Abstract
Human interleukin-12 (IL-12, p70) is an early pro-inflammatory cytokine, comprising two disulfide-linked subunits, p35 and p40. We solved the crystal structures of monomeric human p40 at 2.5 Å and the human p70 complex at 2.8 Å resolution, which reveals that IL-12 is similar to class 1 cytokine–receptor complexes. They also include the first description of an N-terminal immunoglobulin-like domain, found on the p40 subunit. Several charged residues from p35 and p40 intercalate to form a unique interlocking topography, shown by mutagenesis to be critical for p70 formation. A central arginine residue from p35 projects into a deep pocket on p40, which may be an ideal target for a small molecule antagonist of IL-12 formation.
Keywords: cytokine–receptor complex/interleukin-12/structure–function/X-ray crystallography
Introduction
Interleukin (IL)-12 is a potent immunoregulatory cytokine crucial for the generation of cell-mediated immunity to intracellular pathogens (reviewed in Trinchieri, 1998). It is produced by antigen-presenting cells, including dendritic cells, macrophages/monocytes, neutrophils and some B cells (D’Andrea et al., 1992). Among the many biological activities of IL-12 are its abilities to enhance the cytotoxic activity of natural killer (NK) cells and cytotoxic T cells, to stimulate proliferation of activated NK and T cells and to induce production of interferon-γ (IFN-γ) by these cells (Stern et al., 1990; Wolf et al., 1991). IL-12 also plays an important role in immunomodulation by promoting cell-mediated immunity through induction of a class 1 T helper cell (Th1) immune response. Neutralization of IL-12 bioactivity has demonstrated its importance in defense against infection by intracellular bacteria and parasites (Trinchieri, 1998). In addition to its beneficial role in host immunity, IL-12 also contributes to various immunopathological conditions. For example, the severity of disease in IL-12-deficient mice is reduced in the collagen-induced model for rheumatoid arthritis (McIntyre et al., 1996). Therefore, modulating the biological activity of IL-12 may lead to new therapies for this and other autoimmune diseases.
Intact IL-12, also termed p70, is a heterodimeric glycoprotein consisting of 35 and 40 kDa subunits (Gubler et al., 1991; Wolf et al., 1991). It is the only cytokine known to function as a disulfide-linked heterodimer. The primary sequence of the 35 kDa subunit reveals homology to other class 1 cytokines, especially IL-6 and granulocyte-colony stimulating factor (G-CSF) (Merberg et al., 1992). Members of this family are characterized by a four-helix bundle topology. The p40 subunit is not homologous to known cytokines, but is similar in primary sequence to the extracellular domains of the IL-6 receptor α-chain (IL-6Rα) and the ciliary neurotrophic factor receptor (CNTFR), both hematopoietic cytokine receptor family members (Gearing and Cosman, 1991). Like IL-6Rα and CNTFR, p40 contains an N-terminal immunoglobulin (Ig) domain followed by two fibronectin type III domains. The first fibronectin domain contains four conserved cysteines while the second contains the consensus WSXWS box common to all family members. Many cytokine receptors (including IL-6Rα) can be released from cells in a soluble form (which lacks the transmembrane and short cytosolic domains), produced by proteolysis or alternative splicing. Soluble IL-6Rα can bind IL-6 in solution and this complex mediates IL-6 signaling events by binding to cells expressing gp130, the β-chain of the IL-6R signaling complex. Thus, IL-12 can be thought of as a soluble cytokine receptor α-chain (p40) covalently bound to its cytokine (p35), with its cell surface receptors (see below) serving an analogous function to gp130. Although p35 transcripts are found in many cell types, free p35 is not secreted (D’Andrea et al., 1992), suggesting that p35 is unstable in the absence of p40. Interestingly, free p40 is secreted, and in mice can function as an antagonist to IL-12 signaling, either as a monomer or as a disulfide-linked homodimer (Gillessen et al., 1995).
The actions of this p35–p40 heterodimer are mediated by binding to a transmembrane receptor comprised of two subunits (IL-12Rβ1 and IL-12Rβ2) (reviewed in Gately et al., 1998) that are highly homologous to gp130. Each subunit of the receptor is composed of an extracellular ligand-binding domain, a transmembrane domain and a cytosolic domain containing box 1 and box 2 sequences that mediate binding of Janus-family tyrosine kinases. IL-12 binding is believed to result in heterodimerization of β1 and β2 and the generation of a high-affinity receptor complex capable of signal transduction. In this model, dimerization of the receptor leads to juxtaposition of the cytosolic domains and the subsequent tyrosine phosphorylation and activation of the receptor-associated Janus-family kinases, Jak2 and Tyk-2. These activated kinases, in turn, tyrosine phosphorylate and activate several members of the signal transducer and activator of transcription (STAT) family (STAT-1, -3 and -4) (Bacon et al., 1995; Jacobson et al., 1995; Yu et al., 1996). The STATs translocate to the nucleus to activate transcription of several genes, including IFN-γ. The production of IFN-γ has a pleiotropic effect in the cell, stimulating production of molecules important to cell-mediated immunity. In particular, IFN-γ stimulates production of more IL-12 and sets up a positive regulation loop between IL-12 signaling and IFN-γ (Chan et al., 1991). The importance of IL-12 for this loop is demonstrated by IL-12 and STAT-4 knockout mice that are severely compromised in IFN-γ production (Kaplan et al., 1996; Magram et al., 1996).
To understand the unique structural nature of heterodimeric IL-12, we determined the crystal structures of both free (monomeric) p40 and intact IL-12 (p70). These reveal a novel disulfide-linked cytokine, consisting of a long-chain four-helix bundle (p35) bound to a soluble α-chain receptor subunit (p40). They also include an N-terminal Ig-like domain present in p40 and are the first description of this type of domain fold. The p35–p40 binding interface is characterized by an interlocking topography distinct from previous class 1 cytokine–receptor complexes. Mutagenesis of hydrophobic and hydrophilic contacts found at the p35–p40 interface confirms the importance of a central arginine residue that projects from p35 into a deep pocket in p40. Conformational changes in p40 occur upon p70 complex formation, which serve to optimize its interaction with this arginine. Our results indicate that the arginine-binding pocket on p40 may be an ideal target for a small molecule inhibitor of IL-12 formation.
Results
Structure determinations
Crystals of unliganded p40 were grown that diffracted to 2.5 Å, and the structure was solved with multiple isomorphous replacement using mercury- and platinum-derivatized crystals. The resulting experimental map of p40 was of high quality, giving clear density for the entire protein, except for the flexible loop regions 140–144 and 262–265, which were not modeled. The final refined p40 model has an R-factor of 22.7% (R-free 28.1%) (Table I).
