Abstract
Development of cheap, high-throughput, and reliable gene synthesis methods will broadly stimulate progress in biology and biotechnology1. Currently, the reliance on column-synthesized oligonucleotides as a source of DNA limits further cost reductions in gene synthesis2. Oligonucleotides from DNA microchips can reduce costs by at least an order of magnitude3,4,5, yet efforts to scale their use have been largely unsuccessful due to the high error rates and complexity of the oligonucleotide mixtures. Here we use high-fidelity DNA microchips, selective oligonucleotide pool amplification, optimized gene assembly protocols, and enzymatic error correction to develop a highly parallel gene synthesis platform. We tested our platform by assembling 47 genes, including 42 challenging therapeutic antibody sequences, encoding a total of ~35 kilo-basepairs of DNA. These assemblies were performed from a complex background containing 13,000 oligonucleotides encoding ~2.5 megabases of DNA, which is at least 50 times larger than previously published attempts.
The synthesis of DNA encoding regulatory elements, genes, pathways, and entire genomes provides powerful ways to both test biological hypotheses as well as harness biology for our use. For example, since the use of oligonucleotides in deciphering the genetic code6,7 to the recent complete synthesis of a viable bacterial genome8, DNA synthesis has engendered tremendous progress in biology. Currently, almost all DNA synthesis currently relies on the use of phosphoramidite chemistry on controlled-pore glass (CPG) substrates. The synthesis of gene-sized fragments (500–5,000 basepairs) relies on assembling many CPG-oligonucleotides together using a variety of techniques termed gene synthesis2. Technologies to assemble verified gene-sized fragments into much larger synthetic constructs are now fairly mature8,9,10,11,12.
The price of gene synthesis has reduced drastically over the last decade. However, the current commercial price of gene synthesis, ~$0.40–1.00/basepair (bp), has begun to approach the relatively stable cost of the CPG oligonucleotide precursors (~$0.10–0.20/bp)1. At these prices, the construction of large gene libraries and synthetic genomes is out of reach to most. To achieve further cost reductions, many ongoing efforts focus on reducing the cost of the oligonucleotide precursors. For example, microfluidic oligonucleotide synthesis can reduce reagent cost by an order of magnitude and has been used for proof-of-concept gene synthesis13.
Another promising route is to harness existing DNA microchips, which can produce up to a million different oligonucleotides on a single chip, as a source of DNA. Previous efforts have demonstrated the ability to synthesize genes from DNA microchips3,4,5,14. These approaches have thus far failed to scale for at least three reasons. First, the error rates of chip-based oligonucleotides from DNA microchips are higher than traditional column-synthesized oligonucleotides. Second, the assembly of gene fragments becomes increasingly difficult as the diversity of the oligonucleotide mixture becomes larger. Finally, the potential for cross-hybridization between individual assemblies imposes strong constraints on the sequences that can be constructed on an individual microchip.
Recently, LeProust et al. improved the quality of microchip-synthesized oligonucleotides by controlling depurination during the synthesis process15. These arrays produce up to 55,000 200mer oligonucleotides on a single chip and are sold as a ~1–10 picomole pools of oligonucleotides termed OLS (Oligo Library Synthesis). Several groups have used OLS pools for use in DNA capture technologies, promoter analysis, and DNA barcode development16,17,18,19,20. We have previously shown that individual oligonucleotides in a 55,000 150mer OLS pool were evenly distributed18. We reanalyzed this data set to provide a rough estimate of the error rates (see Methods online) and found the error rate of this OLS pool was ~1/500bp both before and after PCR amplification, suggesting that the OLS pools can be used for accurate large-scale gene synthesis (see Supplementary Table 1 online).
