Abstract
Chromatin has a tendency to shift from a relatively decondensed (active) to condensed (inactive) state during cell differentiation due to interactions of specific architectural and/or regulatory proteins with DNA. A promotion of chromatin folding in terminally differentiated avian blood cells requires the presence of either histone H5 in erythrocytes or non-histone protein, myeloid and erythroid nuclear termination stage-specific protein (MENT), in white blood cells (lymphocytes and granulocytes). These highly abundant proteins assist in folding of nucleosome arrays and self-association of chromatin fibers into compacted chromatin structures. Here, we briefly review structural aspects and molecular mode of action by which these unrelated proteins can spread condensed chromatin to form inactivated regions in the genome.
Keywords: Histone H5, MENT, Chromatin condensation, Avian blood cells
Chromatin conformation
Chromatin is not a static macromolecular structure and may adopt a dynamic configuration of chromatin fiber at different chromosomal loci depending on the presence of linker histones (Bustin et al. 2005), non-histone proteins (Eissenberg and Elgin 2000), chromatin remodelers (Corona et al. 2007), and nucleosomal histone posttranslational modifications (Strahl and Allis 2000). A nucleosome consists of a 146-bp DNA duplex wrapped around the octamer of core histones H2A, H2B, H3, and H4 (Luger et al. 1997). The nucleosomes are linked by 20–80-bp linker DNA capable of interacting with linker histones (H1 or H5) to form a nucleosome array. Linear nucleosome strings, also referred to as 10-nm diameter filament, composed of histone octamers bound to DNA represent the first level of hierarchical chromatin structure. Their folding and condensation give rise to a higher-ordered 30-nm-diameter secondary structures (Woodcock and Dimitrov 2001; Hansen 2002) for which two disparate architectural models of the chromatin fiber arrangement have been proposed.
A solenoid model (Finch and Klug 1976) depicts one-start helical structure of the chromatin filaments where the linker DNA is bent so that the adjacent nucleosomes are in a contact with each other to form a helix. A zig zag model (Woodcock et al. 1984), however, represents a two-start helix with a straight linker DNA so that the consecutive nucleosomes are lying at the opposite sites of the fiber axis. Such nucleosomal arrays visible as unique structural motifs that direct chromatin compaction and stabilization have been observed in the intact nuclei (Bednar et al. 1998). In vitro studies of chromatin fiber with force-measuring laser tweezers (Cui and Bustamante 2000) and a calculation of the size distribution of radiation-induced breaks in DNA strands in vivo (Rydberg et al. 1998) seem to favor irregular zig zag conformation of chromatin fiber. Electron microscopy visualization of a loosened nucleosome array obtained following stabilization by cross-linking of histone tail with disulfide compound indicated (Dorigo et al. 2004) that the chromatin fiber contained a parallel arrangement of nucleosome stacks with a zig zag fold in a two-start helix. In addition, using a DiSCO model combined with the Monte Carlo simulations of the array of 12-nucleosome units containing all histone tails, Sun et al. (2005) observed that they adopted a highly irregular conformation at a high ionic strength which transited to the extended “beads-on-a-string” form at a low salt concentration. The salt-dependent condensation is a strong indication of electrostatic interactions including the linker DNA–nucleosome and internucleosomal attractions in addition to the repulsion between linker DNA units. Generally, the nucleosome arrangements with a short nucleosome repeat length (NRL) of 167 bp seem to be stiffer than nucleosomal fibers with 197-bp NRL (Kruithof et al. 2009) as probed by a fiber stretching (at the forces between 0.5 and 3.5 pN) in the presence or absence of linker histones (H1 or H5). The results of single-molecule force spectroscopy (Kruithof et al. 2009) provide strong evidence that the 30-nm chromatin fiber may exist in the form of one-start solenoid. Moreover, it appeared (Kruithof et al. 2009) that linker histones do not affect the length or stiffness of the fiber but rather stabilize its folding. The core histone tails, especially those of H3 (Kan et al. 2007) and H4 (Kan et al. 2009), were responsible for the internucleosome attractions and most of the linker DNA–nucleosome interactions (Sun et al. 2005). In addition to the interaction of an acidic patch on the surface of histone H2A with basic residues in the N-terminal tail of histone H4 (Luger et al. 1997; Kan et al. 2009), the histone H2A C-terminal domain can regulate nucleosome stability and chromatin structure by acting as a recognition module for histone H1 binding to the nucleosome (Vogler et al. 2010). Such histone–histone interactions may affect formation of the 30-nm chromatin fiber whose existence in vivo, irrespective of contentious details of nucleosome arrangement, remains controversial (Maeshima et al. 2010; Fussner et al. 2011). A lack of higher order chromatin structure in high-resolution cryoelectron microscopy images of human mitotic cells persuaded Eltsov et al. (2008) to postulate that nucleosomal fibers form a highly disordered state resembling a polymer melt without folding into 30-nm fiber structure. Although some imperfections in sample preparations for electron microscopy may result in the inability to detect 30-nm fibers in situ, the existence of rigid rodlike structures observed in early experiments using synchrotron radiation X-ray (Bordas et al. 1986) and neutron scattering (Gerchman and Ramakrishnan 1985) in solubilized chromatin was later confirmed using chromatin fiber preparations of 24–25 nm (Dorigo et al. 2004) and 35 nm (Robinson et al. 2006) in a diameter. Similar results were obtained by Ghirlando and Felsenfeld (2008) who identified 33–45-nm rodlike particles with six to seven nucleosomes per 11 nm turn in both constitutive and facultative heterochromatin segments excised from the chicken β-globin locus and released from the nuclei.