Table I. Statistics for data collection, phase determination and refinement.
| Data collection | |||||
|---|---|---|---|---|---|
| p40 |
p70 |
||||
| native | Hg | Pt | native | Hg | |
| Resolution (Å) | 15.0–2.5 | 15.0–2.5 | 15.0–3.0 | 12.0–2.8 | 12.0–3.2 |
| High resolution shell | (2.59–2.50) | (2.59–2.50) | (3.11–3.0) | (2.90–2.80) | (3.31–3.20) |
| No. of crystals | 1 | 2 | 1 | 2 | 1 |
| Unique reflections | 12 537 | 12 656 | 7435 | 21 109 | 14 914 |
| <I/σ(I)> | 35 (7) | 36 (7) | 19 (5) | 25 (6) | 18 (4) |
| Redundancy | 7.2 | 10 | 5.4 | 5.4 | 4.1 |
| Completeness (%) | 97 (79) | 98 (84) | 98 (84) | 96 (96) | 99 (99) |
| Rmerge (%) | 5.4 (16.0) | 4.9 (15.2) | 7.8 (22.9) | 6.6 (34.8) | 7.8 (27.4) |
| Isomorphous differences I (%) |
|
25.1 |
26.3 |
|
15.1 |
| Phasing statisticsa | |||||
| No. of sites | 4 | 5 | 2 | ||
| Phasing power (centrics/acentrics) | |||||
| isomorphous | 1.8/2.4 | 1.3/1.6 | 0.79/0.83 | ||
| anomalous | –/2.3 | –/1.1 | –/0.48 | ||
| Combined figure of merit |
|
0.56/0.56 |
|
0.14/0.19 |
|
| Model refinementb | |||||
| |
|
p40 |
|
p70 |
|
| Resolution range | 15.0–2.5 | 15.0–2.8 | |||
| Refinement data completeness (%) | 95.2 | 95.6 | |||
| Ramachandran plot statistics | |||||
| residues in most favored regions (%) | 88.3 | 78.1 | |||
| residues in additional allowed regions (%) | 10.6 | 16.9 | |||
| residues in generously allowed regions (%) | 0.0 | 3.8 | |||
| residues in disallowed regions (%) | 1.1 | 1.1 | |||
| R-value (%) | 22.7 | 24.1 | |||
| Rfree (%) | 28.1 | 28.4 | |||
| R.m.s.d. (bonds) (Å) | 0.017 | 0.019 | |||
| R.m.s.d. (angles) (°) | 1.88 | 2.2 | |||
| <B-factor> (Å2) protein | 39 | 64 | |||
| R.m.s.d. B-factor main chain bonded atoms (Å2) | 2.2 | 2.2 | |||
aAs reported by SHARP (de La Fortelle and Bricogne, 1997).
bIncludes an anisotropic correction implemented in CNS (Brünger et al., 1998).
Using p40 as a search model, the structure of p70 was solved by molecular replacement with data that extended to 2.8 Å. p35 was subsequently built into solvent-flattened electron density phased by the p40 model and combined with weak phases from a single low-occupancy mercury heavy atom derivative. The structure was refined against 2.8 Å resolution data to an R-factor of 24.1% (Rfree 28.4%) (Table I).
p40 resembles a soluble class 1 cytokine α-receptor
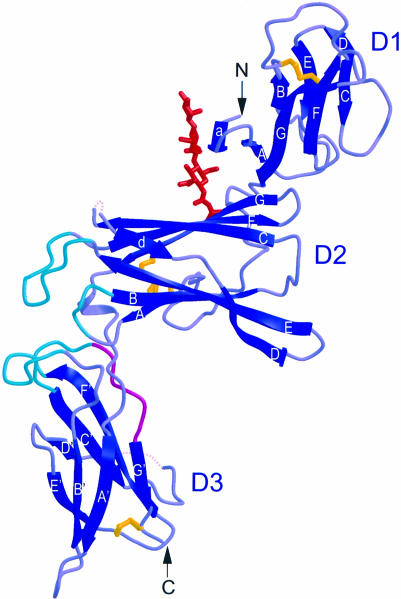
As predicted by sequence similarity, p40 indeed folds like the extracellular domain of other class 1 cytokine receptors, such as growth hormone receptor (GHR). It is an elongated molecule measuring ∼103 × 45 × 25 Å, with three domains, labeled D1, D2 and D3 in Figure 1. D1 folds in a topology most similar to the S-type Ig fold, as classified by Bork et al. (1994), except that strand A switches from the three-strand sheet to join the four-strand sheet. It was suggested that this type of strand switch would be possible, even though it had not been observed previously. This is the first structural description of an N-terminal Ig-like domain commonly present in class 1 cytokine receptors. The similarity of p40 to other class 1 cytokine receptors is apparent in the structures of D2 and D3.
Fig. 1. p40 resembles a soluble class 1 cytokine α-receptor. N- and C-termini are labeled N and C, respectively, and the three domains are labeled D1, D2 and D3. The intrachain disulfide bonds (yellow), an N-linked sugar modification (red) and the WSXWS motif (magenta) are shown. The p40 loops that contact p35 (turquoise) are located at the apex between D2 and D3. Flexible loop regions are indicated by dotted lines. This and all subsequent figures (except Figures 3 and 7) were made using MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997).
D2 adopts a variation of an Ig constant domain 4-on-3 β-sandwich, consistent with Bazan’s original proposal for the topology of cytokine-binding domains (Bazan, 1990). Four conserved cysteine residues form two inter-strand disulfide bonds, which bridge p40 strands A and B (C109–C120) and D and E (C148–C171). An N-linked sugar modification (GlcNAc-GlcNAc-mannose, where GlcNAc is N-acetylglucosamine) is present on D2 and extends from N200 across the D1–D2 interface, stabilizing the relative orientation of these domains through hydrogen bonds and van der Waals contacts.
The third domain, D3, superimposes quite well with the C-terminal cytokine-binding domains of other class 1 cytokine receptor structures, with the exception that p40 contains a unique disulfide bond, bridging strand E′ C278 and the penultimate C-terminal residue from strand G′, C305. It also contains the highly conserved WSXWS motif in strand G′ (WSEWAS in human p40). Structural analysis of a number of WSXWS motifs revealed that WSXWS is part of a β-bulge and π–cation stacking motif, in which positively charged side chains from the adjacent strand F′ stack between aromatic residues from the WSXWS sequence (de Vos et al., 1992; Somers et al., 1994; Livnah et al., 1996).
p70 resembles a soluble class 1 cytokine–receptor complex with a unique interlocking topography
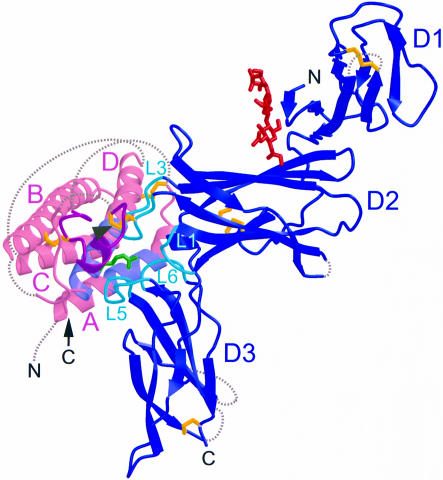
Our structural analysis confirms that IL-12/p70 is formed by the 1:1 complex of p35 and p40 and resembles a soluble class 1 cytokine–receptor complex (Figure 2). In this complex, the mature p35 subunit forms a four-helix bundle ∼60 Å long that adopts a fold characteristic of long-chain class 1 cytokines. In this fold, long loops connect the A and B helices and the C and D helices, permitting an up-up-down-down helical packing. As in other four-helix bundle structures, parts of these long loops are partially disordered. The 11 amino acids C-terminal to C63 are termed the disulfide bond loop (Figure 2). This region contains numerous residues that contact p40, including C74, which forms an interchain disulfide bond with C177 in p40 (Figure 2). Two intrachain disulfide bonds are ordered in this structure, between the A–B loop and helix D (C42–C174), and between the A–B loop and helix B (C63–C101) (Figure 2).