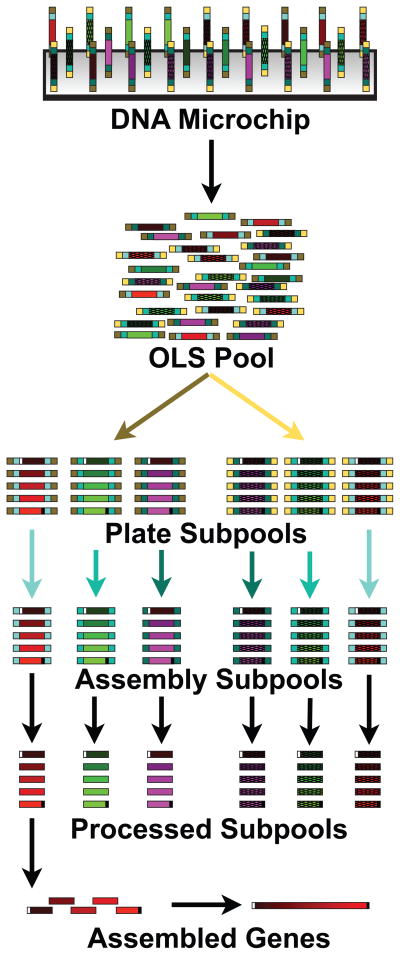
We used these OLS pools to test if they would provide a better starting point for more scalable DNA microchip-based gene synthesis methods. We designed two OLS pools (OLS Pools 1 & 2) of different lengths, each containing ~13,000 130mer or 200mer oligonucleotides respectively. Figure 1 is a general schematic of our methods for utilizing OLS pools in a gene synthesis platform. Briefly, we designed oligonucleotides that were then printed on DNA microchips and recovered as a mixed pool of oligonucleotides (OLS Pool). Next, we took advantage of the long oligonucleotide lengths to independently PCR amplify and process only those oligonucleotides required for a given gene assembly. For the 200mer OLS Pool 2, we first amplified a “plate subpool” that contained DNA to construct up to 96 genes, and then amplified individual “assembly subpools” to separate the oligonucleotides for an individual gene. For the 130mer OLS Pool 1, we directly amplified into assembly subpools, foregoing the plate subpool step. Next, the primers used for these amplification steps were removed by either Type IIS restriction endonucleases to form double-stranded DNA (dsDNA) fragments (OLS Pool 2), or a combination of enzymatic steps to form single-stranded DNA (ssDNA) fragments (OLS Pool 1).
Figure 1. Scalable gene synthesis platform schematic for OLS Pool 2.
Pre-designed oligonucleotides (no distinction is made between dsDNA and ssDNA in the figure) are synthesized on a DNA microchip (a) and then cleaved to make a pool of oligonucleotides (b). Plate-specific primer sequences (yellow or brown) are used to amplify separate Plate Subpools (c) (only two are shown), which contain DNA to assemble different genes (only three are shown for each plate subpool). Assembly specific sequences (shades of blue) are used to amplify assembly subpools (d) that contain only the DNA required to make a single gene. The primer sequences are cleaved (e) using either Type IIS restriction enzymes (resulting in dsDNA) or by DpnII/USER/γ exonuclease processing (producing ssDNA). Construction primers (shown as white and black sites flanking the full assembly) are then used in an assembly PCR reaction to build a gene from each assembly subpool (f). Depending on the downstream application the assembled products are then cloned either before or after an enzymatic error correction step.
Finally, we used PCR assembly to construct full-length genes, performed enzymatic error correction to improve error rates if necessary, and finally cloned and characterized the constructs.
Obtaining subpools of only those DNA fragments required for any particular assembly is crucial for robust gene synthesis in very large DNA backgrounds. In addition, isolating subpools relieves constraints on sequence similarity inherent in past approaches. To facilitate this, we designed 20mer PCR primer sets with low potential cross-hybridization (“orthogonal” primers) derived from a set of 244,000 25mer orthogonal sequences developed for barcoding purposes21. Two separate orthogonal primer sets were constructed for the different OLS pools because of their varying requirements for downstream processing. Both sets were screened for potential cross-hybridization, low secondary structure, and matched melting temperatures to construct large sets of orthogonal PCR primer pairs.
To construct genes from the OLS pools, we developed automated algorithms to split the sequence into overlapping segments with matching melting temperatures such that they could be later assembled by PCR. Genes on OLS Pool 1 and 2 were designed differently to test the effect of different overlap lengths. We designed genes on OLS Pool 1 such that the processed ssDNA pools fully overlapped to form a complete dsDNA sequence. In OLS Pool 2, the processed dsDNA fragments partially overlapped by ~20 bp and can be assembled into a contiguous gene sequence using PCR. We initially constructed a set of fluorescent proteins to test the efficacy of the gene synthesis methods on both OLS Pools.
