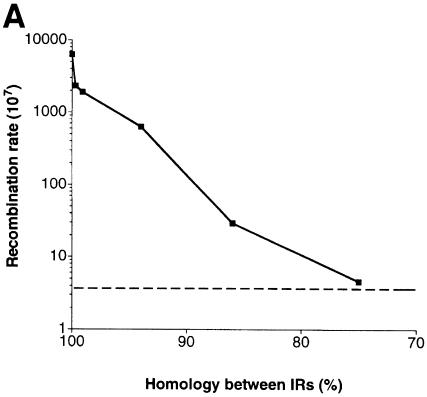
Homologous inverted Alus greatly stimulated ectopic recombination between the lys2 mutant alleles. The rate of interchromosomal recombination in a wild-type strain carrying 100% identical inverted Alu repeats was nearly 2000 times higher than in a strain containing direct homologous repeats (Table I). A single mismatch in the 3′ end of an Alu did not affect the IR-stimulated recombination. In contrast, a single mismatch at the 5′ end (i.e. close to the 12 bp spacer) of an Alu caused an almost 3-fold reduction in the recombination rate relative to the rate for identical IRs. The effects of three mismatches or a single mismatch in the 5′ end of Alu were comparable. Large increases in divergence between the inverted Alus greatly reduced the ability to stimulate recombination: there was a logarithmic decrease in recombination with increased divergence (Table I; Figure 2A). The 94 and 86% identical IRs resulted in 177- and 8-fold increases in recombination, respectively, and an IR composed of repeats with 75% identity failed to induce recombination. Thus, the IR-stimulated recombination requires DNA interactions between the Alus, and these interactions are prevented when the Alu divergence is in the range of 20%.
Table I. Recombination stimulated by inverted Alus in wild-type, msh2 and msh6 strains and effect of sequence divergence.
| Orientation of Alus in LYS2 gene | Homology (%) | Recombination rate (×107) |
||
|---|---|---|---|---|
| Wild type | msh2 | msh6 | ||
| Direct | 100 | 3.6 (2.7–4.5) | 5.4 (3.1–7.2) | 4.0 (2.5–6.7) |
| Inverted | 100 | 6331 (4658–9659) | 1467 (1165–2062) | 6329 (5387–8609) |
| 99.7 (1 mismatch, 3′ end) | 5896 (4203–10390) | 1249 (959–1709) | 5902 (4193–9567) | |
| 99.7 (1 mismatch, 5′ end) | 2311 (1910–3513) | 636 (531–787) | 2030 (1564–3262) | |
| 99.1 (3 mismatches) | 1897 (1523–3253) | 735 (598–1100) | 1411 (1022–3802) | |
| 94 | 634 (417–1402) | 295 (210–595) | 679 (501–1099) | |
| 86 | 29 (25–33) | 17 (15–23) | 20 (19–29) | |
| 75 | 4.6 (4.0–7.6) | 7.8 (4.9–8.5) | 4.8 (3.5–8.1) | |
The inverted and direct Alu repeats were separated by 12 bp. Single mismatches at the 3′ and 5′ ends of the Alu were, respectively, distal and proximal to the spacer region of the IRs (Figure 1). Median rates and 95% confidence intervals (in parentheses) were calculated as stated in Materials and methods.
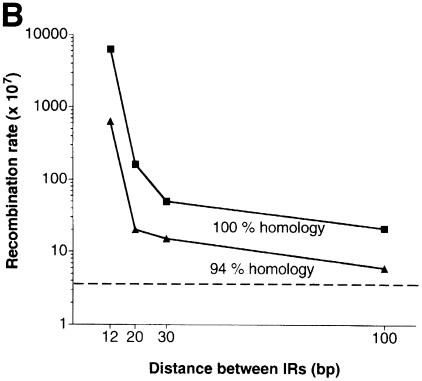
Fig. 2. Effect of DNA divergence and distance between inverted Alu repeats on their ability to stimulate recombination. (A) Effect of sequence divergence. Squares, recombination rates with IRs separated by 12 bp (see Table I). For 99.7% homologous inverted Alu repeats only data for IRs where a single mismatch was located at the 5′ end (i.e. close to the 12 bp spacer) are presented (see Results). Dotted line, level of recombination (4 × 10–7) in a strain containing a direct Alu repeat. (B) Effect of the distance between inverted Alus. Squares and triangles, recombination rates with 100 and 94% identical IRs, respectively. Dotted line, level of recombination (4 × 10–7) in a strain containing a direct Alu repeat.
The IR-stimulated recombination was also strongly affected by distance between the Alu repeats. With an increase of the spacer length from 12 to 20 bp between the 100 or 94% identical inverted Alus, there was a 39- and 31-fold decrease in recombination, respectively (Figure 2B). Further increases in distance from 20 to 100 bp led to a more gradual decline in recombination rates (8- and 3-fold reduction, respectively, for the 100 and 94% identical Alu repeats). An increase in both divergence and distance between IRs caused a synergistic decrease in recombination (Figure 2B). For example, 94% identical inverted Alus stimulated recombination 10 times less frequently than 100% homologous IR, and reducing the spacer from 12 to 100 bp caused a 300-fold decrease in recombination rate. When these factors were combined, there was little if any ability of the IRs to induce recombination (Figure 2B). These observations of dependence on distance and homology further support the view that the stimulation of recombination depends on the ability of the inverted Alu repeats to form hairpin structures.
