Abstract
Protein arginine methylation has been implicated in signal transduction, nuclear transport and transcription regulation. Protein arginine methyltransferases (PRMTs) mediate the AdoMet-dependent methylation of many proteins, including many RNA binding proteins involved in various aspects of RNA processing and/or transport. Here we describe the crystal structure of the rat PRMT3 catalytic core in complex with reaction product AdoHcy, determined at 2.0 Å resolution. The results reveal a two-domain structure: an AdoMet-binding domain and a barrel-like domain. The AdoMet-binding domain is a compact version of the consensus AdoMet-dependent methyltransferase fold. The active site is situated in a cone-shaped pocket between the two domains. The residues that make up the active site are conserved across the PRMT family, consisting of a double-E loop containing two invariant Glu and one His–Asp proton-relay system. The structure suggests a mechanism for the methylation reaction and provides the structural basis for functional characterization of the PRMT family. In addition, crystal packing and solution behavior suggest dimer formation of the PRMT3 core.
Keywords: AdoMet-dependent methyltransferase/PRMT dimer/PRMT3/protein arginine methylation/xenon derivatization
Introduction
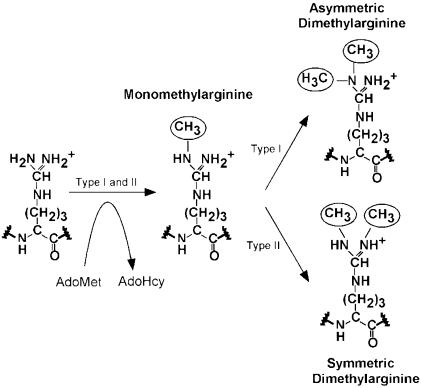
Protein arginine methylation is an abundant post-translational modification. There are two major types of protein arginine methyltransferases (PRMTs) that transfer the methyl group from S-adenosyl-l-methionine (AdoMet) to the guanidino group of arginines in protein substrates, resulting in AdoHcy and methylated proteins (Lee et al., 1977). Both types catalyze the formation of monomethylarginine but differ in the dimethylarginine products: type I PRMTs form asymmetric dimethylarginine; and type II form symmetric dimethylarginine (Figure 1). While type II PRMTs have only one known substrate (myelin basic protein), type I PRMTs have numerous substrates, most of which are RNA binding proteins involved in various aspects of RNA processing and/or transport (Gary and Clarke, 1998). Many of the substrates contain a glycine and arginine rich (GAR) sequence that includes multiple target arginines in RGG or RXR contexts (Gary and Clarke, 1998; Smith et al., 1999; Klein et al., 2000).
Fig. 1. Two major types of protein arginine methylation.
The PRMT genes were cloned almost 30 years after the discovery of protein arginine methylation (Paik and Kim, 1967, 1968). Four mammalian PRMT genes that share extensive homology have been cloned so far: rat and human PRMT1; human PRMT2; rat and human PRMT3; and mouse CARM1 (Lin et al., 1996; Abramovich et al., 1997; Katsanis et al., 1997; Scott et al., 1998; Tang et al., 1998; Chen et al., 1999). The only PRMT homolog from Saccharomyces cerevisiae, termed Rmt1, was also cloned (Gary et al., 1996; Henry and Silver, 1996). This family of PRMTs presumably represents type I enzymes, as type I activity has clearly been demonstrated for PRMT1, PRMT3 and Rmt1. More recently, a protein more distantly related to other PRMTs, Jak kinase binding protein (JBP1), was also found to have protein methyltransferase activity (Pollack et al., 1999).
Despite our knowledge about PRMT genes and substrates, determination of the precise roles of protein arginine methylation is an emerging field. In S.cerevisiae, Rmt1 has been shown to facilitate nuclear export of certain hnRNP proteins and perhaps enhance the efficiency of mRNA export (Shen et al., 1998; Valentini et al., 1999). In mammalian systems, PRMTs have been implicated in receptor-mediated signaling as well as transcription regulation. PRMT1 was cloned independently based on its association with an interferon receptor (Abramovich et al., 1997) and with two antiproliferative immediate-early proteins (Lin et al., 1996). JBP1 was identified through its interaction with the Jak kinase, a family of proteins involved in cytokine signal transduction (Pollack et al., 1999). CARM1, which methylates histone H3 in vitro, was shown to be involved in transcription activation by nuclear hormone receptors (Chen et al., 1999). PRMTs obviously play diverse roles and their significance is just beginning to be appreciated.
Here we present the first structure of a conserved PRMT core. It allows us to propose a mechanism for the methylation reaction and provides the structural basis for functional characterization of the PRMT family.
Results and discussion
A conserved PRMT core
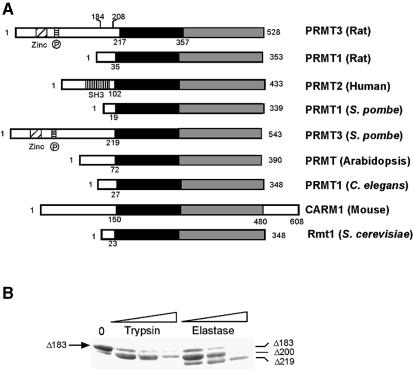
In addition to the S.cerevisiae and four mammalian PRMT genes that have been cloned, complete PRMT genes from Schizosaccharomyces pombe, Arabidopsis and Caenorhabditis elegans have also been identified through genome sequencing projects (Figure 2A). In addition, expressed sequence tags with strong similarity to PRMT1 can be found in Drosophila, Xenopus, zebrafish, sea urchin, rice and tomato, indicating that PRMT is a highly conserved family of proteins in eukaryotes. The PRMT proteins vary in length from 348 (S.cerevisiae Rmt1) to 608 (CARM1) amino acids, but they all contain a conserved ‘core’ region of ∼310 amino acids (Figure 2A). We refer to this conserved region of PRMTs as the ‘PRMT core’.
Fig. 2. Conserved PRMT core region. (A) Schematic representation of nine PRMT homologs. The conserved PRMT core region is shown in black (highly conserved) and in gray (less conserved). The residue number corresponding to the first invariant residue, Y217 in PRMT3, is indicated for each molecule. The first residue of PRMT3 used for crystallization (amino acid 184) and identified by X-ray diffraction (amino acid 208) is also indicated. The human PRMT1 and PRMT3 are omitted because they are >90% identical to the rat proteins. (B) Limited protease assay of PRMT3Δ183. Purified Δ183 (2 µg) was treated with 0.25 µg of trypsin or elastase for 5, 10 or 30 min and separated on a 13% SDS gel. The two fragments generated by elastase cleavage were subjected to N-terminal sequencing.
