Abstract
Importins are members of a family of transport receptors (karyopherins) that mediate the nucleocytoplasmic transport of protein and RNA cargoes. We identified importin-11 as a potential new human member of this family, on the basis of limited similarity to the Saccharomyces cerevisiae protein, Lph2p, and cloned the complete open reading frame. Importin-11 interacts with the Ran GTPase, and constitutively shuttles between the nuclear and cytoplasmic compartments. A yeast dihybrid screen identified UbcM2, an E2-type ubiquitin-conjugating enzyme, as a binding partner and potential transport cargo for importin-11. Importin-11 and UbcM2 interact directly, and the complex is disassembled by Ran:GTP but not by Ran:GDP. UbcM2 is constitutively nuclear and shuttles between the nuclear and cytoplasmic compartments. Nuclear import of UbcM2 requires Ran and importin-11, and is inhibited by wheatgerm agglutinin, energy depletion or dominant interfering mutants of Ran and importin-β. These data establish importin-11 as a new member of the karyopherin family of transport receptors, and identify UbcM2 as a nuclear member of the E2 ubiquitin-conjugating enzyme family.
Keywords: importin/nucleocytoplasmic transport/Ran/ubiquitin
Introduction
Macromolecular traffic between the cytoplasmic and nuclear compartments of eukaryotic cells passes through nuclear pore complexes (Davis, 1995; Stoffler et al., 1999). Pores are assembled from proteins called nucleoporins into structures that span the nuclear envelope to create an aqueous channel with distinct cytoplasmic and nuclear faces (Rout et al., 2000; Wente, 2000). While small molecules diffuse freely through the pores, translocation of nucleic acids and proteins is often mediated by a class of soluble transport receptors (karyopherins) that can associate with the Ran GTPase (Corbett and Silver, 1997; Mattaj and Englmeier, 1998; Melchior and Gerace, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Pemberton et al., 1999). These karyopherins can be classified into several groups, including importins, which facilitate nuclear import, and exportins, which facilitate nuclear export. Several importins have also been termed transportins. Selection of import cargo is accomplished by the recognition of distinct sequence motifs, termed nuclear localization signals (NLSs), present in the cargo molecule (Kalderon et al., 1984). Recognition may be direct or may be mediated by an adaptor protein. For example, the classical basic NLS is recognized by an adaptor called importin-α, which binds to and is transported through the pores by importin-β. Some members of the family, such as importin-β and transportin-1, can bind multiple types of cargo that utilize distinct signals (Jakel and Görlich, 1998), although no example has yet been described of a karyopherin that can act as both an importin and an exportin.
All members of the karyopherin family share an N-terminal region that is predicted, and in several cases has been demonstrated experimentally, to bind to Ran:GTP, whereas the cargo (or adaptor protein) binds to a more C-terminal region (Wozniak et al., 1998). To permit shuttling through the pores, each karyopherin presumably possesses a nucleoporin-binding domain, which in importin-β lies in the central region of the protein (Chi and Adam, 1997). The isolated nucleoporin-binding domain, importin-β(45–465), dominantly interferes with several nuclear import and export pathways, suggesting that there may be common binding sites in the pores for multiple karyopherins (Kutay et al., 1997b), although karyopherin-specific sites also exist (Damelin and Silver, 2000).
A nucleocytoplasmic gradient of Ran:GTP must be maintained to support multiple rounds of nuclear transport. Two factors essential for this gradient are the Ran exchange factor, RCC1 (or RanGEF), and the GTPase activating protein, RanGAP (Cole and Hammell, 1998; Görlich and Kutay, 1999). RCC1 resides in the nucleus. Conversely, RanGAP localizes to the cytoplasm and with the cytoplasmic side of the nuclear pores. The Ran:GTP gradient across the nuclear envelope is used by the karyopherins to provide vectoriality in cargo transport, and to permit transport against a concentration gradient (Corbett and Silver, 1997; Mattaj and Englmeier, 1998; Melchior and Gerace, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Pemberton et al., 1999). Importins bind cargo in the cytoplasm and transit the pores into the nucleus where association with Ran:GTP triggers cargo release. The Ran:GTP–importin complex is then recycled back to the cytoplasm.
