Summary
CREB3L1/OASIS is a cellular transcription factor synthesized as a membrane-bound precursor and activated by regulated intramembrane proteolysis in response to stimuli like ER stress. Comparing gene expression between Huh7 subclones that are permissive for hepatitis C virus (HCV) replication versus the non-permissive parental Huh7 cells, we identified CREB3L1 as a cellular factor that inhibits proliferation of virus-infected cells. Upon infection with diverse DNA and RNA viruses including murine γ-herpesvirus 68, HCV, West Nile virus (WNV) and Sendai virus, CREB3L1 was proteolytically cleaved, allowing its NH2-terminus to enter the nucleus to induce multiple genes encoding inhibitors of the cell cycle to block cell proliferation. Consistent with this, we observed a necessity for CREB3L1 expression to be silenced in proliferating cells that harbor replicons of HCV or WNV. Our results indicate that CREB3L1 may play an important role in limiting virus spread by inhibiting proliferation of virus-infected cells.
Introduction
Hepatitis C virus (HCV), a positive stranded RNA virus within the family Flaviviridae, accounts for most cases of chronic liver disease (Appel et al., 2006; Moradpour et al., 2007). In pioneering studies of HCV replication in cultured cells, Lohmann et al transfected Huh7 cells with a HCV subgenomic replicon that consists of HCV RNA engineered to express a selectable marker gene, neo, in place of a portion of the viral RNA that is not required for viral replication (Lohmann et al., 1999). Following selection with G418, subclones of Huh7 cells were established in which HCV RNA was constantly replicating and producing viral proteins required for its replication such as NS5A (Lohmann et al., 1999). However, only a few cells survived the selection, an observation suggesting that Huh7 cells are not permissive for HCV replication (Lohmann et al., 1999; Blight et al., 2002). Subsequently, Blight et al observed that when HCV RNA was eliminated from the cells harboring the HCV replicon through interferon treatment, the cured cells showed dramatically enhanced permissiveness for HCV RNA replication as demonstrated by the large number of cells that survived G418 selection following re-transfection with the HCV replicon RNA (Blight et al., 2002). The best-characterized line of these cells is Huh7.5 cells (Blight et al., 2002). By comparing the difference between Huh7 cells and Huh7.5 cells, Sumpter et al observed that unlike parental Huh7 cells, Huh7.5 cells failed to produce type 1 interferon in response to viral infection as a result of a dominant negative mutation in the RIG-I gene (Sumpter, Jr. et al., 2005). These studies suggest that comparing the difference between subclones of Huh7 cells that are permissive for HCV replication versus their non-permissive parental Huh7 cells could be a powerful approach to study cellular proteins that defend against viral infection.
In the current study, we identify cAMP Response Element Binding Protein 3-Like 1 (CREB3L1, also known as OASIS) as a cellular transcription factor expressed in parental Huh7 cells but not in Huh7.5 cells and another independent subclone of Huh7 cells highly permissive for HCV replication. CREB3L1 belongs to a family of transcription factors that are synthesized as membrane-bound precursors in the endoplasmic reticulum (ER) and transported to the Golgi where they are activated through regulated intramembrane proteolysis (RIP) (Brown et al., 2000; Murakami et al., 2006). RIP consists of two sequential cleavages mediated by Site-1 protease (S1P) and Site-2 protease (S2P). The S1P-catalyzed cleavage at the luminal side is a prerequisite for the S2P-catalyzed intramembrane cleavage that releases the NH2-terminal domain of the protein from membranes, allowing it to drive transcription of target genes in the nucleus (Brown et al., 2000). In osteoblasts, ER stress triggers RIP of CREB3L1 by S1P and S2P, and the nuclear fragment activates the gene encoding type 1 collagen (Murakami et al., 2009). The function of CREB3L1 in other cells is unknown. Herewe show that CREB3L1 is proteolytically activated in cells infected by HCV or other RNA and DNA viruses to block proliferation of these cells by inducing transcription of genes encoding inhibitors to the cell cycle. As a result, CREB3L1 has to be silenced in proliferating cells that support viral replication.
Results
CREB3L1 inhibits HCV replication
While the mutation in RIG-I helps to render Huh7.5 more susceptible to HCV infection, this mutation may not be sufficient to cause permissiveness for HCV replication. We found that knockdown of RIG-I by RNAi did not enhance replication of HCV in Huh7 cells (Figure S1A). Similar result was also observed previously (Binder et al., 2007). Thus, it is likely that Huh7.5 cells may have altered expression of other genes that limit HCV replication. We sought to identify these genes by comparative microarray analysis of Huh7 and Huh7.5 cells. These experiments were inconclusive due to the large number of genes differentially expressed between these cells. To narrow the candidate genes, we needed an independent line of Huh7 cells also permissive for HCV replication so that we might identify genes with reduced expression in both lines of permissive cells. For this purpose, we treated Huh7-K2040 cells, a line of Huh7 cells that harbor an HCV replicon (Ye et al., 2003), with interferon to obtain a clone of cured Huh7 cells that no longer contained HCV RNA. HCV replicon RNA was then re-transfected into these cells to determine their permissiveness for HCV replication. Similar to Huh7.5 cells, these cells were more permissive for HCV replication than their parental Huh7 cells as measured by the number of colonies that contain the HCV replicon encoding the neo (Figure S1B) or by the activity of luciferase encoded by a HCV replicon RNA that contains the luciferase sequence in lieu of neo (Vrolijk et al., 2003) (Figure S1C). We named the cured Huh7-K2040 cells as HRP (HCV Replication Permissive)-1 cells. Unlike Huh7.5 cells, HRP-1 cells do not contain a mutation in RIG-I and they were not defective in activating interferon-induced genes after infection with Sendai virus (Figure S1D).
Microarray analysis revealed 26 genes whose expression was reduced by more than 10-fold in both HRP-1 and Huh7.5 cells compared to the parental Huh7 cells (Table S1). Among these genes, we chose to focus on CREB3L1, because infection with West Nile Virus (WNV), another flavivirus, was known to reduce the amount of this protein in neurons (van Marle et al., 2007).