Genetic factors that can enhance recombination stimulated by Alu IRs
Recombination between diverged DNAs is prevented by MMR, presumably by blocking annealing of the homologous substrates or the destruction of heteroduplex intermediates (reviewed in Crouse, 1998). Since the MMR system might act on mismatches that would occur in extruded hairpins, we tested whether the MMR system is responsible for the anti-recombinagenic effect of divergence between inverted Alus. If MMR proteins prevent the formation of mispaired hairpins or destroy them after they are formed, the recombination rates in MMR-defective strains would be higher. However, the rates of recombination in Δmsh6, Δmsh2 (Table I), Δmsh3, Δmlh1 and Δpms1 mutants (data not shown) containing diverged inverted Alus were not increased relative to the rates in wild-type strains. For the Δmsh2 (Table I) and Δmsh3 strains (data not shown) there was a 2- to 4-fold reduction in the ability of both homologous and diverged repeats to induce recombination, consistent with the role that these genes play in processing recombination intermediates (Paques and Haber, 1997; Sugawara et al., 1997). We conclude that MMR does not prevent the instability associated with diverged inverted Alus.
We initiated a screen to identify genes that would specifically increase recombination stimulated by inverted Alu repeat motifs. The screen utilized Alu repeats that were 94% homologous and separated by 100 bp, because these have almost no stimulatory effect on their own. Since this motif is present in the human genome (see below), the isolation of mutants that would lead to increased recombination could reveal a novel category of genetic factors and ARMs that are relevant to genome stability. Random gene inactivation was accomplished with a gene disruption library (Ross-Macdonald et al., 1999) and isolates were found that exhibited a hyper-recombination phenotype. Sequencing of DNAs flanking the mTn-3×HA/lacZ insertions identified disruptions of the RAD27 and MMS19 genes. To ascertain that the hyperrec phenotypes were not due to additional mutations and that they were specific to IR-stimulated recombination, these genes were inactivated in strains containing either the 94% homologous Alu IR with a 100 bp spacer or the direct Alu repeat in the LYS2 gene (see Materials and methods). Deletion of the RAD27 gene caused an absolute increase of ∼20 × 10–7 in the recombination rate (from 3.6 to 23.3 × 10–7) for the control strain with the direct Alu repeats (Table II), consistent with other reports of a rad27 hyperrec phenotype (Symington, 1998 and references therein). Disruption of the RAD27 gene in a strain containing the diverged inverted Alu caused a much stronger absolute increase in recombination (104 × 10–7) than that due to the IR or the rad27 mutation alone (i.e. synergistic effect). Since RAD27 is involved in the processing of Okazaki fragments during lagging strand replication (Lieber, 1997), we examined another mutation that has an impact on inverted repeat stability and has been proposed to affect lagging strand replication in yeast: the temperature-sensitive polymerase δ mutation pol3-t (Lobachev et al., 1998 and references therein). It also led to a synergistic increase in IR-induced recombination (Table II).
Table II. Δrad27, pol3-t and Δmms19 mutations increase the recombination potential of distantly separated, diverged inverted Alu repeats.
| Orientation of Alu pairs | Distance (bp) | Homology (%) | Recombination rate (× 107) |
|||
|---|---|---|---|---|---|---|
| wt | Δrad27 | pol3-t | Δmms19 | |||
| Direct | 12 | 100 | 4 (3–5) | 23 (17–38) | 26 (11–37) | 3 (2–5) |
| Inverted | 100 | 94 | 6 (5–12) | 110 (83–164) | 145 (90–223) | 104 (87–123) |
Median rates and 95% confidence intervals (in parentheses) were calculated as described in Materials and methods.
The mms19 mutation differed from the Δrad27 and the pol3-t mutants in that it had a specific effect on IR-stimulated recombination. The recombination rates were comparable for both the wild-type and the Δmms19 strains when one of the lys2 alleles contained a direct Alu repeat. However, the Δmms19 mutation caused a 16-fold increase (∼100 × 10–7 absolute increase) in recombination when the allele contained the inverted diverged Alu repeats separated by 100 bp (Table II).
Analysis of the Alu distribution in the human genome
Based on the above results we analyzed the distribution of Alu elements in the human genome (human sequence database release of September 1999) in order to identify and characterize inverted Alu pairs. Since Alus may be truncated, we identified common (i.e. aligned) regions within each Alu pair that shared sequence homology and all subsequent analyses were based on the aligned regions. The Alus were classified according to their size, distance, extent of homology and orientation relative to each other. Presented in Table III are results for Alu pairs where the region of alignment was >275 bp (i.e. corresponding to approximately full size Alu elements). A complete annotation of all Alus along with a description of inverted and direct pairs has been made (J.E.Stenger, K.S.Lobachev, D.A.Gordenin, T.Darden, J.Jurka and M.A.Resnick, unpublished) and is available on line at http://dir.niehs.nih.gov/ALU.
Table III. Distribution of inverted and direct Alu pairs in the human genome.
| Orientation of Alu pairs | Homology (%) | Number of Alu pairs with aligned regions separated by: |
||
|---|---|---|---|---|
| 0–20 bp | 21–100 bp | 101–500 bp | ||
| Inverted | 60–80 | 127 | 1525 | 4772 |
| 81–100 | 22 | 633 | 2483 | |
| Direct | 60–80 | 2643 | 2317 | 5628 |
| 81–100 | 1518 | 1094 | 2850 | |
Only data for Alu repeats that are >275 bp are presented. A complete analysis of Alu distribution can be found at http://dir.niehs.nih.gov/ALU
We found that highly related, closely spaced Alu pairs are rare in the human genome (Table III). There are similar frequencies of inverted and direct Alu repeats when the separation distance is >20 bp. Among the total inverted plus direct repeats, the frequency of direct repeats is ∼1.2–1.7 that of the IRs for both the 60–80% and the 81–100% homologous Alu pairs. This contrasts with the strong bias in distribution of Alus for distances ≤20 bp. For the 60–80% homologous Alu pairs, the direct repeats are 21 times more frequent than IRs. This is increased to 70 times more frequent when the Alu pairs are more homologous (81–100%). Thus, there is an excluded group of closely spaced highly homologous inverted Alu pairs in the human genome and this corresponds to those IRs that exhibit a high level of instability in yeast.