The sequences beyond the conserved PRMT core region are all N-terminal additions, except that CARM1 also has a C-terminal addition. The size of the N-terminal addition varies from ∼20 amino acids in S.cerevisiae Rmt1 to 200 in PRMT3. Human PRMT1 is known to have three alternatively spliced transcripts that would produce proteins with an N-terminus of 20–40 amino acids (Scott et al., 1998).
Enzymatic activity of the PRMT3 core
The predominant PRMT in mammalian cells, PRMT1, is a mostly nuclear enzyme and exists as an oligomeric complex in rat cells (Tang et al., 1998, 2000). In contrast, PRMT3 is predominantly cytoplasmic and exists as a monomer based on gel filtration analysis (Tang et al., 1998). A fragment of rat PRMT3 containing amino acids 184–528 (termed Δ183) was used for structural determination of the PRMT core. This fragment is similar in size to the full-length PRMT1 and S.cerevisiae Rmt1 (Figure 2A). During crystallization trials, partial proteolysis of the protein was found to correlate with production of crystals of better quality. Limited digestion of the Δ183 by elastase resulted in a stable fragment starting at Ser201 (Figure 2B). A truncated version, PRMT3Δ200, was engineered and yielded better crystals.
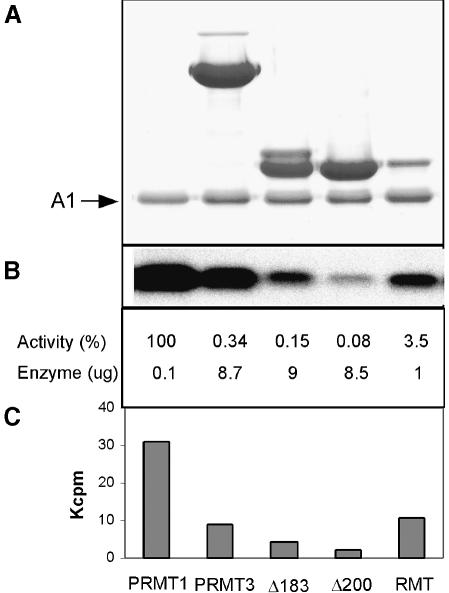
The enzymatic activity of PRMT3Δ183 and PRMT3Δ200 was compared with that of the full-length PRMT3, rat PRMT1 and S.cerevisiae Rmt1, using purified recombinant hnRNPA1 as substrate (Rajpurohit et al., 1994). Figure 3 shows the relative specific activities of various PRMT preparations. The recombinant rat PRMT1 is the most active enzyme (Figure 3, lane 1). The full-length rat PRMT3 has ∼0.3% activity compared with PRMT1 (Figure 3, lane 2). The two deletions containing only the core region, PRMT3Δ183 and PRMT3Δ200, are both active despite a 2- and 4-fold decrease of activity relative to the intact PRMT3 (Figure 3, lanes 3 and 4). This is comparable to the published results using the GAR domain of fibrillarin fused to glutathione S-transferase (GST) as substrate (Tang et al., 1998). As a comparison, S.cerevisiae Rmt1 is also active, with a specific activity of ∼3% of rat PRMT1 (Figure 3, lane 5).
Fig. 3. Relative enzymatic activity of purified PRMTs. The enzymes used were all purified recombinant proteins. The PRMT1 contained a His6 tag, and the PRMT3 and S.cerevisiae Rmt1 were cleaved from GST fusions by thrombin. The reaction mix (20 µl) contained 3 µM hnRNP A1, 40 µM [methyl-3H]AdoMet (0.5 µCi) and various amounts of PRMTs, and was incubated at 37°C for 1 h. (A) Coomassie staining of the reaction mix separated by a 12% SDS gel showing the hnRNP A1 and various amounts of enzymes. (B) Fluorograph showing the amount of [methyl-3H]AdoMet transferred to hnRNP A1. (C) TCA-precipitable counts. The amount of enzyme used in each reaction is indicated, and the relative activity was calculated using the TCA-precipitable counts above the no-enzyme background and the amount of enzyme used.
Structure determination of the PRMT3 core
Crystals of the PRMT3 core were grown in the presence of cofactor AdoHcy. Electron density maps were calculated in P43212 space group using the three-wavelength selenium and the two-wavelength xenon data (Table I). A model consisting of residues 208–528 of PRMT3 was built, and was refined to 2.0 Å resolution with a crystallographic R factor of 0.216 and Rfree value of 0.274. The N-terminal residues (208–214) and the first helix (215–220) are more flexible, as indicated by the discontinuous electron density along the main chain of residues 208–214 and relatively higher crystallographic thermal factors for both regions (∼80 and ∼40 Å2, respectively, compared with an average of ∼20 Å2 for the rest of the molecule). As a result, residues 208–214 were modeled only as alanine. This flexibility might be due to the missing N-terminal segment (amino acids 1–183) or the absence of a substrate. In addition, a 3.3 Å resolution structure of PRMTΔ200 was solved in the space group P21 by molecular replacement (see Materials and methods).
Table I. Summary of X-ray data collection, phasing statistics and model refinement.
| Derivative | Se | Xe | Native | |||
|---|---|---|---|---|---|---|
| Protein | PRMT3Δ183 | PRMT3Δ200 | PRMT3Δ200 | |||
| Cell dimensions (Å) | a = b = 70.9, c = 171.2 | a = b = 70.9, c = 173.4 | a = b = 71.9, c = 175.7 | a = b = 70.91, c = 177.66 | ||
| Wavelength (Å) | 0.983614 | 0.983866 | 0.93 | 1.54 | 1.10 | 1.10 |
| Resolution limit (Å) | 2.85 | 2.85 | 2.85 | 2.8 | 2.3 | 2.03 |
| Completeness (%)a | 96/93 | 96/94 | 95/93 | 99/98 | 96/84 | 90.9/56.9 |
| Rlinear (%)a | 7.3/23.2 | 6.4/22.4 | 6.5/24.3 | 5.0/23.0 | 4.8/23.8 | 5.7/21.0 |
| <I/σ(I)>a | 13/4 | 17/5 | 16/4 | 24/6 | 24/4 | 17/4 |
| Observed reflections | 67 108 | 67 529 | 56 397 | 73 795 | 166 196 | 135 987 |
| Unique reflectionsa | 10 531/469 | 10 536/473 | 10 452/477 | 11 427/1082 | 20 681/1763 | 27 345/1659 |
| Anomalous sites | 5 | 5 | 5 | 1 | 1 | – |
| Phasing power | 1.20 | 1.16 | 0.96 | 2.37 | 2.48 | – |
| R kraut | 0.058 | 0.043 | 0.017 | 0.875 | 1.346 | – |
| Figure of merit | 0.285 | 0.287 | 0.260 | 0.315 | 0.248 | – |
| Overall figure of merit | – | – | 0.543 | – | – | – |
| R valuea | – | – | – | – | – | 0.216/0.281 |
| Rfreea | – | – | – | – | – | 0.274/0.351 |
aThe numbers are given for the whole data set/the highest resolution bin.