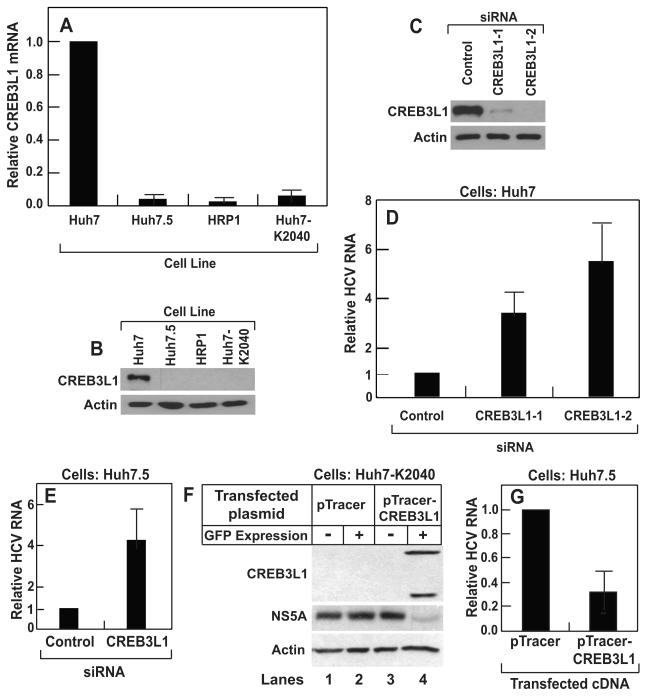
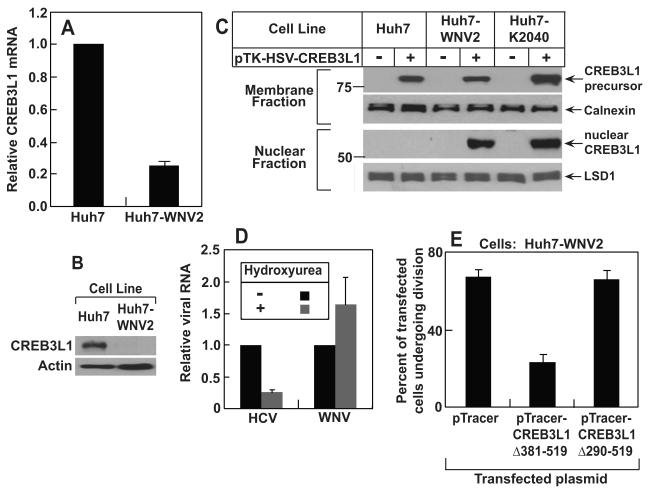
Quantitative real-time PCR (RT-QPCR) analysis showed that the expression of CREB3L1 was reduced by more than 10-fold in both Huh7.5 and HRP-1 compared to the parental Huh7 cells (Figure 1A). Expression of CREB3L1 was also reduced in Huh7-K2040 cells, the progenitor for HRP-1 cells that harbor a HCV replicon (Figure 1A). Immunoblot analysis confirmed the difference in the amount of CREB3L1 protein among these cells (Figure 1B). We then determined whether the lack of expression of CREB3L1 renders cells permissive for HCV replication. CREB3L1 was markedly knocked down by siRNAs targeting two different regions of CREB3L1 mRNA (Figure 1C). Treatment of Huh7 cells with these siRNA raised the amount of HCV replicon RNA replicated in the cells (Figure 1D). To study the replication of live virus, we used the JFH strain of HCV, the only strain of the virus that infects cultured cells (Wakita et al., 2005). We used Huh7.5 instead of Huh7 or HRP-1 cells because the latter cells were not infected by the virion, possibly owing to their intact interferon responses. Although Huh7.5 cells are already permissive for HCV replication and express less CREB3L1 compared to Huh7 cells, knockdown of CREB3L1 by RNAi led to a further increase in HCV RNA after these cells were infected by the HCV virion (Figure 1E).
Figure 1. CREB3L1 inhibits HCV replication.
(A) RT-QPCR quantification of CREB3L1 mRNA in indicated cells with the value in Huh7 cells set to 1.
(B) Immunoblot analysis of CREB3L1 protein in indicated cells.
(C and D) On day 0, Huh7 cells were seeded at 1×105/60 mm dish. On day 1, they were transfected with indicated siRNA (20 pmol/dish). On day 3, these cells were transfected with the HCV replicon RNA (HP, 0.5 μg/dish). On day 4, the cells were harvested and the amount of CREB3L1 protein was determined by immunoblot analysis (C). HCV RNA was quantified by RT-QPCR, with the amount of HCV RNA in cells treated with the control siRNA set to 1 (D).
(E) Huh7.5 cells were seeded and transfected with indicated siRNA as described in (C). On day 3, the cells were infected with the JFH strain of HCV virion. On day 4, the cells were harvested and the amount of HCV RNA was quantified as described in (C).
(F) On day 0, Huh7-K2040 cells were seeded at 7×105/60mm dish. On day 1, they were transfected with 0.5 μg of a plasmid derived from pTracer as indicated. On day 2, the cells were trypsinized and sorted through FACS based on expression of GFP. Lysates of the cells with or without GFP expression were subject to immunoblot analysis with indicated antibodies.
(G) Huh7.5 cells transfected with the indicated plasmid were isolated from untransfected Huh7.5 cells based on GFP expression as described in (D), and seeded at 2×105/60mm dish. They were infected by the JFH strain of HCV virion 24 h later. Following incubation for 3 days, HCV RNA in the cells was quantified by RT-QPCR, with the value in cells transfected with the empty pTracer set at 1.
(A-G), Bar graphs are reported as mean ± S.D. (n=3).
To determine whether overexpression of CREB3L1 inhibits HCV replication, we transfected CREB3L1 into Huh7-K2040 cells. Due to the problem of low transfection efficiency, we subcloned CREB3L1 into a plasmid that also expresses GFP (pTracer). We used a fluorescence activated cell sorter (FACS) to isolate cells that expressed GFP and CREB3L1 (Figure 1F, lane 4). CREB3L1 displayed two bands as a result of proteolytic cleavage (see below). The amount of NS5A, an HCV protein whose expression depends on viral replication (Ye et al., 2003), was markedly reduced in these cells (Figure 1F, lane 4). This effect was not due to the expression of GFP because transfection of the empty pTracer encoding GFP but not CREB3L1 did not affect the amount of NS5A (Figure 1F, lane 2). To determine the effect of CREB3L1 overexpression on infection of the HCV virion, we transfected Huh7.5 cells with pTracer-CREB3L1 or the pTracer vector, isolated transfected cells through FACS, and infected these cells with the HCV virion. As shown in Figure 1G, overexpression of CREB3L1 also reduced the amount of HCV RNA in Huh7.5 cells infected by the HCV virion.