In addition to the classical NLS import pathway mediated by the importin-α/β heterodimer, many pathways mediated by other importins have been identified. In yeast there are eight known importins for which at least one type of cargo has been identified, and it is likely that more exist in metazoans (Wozniak et al., 1998). Why does the cell require a large family of karyopherins? One likely reason is to permit differential regulation of transport, but the regulatory networks remain to be elucidated. Progress has been hampered, particularly in mammalian cells, by the paucity of known cargoes that are transported by the different karyopherins. To gain a full understanding of these regulatory mechanisms, we need to determine the functions of all members of the karyopherin family.
Many nuclear proteins are regulated by ubiquitylation, which, in many cases, targets them for degradation by the proteasome (Varshavsky, 1997). The nuclear localization of these ubiquitin (Ub)-targeted proteins implies that enzymes involved in conjugating Ub may need to be imported into the nucleus. For example, MDM2, an E3-type Ub-ligase that ubiquitylates the tumor suppressor protein, p53, shuttles in and out of the nucleus (Roth et al., 1998; Tao and Levine, 1999). Although the subcellular localization of other ubiquitin enzymes has been examined, the distribution of many remains unknown, and few studies have addressed the mechanisms that control their subcellular location (Hatakeyama et al., 1997).
Here we identify importin-11 as a novel, human member of the karyopherin family that mediates the nuclear import of UbcM2, a murine E2 ubiquitin-conjugating enzyme. This import was inhibited by wheatgerm agglutinin and dominant mutants of Ran and importin-β. In addition, a direct interaction between importin-11 and UbcM2 could be reconstituted in vitro, which was disrupted by addition of Ran:GTP. Collectively, these data establish that importin-11 is a specific import receptor for UbcM2.
Results
Molecular cloning of importin-11
The Saccharomyces cerevisiae genome contains 14 genes that encode members of the karyopherin-β family. Although most of the yeast proteins have been shown to function as nuclear transport receptors (Wozniak et al., 1998), others have so far resisted analysis, and the extent of the functional overlap between yeast and mammalian karyopherins remains to be determined. As one approach to address these issues, we searched the databases for open reading frames related to the uncharacterized S.cerevisiae gene, YPL125W. This gene has also been given the name LPH2. Several ESTs were identified that encode C-terminal fragments of a protein more similar to Lph2p than to other yeast members of the family. The clone encoding one such fragment, ESTAA082435, was completely sequenced. This information was used to generate primers for 5′ rapid amplification of cDNA ends (5′ RACE) from a human brain cDNA library. The complete open reading frame, assembled from the EST and 5′ RACE products, is 2925 bp long and codes for a 975 amino acid residue protein. The Met designated as the initiation codon is in a good Kozak sequence context and matches the expected N-terminus, by comparison with the Lph2p sequence. This gene product was named importin-11 because, as demonstrated below, it imports a protein cargo into the nucleus. It was given the designation ‘11’ because while these studies were in progress an almost identical sequence was deposited in the DDBJ/EMBL/GenBank database and called RanBP11 (accession No. AF111109).