As it was shown in numerous in vitro and in vivo studies (for review, see Hansen 2002), the 30-nm chromatin fiber could be organized into large-scale structural levels, such as fiber segments of ~60–80 and ~100–130 nm in a diameter (Belmont and Bruce 1994) attributed to the heterochromatin states in terminally differentiated cells (Belmont 1999). Extensively compacted chromatin fibers were observed in nuclei of many cell types (Woodcock and Horowitz 1995) where they usually adopted a highly non-uniform structures with non-helical irregular zig zag conformations (Woodcock et al. 1993). Similar patterns of compacted chromatin fibers isolated from distinct blood cells were observed in different ultrastructural studies. Both chicken erythrocyte chromatin imaged by scanning force microscopy (Zlatanova et al. 1994) and chicken granulocyte chromatin visualized by cryoelectron microscopy (Grigoryev et al. 1999) possessed self-associated structures in which chromatin fiber was folded back on itself forming irregular structures thicker than those of 30 nm in a diameter. Whereas the observed folded back structure of 40–50 nm in a diameter was less than that of 60 nm predicted for the two side-by-side aligned 30-nm fibers, the fold back fashion of interfiber interaction needed a reciprocal binding of nucleosomes between laterally arranged fibers (Grigoryev et al. 2006). It seems that these cell-specific condensed chromatin states were created via a common electrostatic mechanism that required neutralization of negatively charged DNA by positively charged proteins, histone H5 in the erythrocytes and MENT in the granulocytes, which were shown to be extra accumulated in the respective cells (Table 1).
Table 1.
A brief characteristics of histone H5 and chromatin protein MENT
| Histone H5 (189 amino acids; 20.5 kDa) | MENT (410 amino acids; 42 kDa) |
|---|---|
| Nuclear localization and functionality | |
| Histone H5 is deposited in terminally differentiated erythrocytes (~1.4 molecule/nucleosome) forming large-scale condensed and repressed heterochromatic regions (Thomas et al. 1992; Bednar et al. 1998; Koutzamani et al. 2002) | MENT is deposited in non-red blood cells (granulocytes ~2 molecules/200 bp of DNA) forming condensed and repressed chromatin (Grigoryev and Woodcock 1998) |
| Domain organization and molecular structure | |
| N-terminal domain (1–21 aa), C-terminal domain (101–189 aa), and globular domain (22–100 aa) containing winged-helix fold consisting of three helix bundles (H1 29–38aa, H2 48–58aa, H3 65–78aa) and two strands of β-ribbon (81–85aa and 93–96aa) (Briand et al. 1980; Ramakrishnan et al. 1993) | M-loop domain (61–91 aa), NLS domain (80–84 aa), and RCL domain (352–379 aa). The molecule adopts α/β fold comprised of nine α-helices (hA–hI) and three β-sheets (A–C) (Grigoryev et al. 1999; McGowan et al. 2006) |
| DNA binding sites | |
| Two DNA binding sites on the globular domain: a primary binding site (Lys69, Arg73, and Lys85), and a secondary binding site (Lys40, Arg42, Lys52, Arg94) (Goytisolo et al. 1996; Duggan and Thomas 2000) | Two DNA binding sites on the M-loop domain: One site around AT-hook motif and the second site around D- and E-helices (McGowan et al. 2006) |
| Mechanism of action | |
| One-step formation of compacted chromatin fibers by cooperative binding of globular domains to DNA with a subsequent dimerization inducing stem-like structures needed for chromatin folding (Bednar et al. 1998; Thomas et al. 1992) | Two-step formation of compacted chromatin fibers by initial binding to DNA and folding the nucleosome array by M-loop domains and further bridging the separate arrays by RCL domains to create self-associated chromatin fibers (Grigoryev 2001; Springhetti et al. 2003) |
Factors involved in chromatin compaction
The pathways of chromatin folding driven by the interactions between nucleosome arrays largely appear to be linked with the neutralization of negatively charged DNA by positively charged tails of core histones. Under certain concentrations of divalent cations, a salt dependent oligomerization (Ausio et al. 1984) and self-association (Schwarz and Hansen 1994) of the nucleosomal arrays have been detected. However, in contrast to divalent cations which are able to induce self-association of nucleosomal structures even at a minimal concentrations, the anions have been found to affect poorly the chromatin fiber oligomerization (Schwarz et al. 1996). Secondary ion mass spectrometry images revealed that both divalent (Mg2+ and Ca2+) and monovalent (Na+ and K+) cations were involved in chromosome condensation in nuclei and isolated mitotic chromosomes through electrostatic neutralization of chromatin components (Strick et al. 2001). Although core histone tails do not function solely as polycations, they mediate self-association of the nucleosome arrays (Garcia-Ramirez et al. 1992; Tse and Hansen 1997) and together with linker histones are engaged in maintaining the solenoidal fiber structure (Allan et al. 1982) in both modified (Jason et al. 2001) and unmodified (Dorigo et al. 2003) forms. By preparing the constructs for nucleosome octamers containing full lenght core histones, Hansen and coworkers (Gordon et al. 2005) revealed that N-terminal tails of all four histones contributed to the salt-dependent oligomerization of the nucleosomal array. Moreover, Fan et al. (2002) detected a core histone-specific oligomerization of chromatin arrays using nucleosomes containing a conserved variant H2A.Z. It appeared that H2A.Z facilitated the intramolecular folding of nucleosomal arrays while simultaneously inhibiting the formation of highly condensed structures that resulted from intermolecular association. This feature of histone H2A.Z may play a fundamental role in creating unique chromatin domains poised for transcriptional activation (Fan et al. 2002).