CREB3L1 is proteolytically activated in cells harboring an HCV replicon
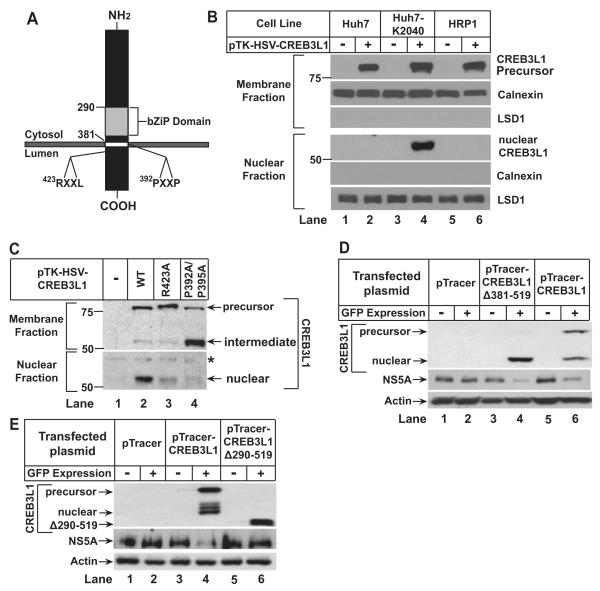
CREB3L1 contains a recognition sequence for S1P (RXXL located within 30 residues from the transmembrane helix) (Hua et al., 1996) and S2P (helix-breaking sequence such as PXXP in the transmembrane helix) (Ye et al., 2000) (Figure 2A). To determine whether RIP of CREB3L1 is induced in hepatoma cells by HCV infection, we transfected a plasmid encoding CREB3L1 tagged at the NH2-terminus with an epitope derived from Herpes Simplex Virus (HSV) into Huh7, HRP-1 and Huh7-K2040 cells. The expression of CREB3L1 encoded by the plasmid was driven by the weak thymidine kinase (TK) promoter to avoid constitutive cleavage of the protein caused by excessive overexpression. The amount of full length and the NH2-terminal fragment of CREB3L1 was detected by anti-HSV, which is sensitive enough to detect transfected proteins driven by the TK promoter in immunoblot analysis (Hua et al., 1996). In Huh7 and HRP-1 cells, which do not contain a HCV replicon, we observed only the full length precursor of CREB3L1 with a molecular weight of ~80 kDa in the membrane fraction (Figure 2B, lanes 2 and 6). Cleaved CREB3L1 was not detectable in the nuclear fraction (Figure 2B, lanes 2 and 6). In Huh7-K2040 cells, which contain an HCV replicon, the cleaved NH2-terminal fragment of CREB3L1 (nuclear form, ~55 kDa) was prominent in the nuclear fraction (Figure 2B, lane 4). In these cells, generation of the nuclear form was reduced when we transfected plasmids encoding CREB3L1 with mutations that disrupt the consensus S1P (R423A) or S2P recognition site (P392A/P395A) (Figure 2C, lanes 2-4). When the S2P recognition site was altered, we observed in the membrane fraction a cleaved fragment with a molecular weight similar to the nuclear form (Figure 2C, lane 4). This fragment is the membrane-bound intermediate form of CREB3L1 that is cleaved by S1P but not by S2P. A similar cleavage intermediate was observed for SREBP-2, a prototype of RIP substrates, when its S2P recognition site was disrupted (Ye et al., 2000). These results suggest that CREB3L1 is activated through RIP in HCV-infected cells.
Figure 2. CREB3L1 undergoes RIP in HCV-infected cells.
(A) Schematic diagram of CREB3L1.
(B and C) On day 0, indicated cells (B) or Huh7-K2040 cells (C) were seeded at 4×105/60 mm dish. On day 1, they were transfected with 50 ng of indicated plasmids. On day 2, the cells were harvested and separated into nuclear and membrane fractions followed by immunoblot analysis with anti-HSV. Asterisk (*) denotes a cross-reactive band. Immunoblots of calnexin and Lysine-specific demethylase 1 (LSD1) served as loading controls for membrane and nuclear fractions, respectively.
(D and E) Huh7-K2040 cells transfected with the indicated plasmid were analyzed as described in Figure 1F.
To determine whether production of the nuclear CREB3L1 is sufficient to inhibit HCV replication, we transfected Huh7-K2040 cells with a plasmid encoding the truncated NH2-terminal domain of CREB3L1 (CREBL1(Δ381-519)), which enters the nucleus without cleavage. Overexpression of CREBL1(Δ381-519) also inhibited HCV replication in Huh7-K2040 cells as indicated by NS5A expression (Figure 2D). This effect was not observed when the DNA-interacting bZIP domain (Figure 2A) was deleted from the nuclear CREB3L1 (CREB3L1(Δ290-519)) (Figure 2E), even though deletion of this domain did not prevent nuclear localization of the protein (Figure S2). These results suggest that the nuclear CREB3L1 is likely to activate transcription of genes that inhibit viral replication.
Nuclear CREB3L1 blocks cell proliferation
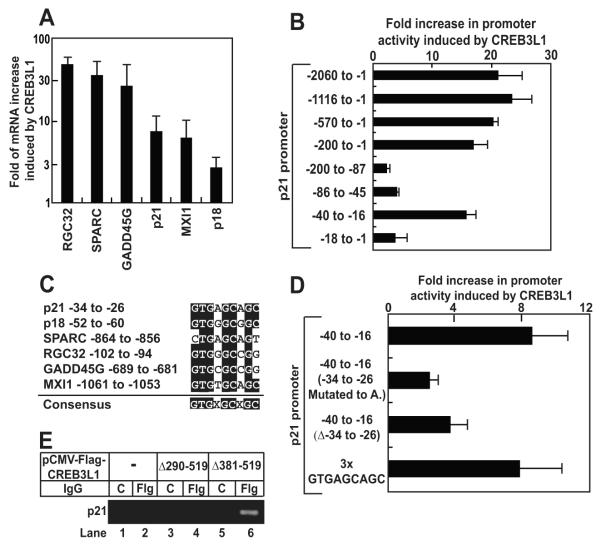
To identify genes activated by CREB3L1, we transfected Huh7-K2040 cells with CREB3L1(Δ381-519) subcloned into pTracer and used FACS to isolate cells that expressed GFP and CREB3L1. Microarray analysis was performed to compare gene expression between these cells and cells transfected with the empty pTracer plasmid. In addition to collagen 1α1, a known target of CREB3L1 (Murakami et al., 2009), CREB3L1(Δ381-519) also activated transcription of a group of genes that inhibit the cell cycle (Table S2). These genes encode cyclin inhibitors p21 and p18 (Sherr and Roberts, 1999), c-myc antagonists Mxi-1 (Hurlin and Huang, 2006), and other proteins known to inhibit cell proliferation such as GADD45γ (Liebermann and Hoffman, 2002), SPARC (Bradshaw and Sage, 2001) and RGC32 (Saigusa et al., 2006). RT-QPCR analysis confirmed that all of these genes were induced by CREB3L1(Δ381-519) (Figure 3A). Despite this induction, none of the 5′-flanking regions of these genes contains the nucleotide sequences reported to bind CREB3L1(Kondo et al., 2005; Murakami et al., 2009). To identify the CREB3L1 response element in these cell cycle-inhibitory genes, we performed reporter assays using a luciferase reporter gene driven by a 2-kb fragment of the 5′-flanking region of p21. Luciferase activity was markedly enhanced (~20-fold) by CREB3L1(Δ381-519) transfected into Huh7-K2040 cells (Figure 3B). Serial deletion analysis indicated that a segment encompassing nucleotide positions −40 to −16 relative to the transcription start site of p21 conveyed CREB3L1 inducibility (Figure 3B). Sequence analysis revealed that position −34 to −26 within this fragment contains a consensus sequence GTGXGCXGC that is conserved in promoters of all genes activated by CREB3L1 (Figure 3C). Deletion of this sequence or changing all nucleotides in the sequence to adenines reduced the ability of CREB3L1 to stimulate luciferase activity (Figure 3D). An artificial promoter containing only the consensus sequence (repeated three times) was activated by CREB3L1 (Figure 3D). Chromatin immunoprecipitation (ChIP) assays revealed that Flag-tagged CREBL1(Δ381-519) bound to the promoter region of endogenous p21 (Figure 3E, lane 6). Deletion of the bZIP domain from the NH2-terminal segment of CREB3L1 (CREB3L1(Δ290-519)) abolished this interaction (Figure 3E, lane 4).