Discussion
Long IRs are a source of genome instability in a variety of prokaryotes and eukaryotes (reviewed in Ehrlich, 1989; Leach, 1994; Gordenin and Resnick, 1998). We reasoned that in addition to such well-characterized unstable sequence motifs in the human genome as triplet repeats, microsatellites and minisatellites, commonly occurring IRs could also be potential threats to genome stability. Alus were investigated because they are expected to be the most common long IRs in the human genome in that they are abundant and tend to form dense tandem clusters. The aim of this study was to determine the structural and genetic parameters that would cause human IRs to be a threat to genome integrity in a model system and to find potentially unstable IRs in human DNA.
Various mechanisms have been proposed to explain IR-induced instability (reviewed in Ehrlich, 1989; Gordenin et al., 1993; Leach, 1994). It is generally accepted that an early step includes the formation of either a hairpin in a single-stranded DNA or a cruciform structure in a double-stranded DNA. Since divergence and distance between Alu repeats are expected to influence the interaction between IRs and the formation of secondary structure, an analysis of these factors will help define the parameters that determine the recombinagenic potential of inverted Alu repeats. We developed a yeast-based system to assess these factors. We have shown that for distances <20 bp, inverted Alus that are >85% identical can efficiently stimulate recombination in a MMR-independent fashion. Guided by results in yeast, we found that highly homologous, closely spaced inverted Alu repeats are rare in the human genome.
Effects of sequence divergence on recombination stimulated by inverted Alus
Sequence divergence can be a barrier to the ability of IRs to stimulate recombination. We observed an exponential decrease in recombination with an increase in divergence between IRs and complete loss of stimulation when homology was reduced to 75% (Table I; Figure 2A). The suppressing effect of sequence divergence could be due to a decreased likelihood of forming a secondary structure. In support of this, a single mismatch reduced the recombinational impact of an IR, but only when it was located proximal to the center of symmetry. In terms of a hairpin model for the initiation of recombination, the generation of the hairpin, whether by extrusion from double-stranded DNA (i.e. a cruciform) or by intrastrand annealing in a single-stranded region, is likely to be more sensitive to mispairs that arise near the center of symmetry.
Multiple mismatches would also reduce the stability of heteroduplex DNA present in a secondary structure. However, this alone does not explain the exponential reduction with decreased homology leading to an ∼220-fold decrease in recombination when the DNAs are diverged by only 14%. A similarly strong barrier is found for recombination between DNAs that are diverged (Datta et al., 1997; Chen and Jinks-Robertson, 1999). Possibly, the impact of multiple mismatches is due to proteins, such as those involved in MMR, that specifically recognize mismatches and either destroy the heteroduplexes or prevent their formation. Numerous in vivo and in vitro studies suggest that MMR proteins can recognize mismatched heteroduplex DNA and prevent recombination between diverged DNAs (reviewed in Crouse, 1998). We found that the recombination stimulated by diverged inverted Alu repeats was not increased in Δmsh2, Δmsh3, Δmsh6, Δmlh1 or Δpms1 mutants (Table I and data not shown). These results argue that the MMR system does not prevent formation of a secondary structure with multiple mismatches and that mismatches in the hairpin stem are not processed by MMR.
There are several explanations for the lack of effect of the MMR system on recombination stimulated by diverged IRs. First, a non-canonical DNA structure (such as a hairpin) containing mismatches may not be recognized by the MMR system. Secondly, it is possible that events leading to initiation of recombination by IRs happen before MMR can recognize and process a hairpin with mismatches. Thirdly, a putative enzyme that binds and cuts secondary structure may cover the stem of the hairpin and thereby protect mismatches from MMR proteins. This was suggested by Nag and Kurst (1997) based on the absence of repair of mismatches in short hairpins during meoisis. Fourthly, results in bacteria (Trinh and Sinden, 1991; Rosche et al., 1995) and yeast (Gordenin et al., 1992, 1993; Ruskin and Fink, 1993) indicate that the formation of a secondary structure by an IR occurs in the lagging template during DNA replication. If there is strand discrimination during MMR in yeast, it is possible that mismatches present in the template strand (i.e. the hairpin) might escape recognition by the MMR system.
The impact of separation on recombination stimulated by inverted Alus
The recombinagenic potential of inverted Alus is strongly dependent on the distance between the repeats, regardless of the level of homology. Increasing the distance between inverted Alus from 12 to 20 bp caused a 39-fold reduction in recombination (Figure 2B), while an additional 80 bp led to only a further 8-fold reduction. It is unlikely that simply the size of single-stranded loops can affect the stability of the expected hairpin. The dramatic effect of distance is more likely to be due to characteristics of extrusion and/or opportunities for protein–DNA interactions. For hairpin extrusion ∼10 bp at the center of symmetry of IRs must unwind; this can occur with a small amount of energy obtainable from negative DNA supercoiling. Much greater energy is needed if there are >10 bp of unique DNA at the center of symmetry (Sinden, 1994). It is also possible that there is a threshold for cell factors that remove secondary structures in single-stranded DNA. For example, the affinity of human replication factor A (RPA) for a 30 bp single-stranded oligonucleotide is 50 times higher than its affinity for a 10 bp oligonucleotide (Wold, 1997). Similar differences might exist for binding of RPA with single-stranded loops in hairpins.