Fig. 2. p70 resembles a soluble class 1 cytokine–receptor complex. Ribbon diagram of the p70 complex: p35 (pink) with helices labeled A–D, p40 (blue) with domains labeled D1–D3, the interchain disulfide bond (yellow, arrowhead), intramolecular disulfide bonds (yellow), an N-linked glycosylation (red) and N- and C-termini labeled N and C. The structural epitope is also highlighted: residues from p40 loops 1, 3, 5 and 6 (turquoise) contact p35 residues from the AD helical face (lavender) and the disulfide bond loop (magenta). The p35 R189 side chain (green) extends from helix D and is buried within the p40 loops.
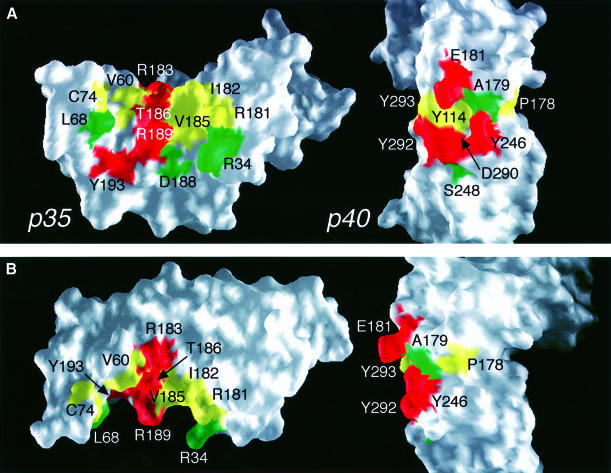
In the p70 complex, there is a large structural epitope between p35 and p40 of ∼1976 Å2 total buried surface area. p35 binds p40 through packing of the p35 AD helical face and disulfide bond loop against p40 loops 1 and 3 from D2 and loops 5 and 6 from D3 (Figure 2). Although p35 is juxtaposed to p40 similarly to growth hormone (GH) (de Vos et al., 1992) and erythropoietin (EPO) (Syed et al., 1998) bound to their receptors, the p35–p40 interface incorporates a much higher degree of polar contacts and forms a unique interlocking topography. Most striking is the binding of R189 from p35 into a deep pocket formed by p40 loops 1, 3, 5 and 6 (Figure 2). Interestingly, this pocket is lined by hydrophobic residues and contains the negatively charged D290 at its base (Figure 3A). Other side chains, such as R34 from p35 and E181 from p40, extend into the interface, in a manner reminiscent of the teeth of a zipper (Figure 3B). These protruding side chains are charged and most are critical to the binding interaction (see below). Figure 3B also shows the complementarity of the binding surfaces of the two subunits. The cupped shape of the p35 binding interface matches the elbow-like bend between D2 and D3 in p40.
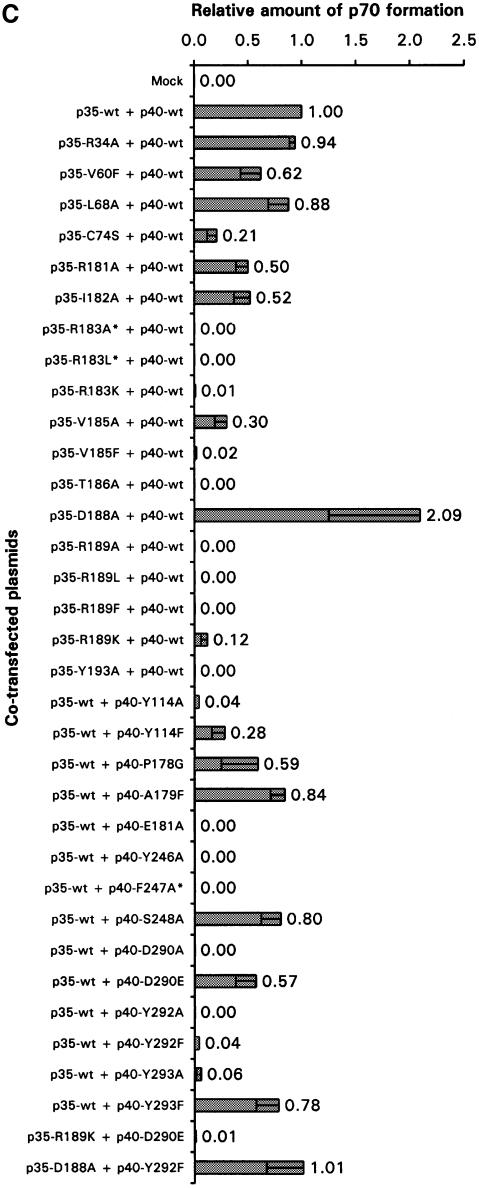
Fig. 3. Interlocking topography of the p35–p40 binding interface with residues important for p70 formation. (A) Open-book view of the p35–p40 interface with mutational effects mapped on the surface of the molecules. Most residues are either critical to (red) or have a partial effect on (yellow) p70 formation when mutated, while a few residues have no effect (green). The centrally located R189 in p35 binds a deep, hydrophobic pocket on p40 containing D290 at its base. (B) Orthogonal views of (A) highlight side chain extension into the interface. (A) and (B) were prepared with GRASP (Nicholls et al., 1991). (C) Mutants were analyzed by transient transfection and ELISA, and p70 formation is reported as a ratio to wild type. Bars represent the means of at least three independent transfections, with error bars indicating standard deviations. Three mutants were not expressed (*).
Identification of the functional epitope
To determine critical residues for the formation of this heterodimeric cytokine (functional epitope), we undertook mutagenesis of the structural epitope. Mutant plasmids were co-transfected into COS cells with a plasmid encoding the wild-type binding partner, and conditioned medium was assayed by ELISA for the p70 heterodimer. A summary of 18 mutations in p35 and 14 mutations in p40 and their effect on p70 formation is shown in Figure 3C. The amount of p70 reported is normalized to wild-type p70 expression levels from the same experiment.