Overall structure
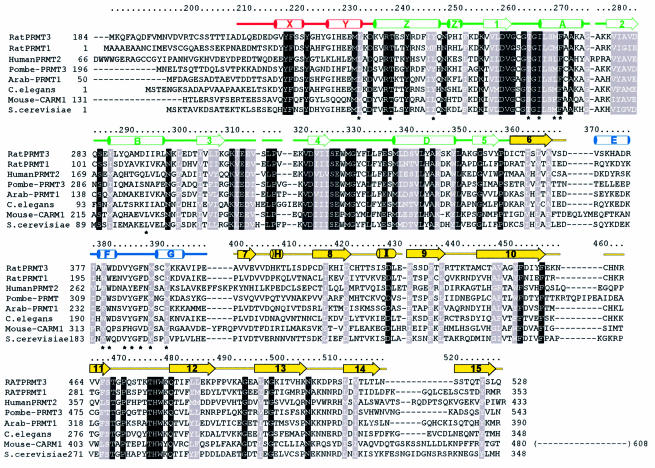
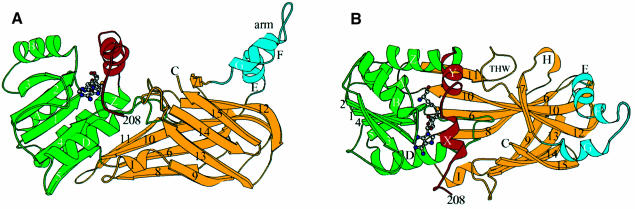
Figure 4 shows the location of structural elements with respect to the amino acid sequence of PRMT3, with the aligned sequences of seven other PRMT cores. Most of the highly conserved residues are concentrated in the N-terminal half of the molecule, between PRMT3 residues 230 and 367. It includes the signature sequences for most classes of MTases, including the GxGxG (residues 260–264) involved in AdoMet binding (Kagan and Clark, 1994; Malone et al., 1995). The C-terminal half of the core is much less conserved except for a short stretch of sequence between residues 466 and 479 of PRMT3. Figure 5 presents two views of the determined PRMT3 core structure. Both Figures 4 and 5 are color coded as follows: red (residues 208–233); green (residues 234–358); blue (residues 370–398); and yellow (residues 359–369 and 399–528). We refer to Figure 5A as the front view and Figure 5B as the top view, where the missing N-terminal segment (residues 1–207) would enter the core structure from the bottom in Figure 5B.
Fig. 4. Sequence alignment of the conserved PRMT core region against the structural elements of PRMT3. The PRMT homologs included are rat PRMT3 (accession No. 3088573), rat PRMT1 (1390025), human PRMT2 (2499805), S.pombe PRMT3 (CAA17825), Arabidopsis thaliana PRMT (CAB79709), C.elegans PRMT1 (CAB54335), mouse CARM1 (AAD41265) and S.cerevisiae Rmt1 (585608). The sequences were initially aligned using Clustal W, and then manually adjusted according to structural elements. The PRMT3 residue numbering is shown above the sequences. Amino acids highlighted are either invariant (white against black) or similar (white against gray) as defined by the following groupings: V, L, I and M; F, Y and W; K and R; E and D; Q and N; S and T; and A, G and P. Residues marked with an asterisk are involved in the dimer interface. The color coding is as described in the text; helices are labeled X–Z before strand 1, and A–I after that; strands are labeled 1–15.
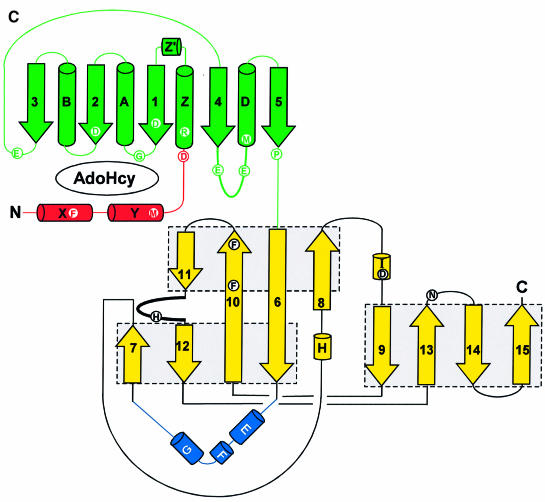
Fig. 5. Schematic Molscript plots (Kraulis, 1991) of the PRMT3 core protein, colored red (two N-terminal helices), green (the AdoMet-binding domain), yellow (barrel structure) and light blue (the three-helix arm). (A) In the front view, the three blue helices represent a high rise arm. (B) In the top view, the green double-E loop is located in the middle of the structure, and the THW loop, connecting strands 11 and 12, is labeled. (C) Topology of the PRMT3 core.
AdoMet-binding domain
As shown in Figure 5A, the enzyme is folded into two domains connected at proline 357 following strand β5. The N-terminal domain consists of a typical Rossmann fold (green) (Rossmann et al., 1974) plus two N-terminal helices αX and αY (red). It also contains the cofactor product AdoHcy, and is thus termed the AdoMet-binding domain. The AdoMet-binding domain of the PRMT3 core is similar to other MTases with two major differences. First, the PRMT3 core contains two N-terminal helices (red), αX and αY, in addition to the α/β Rossmann fold. Secondly, the Rossmann fold (green) in the PRMT core is much smaller than the consensus AdoMet-dependent MTase fold (3↓ 2↓ 1↓ 4↓ 5↓ 7↑ 6↓), as defined by a dozen structurally characterized MTases (Fauman et al., 1999). The PRMT3 core lacks the anti-parallel β-hairpin (7↑ 6↓) that is present in all other MTase structures. Also missing is the short helix (αC) that connects the crossover strands 3 and 4 (Figure 5C).
Residues interacting with AdoHcy come from the three N-terminal helices (αX, αY and αZ), the carboxyl ends of β-strands 1 and 2, and the loops immediately follow strands β1 and β3 (Figure 5C). The 12-residue loop following strand 4 is named the double-E loop, as it contains two invariant amino acids, E326 and E335, and is part of the active site (see below).