Genetic factors destabilizing distantly spaced diverged inverted Alu repeats
We found that diverged Alu repeats separated by 100 bp did not induce recombination. This could be an intrinsic property of this motif; alternatively, the stability could be under genetic control. Using the diverged and distantly separated repeats to identify mutants, we demonstrated that this motif has a high potential for inducing recombination. Three mutants were isolated that could cause the non-recombinagenic pair of inverted Alus to become strong initiators of recombination.
Based on the roles of DNA polymerase δ and FEN1/Rad27 in lagging strand replication (Sugino, 1995; Lieber, 1997), the impact of the pol3-t and Δrad27 mutations could be due to alterations in replication. Gordenin et al. (1992) proposed that pol3-t mutants may lack coordination between leading and lagging strand replication, resulting in extensive or longer lived single-stranded regions during lagging strand synthesis. This would increase the opportunity for secondary structures (i.e. hairpins) between distant IRs, particularly diverged DNAs. The rad27 mutation may have a similar effect by delaying completion of lagging strand replication, which in turn may lead to long single-stranded regions.
The effects of the Δmms19 mutant are different from those of the pol3-t and Δrad27 mutations. It caused a specific increase in IR-induced recombination with no effect on direct repeats, suggesting an impact of MMS19 on secondary structures formed by IRs. The Mms19 protein affects RNA polymerase II transcription, apparently through upstream regulation of the TFIIH complex, which is also involved in nucleotide excision repair (NER) (Lauder et al., 1996; Lombaerts et al., 1997). Possibly, there is increased instability of IRs in a Δmms19 mutant due to weak expression of genes coding proteins that destroy hairpin or cruciform secondary structures. Alternatively, IRs might form secondary structures during transcription of the LYS2 gene and these could be subject to helicase components of TFIIH. The lack of TFIIH activity in a Δmms19 mutant (Lauder et al., 1996) would result in a more stable secondary structure. Another possibility could relate to the role of MMS19 in NER. Inactivation of MMS19 might cause a defect in recognition of secondary structure by components of NER. Consistent with this suggestion, the instability of triplet repeats that form secondary structures was greatly induced in Escherichia coli uvrA– mutants defective in NER (Parniewski et al., 1999).
Implications of human Alu IRs for genome instability
Inverted Alu repeats can have a considerable potential for inducing genome instability. It should be noted that the system we developed detects the potential of inverted Alu repeats to initiate genetic change. While we have measured recombination stimulated by IRs in yeast, other outcomes could result from the initiating event caused by Alu IRs if they occurred in human cells, such as chromosome aberrations and loss of heterozygosity (LOH).
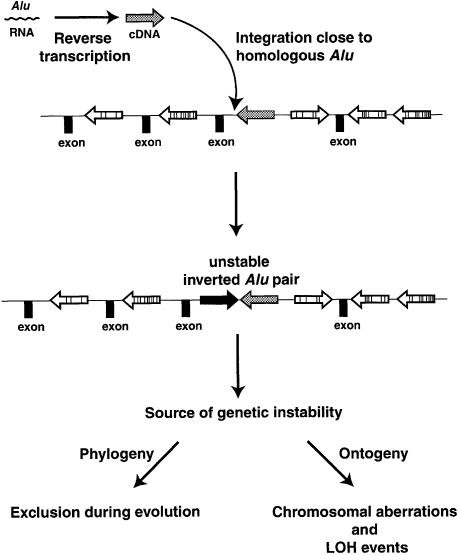
We found that the potential for Alu-induced genetic change was strongly influenced by homology and distance. Even repeats that are only 86% homologous but separated by <20 bp were efficient at inducing recombination. Based on our results in yeast, we developed a new approach to the analysis of the Alu distribution in the human genome. Rather than simply looking at the distance between Alu sequences independently of their size, we analyzed the distribution of Alu pairs according to alignment length of the shared homologous regions, divergence and distance between the aligned regions of the repeats, as well as orientation (direct or inverted). Among all the Alu pairs, direct Alus with closely spaced aligned regions were greatly overrepresented while closely spaced inverted Alu pairs were rare. The results of Jurka (1997) implied that the bias towards closely spaced direct repeats might be due to target preference of an incoming Alu next to an already integrated element. However, this targeting mechanism does not account for the paucity of closely spaced (<20 bp) inverted Alu repeats that are 81–100% identical, especially since the inverted pairs are common when the distance is increased (≥21 bp). These observations along with results in yeast (Table I; Figure 2) lead us to propose that highly homologous, closely spaced inverted Alu repeats are also unstable in the human genome. If such pairs were formed during Alu amplification, they may have been excluded during evolution of the human genome (Figure 3). These results suggest opportunities to track evolutionary changes involving inverted Alu pairs through a comparison of homologous loci from genomes of related primates where Alu repeats are common. It will be interesting to determine whether the exclusion of closely spaced highly homologous IRs observed for the human genome is a general tendency for other complex genomes containing repeats with different types of transposition mechanisms.
Fig. 3. Formation of unstable inverted Alu pairs and proposed consequences to human genome stability. Alu retroposition may lead to an inverted Alu pair. Depending on homology and distance, IRs may initiate genome instability either in wild type or in mutants. Among the outcomes in addition to homologous recombination may be chromosomal aberrations and LOH events. During evolution, inverted Alu pairs that have a destabilizing effect would be excluded from the human genome.