Our hypothesis that R189 is a key binding determinant was borne out by our mutational analysis: R189 and most residues in p40 that comprise its binding pocket were critical to p70 formation (Figures 3 and 4A). First, when we changed R189 to alanine, no p70 was detected in the COS media. Similarly, neither the R189L mutant nor the R189F mutant could bind to p40. The R189K mutant, however, restored partial binding. These data suggest that R189 in p35 is critical, and in particular, its positive charge is essential.
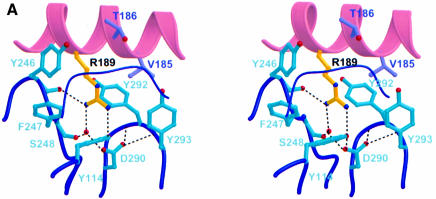
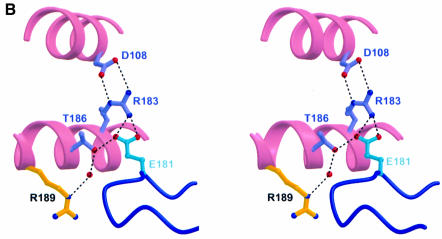
Fig. 4. Important charged interactions in the p35–p40 binding interface. (A) R189 (yellow) from p35 helix D projects into a deep, hydrophobic pocket composed of p40 loops 1, 5, 6 (thick) and 3 (thin). A buried hydrogen bonding network (dashes) connects R189 with D290 from p40 through a bound water (red sphere). (B) E181 in p40 is part of a second important charged interaction. Dashed lines indicate hydrogen bonding of p40 E181 with p35 residues R183 and T186. R183 is further stabilized by electrostatic interactions with p35 D108 in helix B.
The importance of charge in mediating interaction between p35 and p40 is further supported by our mutational analysis of D290 in p40. Buried at the base of the otherwise hydrophobic R189-binding pocket, D290 hydrogen bonds to the hydroxyl group of Y114 and a water molecule that coordinates R189 (Figure 4A). It also forms hydrogen bonds with two amide nitrogen atoms in loop 6 that stabilize the structure of the pocket. When D290 was mutated to alanine, no p70 formation was detected. The D290E mutant, however, displayed partial binding to p35, demonstrating that the charged nature of the interaction between R189 and D290 is important for binding of p35 and p40. Interestingly, we found that most of the other p40 residues forming this deep binding pocket are also essential for p70 formation (Y114, Y246, Y292 and Y293).
In addition to the R189–D290 charged pair, the importance of a second charged interaction involving E181 in p40 loop 3 was highlighted by mutagenesis analysis. The side chain oxygens of E181 form three hydrogen bonds with p35 residues R183 and T186 (Figure 4B). The significance of this particular hydrogen bonding network is supported by mutation of these residues (E181A, R183K and T186A). The hydrophobic interactions of E181 with p35 residues V60 and I182 were not as important, since V60F and I182A could bind p40, albeit at a reduced level. Indeed, mutations in most residues involved in hydrophobic interactions had only partial effects on p70 formation residues (p40 P178G, A179F and p35 V60F, L68A). Thus, we conclude that rather than hydrophobic interactions, charged interactions dominate the formation of p70 and are centered around the R189 side chain buried in a deep hydrophobic pocket on p40.
The interchain disulfide bond is not required for binding
As mentioned earlier, the disulfide bond loop in p35 contains C74, which disulfide bonds to C177 in p40 (Figure 2). This interchain disulfide bond presumably forms in the endoplasmic reticulum and the two subunits are secreted as a covalent complex. We mutated C74 in p35 to a serine and found that intact IL-12 was formed and secreted into the conditioned medium (Figure 3C). These results indicate that the interchain disulfide bond is not necessary for the formation and secretion of IL-12, but instead ensures the stable association of these subunits.
Conformational changes in p40 upon binding of p35
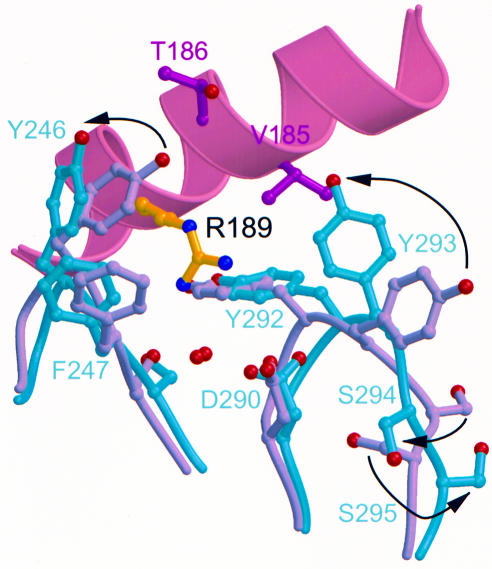
The crystal structures for both free and liganded p40 enabled determination of the conformational changes in this receptor-like subunit upon complex formation. After binding of p35, p40 D3 rotates 14.5° with respect to D2 (not shown), which is similar to changes between the unliganded and liganded forms of the EPO-binding protein (EBP) (Livnah et al., 1999). Furthermore, internal conformational changes occur in p40 loops 3, 5 and 6, which optimize interactions with p35 R189, and involve many residues determined by mutagenesis as critical for p70 formation (Figure 5). For example, rotation of the D290 side chain may serve to anchor the R189 hydrogen bonding network and to stabilize changes in other loop 6 residues, Y293, S294, S295 and S296. It is noted that crystal contacts in the p40 structure in this region may account for some of these differences. However, the interactions seen in the p35–p40 interface are far more extensive than those seen in the p40 crystal lattice.
Fig. 5. Conformational changes in p40 upon p70 complex formation optimize interactions with p35 R189. Internal conformational changes in D3 loops 5 and 6 are depicted as occurring after the D3 domain rotation described in Results. Overlay of unliganded p40 (gray) with p40 bound to p35 (turquoise) highlights movements of individual p40 side chains (arrows). Sixty-seven equivalent Cα atoms from the D3 domain of p40 and p70 were overlayed using lsqkab (CCP4, 1994) to an r.m.s.d. of 0.65 Å. Residues from loop 5 (240–249) and loop 6 (289–297) were excluded from the overlay.
Discussion
The importance of IL-12 in the generation of a Th1-type immune response and its overexpression in various autoimmune diseases make it an attractive therapeutic target. As a first step toward developing small molecule inhibitors of IL-12 formation, we performed a detailed structure–function analysis of this heterodimeric molecule. Solving the crystal structure of IL-12 revealed that although p70 is structurally similar to other class 1 cytokine–receptor complexes, it is distinguished by its unique interlocking topography. Mutating most residues in this structural epitope resulted in partial or complete abolition of p70 complex formation. Our analysis indicates that interactions centered around p35 R189 are critical and may be a good drug target.