Figure 3. Nuclear form of CREB3L1 activates genes that inhibit the cell cycle.
(A) Huh7-K2040 cells were seeded, transfected with pTracer or pTracer-CREB3L1(Δ381-519) as described in Figure 1D. The transfected cells were isolated based on GFP expression as described in Figure 1D. The amount of indicated mRNA was quantified by RT-QPCR. Fold induction of indicated mRNA by CREB3L1(Δ381-519) was determined by setting the amount of the mRNA in cells transfected with the control plasmid pTracer at 1. Result are reported as mean ± S.D. (n=3).
(B and D) On day 0, Huh7-K2040 cells were seed at 7×105/60 mm dish. On day 1, the cells were transfected with a firefly luciferase reporter plasmid containing the indicated region of p21 promoter (1 μg/dish) and a plasmid encoding renilla luciferase driven by the constitutive CMV promoter (0.5 μg/dish) in the absence or presence of cotransfection of pCMV-CREB3L1(Δ381-519) (0.5 μg/dish). On day 2, luciferase activity was measured and the promoter activity was determined by firefly luciferase activity normalized against renilla luciferase activity to control for transfection efficiency. Fold increase in the promoter activity in cells transfected with CREB3L1(Δ381-519) compared to those mock-transfected was presented. Result are reported as mean ± S.D. (n=3).
(C) Alignment of −34 to −26 of the p21 promoter region with promoter sequences from indicated genes.
(E) On day 0, Huh7-K2040 cells were seeded at 2.5×105/60 mm dish. On day 3, they were transfected with 0.5 μg of indicated plasmids. On day 4, the cells were harvested and the cell lysate was immunoprecipitated with control IgG (C) or anti-Flag IgG (Flg) followed by ChIP analysis as described in Experimental Procedures.
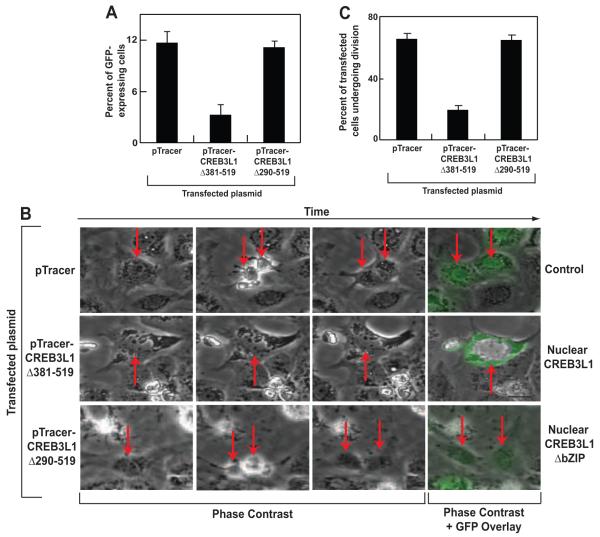
The above results suggest that the nuclear form of CREB3L1 inhibits cell cycle progression by binding to promoters and activating transcription of genes encoding cell cycle inhibitors. To test this hypothesis, we transfected Huh7-K2040 cells with CREB3L1(Δ381-519) subcloned into pTracer, incubated the cells for 3 days to allow these cells to divide, and quantified the number of cells containing the plasmid by FACS. The number of GFP-positive cells was 4-fold less in cells transfected with pTracer-CREB3L1(Δ381-519) than those transfected with the empty pTracer (Figure 4A), suggesting that CREB3L1(Δ381-519) inhibited cell proliferation. This effect was not observed when Huh7-K2040 cells were transfected with the nuclear CREB3L1 in which bZIP domain is deleted (CREB3L1(Δ290-519)) (Figure 4A)
Figure 4. Nuclear form of CREB3L1 inhibits cell proliferation.
(A) Huh7-K2040 cells were seeded and transfected with the indicated plasmid as described in Figure 1D. Three days after the transfection, percentage of the cells expressing GFP encoded by the transfected plasmid was determined through FACS analysis. Results are reported as mean ± S.D. (n=3).
(B) On day 0, Huh7-K2040 cells were seeded at 2.5×105 per well of a 6-well plate. On day 1, they were transfected with 0.25 μg of the indicated plasmid. On day 2, the cells were subjected to time-lapse imaging analysis as described in Experimental Procedures. On day 4 (48 h later), GFP fluorescence images were taken. Representative time-lapse images of these cells were shown, with arrows indicating transfected cells and their daughter cells determined by their expression of GFP at the end of the imaging at 48 h.
(C) Quantification of transfected cells that went through cell divisions in (B). On average, 30 cells transfected with each plasmid were counted in each experiment. Results are reported as mean ± S.D. (n=3).
To more directly access the role of CREB3L1 in cell proliferation, we performed time-lapse imaging analysis of Huh7-K2040 cells transfected with the NH2-terminal fragment of CREB3L1 subcloned into pTracer. At the end of the experiment, GFP fluorescent images were taken to identify cells transfected with the plasmid, and these cells were traced backward to determine if they underwent division events (Movies S1-3). Representative time-lapse images of Huh7-K2040 cells transfected with pTracer-CREB3L1(Δ381-519), pTracer-CREB3L1(Δ290-519) or the pTracer vector were shown in Figure 4B. Quantitative analysis of these images indicated that more than 70% of cells transfected with the empty pTracer went through at least one cycle of cell division during a period of 48 h (Figure 4C). Less than 20% of cells transfected with pTracer-CREB3L1(Δ381-519) divided during the same period (Figure 4C). Deletion of the bZIP domain in CREB3L1 (CREB3L1(Δ290-519)) abolished the inhibition of cell division (Figure 4C).
The data presented above indicate that HCV infection induces the processing of CREB3L1 to its nuclear form, which in turn suppresses cell cycle progression by coordinately activating a host of genes encoding proteins that inhibit the cell cycle. Inasmuch as active division of host cells is known to be required for efficient HCV replication (Pietschmann et al., 2001; Scholle et al., 2004), we conclude that the nuclear form of CREB3L1 suppresses HCV replication by blocking the cell cycle. In order for cells to become permissive for HCV replication, CREB3L1 must be silenced as in Huh7.5 and HRP-1 cells so that these cells are able to divide after they are infected by HCV.