The barrel structure
The C-terminal part of PRMT3 is a barrel-like structure (Figure 5C, yellow) with a three-helix segment (Figure 5C, blue) inserted between strands β6 and β7 of the barrel. Three twisted antiparallel β sheets are formed by (8, 6, 10, 11), (6, 10, 12, 7) and (9, 13, 14, 15) (Figure 5C). The latter two sheets are packed together to form a barrel structure. Strands 6 and 10 are part of, and link together the first two sheets, and strands 11 and 12 are aligned head-to-tail with a loop in between, as if there were a single strand. The loop, termed the THW loop as it contains the most highly conserved stretch of residues in this domain (see Figure 4), is located next to the double-E loop in the AdoMet-binding domain (Figure 5B). These two loops form the active site (see below), which is critically supported and stabilized by the floor formed by the first sheet (8, 6, 10, 11) (Figure 5B), particularly strand 10, which contains two invariant phenylalanines, F452 and F456. F452 packs with M328 and L332 of the double-E loop and W477 of the THW loop, while F456 packs with the proline ring of P357 at the junction between the two domains and Y240 of helix αZ. These residues involved in intramolecular contacts are invariant among all the PRMTs in Figure 4.
The three-helix arm and dimer formation of the PRMT core
A three-helix segment (blue) is inserted between strands 6 and 7 of the barrel, forming a high rise arm (Figure 5A). In the crystal lattice, the arm is stabilized by extensive hydrophobic interactions with the outer surface of the AdoMet-binding domain of a neighboring molecule, resulting in a dimer displaying crystallographic 2-fold symmetry (Figure 6). The elements that comprise the dimer interface are the arm of one protomer (helices αF and αG and the loop in between), and the four helices (αY, αZ, αA and αB) and the loop preceding helix αA of the other.
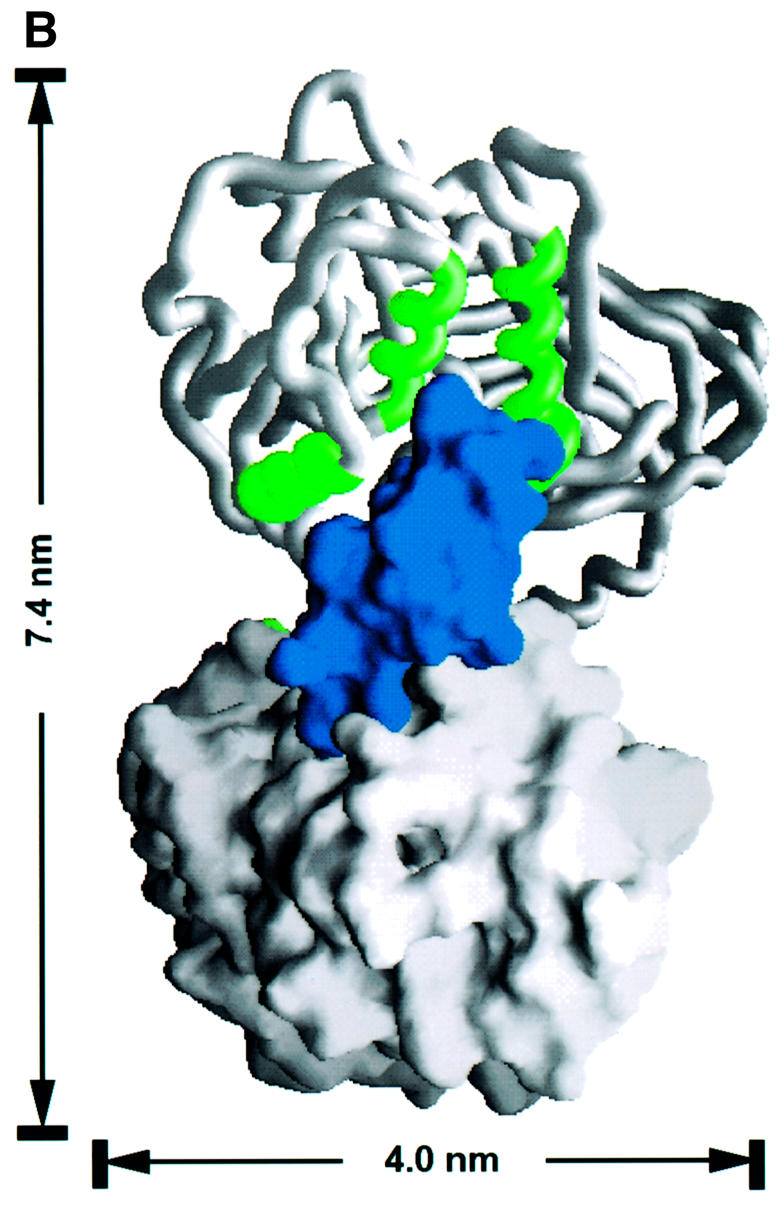
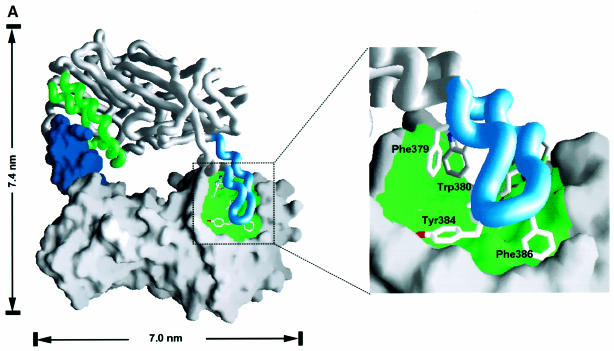
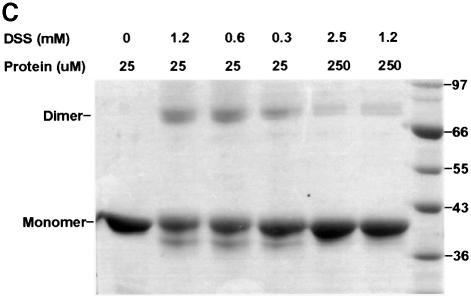
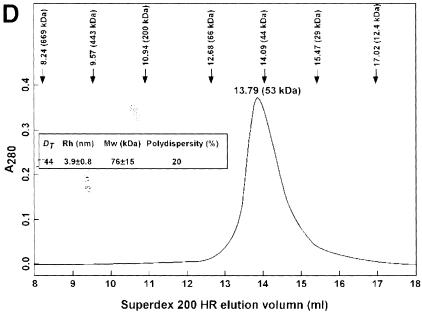
Fig. 6. The dimer interface of the PRMT3 core protein. (A) The crystallographic dimer in space group P43212 is shown with one protomer as a space-filling surface (bottom) and its dimer partner as a worm trace of the polypeptide (top). The view is looking into the hole between the dimer. The F379, W380, Y384 and F386 side chains are shown on the worm representation projecting into surface pockets of the dimer partner. (B) The view is looking from the side. (C) Cross-linking of PRMTΔ200 with disuccinimidyl suberate (DSS). The reaction occurred at room temperature for 30 min in buffer containing 20 mM HEPES pH 7.0, 0.2 M NaCl, 0.1 mM EDTA, 5% glycerol and 0.1% β-mercaptoethanol. Total protein (5 µg) was loaded on a 10% SDS gel. The concentration of protein and cross-linker and the size of the molecular weight marker are indicated. The result of cross-linking with glutaraldehyde is similar and not shown. (D) Elution profile of PRMTΔ200 on a Superdex200HR column (Pharmacia). The column buffer was 20 mM HEPES pH 7.0, 50 mM NaCl, 0.1 mM EDTA, 5% glycerol and 0.1% β-mercaptoethanol, and the protein concentration was ∼1.5 mg/ml. The insert is a summary of the dynamic light scattering experiments using DynaPro Molecular Sizing Instrument, PROTEIN SOLUTIONS, with a protein concentration of 5 mg/ml.