The neutralization of linker DNA charges is also attributed to linker histones (Caterino and Hayes 2011) which in a cation-related manner could bind the DNA through their positive charges and affect nucleosomal spacing and folding of the chromatin fiber (Blank and Becker 1995). Linker histones also influenced chromatin structure especially through their C-terminal and globular domains. While specific subdomains containing S/TPKK motifs in a highly basic C-terminal region (Allan et al. 1986) were critical for the macromolecular events involved in chromatin condensation (Lu and Hansen 2004), the globular domain was capable of reducing nucleosome mobility when bound to a linker DNA at the entry/exit points by forming condensed nucleosome arrays (Allan et al. 1986). A lability of histone H1 binding to chromatin as revealed by fluorescence recovery after photobleaching technique indicates (Raghuram et al. 2009) that a bulk of histone H1 is composed of highly mobile fractions that associate with a majority of the genome while only a small and relatively stable pool is linked with compact regions of the genome. Such a molecular dynamics and intrinsic disorder of C-terminal domains (Hansen et al. 2006) are indicative of the ability of histone H1 to form a varied structural and functional chromatin transitions. The linker histones were, however, able to induce chromatin self-association with a varied efficiency depending on the H1 subtype (Nagaraja et al. 1995; Clausell et al. 2009). It seems that formation of condensed higher-order chromatin structures requires a proper ion environment to ensure a synergistic activity of the core histone tails and linker histones in chromatin array oligomerization, compaction, and stabilization (Carruthers and Hansen 2000; Arya and Schlick 2009).
The core and linker histones are not the sole modifiers of chromatin structure. Numerous studies have demonstrated that diverse chromatin-associated proteins were engaged in the formation of various structural and functional chromatin states (for reviews see Grigoryev 2001 and Adkins et al. 2004) by creating locally loosened transcriptionally active euchromatin regions or condensed transcriptionally inert heterochromatin. Structurally diverse proteins associated with a heterochromatin spreading can usually operate either individually or as a part of multiprotein complexes (Elgin 1996; Henikoff 1997). For example, proteins Sir3 and Sir4 are critical for telomeric silencing in Saccharomyces cerevisiae (Gartenberg 2000), while Su(var)3-7 is a main suppressor of position-effect-variegation in Drosophila (Cléard et al. 1997). The Su(var)3-7 genomic silencing depends upon interaction with a heterochromatic protein 1 (HP1) (Cléard et al. 1997), a highly conserved multifunctional hetrochromatin-associated protein which is present in diverse adult tissues (Nielsen et al. 2001) but absent in the mature nucleated avian erythrocytes (Gilbert et al. 2003). In terminally differentiated human leukocytes, however, the reduced level of HP1 may be balanced by increased accumulation of monocyte and neutrophil elastase inhibitor (MNEI) (Popova et al. 2006), which like the MENT belongs to the serpin family of the proteinase inhibitors (Irving et al. 2000). No MENT homolog, however, is engaged in chromatin condensation in mammalian erythrocytes (Xu et al. 2006). During mammalian erythroid cell maturation, a gradual chromatin condensation is prompted by upregulation of MTB (more than blood), a member of SMC (structural maintenance of chromosome) condensin complex. MTB, a homolog of murine condensin II subunit CAP-G2, promotes downregulation of E-box containing erythroid genes by binding with hematopoietic transcriptional activators, stem cell leukemia (SCL), and E12 bHLH proteins, with a concomitant increase in the level of chromatin compaction (Xu et al. 2006). As reported by Rawlings et al. (2011), a transition from naive to proliferative T cells is accomplished by a conversion of condensed chromatin into decondensed one required for activation of transcription. A proper chromatin compaction in quiescent T cells is maintained by kleisin β, a component of SMC condensin II complex. Surprisingly, none of developmentally regulated architectural proteins is responsible for chromatin condensation in terminally differentiated mouse erythroblasts, in which the high degree of chromatin compaction is well maintained by post-translational modifications of histone proteins (Popova et al. 2009). In the apocentric zone of erythroblast heterochromatin, a decrease in histone acetylation with a concomitant increase in the expression of histone deacetylase HDAC5 and accumulation of methylated histone H3 was observed (Popova et al. 2009).
A highly condensed chromatin structure in avian terminally differentiated blood cells is associated with the presence of developmentally regulated proteins. While the linker histone H5 is bound to the DNA entry/exit points (Travers 1999) of adjacent nucleosomes to keep DNA strands close together by generating stem-like structures in the compacted avian erythrocyte chromatin, the non-histone protein MENT binds to the nucleosomes beyond the entry/exit site (McGowan et al. 2006) connecting neighboring nucleosome arrays in the condensed granulocyte chromatin to form remarkably dense self-associated nucleosome arrays. Below, the structural and functional properties of these chromatin inactivators are briefly discussed.
Histone H5 structure and functionality in erythrocyte chromatin
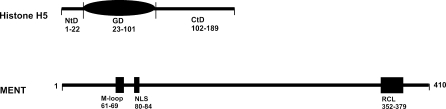
Histone H5, a member of a linker histone family, is a 20.5-kDa protein containing 189 amino acid. This protein is divided into three distinct structural domains (Briand et al. 1980) (Fig. 1). While the N-terminal domain (NtD), residues 1–21, contains most of the hydrophobic residues and all aromatic residues, the strongly basic C-terminal domain (CtD), residues 101–189, possesses on average 50% Lys and Arg residues (Briand et al. 1980). Both H5 terminal tails are separated by a central globular domain (GD), residues 22–100, which has a winged-helix fold consisting of three helix bundles, H1 (residues 29–38), H2 (residues 48–58) and H3 (residues 65–78), and two β-ribbon strands (residues 81–85 and 93–96) (Ramakrishnan et al. 1993).