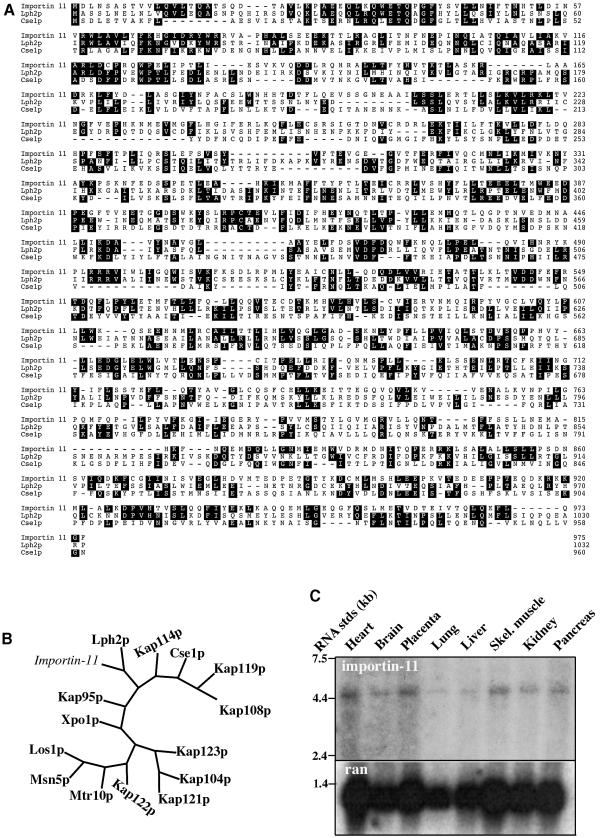
Overall, importin-11 is 24% identical and 44% similar to Lph2p (Figure 1A). The N-terminal region, which constitutes the Ran-binding domain in other karyopherins, is 31% identical to Lph2p and 25% identical to CseIp. Phylogenetic analysis revealed that importin-11 is more similar to Lph2p than to other members of the yeast karyopherin family (Figure 1B), and they belong to a subgroup that includes YGL241W/Kap114p, which transports the TATA-binding protein (Pemberton et al., 1999), and Cse1p/CAS, which is the exportin for importin-α (Kutay et al., 1997a; Kunzler and Hurt, 1998).
Fig. 1. Importin-11 is closely related to Lph2p, and is expressed in many human tissues. (A) Amino acid sequences of importin-11 and S.cerevisiae karyopherins Lph2p and Cse1p were aligned using the Clustal program. Identical residues common to two out of three sequences are shaded. (B) Unrooted phylogenetic tree of importin-11 and the 14 S.cerevisiae karyopherins, generated using PAUP and TreeView. (C) Northern blotting of mRNAs from human tissues was performed with a probe generated using the ESTAA082435 plasmid as a template. The same blot was hybridized with a Ran probe as a loading control.
To determine the tissue distribution of importin-11, a northern blot was performed on mRNAs from multiple human tissues, which revealed a 4.4 kb mRNA for importin-11 in all tissues tested, although detection was low in lung and liver (Figure 1C). Ran was used as a loading control, and was also lower in these tissues. The length of the importin-11 message is consistent with the cDNA sequence.
Importin-11 can bind to the Ran GTPase
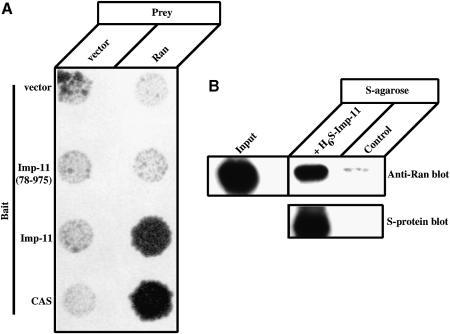
A distinguishing property of karyopherin family members characterized to date is their ability to bind the Ran GTPase in its GTP-bound form. The interaction of importin-11 with Ran was investigated by yeast dihybrid and in vitro binding assays. MATa yeast (HF7c) that expresses importin-11 as a C-terminal fusion to the GAL4 DNA-binding domain (DBD) was mated with the W303 (MATα) strain expressing wild-type Ran as a C-terminal fusion to the VP16 transactivation (TA) domain. After mating, the yeast were replica-plated onto a medium selective for diploids. An interaction between importin-11 and Ran reconstitutes the transcriptional activity necessary for the production of histidine, thus permitting growth on Leu–/Trp–/His– dropout plates. Conversely, lack of growth is interpreted to mean that the two proteins either do not interact or are expressed inefficiently in the yeast nuclei. The results show that importin-11 interacts with Ran, but that importin-11(78–975), an N-terminal truncation lacking the first 77 amino acid residues, does not (Figure 2A). CAS was used as a positive control for Ran binding, and empty vector as the negative control. Failure of importin-11(78–975) to interact with Ran was not due to inadequate expression or to instability of the importin-11(78–975), as indicated by its ability to interact with its import cargo, demonstrated below in a parallel assay (see Figure 4B). Although these data do not demonstrate a direct interaction between importin-11 and Ran:GTP, they are consistent with importin-11 containing an N-terminal Ran-binding domain.