CREB3L1 blocks proliferation of cells infected by viruses other than HCV
To determine the role of CREB3L1 in defending virus other than HCV, we examined the expression of the gene in Huh7-WNV2 cells, a line of a Huh7 cells harboring a replicon of WNV, another flavivirus (Wang et al., 2005). We observed that expression of CREB3L1 is markedly reduced in these cells compared to the parental Huh7 cells as determined by RT-QPCR (Figure 5A) and immunoblot analysis (Figure 5B). CREB3L1 was cleaved to produce the nuclear form upon transfection into Huh7-WNV2 but not Huh7 cells (Figure 5C). Unlike HCV, replication of WNV was not affected by overexpression of the nuclear form of CREB3L1 (Data not shown). This result suggests that unlike HCV, replication of WNV may not depend on active division of host cells. To test this hypothesis, we incubated Huh7 cells harboring a HCV or WNV replicon with hydroxyurea, a drug that inhibits the cell cycle by blocking DNA synthesis (Ye et al., 2003). While reducing the amount of HCV RNA, the drug treatment did not affect the amount of WNV RNA (Figure 5D). However, overexpression of the nuclear form of CREB3L1 still led to suppression of cell division in Huh7-WNV2 cells (Figure 5E). This result might explain why CREB3L1 has to be silenced in Huh7-WNV2 cells as these cells have to proliferate in the presence of persistent replication of WNV genome.
Figure 5. RIP of CREB3L1 inhibits proliferation of cells infected by WNV.
(A) RT-QPCR quantification of CREB3L1 mRNA in indicated cells with the value in Huh7 cells set to 1. Results are reported as mean ± S.D. (n=3).
(B) Immunoblot analysis of CREB3L1 protein precursor in indicated cells
(C) Proteolytic cleavage of CREB3L1 was analyzed as described in Figure 2B.
(D) On day 0, Huh7-K2040 and Huh7-WNV2 cells were seeded at 2×105/60 mm dish. On day 1, the cells were treated with or without 10 μM hydroxyurea. On day 5, cells were harvested for quantification of HCV and WNV RNA in Huh7-K2040 and Huh7-WNV2 cells, respectively, by RT –QPCR, with the values in cells that were not treated with the drug set to 1. Results are reported as mean ± S.D. (n=3).
(E) Quantification of Huh7-WNV2 cells transfected with indicated plasmid that went through cell divisions was carried out as described in Figure 4C.
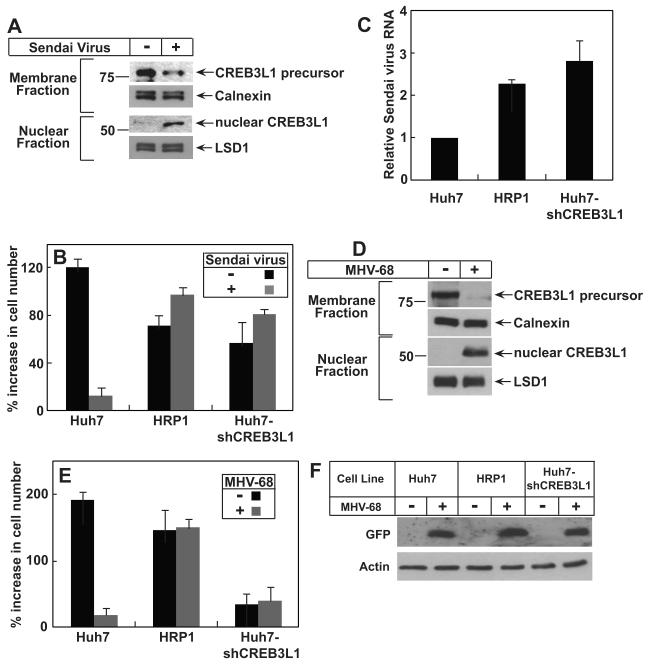
To determine whether the effect of CREB3L1 is restricted to flavivirus, we examined the role of CREB3L1 in inhibiting proliferation of cells infected by Sendai virus, a negative-stranded RNA virus of Paramyxoviridae. Infection of Huh7 cells with Sendai virus stimulated the cleavage of CREB3L1 (Figure 6A). Infection of Sendai virus almost completely blocked proliferation of Huh7 cells, which express CREB3L1 (Figure 6B). The proliferation of HRP-1 cells, which do not express CREB3L1, was not affected by the viral infection (Figure 6B). Huh7-shCREB3L1 cells, which were generated by stably transfecting Huh7 cells with a shRNA targeting CREB3L1, produced much less CREB3L1 mRNA (Figure S3A). Proliferation of these cells was also not affected by the Sendai virus infection (Figure 6B). The failure for Sendai virus to inhibit proliferation of HRP-1 and Huh7-shCREB3L1 cells was not caused by less efficient infection of the cells by the virus, as the amount of viral RNA in these cells was even more than that in Huh7 cells (Figure 6C). Sendai virus did not lyse the cells in the experiments as there was no lactate dehydrogenase activity detected in the culture medium. Similar to WNV, replication of Sendai virus does not require proliferation of host cells, as hydroxyurea treatment did not inhibit replication of the viral RNA (Figure S3B).
Figure 6. CREB3L1 prevents proliferation of cells infected by Sendai Virus and MHV-68.
(A) On day 0, Huh7 cells were seeded at 4×105/60 mm dish. On day 1, the cells were infected with Sendai virus. On day 2, the cells were fractionated into nuclear and membrane fractions and the cleavage of CREB3L1 was determined by immunoblot analysis with anti-CREB3L1.
(B and C) On day 0, Huh7, HRP-1 and Huh7-shCREB3L1 cells were seeded at 2×105/60 mm dish. On day 1, triplicate dishes of the cells were harvested for cell counting. The remaining cells were infected with Sendai virus as indicated. On day 2 (24 h later), triplicate dishes of these cells were harvested and the number of cells in each plate was quantified. The percentage of indicated cells in each plate increased on day 2 compared to that on day 1 is presented (B). Sendai virus RNA in the cells infected by the virus was quantified by RT-QPCR with the value in Huh7 cells set to 1 (C).
(D) Proteolytic cleavage of CREB3L1 was analyzed as in (A) except that the cells were infected by MHV-68 instead of Sendai virus.
(E and F) Indicated cells were seeded and treated as described in (B) except that the cells were infected by MHV-68 instead of Sendai virus. Proliferation of the cells was determined as described in (B). Cellular lysate was subjected to immunoblot analysis with anti-GFP (F).
(A-F) Bar graphs are reported as mean ± S.D. (n=3).