Even though unstable inverted Alu repeats appear to be excluded during evolution, we suggest that Alu retroposition in germinal and somatic tissue may give rise to new unstable pairs (Figure 3). The high level of Alu retroposition (approximately one event per 200 new births; Deininger and Batzer, 1999), the large number of Alus and the preference for integration next to a pre-existing Alu would generate these unstable repeats. Formation of unstable inverted Alu pairs in introns or in intergenic regions might not lead to immediate gene inactivation but could create loci prone to chromosomal changes (such as aberrations and LOH) that may result in disease. These at-risk Alu IRs may be identified in studies of human polymorphisms or through analyses of unstable chromosomal regions.
We identified several inverted Alu pairs in the human genome that are potentially unstable (Table IV). Based on results with wild-type and mutant yeast, two categories of inverted Alus were considered to be at risk: Alu IRs that might be unstable in wild-type human cells (≥85% homologous and separated by <20 bp) and Alu IRs that might induce rearrangements in mutants (≥90% homologous and separated by <100 bp). Only low to moderate levels of instability might be expected in wild-type cells for the Alu repeats presented in Table IV if the effects of divergence and spacer in human cells are similar to those observed in yeast. Additional factors such as chromatin organization, sequence context, transcription level of the Alu repeats and surrounding genes, and distribution of mismatches in the aligned pairs might affect their potential to cause genomic rearrangements. As we have demonstrated in yeast, the instability associated with inverted Alu pairs can be greatly increased in mutants defective in DNA metabolism. This may indicate that reduced levels of these components might increase genome instability. One of the loci identified (the BTF2p44 gene) is frequently deleted in patients with spinal muscular atrophy, although no direct connection with Alu repeats has been described (Burglen et al., 1997; Carter et al., 1997; Wang et al., 1997). It will be interesting to assess the stability of IRs such as those listed in Table IV in human cells and to identify destabilizing genetic and environmental factors. Based on our yeast studies, most inverted Alu pairs in humans are likely to be stable due to high sequence divergence and long spacer distance. Considering the abundance of inverted Alus that are distantly separated (for example, see Table III and Alu database at http://dir.niehs.nih.gov/ALU), factors that would have only a mild destabilizing effect on each inverted Alu pair could dramatically change the integrity of the human genome.
Table IV. Examples of potentially unstable inverted Alu pairs >275 bp in the human genome.
| Position in genome |
Homology (%) | Distance (bp) | Length of aligned regions (bp) | ||
|---|---|---|---|---|---|
| Chromosome | Locus ID | Sequence position | |||
| 17 | AC004771 | 60262–60879 | 86 | 17 | 303 |
| 1q24 | AL021068 | 93086–93681 | 85 | 3 | 297 |
| 17 | AC005274 | 6760–7354 | 85 | 4 | 297 |
| 12p13.3 | AC006057 | 20026–20622 | 85 | 18 | 290 |
| 5q13.1 (BTF2p44 gene) | U80017 | 17753–18403 | 95 | 35 | 311 |
| 21q22.3 | AF064864 | 17714–18401 | 92 | 86 | 302 |
| 22q12.3 | AL008716 | 27276–27976 | 92 | 91 | 306 |
| 17 | AC004584 | 58885–59500 | 91 | 47 | 285 |
| 20q11.1 | AL035071 | 72778–73456 | 91 | 59 | 299 |
| 9 | AC0063120 | 48064–48740 | 91 | 83 | 299 |
Presented are examples of Alu IRs that may be potentially unstable based on structural parameters described in the text. Sequence position indicates the beginning coordinate for the first Alu element and the end coordinate of the locus for the second Alu element in the aligned pair. The Alu pair database is located at http://dir.niehs.nih.gov/ALU
Materials and methods
Strains and plasmids
The CGL strain (MATα, ade5-1, his7-2, leu2-3 112, trp1-289, ura3-Δ) that was used for integration of Alu repeats into the LYS2 gene was an isogenic Lys+ derivative of CG379Δ (Shcherbakova et al., 1996). Plasmids carrying Alu direct and inverted repeats were based on the pFL44S vector, which contains an EcoRI–HindIII fragment of the LYS2 gene in the polylinker site (plasmid p44L2). The Alu repeats were cloned into the unique BamHI site of LYS2 as follows. Four Alu sequences were amplified by PCR using primers that modified the ends of the Alus. The human-specific (HS) Alu consensus sequence from the plasmid pPD39 (Batzer et al., 1994) was amplified using primers 5′-TTATCCATATGCCAAATTGAGGGATCTGAAAAAAGAGCAGGGCAGTTTTTTTTTTTTTTTTTTTTTGAGA-3′ and 5′-CATTGATAGTTGAAATAACATTTGGATCCGTCGACGGCCGGGCGCGGTGGCTCACGCCT-3′. The three Alus [N36, N39 and N14 in Edwards et al. (1990)] with different levels of sequence homology relative to the HS-Alu element were amplified from the human HPRT gene located on a BAC/YAC (Kouprina et al., 1998). These three Alus required two rounds of amplification. For the first round, we used primers to the unique DNA regions flanking the Alu inserts in the HPRT gene. These amplified products were used in the second PCR amplification to modify the ends of Alu elements. The primer sets are described below. Alu N36, first primer set: 5′-CTACGTATTAAGACAAGAAACAGACTG-3′ and 5′-CAAAGCAGTAGTCTATCACATTAGT-3′; second primer set: 5′-TTATCCATATGCCAAATTGAGGGATCTGAAAAAAGAGCAGGGCAGTTTTTTTTTTTTTTGTTTTTTTTTTTGAGACGGAGTCTTGCTC-3′ and 5′-CATTGATAGTTGAAATAACATTTGGATCCGTCGACGGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAG-3′. Alu N39, first primer set: 5′-CCTCTTGAGGTAAGCACTATTATTATC-3′ and 5′-CAGCTTTCTCATCTAAAAAATGGGGATAATAG-3′; second primer set: 5′-TTATCCATATGCCAAATTGAGGGATCTGAAAAAAGAGCAGGGCAGTTTCTTTTTTTTTTTTTTTTTTTTTGACAGTCTTACTCTGTTGCCCAGGCA-3′ and 5′-CATTGATAGTTGAAATAACATTTGGATCCGTCGACGGCCGGGTGCAGTGGTTCGTGCCTGTAATCCC-3′. Alu N14, first primer set: 5′-GGCATGAGCCGCTGCATCAGCCAGCAG-3′ and 5′-CTGGCATGGTGTGGTGGCTCACACTTG-3′; second primer set: 5′-TTATCCATATGCCAAATTGAGGGATCTGAAAAAAGAGCAGGGCAGTTTTTTTTTTTTTTTTTTTTTGAGA-3′ and 5′-CATTGATAGTTGAAATAACATTTGGATCCGTCGACCCGGGGTGTGGTGGCTCACACTTGTAATCCCAGTGCTTT-3′. The N36, N39 and N14 Alus with the PCR modifications are ∼320 bp long and 94, 86 and 75% identical to HS-Alu consensus sequence, respectively.