For OLS Pool 1, we designed two independent “assembly subpools” that encoded for GFPmut3b plus flanking orthogonal primer sequences that are later used for PCR assembly (“construction primers”). The two assembly subpools, GFP43 and GFP35, differed in the average overlap length (43 and 35 bp, respectively), total length (82–90 and 64–78 bases, respectively), and number of oligonucleotides (18 and 22, respectively). We also designed two subpools, Control Subpools 1 and 2, containing ten and five 130mer oligonucleotides, respectively, to test amplification efficacy. The other eight subpools, containing a total of 12,945 130mer sequences, were constructed on the same chip but were not used in this study except to provide potential sources of cross-hybridization. Each of these 12 subpools was flanked with independent orthogonal primer pairs (“assembly-specific primers”). As a control, we used these same algorithms to design a set of shorter CPG oligonucleotides (20 bp average overlap; 35–45 bases in length; and 39 total oligonucleotides) encoding GFPmut3b and obtained them from a commercial provider (IDT). These oligonucleotides were combined to form a third pool that was also tested (“GFP20”). All synthesized oligonucleotides used in the study can be found in the Supplementary Materials online.
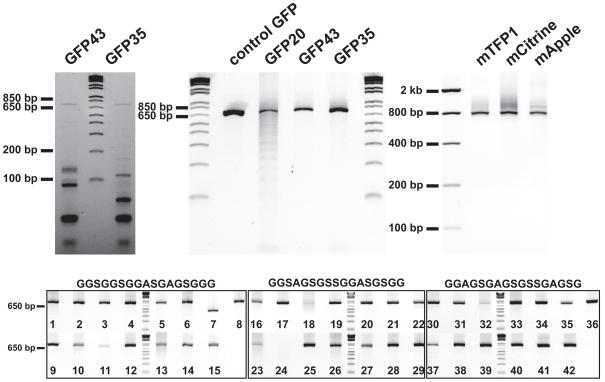
Each of the four subpools (GFP43, GFP35, Control 1, and Control 2) were PCR amplified from the synthesized OLS pool using modified primers that facilitated downstream processing (see Supplementary Figs. 1 and 2a online)18. The oligonucleotides were then processed to remove primer sequences (see Supplementary Figs. 2b and 3 online). Briefly, lambda exonuclease was used to make the PCR products single stranded, and then uracil DNA glycosylase, Endonuclease VIII, and DpnII restriction endonuclease were used to cleave off the assembly-specific primers. The resultant gel shows that while the reaction was efficient, unprocessed oligonucleotide still remained. In addition, we observed spurious cleavage by DpnII that was likely due to the extensive overlap within the subpool that is inherent in the gene synthesis process. We assembled the GFP43, GFP35, and GFP20 subpools using PCR, which resulted in GFP-sized products as well as many incorrect low molecular weight products (Fig. 2a).
Figure 2. Gene synthesis products.
GFPmut3 was PCR assembled (a) from two different assembly subpools (GFP42 and GFP35) that were amplified from OLS Pool 1. Because the majority of the products were of the wrong size, we gel-purified the full-length assemblies and re-amplified them (b). Using the longer oligonucleotides in OLS Pool 2 we were able to develop a PCR assembly protocol that did not require gel-isolation, which we used to build three different fluorescent proteins (c). We then attempted to build 42 scFv regions that contained challenging GC-rich linkers. Of the 42 assemblies (d) 40 resulted in strong bands of the correct size. We gel isolated and re-amplified the two that did not assemble (7 and 24) resulting in bands of the correct size (see Supplementary Fig. 10b online). The antibody that corresponds to each number is given in Supplementary Table 3 online.