Fig. 2. Importin-11 binds Ran directly and this interaction requires the N-terminal 77 residues of importin-11. (A) HF7c (MATa) yeast that express the indicated bait proteins as GAL4 DBD fusions were mated with the W303 (MATα) strain expressing the VP16 TA domain either alone (vector) or as a fusion with Ran. Diploid yeast was selected on Leu–/Trp– plates and replica-plated onto Leu–/Trp–/His– plates. (B) Ran(Q69L)-His6 was mixed with S-protein–agarose beads alone (control) or with beads plus H6-S-Imp-11. Bound proteins were separated by SDS–PAGE and detected by immunoblotting with a monoclonal antibody to Ran and peroxidase-conjugated, anti-mouse secondary, or with peroxidase-conjugated S-protein. A sample representing 2.5% of the Ran(Q69L)-His6 added to each sample is also shown (Input).
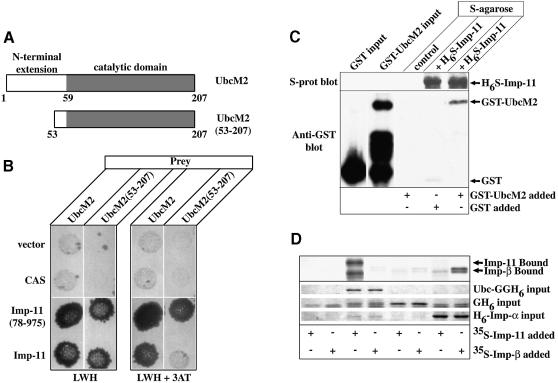
Fig. 4. Importin-11 interacts with an E2-type ubiquitin-conjugating enzyme, UbcM2. (A) Schematic representation of UbcM2, and of a truncation of the enzyme lacking the first 52 amino acid residues [UbcM2(53–207)]. N-terminal extension domain is demarcated by the white box and the catalytic domain by the shaded box. (B) Conjugation assays were carried out as in Figure 2. Diploid yeast were replica-plated onto selective medium (LWH) and similar plates containing 3-AT (LWH+3AT). (C) GST–Ubc or GST was mixed with protein-S–agarose beads plus H6-S-Imp-11 and GST–Ubc was mixed with beads alone (control). Bound proteins were immunoblotted with an anti-GST antibody, or with peroxidase-conjugated S-protein. An aliquot representing 50% of the GST proteins added to each sample is also shown. (D) 35S-labeled importin-11 and importin-β, expressed by in vitro transcription–translation, were incubated with recombinant Ubc-GGH6, GFP-His6 (GH6) or His6-importin-α (H6-Imp-α), immobilized on Ni2+–agarose beads, or with beads alone. Proteins remaining associated with the beads were resolved by SDS–PAGE and detected by staining (Coomassie Brilliant Blue) or fluorography.
To test for direct binding of importin-11 to Ran:GTP we expressed recombinant importin-11 bearing an N-terminal His6-S-peptide tag (H6S-Imp-11) in bacteria. The protein could only be expressed at low levels and slowly precipitated out of solution on storage at 4°C or when concentrated. Precipitation was not prevented by addition of 0.5 M sucrose or 10% glycerol, nor by transfer into other buffers and salt concentrations. Nevertheless, sufficient soluble protein was produced to allow some binding assays to be performed. Ran (Q69L), a mutant of Ran that is defective in GTP hydrolysis, was incubated with either peptide-S–agarose beads alone or beads bound with H6S-Imp-11. The amount of Ran remaining associated with the beads after washing was assessed by immunoblotting. The results of such an experiment (Figure 2B) show that importin-11 can specifically precipitate Ran(Q69L):GTP.