Both the primers used to amplify the HS-Alu sequence and the second set of primers for the Alus from the HPRT gene contained 25–30 nucleotides at the 5′ ends that were identical to sequences at each side of the BamHI site in the LYS2 gene. The four Alu sequences described above were cloned into the BamHI site on a p44L2 vector by the gap repair technique (Oldenburg et al., 1997) through co-transformation of the PCR products and the BamHI-linearized p44L2 plasmid. Recombinant plasmids were rescued into the DH5α strain of E.coli. The HS-Alu sequence on the recombinant plasmid was modified using a QuikChange site-directed mutagenesis kit (Stratagene) to generate Alu sequences that have one or three base pair differences. To change one base pair at the 5′ end of the Alu we used mutation primers 5′-GTAATCCCAGCACTTTGGGAGGCCTAGGCG-3′ and 5′-CGCCTAGGCCTCCCAAAGTGCTGGGATTAC-3′ in the amplification reaction. This leads to the appearance of an AvrII site located 49 bp from the 5′ end of the HS-Alu sequence. To generate a one base pair change in the 3′ end of Alu we used mutation primers 5′-CTGGGCGACAGAGCGAGACTCCGGCTCAAA-3′ and 5′-TTTGAGCCGGAGTCTCGCTCTGTCGCCCAG-3′. This leads to elimination of the Tth111I site located 43 bp from the 3′ end of HS-Alu. The plasmid containing the HS-Alu sequence with the mutation in the 3′ end was used in a subsequent site-directed mutagenesis to introduce an additional change at the 5′ end of the Alu. All cloned and modified Alus were sequenced to confirm the changes. Sequencing of the Alu element with changes at the 5′ and 3′ ends revealed an additional base pair change 121 bp from the 5′ end. The resultant plasmids containing Alu inserts in the LYS2 gene had unique BamHI and SalI sites at the 5′ end of the Alu sequences. To create Alu pairs with different spacer lengths, the Alus cloned into LYS2 were combined with another HS-Alu sequence. The HS-Alu was amplified from pPD39 plasmid where 12, 20, 30 or 100 bp of extra non-Alu DNA and a SalI site were added to one primer and a BamHI site was added to the other. The amplified products were digested with BamHI and SalI and the modified HS-Alu consensus sequences were inserted in inverted or direct orientation next to each of the already cloned Alus on the plasmids using the SalI and BamHI sites. Construction and propagation of plasmids with inverted Alu repeats were carried out in SURE strains of E.coli (Stratagene). The primers used to amplify the HS-Alu sequence from the pPD39 plasmid are described below. For 12 bp spaced direct repeats: 5′-CTGGATCCGGCCGGGCGCGGTGGCTCACGCCT-3′ and 5′-CGTCGACCAACTGGAAAAAAGAGCAGGGCAGT-3′. For 12 bp spaced IRs: 5′-CGTCGACCAACTGGGCCGGGCGCGGTGGCTCACGCCT-3′ and 5′-TGGATCCGAAAAAAGAGCAGGGCAGT-3′. For 20 bp spaced IRs: 5′-AACGGTCGACCAACTGGTTACCATGGCCGGGCGCGGTGGCTCACGCC-3′ and 5′-TGGATCCGAAAAAAGAGCAGGGCAGT-3′. For 30 bp spaced IRs: 5′-AACGGTCGACCAACTGGTTACCATGTTAGGAGGTGGCCGGGCGCGGTGGCTCACGCCT-3′ and 5′-TGGATCCGAAAAAAGAGCAGGGCAGT-3′. For 100 bp spaced IRs: 5′-AACGGTCGACCAACTGGTTACCATGTTAGGAGGTCACATGGAAGATCAGATCCTGGAAAACGGGAAAGGTTCCGTTCAGGACGCTACTTGTGTATAAGAGTCAGGGCCGGGCGCGGTGGCTCACGCCT-3′ and 5′-TGGATCCGAAAAAAGAGCAGGGCAGT-3′. Finally, the ClaI fragment containing the ARSCEN cassette was cut out from the resulting plasmids to use them for integrative transformation. The CGL strain was transformed with HpaI-digested vectors and inverted and direct Alu repeats were transferred into the chromosomal LYS2 gene using a two-step replacement procedure. All replacements were confirmed by Southern blotting.