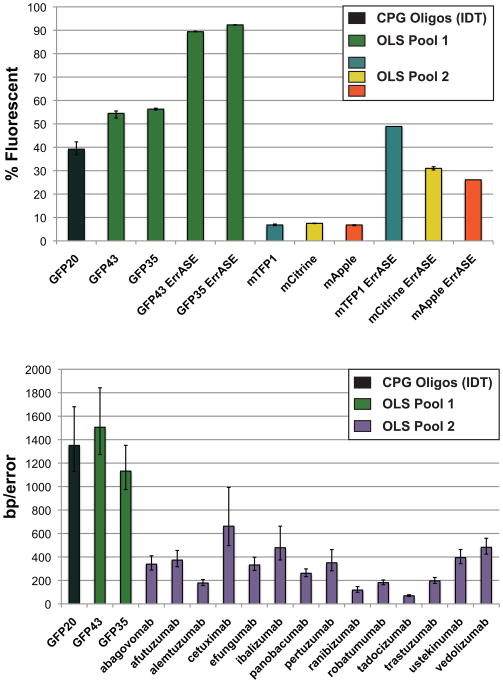
We gel isolated, digested, and then cloned the assembly products into an expression vector (Fig. 2b and Supplementary Fig. 4 online). We used flow cytometry tests, manual colony counts, and sequencing of individual clones to measure the error rates (see Supplementary Figs. 5a and 5b online). All three of the assays correlated well, and the error rates determined through sequencing were 1/1,500 bp, 1/1,130 bp, and 1/1,350 bp for the GFP43, GFP35, and GFP20 synthesis reactions, respectively (See Fig. 3 and Supplementary Table 2 online).
Figure 3. Characterization of products from OLS Pools 1 and 2.
The percentage of fluorescent cells resulting from synthesis products derived from column-synthesized oligonucleotides (black), OLS Chip 1 subpools GFP43 and GFP35 (green) and the three fluorescent proteins produced on OLS Chip 2 with and without ErrASE treatment (blue, yellow, and orange) are shown (a). The error bars correspond to the range of replicates from separate ligations. The error rates (average bp of correct sequence per error) from various synthesis products are shown (b). Error bars show the expected Poisson error based on the number of errors found (±√n). Deletions of more than 2 consecutive bases are counted as a single error (no such errors were found in OLS Pool 1).
These results demonstrated a number of important findings. First, our subpool assembly primers were sufficiently well-designed to provide stringent subpool amplification of as few as 5 oligonucleotides out of a 12,995 oligonucleotide background. Second, the relative quantities of the oligonucleotides in the assembly subpools were sufficient to allow PCR assembly. Third, the error rates from 130mer OLS pools are sufficient to construct gene-sized fragments (717 bp) such that >50% of the sequences will be perfect. In fact, the error rates from both the GFP43 and GFP35 assemblies were indistinguishable from the column-synthesized GFP20 assemblies. Fourth, our data showed that the level of fluorescence of our gene assemblies correlated with the number of constructs with perfect sequence, providing a useful screen to test fluorescent gene assemblies in OLS Pool 2 (see Supplementary Fig. 6 online). Finally, while PCR assembly was able to generate full-length product, many smaller misassembled products were also formed, requiring the use of difficult-to-automate gel isolation steps.
In OLS Pool 2, we designed 836 assembly subpools split into 11 plate subpools, encoding 2,456,706 bases of oligonucleotides that could potentially result in 869,125bp of final assembled sequence. We first constructed three fluorescent proteins to test assembly protocols in OLS Pool 2: mTFP1, mCitrine, and mApple. We found that the PCR assembly protocols developed for ssDNA subpools in OLS Pool 1 only produced short (<200 bp) misassemblies when applied to the dsDNA subpools in OLS Pool 2. We tested over 1,000 assembly PCR conditions by varying parameters such as DNA concentration, annealing temperatures, cycle numbers, polymerase choice, and buffer conditions. Using the best protocol (see Supplementary Note 1), we assembled the three genes with no detectable misassemblies, thereby removing the need for gel isolation (Fig. 2c and Supplementary Figs. 7a and 7b). Cloning followed by flow cytometry screening showed that 6.8%, 7.5%, and 6.8% of the cells were fluorescent for mTFP1, mCitrine, and mApple assemblies, respectively (see Fig. 3a).