There are several indications that this crystallographic dimer may also exist in solution. (i) The outer surface of the three-helix arm is highly hydrophobic and would not be very stable if exposed to solvent as a monomer. (ii) The molecular contacts observed in the crystallographic dimer include multiple hydrophobic contacts between one molecule (L231, T237, Y245, T263, I265, M268, F269 and I293) and its partner (F379, W380, V383, Y384, F386, M388, C390 and M391). As shown in Figure 4, these hydrophobic residues are conserved among all family members, suggesting that the dimer interaction can be conserved. (iii) This dimer contact is preserved in another crystal form of PRMT3Δ200 in space group P21. In this case, the dimer is non-crystallographic and two copies of it were present in each asymmetric unit. In addition, the same crystallographic dimer interaction is also observed in the structure of rat PRMT1 crystallized in a different space group under a very different condition (X.Zhang, L.Zhou and X.Cheng, manuscript in preparation). (iv) Dynamic light scattering measurement suggested that the hydrodynamic radius of PRMT3Δ200 is 3.9 nm (Figure 6D insert). This matches quite well to the longer dimensions of the dimer (7.4 × 7.0 × 4.0 nm, Figure 6B), and would correspond to an 80 kDa globular protein. (v) A portion of PRMT3Δ200 formed a covalent dimer after cross-linking by glutaraldehyde or disuccinimidyl suberate (Figure 6C).
However, gel filtration suggests that PRMT3 has an apparent molecular weight of a monomer. On a Superdex200 column (Pharmacia), PRMT3Δ200 eluted as an ∼53 kDa globular protein, slightly larger than the calculated molecular weight (36.8 kDa) (Figure 6D). This is similar to the full-length PRMT3 (with a calculated molecular weight of 59.4 kDa) in rat cell extract, which eluted as a 64 kDa protein (Tang et al., 1998). These data suggest that the dimers seen in the crystal structures of the PRMT3 core could also exist in solution, perhaps in equilibrium with the monomer. This is probably true for other PRMT family members as well.
The function of the PRMT dimer is not yet clear. We note that one segment that is involved in the dimer interaction, helix αA and the loop immediately preceding it, also contains many side chains that interact with AdoHcy (see below) on the opposite side of the dimer interface. One possibility is that the dimer interaction could influence the cofactor AdoMet binding and regulate PRMT activity. For example, glycine-N-MTase is a multimeric enzyme that shows cooperative binding of AdoMet (Ogawa et al., 1998). Another consequence of the dimer formation is the enclosure of the active sites (see below) into a hole between the two molecules. This may place a restriction on the substrate that can enter the active site. Consistent with this notion, we have observed that PRMT3 has a much higher activity against a small peptide (∼100-fold) than protein substrates (data not shown).
Substrate-binding surface
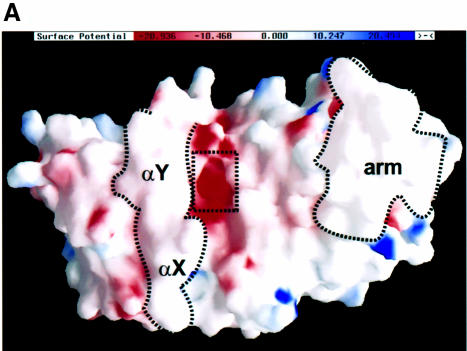
Most known PRMT substrates contain multiple arginines in the context of RG repeats or RXR clusters (Najbauer et al., 1993; Smith et al., 1999). Therefore, one would expect the substrate-binding surface to be negatively charged. Figure 7A shows a molecular surface representation of the PRMT3 core showing the surface charge distribution. The top surface of the PRMT core is enriched in acidic residues. A highly acidic groove runs almost parallel to the two N-terminal helices (αX and αY), with a deep acidic pocket in the middle. We think that the pocket is the active site where the target arginine will be inserted, and that the acidic groove represents a substrate-binding surface that interacts with other arginines surrounding the target.
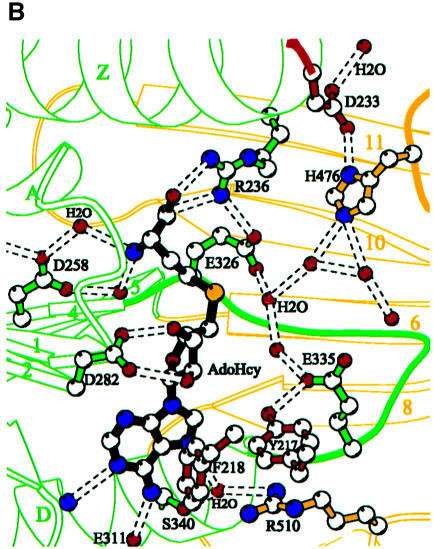
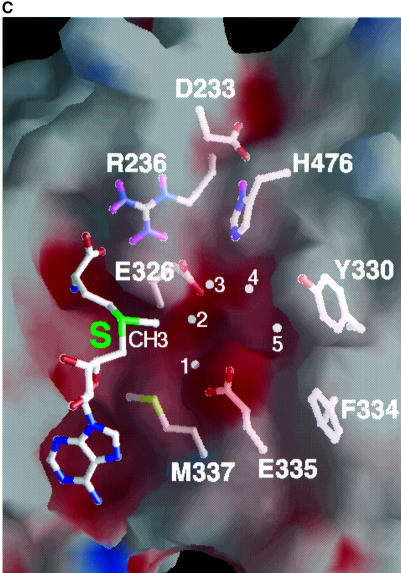
Fig. 7. The active site pocket. (A) Solvent accessible molecular GRASP surface (Nicholls et al., 1991) of the electrostatic potential of the PRMT3 core. The surface is colored blue for positive (20 kBT), red for negative (–20 kBT) and white for neutral, where kB is the Boltzmann’s constant and T is the temperature. (B) Detailed Molscript plots (Kraulis, 1991) of a network of interactions center on the active site and AdoHcy (with the transferable methyl group modeled onto its sulfur atom). (C) A close-up view of the active site pocket.