Comparison with other structures
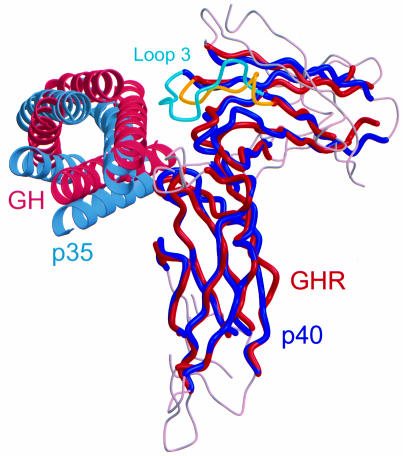
The crystal structure of IL-12 confirmed that this heterodimeric cytokine is similar to a class 1 cytokine–receptor complex in its overall architecture. The cupped epitope on p35 wraps around the receptor loops, as is seen in other complexes. Furthermore, the arrangement of p35 relative to p40 resembles the binding of GH to GHR (de Vos et al., 1992) (Figure 6). However, due to the increased size and prominence of p40 loop 3 compared with the equivalent loop in GHR, p35 shifts ∼5 Å towards p40 D3 relative to GH. The same is true when comparing p40 loop 3 with the equivalent loop found in prolactin receptor (PRLR) (Somers et al., 1994) and EBP (Syed et al., 1998) as well.
Fig. 6. Optimal superposition of IL-12 with growth hormone–growth hormone receptor (GH–GHR) complex. Equivalent Cα atoms are colored blue (p40) and red (GHR), whereas non-equivalent regions in p40 and GHR are gray. p35 (light blue) shifts ∼5 Å toward p40 D3 relative to GH (pink), due to the increased size of p40 loop 3 (turquoise) compared with the equivalent loop in GHR (yellow). Inter-helical loops in p35 and GH are omitted for clarity. The overlay was initiated by manual placement of the high-affinity portion of 1:2 GH–GHR (PDB code 3hhr) onto p70 at the D2–D3 helical linker, then optimized and equivalent pairs defined by LSQMAN (Kleywegt and Jones, 1994). Pairs of Cα atoms were included if their separation was <3.5 Å and they formed a contiguous stretch of at least five residues. A total of 129 residue pairs match to an overall r.m.s.d. of 1.9 Å.
The most striking difference, however, is the unique zipper-like binding interface found in IL-12. In the GH–GHR complex, two key tryptophan residues (Clackson and Wells, 1995) pack against one another and a nearby arginine, and this triad protrudes into an elongated groove on the hormone. In IL-12, on the other hand, the invagination is formed in this area on the ‘receptor-side’ instead, through the clustering of the p40 loops around the deeply embedded p35 R189. Other residues, such as p35 R34 and p40 E181, protrude into the interface and complete the interlocking topography.
Although this study did not include binding analysis of each mutant, one would predict that the R189 region will comprise a ‘hot spot’ of binding energy (Clackson and Wells, 1995). The salient features of hot spots are characteristic of the R189 region in IL-12: location at the center of the binding interface allowing exclusion of bulk solvent, complementarity between the binding partners and the high occurrence of tyrosine and arginine due to their ability to make multiple types of favorable interactions (Bogan and Thorn, 1998).
The importance of charged residues and polar contacts in the p35–p40 binding interaction is similar to results obtained in studies on G-CSF and IL-4 binding their respective receptors (Layton et al., 1997; Wang et al., 1997; Young et al., 1997). In both cases, the polar residues identified by mutagenesis formed critical contacts in the cytokine–receptor complexes (Aritomi et al., 1999; Hage et al., 1999). However, that charged residues are important to cytokine–receptor interactions may be unexpected due to earlier studies on the GH–GHR and EPO–EBP complexes, which showed that the majority of the binding energy can be attributed to a few hydrophobic residues (Cunningham and Wells, 1993; Clackson and Wells, 1995; Middleton et al., 1996). Based on these data, two distinct situations seem to exist for cytokine–receptor binding: one dominated by hydrophobic residues, as for GH and EPO, and a second dominated by charged and polar contacts, as for IL-4, G-CSF and now IL-12. Moreover, the p35–p40 interface in IL-12 may be further distinguished from these complexes by its unique interlocking topography, containing a charged residue buried in a deep hydrophobic pocket, and defines a novel mode of cytokine–receptor recognition.
The role of the interchain disulfide bond in the formation of IL-12
Our analysis of the p35 C74S mutant is the first direct evidence that the interchain disulfide bond is not required for IL-12 formation or secretion. This possibility had been suggested by experiments on EBI3, a p40 homolog induced in B cells upon Epstein–Barr virus infection. EBI3 can form a heterodimer with p35 in co-transfection experiments even though it lacks a p40 C177 disulfide-bonding equivalent (Devergne et al., 1997). The IL-12 disulfide bond probably evolved due to the inherent instability of the p35 subunit, since p35 protein never appears to be secreted in the absence of p40 even though its transcripts are present in many cell types (D’Andrea et al., 1992). Other interchain disulfide bonds not required for oligomerization include those in chicken acetylcholinesterase (Gough and Randall, 1995) and platelet-derived growth factor-B (PDGF-B) (Prestrelski et al., 1994). In PDGF-B, the interchain disulfide bond was required for protein stability and resistance to denaturation. Similarly, we might expect the disulfide bond in IL-12 to function as a tether, ensuring stability of the heterodimer.
IL-12 signaling model
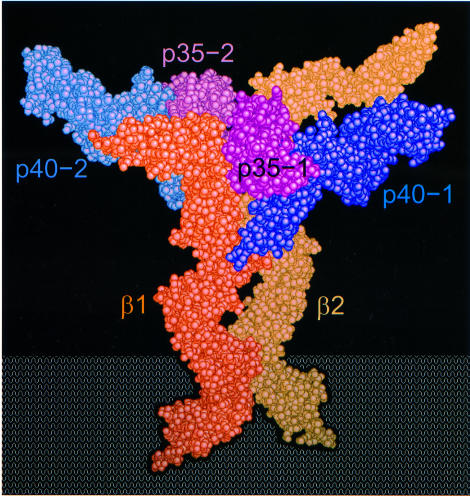
Both IL-12R subunits are required for efficient signaling and together form a high-affinity signaling complex when bound to IL-12 (Gately et al., 1998). The structural and sequence similarity of IL-12 with the IL-6/gp130 system suggests that IL-12 signaling may be mediated by a hexameric complex similar to that proposed for IL-6 (Ward et al., 1994; Paonessa et al., 1995). In our model, either the IL-12Rβ1 or the IL-12Rβ2 would bind to p70 along the p35 AC helical face. Two of these complexes would then interact to form a pseudo-symmetric heteroligomer containing two p70s, and one each of the IL-12Rβ1 and IL-12Rβ2 subunits (Figure 7). Binding of the initial p70 to IL-12Rβ1 may cause conformational changes that might enhance the affinity of the resulting complex for an incoming p70–IL-12Rβ2 complex. Alternatively, the first p70–IL-12Rβ1 complex may bind the second p70, and the resulting complex would then present a preformed binding site with high affinity for IL-12Rβ2. We expect this complex would be further stabilized by interactions between IL-12Rβ1 and p40 (p40-2 in Figure 7), since it has been observed that p40 has a measurable affinity for IL-12Rβ1 (Y.Zhang and J.F.Tobin, unpublished data; Presky et al., 1998). In either scenario, IL-12Rβ1 would act as an affinity converter to facilitate formation of a higher affinity IL-12 hexameric signaling complex. Further experiments, including mutagenesis of residues in the p35 AC helical face and identification of partially formed signaling complexes, will have to be conducted to determine the validity of this model.