Assuming 6% correct sequence per construct and no selection against errors in the assembly process, the error rate was ~1/250 bp for 200mer OLS Pool 2, significantly above that of the estimates for 130mer OLS Pool 1 (~1/1000 bp) and the sequenced 55K 150mer OLS pool (~1/500 bp). This is not completely unexpected, as the amount of depurination is dependent upon the number of deprotection steps during synthesis and thus the oligonucleotide length. Despite the higher error rate, there were several advantages to the 200mer OLS Pool 2. First, the extensive overlaps designed in OLS Pool 1 caused spurious processing of the primers from the assembly subpools. The use of Type IIs restriction endonucleases to process primers to form dsDNA resulted in more robust processing. Second, the use of two amplification steps conserves chip-eluted DNA to allow for future scaling of the gene synthesis process (See Supplementary Note 2). Third, the assemblies of OLS Pool 1 produced many smaller bands and required lower-throughput gel isolation procedures. This could be due to mispriming during PCR assembly because of the long overlap lengths used in the design process. The assemblies in OLS Pool 2 used much shorter overlap lengths, and resulted in no smaller molecular weight misassembled products.
In order to improve the error rates of the genes assembled from OLS Pool 2, we used ErrASE, a new commercially-available enzyme cocktail, to remove errors in the assembled fluorescent proteins. For each gene, we applied ErrASE at six different stringencies, re-amplified the constructs, cloned the PCR products, and re-screened the cloned genes using flow cytometry. Improvement of the level of fluorescence progressively increased with increased ErrASE stringency. At the highest levels of error correction, the fluorescence levels were 31%, 49%, and 26% for mTFP1, mCitrine, and mApple respectively (see Fig. 3a and Supplementary Fig. 8 online). We also performed the ErrASE procedure on our GFP43 and GFP35 pools from OLS Pool 1, resulting in fluorescence levels of 89% and 92% respectively (Fig. 3a and Supplementary Fig. 5c). We sequenced clones of GFP43 and GFP35 and found 3 errors in 21,510 (1/7170 bp) and 4 errors in 20,076 (1/5019 bp) sequenced bases, respectively.
As a more challenging test for our DNA synthesis technology, we designed and synthesized oligonucleotides for 42 genes encoding single-chain Fv (scFv) regions corresponding to a number of well-known antibodies in OLS Pool 2. We have previously had trouble synthesizing these genes using commercial gene synthesis companies. This might be partly due to the prototype (Gly4Ser)3 linker, which is designed to maximize flexibility and allow the heavy and light V regions to assemble22. The repetitive nature and high GC content of the linker-encoding sequences often represents a challenge for accurate DNA synthesis. We therefore tested three different linker sequences that varied in GC content and repetitive character of the linker encoding sequence. In addition, the presence of high sequence homology in the antibody backbones and linkers represented a potential source of cross-hybridization that could interfere with assembly.
As expected, the antibody sequences did not assemble as robustly as the fluorescent proteins and thus we further optimized the conditions during pre-and post-assembly (see Supplementary Figs. 7c, 9, and 10a online). Under the best protocol, 40of the 42 constructs assembled to the correct size (see Fig. 2d and Supplementary Table 3 online). The two misassembled genes displayed faint bands at the correct size, which were gel isolated and reamplified to produce strong bands of the correct size. We sequenced 15 antibodies including representatives from all three linker types. We performed enzymatic error correction using ErrASE, gel isolated the product and finally cloned the constructs into an expression vector. One of the 15 antibodies did not clone, and another had a deleted linker region in all 21 sequenced clones. Both of these antibodies were encoded with the highest GC content linker. The average error rate of the 14 antibodies that did clone was1/315 bp (see Fig. 3b and Supplementary Table 2 online); this was significantly higher than the GFP assemblies, but still sufficient for construction of genes of this size (~10% clones should be perfect on average). In addition, sequence analysis showed no instances of subpool cross-contamination during the assembly process.