AdoHcy binding
Figure 7B shows a network of polar and hydrophobic interactions with AdoHcy. The interactions can be grouped according to the three moieties of AdoHcy. (i) For the methionine moiety, R236 of helix αZ interacts with the carboxylate, while D258 of strand β1 interacts with the amino group via two water molecules. (ii) For the adenosine ribose moiety, D282 at the end of strand β2 interacts with both hydroxyls, which is an interaction highly conserved among AdoMet-dependent MTases (Fauman et al., 1999). The Cα of G260 of the loop between β1 and αA interacts with the O4′ of the ribose ring. (iii) For the adenine ring moiety, the nitrogens N6, N1 and N7 interact, respectively, with E311 in the loop after strand β3, the main-chain nitrogen atom of I310, and a water molecule. The water molecule in turn interacts with S340 of helix αD and R510 in the loop following strand β13. The phenyl ring of F218 from helix αX packs against the face of the adenine ring in a perpendicular fashion.
The hydrophobic force of F218 is important to lock the AdoHcy in position and almost completely buries the AdoHcy. In other substrate-free MTase–cofactor complexes, the AdoHcy is much more exposed because the corresponding phenyl ring comes from within the Rossmann fold structure, i.e. the loop between strands 1 and 2 in HhaI DNA MTase or helix αD in TaqI DNA MTase (Schluckebier et al., 1995). This would suggest that the N-terminal helix αX of PRMT3 has to move during the methylation reaction to let AdoMet into and AdoHcy out of the binding site. The high flexibility of helix αX observed in the structure is consistent with such movement. In fact, helix αX is disordered in the P21 space group while the AdoHcy molecule is largely flexible.
Putative arginine binding pocket
The active site is likely to be situated in the pocket along the groove between the two domains. The pocket is cone shaped with an opening of ∼8 × 6 Å and a depth of ∼8 Å. This dimension would accommodate the side chain of an arginine so that the terminal amino group could reach the AdoMet. The hairpin loop contributes most of the residues (E326, Y330, F334, E335, M337) in the active site pocket, except that H476 is part of the THW loop. These residues and the AdoHcy sulfonium atom line the sides and the bottom of the pocket (Figure 7C). The residues lining the pocket are negatively charged (E326 and E335) at the bottom and hydrophobic (F334 and Y330) on the side. This fits well with the asymmetric polarity of an arginine, which consists of three hydrophobic methylene groups and the basic guanidino group. In the absence of the target arginine, the active site is occupied by five ordered water molecules, three of which (sites 1, 2 and 3) directly interact with E326 and E335, and H476 (Figure 7B).
A proposed catalytic mechanism for protein arginine amino methylation
We propose a catalytic mechanism for PRMT based on several features of the active site. (i) The two terminal amino groups (Nη1 and Nη2) of the substrate arginine would occupy water sites 1 and 2, and interact with the side chains of E326 and E335, respectively. The positions of E326 and E335 and the hydrogen-bonding distance between water sites 1 and 2 are consistent with this arrangement. (ii) The Nη2 at site 2 is in a position to become the nucleophile attacking the methyl group of AdoMet. It is only 2 Å away from the methyl group modeled onto the sulfur atom of AdoHcy. It is in line with the methyl group and the sulfur atom of AdoMet; such linear arrangement of the nucleophile, the methyl group and the leaving thioester group in transition state is required by the classic SN2 reaction mechanism used by most (if not all) other MTases (Coward, 1977). (iii) Through interactions with active site residues, the positive charge on the guanidino group is re-distributed towards Nη1. This leaves a lone pair of electrons on the nucleophilic Nη2 to attack the nearby methyl group of AdoMet. The most critical interaction that distinguishes Nη2 from Nη1 involves the negatively charged E335 that stabilizes the positive charge on the Nη1 atom. In contrast, the negative charge of E226 that interacts with the Nη2 atom is effectively neutralized by an interaction with R236, leaving only one carboxylate oxygen to position the target amino group (Nη2) at site 2 through hydrogen bonding. (iv) The proton elimination step after methyl transfer is likely to be accomplished through a His–Asp proton relay system, similar to the one found in many serine proteases (Fersht and Sperling, 1973). The carboxylate oxygen of D233 forms a hydrogen bond to H476, which is in a position to accept a proton from the target amino group and transfer the proton to D233, and eventually to the solvent. Alternatively, this His–Asp pair could deprotonate the target arginine nitrogen atom in concert with or prior to the methyl transfer.
All residues involved in this mechanism are invariant among all PRMTs (Figure 4), suggesting a common mechanism. The most critical catalytic residues are E236 and E335, which make the Nη2 an effective nucleophile. Replacing the equivalent E144 or E153 in PRMT1 with a glutamine essentially abolishes the catalytic activity of the enzyme (X.Zhang, L.Zhou and X.Cheng, manuscript in preparation).
One characteristic feature of this family of PRMTs is that they only form mono- or asymmetric dimethylarginine, but not symmetric dimethylarginine (Figure 1). This specificity could be achieved by excluding any mono-methylated amino group from occupying site 1. The sulfur atom of M337 (an invariant amino acid) of helix αD is only 3.6 Å away from site 1 and would pose a steric block to an extra methyl group on the Nη1. In contrast, site 2 has space to accommodate a mono-methylated amino group and would allow the synthesis of asymmetric dimethylarginine.
Conclusions
We have described the crystallographic structure of the conserved core of PRMT3. This structure reveals that highly conserved residues of PRMT are involved in cofactor binding, catalysis, intramolecular contacts between the two domains, as well as the putative dimer interface, suggesting a common fold and catalytic mechanism.