Fig. 7. Hexameric model of the IL-12 signaling complex. The model is composed of two p70 heterodimers juxtaposed in ∼2-fold symmetry bound to one each of IL-12Rβ1 and IL-12Rβ2 extracellular domains in a position analogous to the low-affinity binding sites on GHR and EBP. The IL-12Rβ1 and IL-12Rβ2 p70-binding regions were modeled using gp130 coordinates (PDB code 1BQU). The other domains in IL-12Rβ1and IL-12Rβ2 were made by duplication and manual placement of fibronectin domains from gp130, with the exception of an N-terminal Ig-like domain in IL-12Rβ2, which was constructed using p40 D1. This figure was made using QUANTA and GRASP (Nicholls et al., 1991).
Designing a small molecule inhibitor of IL-12 formation
The heterodimeric nature of IL-12 offers a unique opportunity to develop a small molecule inhibitor of cytokine formation as a potential therapeutic. Because the R189 interaction is a key determinant in p70 formation and requires burying charge within a deep hydrophobic pocket, the R189-binding site on p40 may be an ideal target for a small molecule drug. Interestingly, an arginine from a neighboring p40 molecule binds to this pocket in the unliganded p40 crystal lattice, much like R189 from p35. Since there are two unrelated examples of arginine binding in this site, one could imagine screening for arginine mimetics that bind this pocket. Optimizing interactions of the small molecule with other nearby residues in p40 would also benefit from consideration of the unliganded and liganded structures of p40. We would expect that the R189-binding site on p40 may be amenable to small molecule binding, since both GHR and EBP are able to use similar regions to bind multiple ligands (Livnah et al., 1996; Wells and de Vos, 1996; Syed et al., 1998). Finally, mutagenesis of the interface between p35 and the IL-12R subunits to test the validity of the hexameric model described above may reveal additional drug targets.
Materials and methods
Purification and crystallization
Recombinant p40 was expressed in Chinese hamster ovary (CHO) cells and purified using a combination of anion-exchange, hydrophobic and size exclusion chromatography. The protein was treated with PNGaseF, and to prevent aggregation, the two free cysteines in p40, C177 and C252, were modified with 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB). For crystallization, p40 protein (4 mg/ml in 10 mM Tris pH 8.0) was combined with an equal volume of well solution (18% PEG 8000, 0.2 M calcium acetate and 0.1 M sodium cacodylate pH 6.5) at 18°C in a hanging drop vapor diffusion experiment. These crystals, measuring up to 0.3 × 0.3 × 0.1 mm, were stabilized in a solution of 18% PEG 8000, 0.2 M calcium acetate, 0.1 M sodium cacodylate pH 6.5, 10 mM Tris pH 8.0 and 18% ethyleneglycol, and then rapidly cooled by plunging into liquid propane at close to liquid nitrogen temperatures. The p40 crystals belong to the space group P212121 with the cell dimensions a = 34.0, b = 55.0, c = 189.2 Å and have a single molecule in the asymmetric unit.
p70 was also expressed in CHO cells and purified by using Toyopearl Q, CM Sepharose, phenyl, heparin, mono Q and size exclusion columns. A variety of endoglycosidases (PNGaseF and O-glycosidase) and exoglycosidases (neuraminidase, β-galactosidase, α1-2,3-mannosidase, α1-2,3-galactosidase and α1-2-fucosidase) were used for partial deglycosylation during the purification process, which appeared primarily to affect the p35 subunit as analyzed by SDS–PAGE. p70 protein (10 mg/ml in 10 mM Tris pH 7.4 and 100 mM sodium chloride) was also crystallized in hanging drops at 18°C by combining it with an equal volume of well solution [12% (v/v) 2-methyl-2,4-pentanediol (MPD), 550 mM sodium chloride and 100 mM sodium acetate pH 5.0]. Crystals measuring up to 0.25 × 0.25 × 0.15 mm were stabilized in 13% MPD, 600 mM sodium chloride, 100 mM sodium acetate and 5% glycerol, and cooled to liquid nitrogen temperatures by plunging into liquid propane. The p70 crystals belong to the space group C2221, a = 112.9, b = 154.2, c = 101.7 Å, with one molecule per asymmetric unit.
Data collection and processing
All p40 data were collected using a Rigaku R-axis II image plate on an RU-200 X-ray generator with mirror focusing optics on crystals maintained at –170°C. The diffraction was anisotropic, extending to 2.3 Å along the c axis but only 2.7 Å along a and b. Heavy atom derivative data were collected on crystals soaked in 2 mM mersalyl acid for 2 days and on a platinum derivative using 1 mM K2PtCl4.
All data for the p70 crystals were collected at beamline X4A at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. These crystals suffered from radiation decay so data from two native p70 crystals were merged into one data set extending to 2.8 Å resolution. A heavy atom derivative data set was collected at a wavelength of 0.99987 Å from a crystal soaked in 2.5 mM mercury acetate for 2 days. All p40 and p70 data were processed and scaled using DENZO/SCALEPACK (Otwinowski, 1993).
Structure determination and refinement
The p40 structure was solved using multiple isomorphous replacement. Heavy atom sites for mercury and platinum data sets were identified by Patterson and difference Fourier analysis, initially refined with MLPHARE (CCP4, 1994), then further refined in SHARP (de La Fortelle and Bricogne, 1997). Two isomorphous difference peaks were present from the mersalyl acid derivative, which, unlike the Hg sites, lacked equivalent peaks in anomalous difference Fourier maps; these sites were refined as sodium. In total, four Hg, five Pt and two Na sites were refined in SHARP, which, after solvent flattening of the electron density with Solomon (CCP4, 1994), resulted in maps with continuous electron density for most of p40. The p40 model was built using QUANTA (Biosym/Molecular Simulations) and refined with XPLOR (Brünger et al., 1987), then with CNS (Brünger et al., 1998). The progress of refinement was monitored using an Rfree set (Brünger, 1992) constructed by a random selection of 5% of the reflection data. Owing to the severe anisotropy, reflections with an I/σ(I) of <2 were not used in the refinement. The final p40 model has an R-factor of 22.7% (Rfree 28.1%), reasonable stereochemistry and geometry (r.m.s.d.: bonds 0.017 Å, angles 1.88°) with 88% of residues in the most favored region of the Ramachandran plot (PROCHECK; Laskowski et al., 1993) and an average B-factor of 39 Å2 (Table I). The final model contains the residues 1–139, 145–261 and 266–306, 57 water molecules and one carbohydrate comprised of two GlcNAc moieties and one mannose moiety. Both DTNB cysteine modifications were visible and included in the final model. The side chains of K99, E100, E102, N103, K104, K135, K225 and R279 were not observed and were therefore modeled as alanine.