Our results show for the first time the assembly of gene-sized DNA fragments totaling ~35,000 bp from oligonucleotide pools of more than >50 kilobases. A number of key features are important to make the process work including the use of low-error starting material, well-chosen orthogonal primers, subpool amplification of individual assemblies, optimized assembly methods, and enzymatic error correction. We describe two separate OLS pool lengths and assembly methods, which have their own advantages and disadvantages (see Supplementary Fig. 1 online). The shorter, 130mer OLS Pool 1 assemblies have lower error rates, but because there are no plate amplifications, will be harder to scale as we begin to utilize larger OLS pools. The longer 200mer OLS Pool 2 is easier to scale, but contained higher error rates. The costs of oligonucleotides in both processes are less than $0.01/bp of final synthesized sequence, and thus the dominant costs are enzymatic processing, cloning, and sequence verification. Future work on lowering cost of perfect sequence will focus on the ability to lower sequencing costs such as by using cheaper next-generation sequencing technologies, or by incorporating other error-correction techniques such as PAGE selection of oligonucleotide pools or mutS-based error filtration3,23.
Online Methods
Reanalysis of OLS Pool Error Rates
We reanalyzed a previously published data set for determining sequencing error rates24. Briefly, the dataset was derived from high-throughput sequencing using the Illumina Genome Analyzer platform of a 53,777 150mer OLS pool. Two sequencing runs were performed; the first before any amplification, and the second after two rounds of ten cycles of PCR (20 cycles total). As our previous analyses were mostly looking for distribution effects, we reanalyzed this existing data to get an estimate of error rates pre-and post-PCR amplification. We realigned the dataset using Exonerate to allow for gapped alignments and analysis of indels25. Specifically, we used an affine local alignment model that is equivalent to the classic Smith-Waterman-Gotoh alignment, a gap extension penalty of -5, and used the full refine option to allow for dynamic programming based optimization of the alignment. These reads were solely mapped on base calls by the Illumina platform. We used these alignments to count mismatches, deletions, and insertions as compared to the designed sequences. However, since base-calling can be more error prone on next generation platforms than traditional Sanger-based approaches, we filtered the results based only on high-quality base-calls (Phred scores of 30 or above or >99.9% accuracy). This was accomplished by converting Illumina quality scores to Phred values using the Maq utility sol2sanger25 and only using statistics from base calls of Phred 30 or higher. All error rate analysis scripts were implemented in Python and are available upon request. While this method provides an estimate for error rates, unmapped reads may have higher error rates and thus underestimating the total average error rate. In addition, base-calling errors might still overestimate the error rate. Finally, using only high-quality base calls, which usually occur only in the first 10 bases of a read, might only reflect error rates on the 5′ end of the synthesized oligonucleotide.
Design and Synthesis of OLS Pools
The 13,000 oligos in the first OLS library (“OLS Pool 1”) were broken up into 12 separately amplifiable subpools (“assembly subpools). Each assembly subpool was defined by unique 20 bp priming sites that flanked each of the oligos in the pool. The priming sites were designed to minimize amplification of oligos not in the particular assembly subpool. This was done by designing set of orthogonal 20-mers (“assembly-specific primers”) using a set of 240,000 orthogonal 25-mers designed by Xu et al.21 as a seed. From these sequences we selected 20-mers with 3′ sequence ending in thymidine or ‘GATC’ for the forward and reverse primers respectively. We screened for melting temperatures between 62–64 °C and low primer secondary structure. After the additional filtering, 12 pairs of forward and reverse primers were chosen to be the assembly-specific primers. The 13,000 oligos in the second OLS library (“OLS Pool 2”) were broken up into 11 subpools corresponding to 11 sets of up to 96 assemblies (“plate subpools”), which were further divided into a total of 836 assembly subpools. A new set of orthogonal primers were designed similarly to the previous set (without the GATC and thymidine constraints) but further filtered to remove Type IIS restriction sites, secondary structure, primer dimers, and self-dimers. The final set of primer pairs was distributed among the plate-specific primers, assembly-specific primers, and construction primers. See Supplementary Methods online for more detailed design information and primer sequences.
OLS pools were synthesized by Agilent Technologies, and are available upon signing a Collaborative Technology Development agreement with Agilent. Costsof OLS pools area function of the number of unique oligos synthesized and of the length of the oligos (<$0.01 per final assembled base-pair for all scales used in this study). OLS Pools 1 and 2 were independently synthesized, cleaved, and delivered as lyophilized ~1–10 picomole pools.