The EcoRI–HindIII fragment containing the lys2-8 allele (Lobachev et al., 1998) was cloned into the pRS305 integrative plasmid to generate p305L28. This plasmid was used to create an interchromosomal lys2 duplication. Strains containing Alu repeats in the LYS2 gene were transformed with SfoI-digested p305L28 plasmid to target lys2-8 allele to the LEU2 locus. Transformants were analyzed by Southern blotting.
Deletions of the MSH2, MSH3, MSH6, PMS1, MLH1, RAD27 and MMS19 genes were made by one-step replacement using a PCR disruption technique with a kanMX module (Wach et al., 1994) or disruption plasmids. Nucleotide sequences of the primers used to make and to confirm gene deletions and the disruption plasmids are available upon request. The pol3-t allele was introduced into the Alu-containing strains using the p171 plasmid (Tran et al., 1999a).
Genetic and molecular procedures
Genetic and molecular procedures were described previously (Gordenin et al., 1992, 1993). Rates of homologous recombination and 95% confidence intervals were determined in fluctuation tests using at least 14 independent cultures (Lobachev et al., 1998). The mTn-3×HA/lacZ disruption library used for screening of hyperrec mutants was obtained from Yale Genome Analysis Center (Ross-Macdonald et al., 1999). Screening of mutants was performed as recommended at http://ycmi.med.yale.edu/YGAC/protocol.html and loci containing inserts were determined as described previously (Tran et al., 1999b). The junction regions between the yeast genome and mTn-3×HA/lacZ insertions were amplified using primers lacZ 5′-GCGGGCCTCTTCGCTATTACG-3′, and lacZ-2 5′-TGAATGGCGAATGGCGCTTTG-3′ and, subsequently, sequenced using HAT primer 5′-TTCAATGGCCGCCTTAACGT-3′.
Computer analysis of the human genome database
The computational methods used to analyze the distribution of Alus in the human genome are available on line at http://dir.niehs.nih.gov/ALU. Alu sequences were obtained from the GenBank database (release 112.0 of September 1999, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD). A total of 153 645 Alu sequences (corresponding to ∼15–30% of the total Alus in the human genome) were analyzed.
Acknowledgments
Acknowledgements
We are grateful to J.Sterling and N.Degtyareva for assistance in experiments; to S.Prakash for pMB219 plasmid; to P.Klonowski and T.Darden for help in analyzing the human genome database and insightful comments; to R.Slebos, Y.Pavlov and M.Edgell for their critical readings of the manuscript.
References
- Akgun E., Zahn,J., Baumes,S., Brown,G., Liang,F., Romanienko,P.J., Lewis,S. and Jasin,M. (1997) Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol., 17, 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer M.A., Alegria-Hartman,M. and Deininger,P.L. (1994) A consensus Alu repeat probe for physical mapping. Genet. Anal. Tech. Appl., 11, 34–38. [DOI] [PubMed] [Google Scholar]
- Burglen L., Seroz,T., Miniou,P., Lefebvre,S., Burlet,P., Munnich,A., Pequignot,E.V., Egly,J.M. and Melki,J. (1997) The gene encoding p44, a subunit of the transcription factor TFIIH, is involved in large-scale deletions associated with Werdnig–Hoffmann disease. Am. J. Hum. Genet., 60, 72–79. [PMC free article] [PubMed] [Google Scholar]
- Carter T.A. et al. (1997) A multicopy transcription-repair gene, BTF2p44, maps to the SMA region and demonstrates SMA associated deletions. Hum. Mol. Genet., 6, 229–236. [DOI] [PubMed] [Google Scholar]
- Chen W. and Jinks-Robertson,S. (1999) The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics, 151, 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collick A., Drew,J., Penberth,J., Bois,P., Luckett,J., Scaerou,F., Jeffreys,A. and Reik,W. (1996) Instability of long inverted repeats within mouse transgenes. EMBO J., 15, 1163–1171. [PMC free article] [PubMed] [Google Scholar]
- Collins J., Volckaert,G. and Nevers,P. (1982) Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene, 19, 139–146. [DOI] [PubMed] [Google Scholar]
- Connelly J.C. and Leach,D.R. (1996) The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells, 1, 285–291. [DOI] [PubMed] [Google Scholar]
- Crouse G.F. (1998) Mismatch repair systems in Saccharomyces cerevisiae. In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Repair in Prokaryotes and Lower Eukaryotes. Vol. 1. Humana Press, Totowa, NJ, pp. 411–448. [Google Scholar]
- Datta A., Hendrix,M., Lipsitch,M. and Jinks-Robertson,S. (1997) Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl Acad. Sci. USA, 94, 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P.L. and Batzer,M.A. (1999) Alu repeats and human disease. Mol. Genet. Metab., 67, 183–193. [DOI] [PubMed] [Google Scholar]
- Deininger P.L. and Schmid,C.W. (1976) An electron microscope study of the DNA sequence organization of the human genome. J. Mol. Biol., 106, 773–790. [DOI] [PubMed] [Google Scholar]
- Edwards A. et al. (1990) Automated DNA sequencing of the human HPRT locus. Genomics, 6, 593–608. [DOI] [PubMed] [Google Scholar]
- Egner C. and Berg,D.E. (1981) Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc. Natl Acad. Sci. USA, 78, 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich D.S. (1989) Illegitimate recombination in bacteria. In Berg,D.E. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 799–832. [Google Scholar]
- Foster T.J., Lundblad,V., Hanley-Way,S., Halling,S.M. and Kleckner,N. (1981) Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell, 23, 215–227. [DOI] [PubMed] [Google Scholar]
- Gordenin D.A. and Resnick,M.A. (1998) Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat. Res., 400, 45–58. [DOI] [PubMed] [Google Scholar]