Fig. 1.
A domain structure for histone H5 and MENT molecules. While the histone H5 is composed of a globular domain (GD, residues 23–101) flanked by an N-terminal domain (NtD, residues 1–22) and C-terminal domain (CtD, residues 102–189), the MENT possesses an M-loop domain (M-loop, residues 61–91), nuclear localization signal domain (NLS, residues 80–84), and reactive center loop domain (RCL, residues 352–379)
It has been revealed (Ramakrishnan et al. 1993; Duggan and Thomas 2000) that two putative DNA-binding sites were arranged as two clusters of positively charged residues located on the opposite sites on the histone H5 globular domain. The first cluster, referred to as “primary binding site,” comprised Lys69, Arg73 and Lys85 and the second one, known as “secondary binding site” contains Lys40, Arg42, Lys52, and Arg94. The site-directed mutagenesis and chromatin binding assays have revealed (Goytisolo et al. 1996) that both DNA-binding sites in the histone H5 globular domain are required for correct binding to the nucleosome. The H5 globular domain can interact with the nucleosome by connecting one terminus of chromatosomal DNA with the site located close to the dyad (Lambert et al. 1991). An asymmetrical mode of histone H5 binding was supported by Zhou and coworkers (1998) who, by using protein-DNA cross-linking experiments, mapped Ser 71 close to Arg 73 to the one terminal end of the chromatosomal DNA in the primary binding site, and Ser 41 between Lys 40 and Arg 42 to a site close to the midpoint in the secondary binding site, so that histone H5 globular domain created a bridge between one arm of the DNA and the nucleosome dyad resulting in a simultaneous connection of two gyres of the DNA. A new model of histone H5 interaction with nucleosome revealed, however, three potential sites through which the H5 globular domain contacted the DNA (Fan and Roberts 2006). The primary protein–DNA interactions occur at the site II through the insertion of Lys 85 into the major groove of DNA accompanied by additional contacts at the site I (the interaction of the Arg 47, Lys 69, and Arg 74 with the DNA major groove), and at the site III with the DNA backbone located between Lys 40 and Lys 97. The implication is that the H5 globular domain may interact with both the DNA dyad and with the DNA arms which, as authors suggested, agree with a symmetrical mode of the binding of histone H5 to the nucleosome (Simpson 1978). According to the all–atom model of chromatin fiber with linker histones (Wong et al. 2007), the globular domain of histone H5 can be positioned within a chromatosome depending on the nucleosome repeat length and could be placed either on the dyad axis in between two DNA linkers in the nucleosomes with short NRLs or located in a pocket made by longer linkers at the entry/exit point to bridge both DNA linkers and the nucleosomal DNA at the dyad in the nucleosome arrays containing longer NRLs. For chromatosomes with a relatively short NRL (177 bp), the DNA joining consecutive nucleosomes is very short (10 bp in this case) and seems to be lacked binding sites for H5 tails, whereas the chromatosomes with longer NRLs can make contacts with H5 tails, the number of which increase by 1 with every increment of 10 bp above the 177 bp NRL. In the latter cases, the linker histones induce DNA kinks with a consequent change in the direction of DNA linker at the exit site.
Despite a controversial view of how the structured histone H5 globular domain binds to the nucleosome, its role in DNA stabilization in the nucleosome core has been established in different experiments (Buckle et al. 1992 and references therein). Moreover, the histone H5 C-terminal domain is equally important in the determination of folded states involved in chromatin condensation (Vila et al. 2000; Lu and Hansen 2004). This domain, unstructured in aqueous solutions (Allan et al. 1986), may adopt α-helical conformation when bound to the DNA (Clark et al. 1988). The α-helix formed by intra-helix hydrogen bonding and neutralization of positive charges of the lysine and arginine residues was divided into the segments demarcated by proline residues which resulted in the bending of the α-helical region. Thus, the α-helical segment in the C-terminal domain may twist around the DNA phosphate backbone exerting a stabilizing effect on the linker DNA in the 30-nm chromatin fiber. In addition, when chromatin fiber is folded into a higher order structure, the histone H5 C-terminal domains remain close enough between adjacent nucleosomes to be linked with a cross-linking agent (Lennard and Thomas 1985).