Amplification and processing of OLS subpools
Lyophilized DNA from OLS Pools 1 and 2 were resuspended in 500 μL TE. Assembly subpools were amplified from 1 μL of OLS Pool 1 in a 50 μL qPCR reaction using the KAPA SYBR FAST qPCR kit (Kapa Biosystems). A secondary 20 mL PCR amplification using Taq polymerase was performed from the primary amplification product. The barcode primer sites were removed using a technique previously described20. In brief, the forward primers contained a phosphorothioate bond at the 5′ end and the last nucleotide on the 3′ end was a deoxyuridine; the reverse primers contained a DpnII recognition site (‘GATC’) at the 3′ end and a phosphorylated 5′ end. PCR amplification was followed by γ exonuclease digestion of 5′ phosphorylated strands, hybridization of the 3′ primer site to its complement, and cleavage of the 5′ and 3′ primer sites using USER enzyme mix and DpnII (New England Biolabs), respectively. Plate subpools were amplified from1 μL of OLS Pool 2 in 50 μL Phusion polymerase PCR reactions. Assembly subpools were amplified from the plate subpools by 100μL Phusion polymerase PCR reactions. A BtsI digest removed the forward and reverse primer sites. See the Supplementary Methods online for more detailed protocols.
Assembly of fluorescent proteins
GFPmut326 was assembled from the OLS Pool 1 assembly subpools by PCR. The GFP43 and GFP35 subpools were designed such there was full overlap between neighboring oligos during assembly, with average overlaps of 43 bp and 35 bp for GFP43 and GFP35, respectively. For the first set of assemblies, 330 pg of the GF43 subpool or 40 pg of the GFP35 subpool were used per 20 μL Phusion polymerase PCR assembly. The full-length product was gel-isolated, amplified using Phusion polymerase, and cloned into pZE21 after a HindIII/KpnI digest. The second set of assemblies was built using a similar procedure, except that the assembly PCR used 170 pg or 190 pg of GFP43 and GFP35 subpools, respectively; and the gel-isolated product was not re-amplified prior to cloning.
Oligonucleotides for mTFP1, mCitrine, and mApple were designed such that there was on average a 20 bp overlap between adjacent oligonucleotides. The proteins were built from OLS Pool 2 assembly subpools by first performing a KOD polymerase pre-assembly reaction that was done in the absence of construction primers followed by a KOD polymerase assembly PCR in which the construction primers were included. ErrASE error correction was then performed on aliquots of the synthesis products following the manufacturer’s instructions. The assembled product was digested with HindIII and KpnI and cloned into pZE21. Sequencing of clones was performed by Beckman Coulter Genomics. See the Supplementary Methods online for more detailed protocols.
ErrASE
ErrASE is an enzyme cocktail designed to remove errors in synthetically assembled genes (Novici Biotech, Vacaville CA). Assembled genes are denatured and re-annealed to allow for the formation of hetero-duplexes. A resolvase enzyme in ErrASE then recognizes and cuts at mismatched positions. Other enzymes in the cocktail remove these cut mismatched positions. The products could then be reamplified by PCR to reassemble the full-length gene.
Specifically, sixaliquots of 10–50 ng of each assembled gene was added to 10 μL of PCR buffer (we have also tested the effects of including betaine in the buffer see Supplementary Fig. 11). Hetero-duplexes were formed by denaturing at 95°C and slowly cooling to room temperature. Each aliquot was then used to resuspend six different lyophilized ErrASE mixtures of increasing stringency provided by the manufacturer. After a 1–2 hour room temperature incubation, the assemblies were re-amplified and visualized on an agarose gel. Of the reactions that resulted in a correctly-sized band, the one that used the most stringent ErrASE protocol was selected for cloning.
Flow cytometry
Fluorescent cell fractions of the cloned libraries of assembly products were quantified using a BD LSR Fortessa flow cytometer either a 488 nm laser with a 530 nm filter (30 nm bandpass) or a 561 nm laser with a 610 nm filter (20 nm bandpass).