- Gordenin D.A., Malkova,A.L., Peterzen,A., Kulikov,V.N., Pavlov,Y.I., Perkins,E. and Resnick,M.A. (1992) Transposon Tn5 excision in yeast: influence of DNA polymerases α, δ and ε and repair genes. Proc. Natl Acad. Sci. USA, 89, 3785–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin D.A., Lobachev,K.S., Degtyareva,N.P., Malkova,A.L., Perkins,E. and Resnick,M.A. (1993) Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol., 13, 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan C.E. and Warren,G.J. (1983) Viability of palindromic DNA is restored by deletions occurring at low but variable frequency in plasmids of Escherichia coli. Gene, 24, 317–326. [DOI] [PubMed] [Google Scholar]
- Henderson S.T. and Petes,T.D. (1993) Instability of a plasmid borne inverted repeat in Saccharomyces cerevisiae. Genetics, 133, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. (1997) Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA, 94, 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N., Annab,L., Graves,J., Afshari,C., Barrett,J.C., Resnick,M.A. and Larionov,V. (1998) Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3′ end target sequence. Proc. Natl Acad. Sci. USA, 95, 4469–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder S., Bankmann,M., Guzder,S.N., Sung,P., Prakash,L. and Prakash,S. (1996) Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol. Cell. Biol., 16, 6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.R. (1994) Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays, 16, 893–900. [DOI] [PubMed] [Google Scholar]
- Leach D.R., Okely,E.A. and Pinder,D.J. (1997) Repair by recombination of DNA containing a palindromic sequence. Mol. Microbiol., 26, 597–606. [DOI] [PubMed] [Google Scholar]
- Lewis S.M. (1999) Palindromy is eliminated through a structure-specific recombination process in rodent cells. Nucleic Acids Res., 27, 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S., Akgun,E. and Jasin,M. (1999) Palindromic DNA and genome stability. Further studies. Ann. N Y Acad. Sci., 870, 45–57. [DOI] [PubMed] [Google Scholar]
- Lieber M.R. (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- Lobachev K.S., Shor,B.M., Tran,H.T., Taylor,W., Keen,J.D., Resnick,M.A. and Gordenin,D.A. (1998) Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics, 148, 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaerts M., Tijsterman,M., Verhage,R.A. and Brouwer,J. (1997) Saccharomyces cerevisiae mms19 mutants are deficient in transcription-coupled and global nucleotide excision repair. Nucleic Acids Res., 25, 3974–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D.K. and Kurst,A. (1997) A 140 bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics, 146, 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg K.R., Vo,K.T., Michaelis,S. and Paddon,C. (1997) Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res., 25, 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1997) Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniewski P., Bacolla,A., Jaworski,A. and Wells,R.D. (1999) Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res., 27, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W.A., Trinh,T.Q. and Sinden,R.R. (1995) Differential DNA secondary structure-mediated deletion mutation in the leading and lagging strands. J. Bacteriol., 177, 4385–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- Ruskin B. and Fink,G.R. (1993) Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics, 134, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C.W. (1996) Alu: structure, origin, evolution, significance and function of one-tenth of human DNA. Prog. Nucleic Acid Res. Mol. Biol., 53, 283–319. [DOI] [PubMed] [Google Scholar]
- Shcherbakova P.V., Noskov,V.N., Pshenichnov,M.R. and Pavlov,Y.I. (1996) Base analog 6-N-hydroxylaminopurine mutagenesis in the yeast Saccharomyces cerevisiae is controlled by replicative DNA polymerases. Mutat. Res., 369, 33–44. [DOI] [PubMed] [Google Scholar]
- Shen M.R., Batzer,M.A. and Deininger,P.L. (1991) Evolution of the master Alu gene(s). J. Mol. Evol., 33, 311–320. [DOI] [PubMed] [Google Scholar]
- Sinden R.R. (1994) DNA structure and Function. Academic Press, San Diego, CA. [Google Scholar]
- Sugawara N., Paques,F., Colaiacovo,M. and Haber,J.E. (1997) Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl Acad. Sci. USA, 94, 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A. (1995) Yeast DNA polymerases and their role at the replication fork. Trends Biochem. Sci., 20, 319–323. [DOI] [PubMed] [Google Scholar]
- Symington L.S. (1998) Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res., 26, 5589–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.T., Degtyareva,N.P., Gordenin,D.A. and Resnick,M.A. (1999a) Genetic factors affecting the impact of DNA polymerase δ proofreading activity on mutation avoidance in yeast. Genetics, 152, 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.T., Gordenin,D.A. and Resnick,M.A. (1999b) The 3′→5′ exonucleases of DNA polymerase δ and ε and the 5′-3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2000–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh T.Q. and Sinden,R.R. (1991) Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E.coli. Nature, 352, 544–547. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Carter,T.A., Das,K., Xu,J., Ross,B.M., Penchaszadeh,G.K. and Gilliam,T.C. (1997) Extensive DNA deletion associated with severe disease alleles on spinal muscular atrophy homologues. Ann. Neurol., 42, 41–49. [DOI] [PubMed] [Google Scholar]
- Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- Yoshimura H., Yoshino,T., Hirose,T., Nakamura,Y., Higashi,M., Hase,T., Yamaguchi,K., Hirokawa,H. and Masamune,Y. (1986) Biological characteristics of palindromic DNA (II). J. Gen. Appl. Microbiol., 32, 393–404. [Google Scholar]