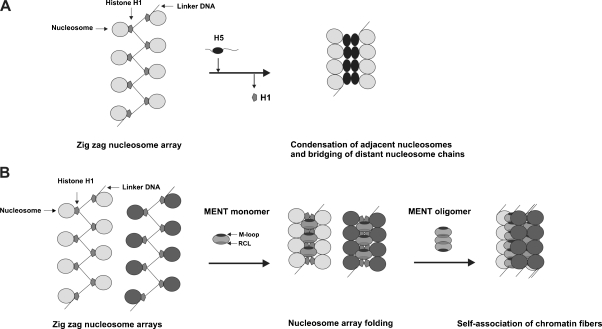
Histone H5 replacing histone H1 in mature erythrocytes constituted about 60% of the total amount of linker histones (Koutzamani et al. 2002). Exchange of H1 by H5 prevented the mobility of nucleosomes (Pennings et al. 1994) resulting in a greater stability of chromatin (Bates and Thomas 1981) so that the histone H5 is regarded to be more potent factor in producing extensively repressed regions in the erythroid genome. A microinjection of H5 into proliferating myoblasts generated a densely compacted chromatin in the injected cells compatible with the inhibition of transcription and replication (Bergman et al. 1988). A similar repressive effect was observed after expression of histone H5 in rat sarcoma cells transfected with MMTV-H5 constructs (Sun et al. 1990). A higher binding affinity of histone H5 compared to H1 subtypes (Orrego et al. 2007) could explain a greater ability of the H5 to produce more compacted chromatin fibers in transfected rat sarcoma cells (Sun et al. 1990). Such effects may result from a better neutralization of DNA charges by the H5 due to its higher content of Arg residues. The efficiently bound nucleosomes tended to form axially stable structures similar to those in mature erythrocyte chromatin (Sun et al. 1990). Unlike the rest of linker histones, the histone H5 has a tendency to form dimers in solution by specific self-contacts within the globular domains (Carter and van Holde 1998). Using a computational docking tool for evaluation of the interaction between histone H5 globular domains, Fan and Roberts (2006) corroborated that the nucleosome dimerization was triggered by the interaction of aromatic residues between the H5 monomers (Ramakrishnan et al. 1993) which induced greater chromatin compaction by bringing the adjacent nucleosomes closer together to form zig zag structures (Rydberg et al. 1998; Bednar et al. 1998) and by connecting distant nucleosome segments to create a uniformly thick nucleosomal array (Fan and Roberts 2006) (Fig. 2a). These findings are consistent with the linker histone-induced stem formation required for chromatin folding (Bednar et al. 1998) as well as with a continuous fiber model obtained by successively stacking of tetranucleosomes one on another (Schalch et al. 2005). Moreover, the avian erythrocyte histone H5 is gradually phosphorylated after synthesis and then dephosphorylated to become fully devoid of phosphate at the terminal stage of development (Sung et al. 1977). Dephosphorylation is correlated with a strong increase in the affinity of histone H5 for DNA resulting in a sharp shift of chromatin to fully condensed state (Aubert et al. 1991).
Fig. 2.
Models for the formation of compacted chromatin fibers. a A formation of dense chromatin structure by histone H5-mediated association of adjacent nucleosomes and bridging of distant nucleosome chains. A side-by-side self-association of compacted neighboring poly-nucleosome arrays may facilitate further chromatin condensation. b Two-step formation of compacted chromatin fibers by initial binding of MENT-monomers to DNA and folding the nucleosome arrays and subsequent self-association of the chromatin fibers by MENT-oligomers
A developmentally regulated histone H5 is a main factor mediating the stability of erythrocyte chromatin. The neutralization of DNA negative charges by a deposition of positively charged histone H5 molecules (Blank and Becker 1995) resulted in a formation of condensed chromatin structure. Histone H5 can produce a folded chromatin independently of other chromatin-modifying proteins (Sun et al. 1990) without significant alterations in the arrangement of the nucleosomal arrays (Bednar et al. 1998). The histone H5-induced repressive effects mainly depend on the interactions between H5 globular domains themselves (Thomas et al. 1992) placed at axial positions (Thoma et al. 1979) because spatial juxtaposition of globular histone H5 domains promotes their self-association and tends to stabilize chromatin fiber (Maman et al. 1994). By forming stable stem-like structures (Hamiche et al. 1996), the histone H5 may facilitate partial interdigitating of the nucleosome arrays to create structurally heteromorphic regions along chromatin fibers (Horowitz et al. 1994; Grigoryev et al. 2009).
MENT structure and functionality in granulocyte chromatin
MENT, a basic 42-kDa protein containing 410 amino acids, is classified as a member of serpin family due to the presence of serpin-related conserved reactive center loop (RCL) domain (residues 352–379) engaged in the formation of disordered conformations in the proteases (Huntington et al. 2000; Irving et al. 2000). But unlike the other serpins, the MENT possesses additional M-loop domain (residues 61–91) which is able to interact with DNA and a nuclear localization signal (NLS) domain (residues 80–84) responsible for the nuclear targeting (Grigoryev and Woodcock 1998; Grigoryev et al. 1999). These protein domains are made up of three β-sheets (A–C) surrounded by nine α-helices (hA–hI) (McGowan et al. 2006) (Table 1) forming a charged patch centered on D- and E-helices with a potential for DNA-binding. While the RCL extension is connecting two strands of the four stranded C β-sheet around which the neutral and acidic amino acids are arranged, the M-loop harboring AT hook motif exposed on the surface of the MENT as well as D- and E-helices containing positively charged amino acids contribute to the formation of DNA binding sites (McGowan et al. 2006). One DNA binding site is encompassing a positively charged segment around the D- and E-helices, while the second one corresponds to a short sequence related to the AT-hook motif (McGowan et al. 2006) which has been recognized as a typical motif in nuclear proteins known to interact with the DNA (Aravind and Landsman 1998).