Synthesis of Antibodies
125 ng of each antibody assembly pool was pre-assembled in 20 μL KOD pre-assembly reactions. We then tested 9 amplification protocols for the ability to amplify the 42 antibody pre-assemblies into full-length genes. We attempted to clone 8 constructs from the best assembly protocol (afutuzumab, efungumab, ibalizumab, oportuzumab, panobacumab, robatumumab, ustekinumab, and vedolizumab; see Supplementary Fig. 10a and Supplementary Table 3). The 8 assemblies were error-corrected using ErrASE, gel-isolated, re-amplified using Phusion polymerase, gel-isolated again, and cloned into pSecTag2A after an ApaI/SfiI digest. Sequencing was performed by Genewiz. All but oportuzumab cloned successfully. We then repeated the experiment, increasing the amount of assembly pool DNA in the pre-assembly reaction to 400 ng. We selected a different set of 8 constructs from this second set of assemblies for cloning (abagovomab, alemtuzumab, ranibizumab, cetuximab, efungumab, pertuzumab, tadocizumab, and trastuzumab; see Fig. 2d and Supplementary Table 3). Using the same methods as with the first set of cloned antibodies, this second set was error-corrected, gel-isolated, cloned, and sequenced. See the Supplementary Methods online for more detailed protocols.
Supplementary Material
Acknowledgments
This work was supported by the US ONR (N000141010144), NIHGRI Center for Excellence in Genomics Science (P50 HG003170), DOE Genomes to Life (DE-FG02-02ER63445), DARPA (W911NF-08-1-0254), and the Wyss Institute for Biologically Inspired Engineering (all to G.M.C.).We thank H.Padgett for providing ErrASE and expertise during optimization and J. Boeke for advice on gene assembly protocols. We also thank S. Raman, F. Vigneault, and F. Zhang for critical readings of the manuscript, G. Dantas for pZE21, F. Isaacs for pZE21G, and J.S. Workman for pSecTag2A.
Footnotes
Author Contributions
S.K. and N.E. wrote the paper with contributions from all authors; S.K. and G.M.C. conceived the study; S.K. wrote all algorithms and designed all sequences; S.K. and N.E. designed and performed all experiments; E.L. provided the oligonucleotides libraries; M.S. and J.F. designed the single-chained versions of commercial antibodies; J.B.L. performed the OLS high-throughput sequencing experiment and provided critical advice on the processing of subpools.
Competing Financial Interests
E.M.L. is an employee of Agilent Technologies, the commercial provider of OLS pools. G.M.C. is a co-founder of an early-stage startup company involved in gene synthesis. S.K., N.E., and G.M.C. are named inventors on a patent application on technologies described in this article. S.K. is a post-doctoral fellow whose future employment prospects depend upon refereed publications.
References
- 1.Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 2.Tian J, Ma K, Saaem I. Advancing high-throughput gene synthesis technology. Mol BioSyst. 2009;5:714–722. doi: 10.1039/b822268c. [DOI] [PubMed] [Google Scholar]
- 3.Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 4.Richmond KE, et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 2004;32:5011–5018. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, et al. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences. Nucleic Acids Res. 2004;32:5409–5417. doi: 10.1093/nar/gkh879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. P Natl Acad Sci USA. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söll D, et al. Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA’s to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. P Natl Acad Sci USA. 1965;54:1378–1385. doi: 10.1073/pnas.54.5.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 9.Gibson DG. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009;37:6984–6990. doi: 10.1093/nar/gkp687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 11.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 12.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C-C, Snyder TM, Quake SR. A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 2010;38:2514–2521. doi: 10.1093/nar/gkq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim C, et al. Progress in gene assembly from a MAS-driven DNA microarray. Microelectronic Eng. 2006;83:1613–1616. [Google Scholar]
- 15.Leproust EM, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nature Biotech. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlabach MR, et al. Synthetic design of strong promoters. P Natl Acad Sci USA. 2010;107:2538–2543. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JB, et al. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res. 2009;19:1606–1615. doi: 10.1101/gr.092213.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 20.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 21.Xu Q, et al. Design of 240,000 orthogonal 25mer DNA barcode probes. P Natl Acad Sci USA. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huston JS, et al. Medical applications of single-chain antibodies. Int Rev Immunol. 1993;10:195–217. doi: 10.3109/08830189309061696. [DOI] [PubMed] [Google Scholar]
- 23.Carr PA, et al. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H. Maq: mapping and assembly with qualities. Welcome Trust Sanger Institute; 2010. Available at: http://maq.sourceforge.net. [Google Scholar]
- 26.Carmack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.