We still do not understand the function of two invariant residues in the less conserved barrel domain: D428 of helix αI and N505 following strand β13 (Figure 4). Both residues are exposed and face the relatively flexible N-terminus in the current structure. The sequence at the N-terminus is variable among different PRMTs as well as different splicing variants of PRMT1 (Scott et al., 1998). We think that the variable N-terminus probably determines the different substrate specificity of PRMTs by interacting with regions of the substrates outside the target arginine. A mutation near the N-terminus of S.cerevisiae Rmt1 has indeed been shown to alter its substrate specificity (McBride et al., 2000). Being in the same vicinity as the N-terminus, D428 and N505 perhaps play a role in the substrate recognition process. A detailed account of how PRMTs confer substrate specificity must await a high-resolution crystal structure of a PRMT–substrate complex.
Materials and methods
Overexpression and purification of the PRMT3 core
The core domain containing amino acids 184–528 (Δ183) or 201–528 (Δ200) was expressed as GST fusions (Tang et al., 1998). The proteins were purified using glutathione–agarose, Mono Q and gel filtration columns. The GST tag was cleaved by applying thrombin to fusion proteins bound to the glutathione–agarose column. The proteins were incubated with cofactor analog AdoHcy prior to the Mono Q column.
Assay of PRMT activity
We used recombinant hnRNP A1, a well-characterized substrate for PRMT (Rajpurohit et al., 1994), to measure the activity of purified PRMTs. The published procedure for the purification of hnRNP A1 (Mayeda and Krainer, 1992) was modified by replacing the ultracentrifugation step with ion-exchange and gel filtration columns. PRMT reactions (20 µl) contained 3 µM hnRNP A1, 40 µM [methyl-3H]AdoMet (0.5 µCi) and various amounts of PRMTs in 100 mM Tris pH 8.0, 200 mM NaCl, 2 mM EDTA and 1 mM dithiothreitol. The reactions were carried out at 37°C for 1 h. Methylation was analyzed either by SDS–PAGE and fluorography as described previously (Lin et al., 1996), or by precipitation with 10% trichloroacetic acid (TCA), filtering and washing through a GF/F filter (Millipore), and liquid scintillation counting. Under these conditions, the TCA-precipitable counts are linear to the amount of PRMT1 enzyme used (data not shown).
Crystallography in the P43212 space group
Crystals were obtained for PRMT3Δ183 or PRMT3Δ200 using mother liquor containing 100 mM 2-morpholinoethane sulfonic acid (MES) pH 6.3, 16.5–18% polyethylene glycol (PEG) 4000 and 0.2 M ammonium acetate. In some cases, cryoprotectant such as 16% glycerol or 10–15% 2-methyl-2,4-pentanediol (MPD) was also included in the mother liquor. The crystal belongs to primitive tetragonal space group P43212 and each asymmetric unit contains one molecule. Se-containing PRMT3Δ183 (with 10 methionines) was expressed in a methionine auxotroph strain B834 grown in the presence of Se-methionine. To increase the crystal size and reduce precipitation, the Se-protein was allowed to degrade completely to a size similar to that of PRMT3Δ200 (containing eight methionines) by leaving out phenylmethylsulfonyl fluoride after thrombin cleavage. Se-containing crystals were obtained under slightly different conditions from the native protein (100 mM MES pH 6.5, 18–19.5% PEG 4000, 0.2 M ammonium acetate, and 16% glycerol as cryoprotectant).
Complete data sets were collected from crystals of both PRMT3Δ183 (native and SeMet-incorporated) and Xe-incorporated PRMT3Δ200 (Table I). The xenon derivative was obtained by exposing a single crystal (after transferring to the cryobuffer) to 100–250 p.s.i. of the noble gas xenon (Nova Gas Technologies, Cryogenic Rare Gas Laboratories Inc.) for 15 min and flash freezing in liquid carbon tetrafluoride without depressurization, using the Cryo-Xe-Siter (Molecular Structure Corporation). One xenon molecule is bound inside the barrel and makes van der Waals interactions with side chains of three leucines (amino acids 484 of strand β12, and 516 and 518 of strand β14) and the Cβ atom of Gln523 of strand β15. The X-ray data were collected at our home X-ray facility (wavelength 1.54 Å, RAXIS-IV) and at the beamlines X26C (wavelength 1.1 Å, ADSC Q4) and X12C (wavelengths 0.93 Å and at near the Se-absorption edge 0.98 Å, Brandeis B2) of the National Synchrotron Light Source, Brookhaven National Laboratory.
SOLVE (Terwilliger and Berendzen, 1999) first determined the positions of six selenium atoms and five of them were confirmed by Xe-phased difference Fourier methods. Since the cell dimension in the c axis varies slightly from crystal to crystal, we decided to explore only the anomalous diffraction data on the selenium and xenon derivatives (Table I). The multiple anomalous data were used to generate initial protein phases, using PHASES (Furey and Swaminathan, 1997). The resulting initial phases at 2.85 Å were improved by density modification with SOLOMON (Abrahams and Leslie, 1996). The solvent-flattened map was of sufficient quality to place amino acids 221–528 of PRMT3, with the indications from the positions of selenium atoms, into the recognizable densities using the molecular modeling program O (Jones and Kjeldgaard, 1997). The resultant model was refined against a higher resolution data set (up to 2.0 Å) collected from a native PRMT3Δ183 crystal in the resolution range 30.0–2.0 Å by simulated annealing and least-square minimization using X-PLOR (Brünger, 1992). Amino acids 208–220 of PRMT3 and a molecule of AdoHcy were placed in the final model, which contains 2673 protein atoms, 149 water molecules and 26 AdoHcy atoms. The Protein Data Bank coordinates code is 1F3L.
Molecular replacement in the P21 space group
A different crystal form was obtained under conditions similar to those for the crystals of the P43212 space group, except that 16% glycerol or 5–12.5% MPD was added as cryoprotectant. The crystal belongs to the monoclinic space group P21 with cell dimensions of a = 108.5 Å, b = 123.1 Å, c = 71.7 Å and β = 102.7° and each asymmetric unit contains four molecules. The crystal diffracted to 3.3 Å resolution (26 959 unique reflections out of 76 712 observations, 95.7% completeness, Rsym = 0.111 with <I/σ> = 8). The structure was solved by molecular replacement using the refined monomer PRMT3 core structure as the search model, using AMoRe (Navaza and Saludjian, 1997); four solutions were found, with the correlation coefficient of 0.75 and R factor of 0.29, respectively, and the resulting model was refined.