p70 was solved by molecular replacement using AMoRe (Navaza, 1994). Model-phased, SIGMAA-weighted (CCP4, 1994) electron density maps were solvent flattened in DM (Cowtan, 1994) and revealed electron density adjacent to p40 consistent with a four-helix bundle. These maps were used for initial model building although a heavy atom data set was subsequently collected from crystals soaked in mercury acetate. The model-phased difference Fourier indicated a mercury site adjacent to the only free cysteine and was also consistent with observed difference Patterson peaks. The mercury-phased electron density maps were poor but showed features consistent with p70. Iterative rebuilding continued using phase-combined maps. Using similar techniques to the p40 refinement, the final p70 refinement was completed using all the data to 2.8 Å and resulted in an R-factor of 24.1% (Rfree 28.4%) (Table I). The p70 model has reasonable stereochemistry and geometry (r.m.s.d.: bonds 0.019 Å, angles 2.2°) with 78% of residues found in the most favored region of the Ramachandran plot (PROCHECK; Laskowski et al., 1993) and an average B-factor of 64 Å2. The model contains p40 residues 1–68, 76–156, 164–256, 264–279 and 283–306, two GlcNAc moieties and one mannose moiety, p35 residues 20–42, 58–74, 79–86, 95–148 and 167–197, and 27 water molecules. The side chains of E33, K99, E100, E102, K135 and R279 were modeled as alanine.
Atomic coordinates
Atomic coordinates for p40 (PDB code 1F42) and p70 (PDB code 1F45) have been deposited in the Protein Data Bank.
COS transfection and ELISA
Site-directed mutants were generated by PCR in pEMC (Kaufman et al., 1991) and sequenced. p35 was generated as IgG1 heavy chain (Fc) chimeras to aid in secretion and detection of this subunit. The presence of the Fc did not interfere with p35 as indicated by the ability of p35–Fc to bind to p40 using BIA-CORE (Pharmacia) (data not shown).
Each plasmid (4 µg) (p35–Fc + p40) was transfected into COS M6 cells using LipofectAmine (Life Technologies) according to the manufacturer’s instructions. Cells were incubated at 37°C for 65 h. Conditioned media were assayed for p70 formation by ELISA using mouse anti-human IL-12/p70 monoclonal antibody (mAb) (MAB611; R&D Systems) for capture and biotinylated mouse anti-human p40 mAb (C11.5.14; gift of Denise O’Hara, Wyeth Research) and alkaline phosphatase-conjugated streptavidin (Caltag) for detection. Each mutant was tested at least three times and results were normalized to wild-type p70 levels in the same experiment. The range in wild-type p70 formation varied from 0.6 to 4.9 µg/ml for 10 ml of media collected per 10 cm plate. Control experiments confirmed that the relative ratio (mutant to wild type) remains constant over this range of expression (data not shown), thus comparisons could be made between samples tested on different days. Co-culturing cells transfected with each subunit separately indicated that the amount of p70 detected in the conditioned medium was produced by secretion of the heterodimer rather than association of the two subunits outside the cell (data not shown).
Subunit-specific ELISAs confirmed that mutant proteins were translated and secreted properly. For the p35 ELISA, the presence of the Fc was assayed using goat anti-human IgG polyclonal antibody (Southern Biotechnology) for capture and alkaline phosphatase-conjugated goat anti-human IgG polyclonal antibody (Southern Biotechnology) for detection. For the p40 ELISA, mouse anti-human p40 mAb (C11.79.15; gift of Denise O’Hara, Wyeth Research) was used for capture and biotinylated mouse anti-human p40 mAb (C11.5.14) and alkaline phosphatase-conjugated streptavidin for detection. Three of the 34 mutants generated were not expressed (p35-R183A, -R183L and p40-F247A).
Acknowledgments
Acknowledgements
We thank Brian Hubbard and Jeffrey Robinson for protein expression and purification help, Craig Ogata for assistance with crystallographic data collection, Gérard Bricogne and Pietro Roversi for help with SHARP/BUSTER, Susan Benard for sequencing, and Denise O’Hara for the p40 antibodies. We are grateful to Deirdre Crommie and David Erbe for helpful comments on the manuscript.
References
- Aritomi M., Kunishima,N., Okamoto,T., Kuroki,R., Ota,Y. and Morikawa,K. (1999) Atomic structure of the GCSF–receptor complex showing a new cytokine–receptor recognition scheme. Nature, 401, 713–717. [DOI] [PubMed] [Google Scholar]
- Bacon C.M., Petricoin,E.F.,III, Ortaldo,J.R., Rees,R.C., Larner,A.C., Johnston,J.A. and O’Shea,J.J. (1995) Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc. Natl Acad. Sci. USA, 92, 7307–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J.F. (1990) Structural design and molecular evolution of a cytokine receptor superfamily. Proc. Natl Acad. Sci. USA, 87, 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan A.A. and Thorn,K.S. (1998) Anatomy of hot spots in protein interfaces. J. Mol. Biol., 280, 1–9. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm,L. and Sander,C. (1994) The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol., 242, 309–320. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–474. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Kuriyan,J. and Karplus,M. (1987) Crystallographic R factor refinement by molecular dynamics. Science, 235, 458–460. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chan S.H. et al. (1991) Induction of interferon γ production by natural killer cell stimulatory factor: characterization of the responder cells and synergy with other inducers. J. Exp. Med., 173, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T. and Wells,J.A. (1995) A hot spot of binding energy in a hormone–receptor interface. Science, 267, 383–386. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K.D. (1994) ‘dm’: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Cunningham B.C. and Wells,J.A. (1993) Comparison of a structural and a functional epitope. J. Mol. Biol., 234, 554–563. [DOI] [PubMed] [Google Scholar]
- D’Andrea A. et al. (1992) Production of natural killer cell stimulatory factor (interleukin 12) by peripheral blood mononuclear cells. J. Exp. Med., 176, 1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Devergne O., Birkenbach,M. and Kieff,E. (1997) Epstein–Barr virus-induced gene 3 and the p35 subunit of interleukin 12 form a novel heterodimeric hematopoietin. Proc. Natl Acad. Sci. USA, 94, 12041–12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos A.M., Ultsch,M. and Kossiakoff,A.A. (1992) Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science, 255, 306–312. [DOI] [PubMed] [Google Scholar]