To investigate the role of CREB3L1 in defending against infection of DNA virus, we infected Huh7 cells with murine γ-herpesvirus 68 (MHV-68). The genome of MHV-68 we used also encodes GFP that serves as a marker for viral infection (Dong et al., 2010). Similar to Sendai virus, infection of Huh7 cells with MHV-68 also triggered the cleavage of CREB3L1 (Figure 6D). Infection with MHV-68 blocked the proliferation of Huh7 cells (Figure 6E). Proliferation of HRP-1 and Huh7-shCREB3L1 cells, which express much less CREB3L1 compared to the parental Huh7 cells, was not affected by the viral infection (Figure 6E). The lack of CREB3L1 did not affect viral infection, as expression of GFP encoded by MHV-68 was the same in Huh7, HRP-1 and Huh7-shCREB3L1 cells (Figure 6F).
To determine the function of CREB3L1 in cells that are not derived from Huh7 cells, we infected SV589 cells, an immortalized line of human fibroblasts (Yamamoto et al., 1984), with Sendai virus. The viral infection stimulated the cleavage of CREB3L1 in SV589 cells (Figure S3C). Sendai virus infection reduced the rate of proliferation of SV589 cells transfected with the control siRNA but not those transfected with the siRNA targeting CREB3L1 (Figure S3D), which decreased the amount of CREB3L1 mRNA by ~90% (Figure S3E). Knockdown of CREB3L1 did not alter the amount of viral RNA in SV589, suggesting that the failure for Sendai virus to inhibit proliferation of SV589 cells in which CREB3L1 was knocked down was not caused by less efficient infection of the cells by the virus (Figure S3F).
Discussion
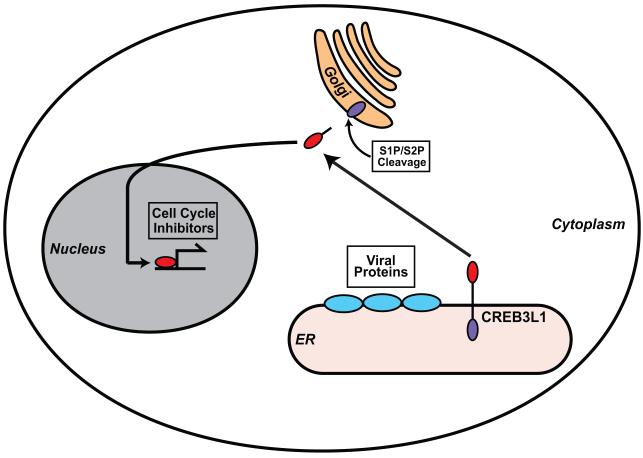
The results presented above support a model shown in Figure 7. CREB3L1 is synthesized as a membrane-bound precursor. In cells infected by a DNA virus such as MHV-68, negative stranded-RNA virus such as Sendai virus, or positive stranded-RNA viruses such as HCV and WNV, CREB3L1 is activated by RIP catalyzed by S1P and S2P. As an analogy to other well-characterized transcription factors activated through RIP, we assume that the viral infection stimulates the translocation of CREB3L1 precursor from the ER to Golgi complex in which S1P and S2P reside (DeBose-Boyd et al., 1999; Shen et al., 2002). Following the cleavage, the NH2-terminal domain of CREB3L1 enters the nucleus, where it activates transcription of genes encoding cell cycle inhibitors to block proliferation of the virus-infected cells. For most of the viruses we analyzed, inhibition of proliferation of their host cells did not affect their replication. Thus, rather than directly inhibiting viral replication, CREB3L1 may play an important role to prevent virus spread by limiting proliferation of virus-infected cells. Among the viruses we studied, HCV is unique in that its replication in Huh7-derived cells requires active division of the host cells (Pietschmann et al., 2001; Scholle et al., 2004). As a result, CREB3L1-mediated signaling not only blocks proliferation of virus-infected cells but also inhibits viral replication. However, the in vivo relevance of the observation remains to be determined. Although hepatocytes do not enter the cell cycle in healthy livers, they proliferate rapidly to repair liver injuries induced by HCV infection. Thus, further studies are required to determine whether CREB3L1 inhibits division of virus-infected hepatocytes in vivo to limit HCV replication at a low level and to prevent the spread of the virus.
Figure 7. A model illustrating the role of CREB3L1 in limiting HCV infection.
CREB3L1 is synthesized as a membrane-bound precursor. In cells infected by virus, CREB3L1 is activated through RIP, presumably through ER stress caused by expression of ER-associated viral proteins. The proteolytic cleavage allows the NH2-terminal domain of the protein to enter the nucleus, where it activates transcription of genes encoding cell cycle inhibitors to block proliferation of the cells infected by the virus.
It was reported previously that ER stress triggers cleavage of CREB3L1 (Murakami et al., 2006). ER stress is also known to be induced by massive synthesis of ER-associated viral proteins in cells infected by viruses (He, 2006). These viruses include those used in the current study such as HCV (Tardif et al., 2002; von dem Bussche et al., 2010) and WNV (Medigeshi et al., 2007)). Thus, it is conceivable that viral infection triggers cleavage of CREB3L1 through ER stress. If this is the case, CREB3L1 may belong to a growing list of proteins that defend against viral infection through ER stress (Martinon and Glimcher, 2010).
In the current study, we show that both Huh7.5 and HRP-1 cells are highly permissive for replication of HCV subgenomic replicons derived from the genotype 1 HCV owing to the lack of expression of CREB3L1. However, only Huh7.5 but not HRP-1 cells can be infected by the JFH1 strain of the genotype 2 HCV. Unlike genotype 1 HCV, genotype 2 HCV is much more sensitive to interferon (Zein, 2000). This difference between Huh7.5 and HRP-1 cells might be explained by a mutation in RIG-I found in Huh7.5 (Sumpter, Jr. et al., 2005) but not in HRP-1 cells, which makes Huh7.5 but not HRP-1 cells defective to produce interferon in response to viral infection.
Inasmuch as CREB3L1 inhibits cell proliferation by activating genes encoding proteins that inhibit the cell cycle, it may be considered as a tumor suppressor gene. Indeed, inactivation of CREB3L1 through chromosome fusion is associated with development of low-grade fibromyxoid sarcoma (Mertens et al., 2005). Considering that CREB3L1 is proteolytically activated in virus-infected cells, the protein may play an important role in preventing virus-induced tumorigenesis. Notably, the ability of CREB3L1 to inhibit cell proliferation has not been observed in previous studies that analyze genes activated by the protein (Kondo et al., 2005; Murakami et al., 2009; Vellanki et al., 2010; Fox et al., 2010). Unlike the current study, these analyses were not performed in virus-infected cells. We observed that the nuclear form of CREB3L1 activated genes that suppress cell proliferation much more profoundly in Huh7-K2040 cells that harbor an HCV replicon than the naïve Huh7 cells (data not shown). Thus, it is likely that another factor generated in virus-infected cells cooperates with the nuclear form of CREB3L1 to activate these genes.