Springhetti et al. (2003) reported the effects of M-loop and RCL segments on the initiation and maintaining of compacted chromatin at terminal stages of granulocyte differentiation. The electrophoretic mobility shift assays and electron microscopy image analysis of a series of MENT mutants with fully swapped RCL domain by the inactive one from ovalbumin or the mutants with deleted M-loop residues as well as with a single point mutation (T → R) in the RCL hinge region have shown that all these mutations influenced the interaction of MENT with the chromatin. Because the mutated forms differently interacted with both naked DNA and reconstituted nucleosome arrays, the activity of both M-loop and RCL domain in wild-type MENT was necessary for proper chromatin condensation. Interestingly, these two domains fulfilled distinct functions in the course of MENT-induced chromatin transition to more compacted states. The wild-type MENT and inactivated RCL mutants formed juxtaposed parallel structures of folded DNA, while deletion of M-loop prevented their formation. Moreover, the M-loop inactivating mutants prevented a cooperative interaction of the MENT with DNA. The folding and self-oligomerization were apparent after the reconstitution of nucleosome arrays with the wild-type MENT but not with the M-loop mutants. The loss-of-function RCL mutant exhibited an impaired capacity for self-association of folded nucleosome arrays. Therefore, the MENT-dependent chromatin condensation requires both a cooperative binding and folding of DNA associated with the M-loop domain and the RCL-dependent MENT oligomerization which in cooperation with linker histones is needed to form self-associated chromatin fibers (Springhetti et al. 2003). A bipartite mechanism leading to repression of granulocyte chromatin is initiated by the binding of the MENT to DNA linker entry–exit segments bringing them to close apposition in a linker region (Grigoryev 2001) followed by the RCL β-strand interactions between adjoining molecules (McGowan et al. 2006) facilitating the protein oligomerization. The MENT oligomers are engaged in bridging laterally self-associated chromatin fibers and forming strongly compacted chromatin states (Fig. 2b). As mentioned above, the MENT-promoted chromatin condensation might be locally enhanced by other proteins, especially linker and core histones. Since MENT reconstitution with chicken oligonucleosomes containing linker histones resulted in a higher rate of chromatin self-association compared to a weaker chromatin bridging when linker histones were absent (Springhetti et al. 2003), it seems that the heterochromatin spreading depends on the changing levels of the MENT (Grigoryev and Woodcock 1998) or the linker histone composition (Koutzamani et al. 2002) in distinct chromatin regions. In addition, a cooperation between dimethylated Lys9 in the N-terminal domain of histone H3 and the MENT RCL domain has been found to result in promoting facultative chromatin condensation (Istomina et al. 2003).
It seems that exact function of the MENT in a compact peripheral heterochromatin in avian blood cells (Grigoryev and Woodcock 1998), where it selectively interacts with silent genetic loci (Grigoryev 2001), probably does not rely on a direct formation of condensed chromatin via binding to a specific DNA sequence but rather on the maintenance of already inactivated portion of the genome by holding together self-associated chromatin fibers which give rise to large-scale repressed area of chromatin.
Conclusions
Linker histone H5 highly expressed in avian erythrocytes and non-histone chromatin protein MENT abundant in avian granulocytes represent chromatin-condensing proteins. The remodeling factor MENT acts by gluing together nucleosome arrays and promoting self-association of chromatin fibers. The establishment and development of compacted chromatin regions seem to follow a similar course driven by structurally unrelated domains of histone H5 and MENT. While compaction of erythrocyte chromatin may be mainly attributed to the histone H5 globular domain which due to a tendency toward dimerization can induce the aggregation of nucleosomes and maintain self-associatied chromatin fibers, the protein MENT is engaged in the folding of nucleosome arrays through its M-loop domain and subsequent RCL domain-dependent protein oligomerization to hold self-associated chromatin fibers via protein bridges. The condensation and subsequent repression of chromatin may arise from the electrostatic attractions between negative charges of DNA and positive charges of the protein domains which in common with counter-ions could enable compaction and self-association of nucleosome arrays. Therefore, the high level of compaction may require either positively charged histone H5 globular domain or a positively charged amino acid cluster around the MENT M-loop domain which is engaged in neutralization of DNA charges. In addition, a negatively charged surface around the MENT R-loop domain is needed for the interactions with other positively charged proteins. The main difference in the effectiveness of H5 and MENT seems to concern the extend of chromatin compactness through the neutralization of the DNA charge. A presence of greater number of positively charged residues in granulocyte MENT compared with erythrocyte histone H5 might induce a stronger compaction and repressive effect in granulocytes. However, regardless of the level of the inert chromatin generated in different blood cells, it seems that chromatin inactivation may be driven by dissimilar structural domains in distinct proteins which can adopt similar mechanisms which allow performing a related function.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CtD
C-terminal domain
- GD
Globular domain
- HDAC5
Histone deacetylase 5
- HP1
Heterochromatic protein 1
- MNEI
Monocyte and neutrophil elastase inhibitor
- MENT
Myeloid and erythroid nuclear termination stage-specific protein
- MTB
More than blood
- NLS
Nuclear localization signal
- NRL
Nucleosome repeat length
- NtD
N-terminal domain
- RCL
Reactive center loop
- SLC
Stem leukemia cell
- SMC
Structural maintenance of chromosome
References