- Gately M.K., Renzetti,L.M., Magram,J., Stern,A.S., Adorini,L., Gubler,U. and Presky,D.H. (1998) The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu. Rev. Immunol., 16, 495–521. [DOI] [PubMed] [Google Scholar]
- Gearing D.P. and Cosman,D. (1991) Homology of the p40 subunit of natural killer cell stimulatory factor (NKSF) with the extracellular domain of the interleukin-6 receptor. Cell, 66, 9–10. [DOI] [PubMed] [Google Scholar]
- Gillessen S. et al. (1995) Mouse interleukin-12 (IL-12) p40 homodimer: a potent IL-12 antagonist. Eur. J. Immunol., 25, 200–206. [DOI] [PubMed] [Google Scholar]
- Gough N.R. and Randall,W.R. (1995) Oligomerization of chicken acetylcholinesterase does not require intersubunit disulfide bonds. J. Neurochem., 65, 2734–2741. [DOI] [PubMed] [Google Scholar]
- Gubler U. et al. (1991) Coexpression of two distinct genes is required to generate secreted bioactive cytotoxic lymphocyte maturation factor. Proc. Natl Acad. Sci. USA, 88, 4143–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage T., Sebald,W. and Reinemer,P. (1999) Crystal structure of the interleukin-4/receptor α chain complex reveals a mosaic binding interface. Cell, 97, 271–281. [DOI] [PubMed] [Google Scholar]
- Jacobson N.G., Szabo,S.J., Weber-Nordt,R.M., Zhong,Z., Schreiber,R.D., Darnell,J.E.,Jr and Murphy,K.M. (1995) Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med., 181, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.H., Sun,Y.L., Hoey,T. and Grusby,M.J. (1996) Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature, 382, 174–177. [DOI] [PubMed] [Google Scholar]
- Kaufman R.J., Davies,M.V., Wasley,L.C. and Michnick,D. (1991) Improved vectors for stable expression of foreign genes in mammalian cells by use of the untranslated leader sequence from EMC virus. Nucleic Acids Res., 19, 4485–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) A super position. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 9–14. [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Layton J.E., Iaria,J., Smith,D.K. and Treutlein,H.R. (1997) Identification of a ligand-binding site on the granulocyte colony-stimulating factor receptor by molecular modeling and mutagenesis. J. Biol. Chem., 272, 29735–29741. [DOI] [PubMed] [Google Scholar]
- Livnah O., Stura,E.A., Johnson,D.L., Middleton,S.A., Mulcahy,L.S., Wrighton,N.C., Dower,W.J., Jolliffe,L.K. and Wilson,I.A. (1996) Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 Å. Science, 273, 464–471. [DOI] [PubMed] [Google Scholar]
- Livnah O., Stura,E.A., Middleton,S.A., Johnson,D.L., Jolliffe,L.K. and Wilson,I.A. (1999) Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science, 283, 987–990. [DOI] [PubMed] [Google Scholar]
- Magram J. et al. (1996) IL-12-deficient mice are defective in IFN γ production and type 1 cytokine responses. Immunity, 4, 471–481. [DOI] [PubMed] [Google Scholar]
- McIntyre K.W., Shuster,D.J., Gillooly,K.M., Warrier,R.R., Connaughton,S.E., Hall,L.B., Arp,L.H., Gately,M.K. and Magram,J. (1996) Reduced incidence and severity of collagen-induced arthritis in interleukin-12-deficient mice. Eur. J. Immunol., 26, 2933–2938. [DOI] [PubMed] [Google Scholar]
- Merberg D.M., Wolf,S.F. and Clark,S.C. (1992) Sequence similarity between NKSF and the IL-6/G-CSF family. Immunol. Today, 13, 77–78. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Middleton S.A., Johnson,D.L., Jin,R., McMahon,F.J., Collins,A., Tullai,J., Gruninger,R.H., Jolliffe,L.K. and Mulcahy,L.S. (1996) Identification of a critical ligand binding determinant of the human erythropoietin receptor. Evidence for common ligand binding motifs in the cytokine receptor family. J. Biol. Chem., 271, 14045–14054. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) Oscillation data reduction program. In Sawyer,L., Isaacs,N. and Bailey,S. (eds), Data Collection and Processing: Proceedings of the CCP4 Study Weekend. SERC Daresbury Laboratory, Warrington, UK. [Google Scholar]
- Paonessa G., Graziani,R., De Serio,A., Savino,R., Ciapponi,L., Lahm,A., Salvati,A.L., Toniatti,C. and Ciliberto,G. (1995) Two distinct and independent sites on IL-6 trigger gp 130 dimer formation and signalling. EMBO J., 14, 1942–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presky D.H., Minetti,L.J., Gillessen,S., Wilkinson,V.L., Wu,C.Y., Gubler,U., Chizzonite,R. and Gately,M.K. (1998) Analysis of the multiple interactions between IL-12 and the high affinity IL-12 receptor complex. J. Immunol., 160, 2174–2179. [PubMed] [Google Scholar]
- Prestrelski S.J., Arakawa,T., Duker,K., Kenney,W.C. and Narhi,L.O. (1994) The conformational stability of a non-covalent dimer of a platelet-derived growth factor-B mutant lacking the two cysteines involved in interchain disulfide bonds. Int. J. Pept. Protein Res., 44, 357–363. [DOI] [PubMed] [Google Scholar]
- Somers W., Ultsch,M., De Vos,A.M. and Kossiakoff,A.A. (1994) The X-ray structure of a growth hormone–prolactin receptor complex. Nature, 372, 478–481. [DOI] [PubMed] [Google Scholar]
- Stern A.S. et al. (1990) Purification to homogeneity and partial characterization of cytotoxic lymphocyte maturation factor from human B-lymphoblastoid cells. Proc. Natl Acad. Sci. USA, 87, 6808–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed R.S. et al. (1998) Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature, 395, 511–516. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. (1998) Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv. Immunol., 70, 83–243. [DOI] [PubMed] [Google Scholar]
- Wang Y., Shen,B.J. and Sebald,W. (1997) A mixed-charge pair in human interleukin 4 dominates high-affinity interaction with the receptor α chain. Proc. Natl Acad. Sci. USA, 94, 1657–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L.D., Howlett,G.J., Discolo,G., Yasukawa,K., Hammacher,A., Moritz,R.L. and Simpson,R.J. (1994) High affinity interleukin-6 receptor is a hexameric complex consisting of two molecules each of interleukin-6, interleukin-6 receptor and gp-130. J. Biol. Chem., 269, 23286–23289. [PubMed] [Google Scholar]
- Wells J.A. and de Vos,A.M. (1996) Hematopoietic receptor complexes. Annu. Rev. Biochem., 65, 609–634. [DOI] [PubMed] [Google Scholar]
- Wolf S.F. et al. (1991) Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. J. Immunol., 146, 3074–3081. [PubMed] [Google Scholar]
- Young D.C., Zhan,H., Cheng,Q.L., Hou,J. and Matthews,D.J. (1997) Characterization of the receptor binding determinants of granulocyte colony stimulating factor. Protein Sci., 6, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.R., Lin,J.X., Fink,D.W., Akira,S., Bloom,E.T. and Yamauchi,A. (1996) Differential utilization of Janus kinase-signal transducer activator of transcription signaling pathways in the stimulation of human natural killer cells by IL-2, IL-12 and IFN-α. J. Immunol., 157, 126–137. [PubMed] [Google Scholar]