In addition to genes encoding proteins that inhibit the cell cycle, CREB3L1 also activates transcription of type 1 collagen and genes involved in assembly of collagen matrix as previously reported (Murakami et al., 2009; Vellanki et al., 2010). This observation suggests that in addition to preventing proliferation of virus-infected cells, CREB3L1 may also limit the spread of the virus by activating production of small amounts of collagen in virus-infected cells to segregate these cells from their environment. Further study will be required to determine whether chronic deposition of collagen produced in virus-infected cells contributes to fibrosis induced by viral infection.
Experimental Procedures
Antibodies
We obtained rabbit anti-actin and anti-GFP from Abcam; mouse anti-HSV from Novagen; rabbit anti-LSD1 from cell signaling; mouse anti-calnexin from Enzo Life Sciences; peroxidase-conjugated secondary antibodies from Jackson ImmunoResearch. Rabbit anti-NS5A was described previously (Huang et al., 2007). A rabbit polyclonal antibody against human CREB3L1 was generated by immunizing rabbits with a protein consisting of the NH2-terminal 290 amino acid residues of human CREB3L1.
Cell culture
Huh7, Huh7.5, HRP-1 and SV589 cells were maintained in medium A (Dulbecco’s modified Eagle medium with 4.5 g/L glucose, 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, and 10% fetal calf serum). Huh7-K2040 cells were maintained in medium A supplemented with 200 μg/ml G418. Huh7-GL cells, a line of Huh7 cells that contain a chromosomally integrated JFH strain of HCV cDNA and constitutively produce infectious virus (Cai et al., 2005), were maintained in medium A supplemented with 5 μg/ml blasticidine. Huh7-shCREB3L1 cells were generated by stably transfecting a shRNA targeting CREB3L1 (SA Biosciences, Clone ID 2) into Huh7 cells. The cells were maintained in medium A supplemented with 10 μg/ml puromycin. All cells were cultured in monolayer at 37 °C in 5% CO2.
Plasmids
pTracer-CMV (Invitrogen) is a plasmid that allows detection of transfected cells through expression of GFP encoded by the plasmid. pTracer-CREB3L1, pTracer-CREB3L1(Δ381-519), pTracer-CREB3L1(Δ290-519) encode CREB3L1 and its deletion mutants. pTK-HSV-CREB3L1 encodes full length CREB3L1 preceded with two copies of the HSV epitope tag (QPELAPEDPED) at the NH2-terminus under the control of the thymidine kinase (TK) promoter. pTK-HSV-CREB3L1(R423A) and pTK-HSV-CREB3L1(P392A/P395A) were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) with pTK-HSV-CREB3L1 as the template. pCMV-CREB3L1(Δ381-519) encodes a deletion mutant of CREB3L1 under the control of the CMV promoter. pCMV-Flag-CREB3L1(Δ290-519) and pCMV-Flag-CREB3L1(Δ381-519) encodes deletion mutants of CREB3L1 preceded with a Flag epitope tag (DYKDDDDK) under control of a CMV promoter. For luciferase reporter experiments shown in Figures 3B and 3D, indicated regions of p21 promoter were cloned into pGL3 vector (Genescript).
RT-QPCR
RT-QPCR was performed as previously described (Ye et al., 2003; Liang et al., 2002). Briefly, total RNA prepared from cells was treated extensively with DNase I (DNA-free, Ambion, Austin, TX). First-strand cDNA was synthesized from the DNA-free RNA by using random hexamer primers and the ABI cDNA synthesis kit (Applied Biosystems). cDNA was mixed with SYBR Green PCR Master Mix (Applied Biosystems) and sets of forward and reverse primers specific for the RNA subjected to the measurement or human 36B4 mRNA and analyzed by real-time quantitative PCR with the ABI PRISM 7900HT sequence detection system (Applied Biosystems). All reactions were performed in triplicate. The relative amounts of RNAs were calculated through the comparative cycle threshold method by using human 36B4 mRNA as the invariant control.
Virus infection
The JFH1 strain of HCV was produced from Huh7-GL cells, a gift from Dr. Guangxiang Luo (University of Kentucky), as previously described (Huang et al., 2007). The HCV HP or con1 replicon RNA was in vitro transcribed as previously described (Sumpter, Jr. et al., 2004) and transfected into Huh7 cells using Transmessenger reagent (Qiagen). Sendai virus (Cantrell strain) was purchased from Charles River Labs. The virus was added to cells after it was diluted 50-fold in cell culture medium (3 ml/60 mm plate). MHV-68, a gift from Dr. Pinghui Feng (UT Southwestern Medical Center), was used to infect the cells with a multiplicity of infection at 2 through spin infection by spinning the cells at 1000 g for 30 min.
RNA interference
Duplexes of siRNA were synthesized by Dharmacon Research. The two siRNA sequences targeting human CREB3L1 (NCBI Accession number NM_052854) are at nucleotide positions (relative to the codon for the initiation methionine) 131 - 149 and 884 - 902 for CREB3L1-1 and CREB3L1-2, respectively. The control siRNA targeting GFP was reported previously (Adams et al., 2004). Cells were transfected with siRNA using Lipofectamine™ RNAiMAX reagent (Invitrogen) as described by the manufacturer, after which the cells were used for experiments as described in the figure legends.
Immunoblot analyses to determine RIP of CREB3L1
Cells were harvested and separated into nuclear and membrane fractions as described (Sakai et al., 1996). 4 μg of nuclear fractions and 10 μg of membrane fractions were analyzed by 10% SDS-PAGE followed by immunoblot analysis with anti-HSV (0.25 μg/ml). Bound antibodies were visualized with a peroxidase-conjugated secondary antibody using the SuperSignal ECL-HRP Substrate System (Pierce).
Isolation of transfected cells through FACS
Cells were trypsinized and sorted on a BD FACSAria Flow Cytometer (Beckin-Dickenson) based on expression of GFP by the UT Southwestern Flow Core Facility. Sorted cells with or without GFP expression were then used for assays described in figure legends.
Luciferase assays
Luciferase activity in the cell lysate was assayed with the Dual Luciferase Reporter Assay system (Promega) using the Synergy 4 plate reader and Gen5 1.10 Software (Biotek). Promoter activity was determined by firefly luciferase activity normalized against renilla luciferase activity to control for transfection efficiency.
ChIP analyses
ChIP was performed using the SimpleChiP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s protocol. Genomic DNA was used for PCR with the AccuPrime Pfx Supermix Kit (Invitrogen) using primers 5′-AACTCGGCCAGGCTCAGCTG-3′ and 5′-GCTCCACAAGGAACTGACTTCGGCAG-3′ to amplify a 200bp segment of the p21 promoter. PCR products were run on a 2% agarose gel and imaged with the Eagle Eye II gel dock system with EagleSight Software (Stratagene).