- Adkins NL, Watts M, Georgel PT. To the 30-nm chromatin fiber and beyond. Biochim Biophys Acta. 2004;1677:12–23. doi: 10.1016/j.bbaexp.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Allan J, Harborne N, Rau DC, Gould H. Participation of core histone “tails” in the stabilization of the chromatin solenoid. J Cell Biol. 1982;93:285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya G, Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J Phys Chem A. 2009;16:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D, Garcia M, Benchaibi M, Poncet D, Chebloune Y, Verdier G, Nigon V, Samarut J, Mura CV. Inhibition of proliferation of primary avian fibroblasts through expression of histone H5 depends on the degree of phosphorylation of the protein. J Cell Biol. 1991;113:479–506. doi: 10.1083/jcb.113.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J, Seger D, Eisenberg H. Nucleosome core particle stability and conformational change. Effect of temperature, particle and NaCl concentrations, and crosslinking of histone H3 sulfhydryl groups. J Mol Biol. 1984;176:77–104. doi: 10.1016/0022-2836(84)90383-8. [DOI] [PubMed] [Google Scholar]
- Bates DL, Thomas JO. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman MG, Wawra E, Winge M. Chicken histone H5 inhibits transcription and replication when introduced into proliferating cells by microinjection. J Cell Sci. 1988;91:201–209. doi: 10.1242/jcs.91.2.201. [DOI] [PubMed] [Google Scholar]
- Blank TA, Becker PB. Electrostatic mechanism of nucleosome spacing. J Mol Biol. 1995;252:305–313. doi: 10.1006/jmbi.1995.0498. [DOI] [PubMed] [Google Scholar]
- Bordas J, Perez-Grau L, Koch MHJ, Vega MC, Nave C. The superstructure of chromatin and its condensation mechanism. I. Synchrotron radiation X-ray scattering results. Eur Biophys J. 1986;13:157–173. doi: 10.1007/BF00542560. [DOI] [PubMed] [Google Scholar]
- Briand G, Kmiecik D, Sautiere P, Wouters D, Borie-Loy O, Biserte G, Mazen A, Champagne M. Chicken erythrocyte histone H5. IV. Sequence of the carboxy-termined half of the molecule (96 residues) and complete sequence. FEBS Lett. 1980;112:147–151. doi: 10.1016/0014-5793(80)80167-0. [DOI] [PubMed] [Google Scholar]
- Buckle RS, Maman JD, Allan J. Site-directed mutagenesis studies on the binding of the globular domain of linker histone H5 to the nucleosome. J Mol Biol. 1992;223:651–659. doi: 10.1016/0022-2836(92)90981-o. [DOI] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim J-H. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Hansen JC. The core histone N termini function independently of linker histones during chromatin condensation. J Biol Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- Carter GJ, van Holde K. Self-association of linker histone H5 and of its globular domain: evidence for specific self-contacts. Biochemistry. 1998;37:12477–12488. doi: 10.1021/bi980716v. [DOI] [PubMed] [Google Scholar]
- Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DJ, Hill CS, Martin SR, Thomas JO. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988;7:69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS ONE. 2009;4:e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléard F, Delattre M, Spierer P. SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DFV, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:2011–2021. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- Duggan MM, Thomas JO. Two DNA-binding sites on the globular domain of histone H5 are required for binding to both bulk and 5S reconstituted nucleosomes. J Mol Biol. 2000;304:21–33. doi: 10.1006/jmbi.2000.4205. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Elgin SCR. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existance of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Roberts VA. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc Natl Acad Sci USA. 2006;103:8384–8389. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussner E, Ching RW, Bazett-Jones DP. Living without 30 nm chromatin fibers. Trends Biochem Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M, Dong F, Ausio J. Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- Gartenberg MR. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr Opin Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- Gerchman SE, Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci USA. 1985;84:7082–7086. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. Hydrodynamic studies of defined heterochromatin fragments support a 30 nm fiber having 6 nucleosomes per turn. J Mol Biol. 2008;376:1417–14275. doi: 10.1016/j.jmb.2007.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Sutherland H, De Las Heras J, Allan J, Jenuwein T, Bickmore WA. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem. 2005;280:3371–3376. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- Goytisolo FA, Gerchman SE, Yu X, Rees C, Graziano V, Ramakrishnan V, Thomas JO. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- Grigoryev SA. Higher-order folding of heterochromatin: protein bridges span the nucleosome arrays. Biochem Cell Biol. 2001;79:227–241. [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL. Chromatin structure in granulocytes. A link between tight compaction and accumulation of a heterochromatin-associated protein (MENT) J Biol Chem. 1998;273:3082–3089. doi: 10.1074/jbc.273.5.3082. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Bednar J, Woodcock CL. MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J Biol Chem. 1999;274:5626–5636. doi: 10.1074/jbc.274.9.5626. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Bulynko YA, Popova EY. The end adjusts the means: heterochromatin remodelling during terminal cell differentiation. Chromosome Res. 2006;14:53–69. doi: 10.1007/s10577-005-1021-6. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Xu L, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington JA, Read RJ, Carrell RW. Structure of a serpin–protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–1864. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- Istomina NE, Shushanov SS, Springhetti EM, Karpov VL, Krasheninnikov IA, Stevens K, Zaret KS, Sinhg PB, Grigoryev SA. Insulation of the chicken β-globin chromosomal domain from a chromatin-condensing protein. MENT Mol Cell Biol. 2003;23:6455–6468. doi: 10.1128/MCB.23.18.6455-6468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason LJ, Moore SC, Ausio J, Lindsey G. Magnesium dependent association and folding of oilgonucleosomes reconstituted with ubiquinated H2A. J Biol Chem. 2001;276:14597–14601. doi: 10.1074/jbc.M011153200. [DOI] [PubMed] [Google Scholar]
- Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol Cell Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutzamani E, Laborg H, Sarg B, Lindner HH, Rundquist I. Linker histone subtype composition and affinity for chromatin in situ in nucleated mature erythrocytes. J Biol Chem. 2002;277:44688–44694. doi: 10.1074/jbc.M203533200. [DOI] [PubMed] [Google Scholar]