Acknowledgments
Acknowledgements
We thank Dr Harvey R.Herschman (UCLA) for providing the construct to overexpress the GST–Δ183 fusion, Dr Adrian Krainer (Cold Spring Harbor Laboratory) for providing the construct to express hnRNP A1, Susan Sunay for technical assistance during protein purification and crystallization, Paul Kearney for mutagenesis, Aiping Dong for help with X-ray data collection and xenon derivative preparation, Drs Robert M.Sweet and Dieter Schneider (Brookhaven National Laboratory) for help with X-ray data collection at beamlines X12-C and X26-C in the National Synchrotron Light Source, and Dr Robert M.Blumenthal (Medical College of Ohio) for critical comments on the manuscript. These studies were supported in part by the American Cancer Society Institutional Research Grant (IRG-182), the University Research Committee of Emory University, the Georgia Research Alliance and the National Institutes of Health GM61355 to X.C. and X.Z.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determinaiton of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Abramovich C., Yakobson,B., Chebath,J. and Revel,M. (1997) A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J., 16, 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR. A system for X-ray crystallography and NMR, version 3.1. Yale University, New Haven, CT. [Google Scholar]
- Chen D., Ma,H., Hong,H., Koh,S.S., Huang,S.-M., Schurter,B.T., Aswad,D.W. and Stallcup,M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. [DOI] [PubMed] [Google Scholar]
- Coward J.K. (1977) Chemical mechanisms of methyl transfer reactions: comparison of methylases with nonenzymic ‘model reactions’. In Salvatore,F., Borek,E., Zappia,V., Williams-Ashman,H.G. and Schlenk,F. (eds), The Biochemistry of Adenosylmethionine. Columbia University Press, New York, NY, pp. 127–144. [Google Scholar]
- Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of Adomet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, pp. 1–38. [Google Scholar]
- Fersht A.R. and Sperling,J. (1973) The charge relay system in chymotrypsin and chymotrypsinogen. J. Mol. Biol., 74, 137–149. [DOI] [PubMed] [Google Scholar]
- Furey W. and Swaminathan,S. (1997) PHASES-95: A program package for processing and analyzing diffraction data from macromolecules. Methods Enzymol., 277, 590–620. [DOI] [PubMed] [Google Scholar]
- Gary J.D. and Clarke,S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol., 61, 65–131. [DOI] [PubMed] [Google Scholar]
- Gary J.D., Lin,W.J., Yang,M.C., Herschman,H.R. and Clarke,S. (1996) The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem., 271, 12585–12594. [DOI] [PubMed] [Google Scholar]
- Henry M.F. and Silver,P.A. (1996) A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol., 16, 3668–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A. and Kjeldgaard,M. (1997) Electron-density interpretation. Methods Enzymol., 277, 173–208. [DOI] [PubMed] [Google Scholar]
- Kagan R.M. and Clarke,S. (1994) Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes [published erratum appears in Arch. Biochem. Biophys. (1995) 316, 657]. Arch. Biochem. Biophys., 310, 417–427. [DOI] [PubMed] [Google Scholar]
- Katsanis N., Yaspo,M.L. and Fisher,E.M. (1997) Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mamm. Genome, 8, 526–529. [DOI] [PubMed] [Google Scholar]
- Klein S. et al. (2000) Biochemical analysis of the arginine methylation of high molecular weight fibroblast growth factor-2. J. Biol. Chem., 275, 3150–3157. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lee H.W., Kim,S. and Paik,W.K. (1977) S-adenosylmethionine: protein-arginine methyltransferase. Purification and mechanism of the enzyme. Biochemistry, 16, 78–85. [DOI] [PubMed] [Google Scholar]
- Lin W.J., Gary,J.D., Yang,M.C., Clarke,S. and Herschman,H.R. (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem., 271, 15034–15044. [DOI] [PubMed] [Google Scholar]
- Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- McBride A.E., Weiss,V.H., Kim,H.K., Hogle,J.M. and Silver,P.A. (2000) Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem., 275, 3128–3136. [DOI] [PubMed] [Google Scholar]
- Najbauer J., Johnson,B.A., Young,A.L. and Aswad,D.W. (1993) Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem., 268, 10501–10509. [PubMed] [Google Scholar]
- Navaza J. and Saludjian,P. (1997) AMoRe: an automated molecular replacement program package. Methods Enzymol., 276, 581–594. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Gomi,T., Takusagawa,F. and Fujioka,M. (1998) Structure, function and physiological role of glycine N-methyltransferase. Int. J. Biochem. Cell Biol., 30, 13–26. [DOI] [PubMed] [Google Scholar]
- Paik W.K. and Kim,S. (1967) Enzymatic methylation of protein fractions from calf thymus nuclei. Biochem. Biophys. Res. Commun., 29, 14–20. [DOI] [PubMed] [Google Scholar]
- Paik W.K. and Kim,S. (1968) Protein methylase I. Purification and properties of the enzyme. J. Biol. Chem., 243, 2108–2114. [PubMed] [Google Scholar]
- Pollack B.P., Kotenko,S.V., He,W., Izotova,L.S., Barnoski,B.L. and Pestka,S. (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem., 274, 31531–31542. [DOI] [PubMed] [Google Scholar]
- Rajpurohit R., Lee,S.O., Park,J.O., Paik,W.K. and Kim,S. (1994) Enzymatic methylation of recombinant heterogeneous nuclear RNP protein A1. Dual substrate specificity for S-adenosylmethionine: histone-arginine N-methyltransferase. J. Biol. Chem., 269, 1075– 1082. [PubMed] [Google Scholar]
- Rossmann M.G., Moras,D. and Olsen,K. (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature, 250, 194–199. [DOI] [PubMed] [Google Scholar]
- Schluckebier G., O'Gara,M. Saenger,W. and Cheng,X. (1995) Universal catalytic domain structure of AdoMet-dependent methyltransferase. J. Mol. Biol., 247, 16–20. [DOI] [PubMed] [Google Scholar]
- Scott H.S., Antonarakis,S.E., Lalioti,M.D., Rossier,C., Silver,P.A. and Henry,M.F. (1998) Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2). Genomics, 48, 330–340. [DOI] [PubMed] [Google Scholar]
- Shen E.C., Henry,M.F., Weiss,V.H., Valentini,S.R., Silver,P.A. and Lee,M.S. (1998) Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev., 12, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Rucknagel,K.P., Schierhorn,A., Tang,J., Nemeth,A., Linder,M., Herschman,H.R. and Wahle,E. (1999) Unusual sites of arginine methylation in poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J. Biol. Chem., 274, 13229–13234. [DOI] [PubMed] [Google Scholar]
- Tang J., Gary,J.D., Clarke,S. and Herschman,H.R. (1998) PRMT3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity and regulation. J. Biol. Chem., 273, 16935–16945. [DOI] [PubMed] [Google Scholar]
- Tang J., Frankel,A., Cook,R.J., Kim,S., Paik,W.K., Williams,K.R., Clarke,S. and Herschman,H.R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem., 275, 7723–7730. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini S.R., Weiss,V.H. and Silver,P.A. (1999) Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA, 5, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]