Time-lapse imaging
Cells were changed into CO2-Independent Medium (Invitrogen) supplemented with 10% FCS, 1% penicillin/streptomycin, 2 mM GlutaMAX (Invitrogen), and 1 mM sodium pyruvate for the imaging analysis. Phase contrast time-lapse imaging was performed with a Axiovert 200M (Ziess) microscope in a 37°C chamber and images were taken every ten minutes for 48 h in at least four different fields per well using MetaMorph Software (Molecular Devices). GFP fluorescence images were taken at the end of the experiment at 48 h. Cells transfected with different plasmids were imaged in parallel using a motorized stage (Marzhauser). Cells expressing GFP at 48 h were traced backward to determine if these cells underwent division events.
Microarray Analysis
Microarray analysis was performed exactly as previously described (Horton et al., 2003). Microarray analyses comparing gene expression between Huh7 versus Huh7.5 and HRP-1 cells, and Huh7-K2040 cells transfected with CREB3L1(Δ381-519) versus those mock transfected were deposited at Gene Expression Omnibus (GEO) with accession number GSE25156, and GSE25157, respectively.
Supplementary Material
Highlight.
Viral infection triggers proteolytic cleavage of membrane-bound CREB3L1.
The cleaved NH2-terminal fragment of CREB3L1 enters the nucleus.
Nuclear CREB3L1 activates genes inhibiting proliferation of virus-infected cells.
Acknowledgement
We thank Drs. Michael S. Brown and Joseph L. Goldstein for their support and advice; Dr. Michael Gale for his gifts of Huh7-K2040 and Huh7-WNV2 cells; Charlie Rice, Guangxiang Luo and Pinghui Feng for their gifts of Huh7.5 cells, Huh7-GL cells and MHV-68, respectively; Takaji Wakita for the JFH technology; Lisa Beatty, Angela Carroll, Shomanike Head and Ijeoma Onwuneme for help with tissue culture; and Saada Abdalla for technical assistance. This work was supported by grants from the National Institutes of Health (AI 090119) and Perot Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information Supplementary information including supplementary figures, tables and movies can be found with this article online at__________.
Reference
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- Appel N, Schaller T, Penin F, Bartenschlager R. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 2006;281:9833–9836. doi: 10.1074/jbc.R500026200. [DOI] [PubMed] [Google Scholar]
- Binder M, Kochs G, Bartenschlager R, Lohmann V. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology. 2007;46:1365–1374. doi: 10.1002/hep.21829. [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 2005;79:13963–13973. doi: 10.1128/JVI.79.22.13963-13973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of Site-1 Protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Dong X, Feng H, Sun Q, Li H, Wu TT, Sun R, Tibbetts SA, Chen ZJ, Feng P. Murine Gamma-Herpesvirus 68 hijacks MAVS and IKK+¦ to initiate lytic replication. PLoS Pathog. 2010;6:e1001001. doi: 10.1371/journal.ppat.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J. Cell Biol. 2010;191:479–492. doi: 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr., Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Sem. Cancer Biol. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, Iseki K, Wanaka A, Imaizumi K. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol. 2005;7:186–194. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS. Diminished hepatic response to fasting/refeeding and Liver X Receptor agonists in mice with selective deficiency of Sterol Regulatory Element-binding Protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- Liebermann DA, Hoffman B. Myeloid Differentiation (MyD)/Growth Arrest DNA Damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia. 2002;16:527–541. doi: 10.1038/sj.leu.2402477. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Martinon F, Glimcher LH. Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr. Opin. Immunol. 2010;23:1–6. doi: 10.1016/j.coi.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, DeFilippis V, Fruh K, Mason PW, Nikolich-Zugich J, Nelson JA. West Nile Virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J. Virol. 2007;81:10849–10860. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens F, Fletcher CDM, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, Reid R, Rosai J, Sciot R, Mandahl N, Panagopoulos I. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS//CREB3L1 fusion gene. Lab Invest. 2005;85:408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat. Rev. Micro. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Murakami T, Kondo S, Ogata M, Kanemoto S, Saito A, Wanaka A, Imaizumi K. Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J. Neurochem. 2006;96:1090–1100. doi: 10.1111/j.1471-4159.2005.03596.x. [DOI] [PubMed] [Google Scholar]
- Murakami T, Saito A, Hino S.i., Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat. Cell Biol. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigusa K, Imoto I, Tanikawa C, Aoyagi M, Ohno K, Nakamura Y, Inazawa J. RGC32, a novel p53-inducible gene, is located on centrosomes during mitosis and results in G2//M arrest. Oncogene. 2006;26:1110–1121. doi: 10.1038/sj.onc.1210148. [DOI] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. Virus-host cell interactions during hepatitis C virus RNA replication: Impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 2004;78:1513–1524. doi: 10.1128/JVI.78.3.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr., Wang C, Foy E, Loo YM, Gale M., Jr. Viral evolution and interferon resistance of Hepatitis C virus RNA replication in a cell culture model. J. Virol. 2004;78:11591–11604. doi: 10.1128/JVI.78.21.11591-11604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif KD, Mori K, Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 2002;76:7453–7459. doi: 10.1128/JVI.76.15.7453-7459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G, Antony J, Ostermann H, Dunham C, Hunt T, Halliday W, Maingat F, Urbanowski MD, Hobman T, Peeling J, Power C. West Nile Virus-induced neuroinflammation: glial infection and capsid protein-mediated neurovirulence. J. Virol. 2007;81:10933–10949. doi: 10.1128/JVI.02422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanki RN, Zhang L, Guney MA, Rocheleau JV, Gannon M, Volchuk A. OASIS/CREB3L1 induces expression of genes involved in extracellular matrix production but not classical endoplasmic reticulum stress response genes in pancreatic β-cells. Endocrinology. 2010;151:4146–4157. doi: 10.1210/en.2010-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von dem Bussche A, Machida R, Li K, Loevinsohn G, Khander A, Wang J, Wakita T, Wands JR, Li J. Hepatitis C virus NS2 protein triggers endoplasmic reticulum stress and suppresses its own viral replication. J. Hepatol. 2010;53:797–804. doi: 10.1016/j.jhep.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods. 2003;110:201–209. doi: 10.1016/s0166-0934(03)00134-4. [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gale J, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Davis CG, Brown MS, Schneider WJ, Casey ML, Goldstein JL, Russell DW. The human LDL receptor: A cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 1984;39:27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]
- Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease. Proc. Natl. Acad. Sci. USA. 2000;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Wang C, Sumpter R, Jr., Brown MS, Goldstein JL, Gale M., Jr. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA. 2003;100:15865–15870. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zein NN. Clinical significance of Hepatitis C Virus genotypes. Clin. Microbiol. Rev. 2000;13:223–235. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.