- Kruithof M, Chien F-T, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- Lambert S, Muyldermans S, Baldwin J, Kilner J, Ibel K, Wijns L. Neutron scattering studies of chromatosomes. Biochem Biophys Res Commun. 1991;179:810–816. doi: 10.1016/0006-291x(91)91889-k. [DOI] [PubMed] [Google Scholar]
- Lennard AC, Thomas JO. The arrangement of H5 molecules in extended and condensed chicken erythrocyte chromatin. EMBO J. 1985;4:3455–3462. doi: 10.1002/j.1460-2075.1985.tb04104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Maman JD, Yager TD, Allan J. Self-association of the globular domain of histone H5. Biochemistry. 1994;33:1300–1310. doi: 10.1021/bi00172a003. [DOI] [PubMed] [Google Scholar]
- McGowan S, Buckle AM, Irving JA, Ong PC, Bashtannyk-Puhalovich TA, et al. X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation. EMBO J. 2006;25:3144–3155. doi: 10.1038/sj.emboj.7601201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja S, Delcuve GP, Davie JR. Differential compaction of transcriptionally competent and repressed chromatin reconstituted with histone H1 subtypes. Biochem Biophys Acta. 1995;1260:207–214. doi: 10.1016/0167-4781(94)00201-d. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- Orrego M, Ponte I, Roque A, Buschati N, Mora X, Suau P. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 2007;5:22. doi: 10.1186/1741-7007-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings S, Meerseman G, Bradbury EM. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci USA. 1994;91:10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Claxton DF, Lukasova E, Bird PI, Grigoryev SA. Epigenetic heterochromatin markers distinguish terminally differentiated leukocytes from incompletely differentiated leukemia cells in human blood. Exp Hematol. 2006;34:453–462. doi: 10.1016/j.exphem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Popova EY, Krauss SW, Short SA, Lee G, Villalobos J, Etzell J, Koury MJ, Ney PA, Chasis JA, Grigoryev SA. Chromatin condensation in terminally differentiating mouse erythroblasts does not involve special architectural proteins but depends on histone deacetylation. Chromosome Res. 2009;17:47–64. doi: 10.1007/s10577-008-9005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram N, Carrero G, Th’ng J, Hendzel MJ. Molecular dynamics of histone H1. Biochem Cell Biol. 2009;87:189–206. doi: 10.1139/O08-127. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Rawlings JS, Gatzka M, Thomas PG, Ihle JN. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 2011;30:263–276. doi: 10.1038/emboj.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJJ, Fairall H, Huynh VAT, Rhodes D. EM measurements define the dimensions of the “30-nm” fiber: evidence for a compact interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Holley W, Mian I, Chatterjee A. Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J Mol Biol. 1998;284:71–84. doi: 10.1006/jmbi.1998.2150. [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- Schwarz PM, Hansen JC. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J Biol Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Springhetti EM, Istomina NE, Whisstock JC, Nikitina T, Woodcock CL, Grigoryev SA. Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J Biol Chem. 2003;278:43384–43393. doi: 10.1074/jbc.M307635200. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis C. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strick R, Strissel PL, Gavrilov K, Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JM, Ali Z, Lurz R, Ruiz-Carrillo A. Replacement of histone H1 by H5 in vivo does not change the nucleosome repeat length of chromatin but increases its stability. EMBO J. 1990;9:1651–1658. doi: 10.1002/j.1460-2075.1990.tb08285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang Q, Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc Natl Acad Sci USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MT, Wagner TE, Hartford JB, Serra M, Vandegrift V. Phosphorylation and dephosphorylation of histone V (H5): controlled condensation of avian erythrocyte chromatin. Biochemistry. 1977;16:286–290. doi: 10.1021/bi00621a020. [DOI] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Rees C, Finch JT. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 1992;20:187–194. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. The location of the linker histone on the nucleosome. Trends Biochem Sci. 1999;24:4–7. doi: 10.1016/s0968-0004(98)01339-5. [DOI] [PubMed] [Google Scholar]
- Tse C, Hansen JC. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- Vila R, Ponte I, Jimenez MA, Rico M, Suau P. A helix-turn motif in the C-terminal domain of histone H1. Protein Sci. 2000;9:627–636. doi: 10.1110/ps.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler C, Huber C, Waldmann T, Ettig R, Braun L, Izzo A, Daujat S, Chassignet I, Lopez-Contreras AJ, Fernandez-Capetillo O, Dundr M, Rippe K, Langst G, Schneider R. Histone H2A C-terminus regulates chromatin dynamics, remodeling and histone H1 binding. PLoS Genet. 2010;6:1–12. doi: 10.1371/journal.pgen.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Victor J-M, Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosome repeat length. PLoS ONE. 2007;9:1–8. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S. Higher order structure of chromatin and chromosomes. Curr Opin Genet Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Horowitz RA. Chromatin organization re-viewed. Trends Cell Biol. 1995;5:272–277. doi: 10.1016/s0962-8924(00)89038-8. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N. A chromatin folding model that incorporates linker variability generated fibers resembling the native structures. Proc Natl Acad Sci USA. 1993;90:9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Leung CG, Lee DC, Kennedy BK, Crispino JD. MTB, the murine homolog of condensin II subunit CAP-G2, represses transcription and promotes erythroid cell differentiation. Leukemia. 2006;20:1261–1269. doi: 10.1038/sj.leu.2404252. [DOI] [PubMed] [Google Scholar]
- Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Leuba SH, Yang G, Bustamante C, van Holde K. Linker DNA accessibility in chromatin fibers of different conformations: a reevaluation. Proc Natl Acad Sci USA. 1994;91:5277–5280. doi: 10.1073/pnas.91.12.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]