Importin-11 shuttles constitutively between the nucleus and cytoplasm
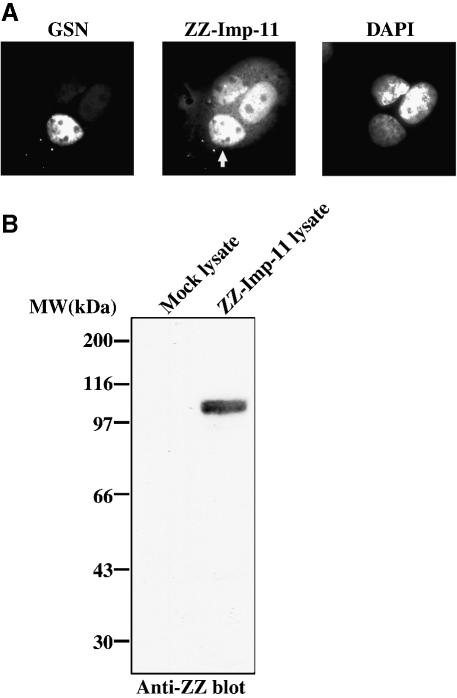
A common characteristic of karyopherins is their ability to shuttle through nuclear pores as they transport cargo between the cytoplasmic and nuclear compartments. When ectopically expressed, importin-11 has a predominantly nuclear distribution (Figure 3A, center panel). We used a heterokaryon fusion assay to determine whether the protein resides permanently within the nucleus or shuttles between the nucleus and cytoplasm (Borer et al., 1989). In this assay, donor cells expressing the protein of interest are mixed with acceptor cells that do not express the protein, in the presence of a fusogen. Equilibration of the protein between the nuclei of a fused cell (in the absence of new protein synthesis) demonstrates that the protein has exited one nucleus and entered the other.
Fig. 3. Importin-11 shuttles in and out of the nucleus. (A) Transfected donor BHK cells, expressing zz-tagged importin-11 (ZZ-Imp-11), were fused to GSN2 acceptor cells. The fused cells were grown in medium containing cycloheximide, then fixed, permeabilized and immunostained with a Texas Red-conjugated antibody to detect ZZ-Imp-11. DNA was stained with DAPI. Left panel shows the green channel (GSN); middle panel shows the red channel (ZZ-Imp-11); right panel shows the DNA staining. The white arrow in the middle panel denotes the acceptor nucleus within the fused cell. (B) Immunoblot of lysates prepared from mock-transfected and ZZ-Imp-11-transfected BHK cells. The blot was probed with an HRP-conjugated, rabbit anti-goat antibody.
Baby hamster kidney (BHK) cells expressing zz-tagged importin-11 were fused with GSN2 cells, a stably transformed HeLa cell line expressing a fusion protein consisting of green fluorescent protein (GFP)-streptavidin and the SV-40 large-T-antigen NLS (Holaska and Paschal, 1998). This fusion protein does not possess a nuclear export signal and is too large to diffuse out through the nuclear pores. It therefore functions as a marker of the acceptor nucleus and as a negative control for shuttling. After fusion, the cells were incubated with cycloheximide to inhibit further protein synthesis, then fixed and processed to detect zz-importin-11. As shown in Figure 3A, the zz-importin-11 did equilibrate between the donor and acceptor nuclei of fused cells, while the GSN protein did not. The movement of the zz-importin-11 was not attributable to passive diffusion of immunoreactive proteolytic fragments (Figure 3B), and the zz tag itself did not promote shuttling when attached to a resident nuclear protein such as RCC1 (data not shown). These data demonstrate that importin-11 can shuttle constitutively between the nuclear and cytoplasmic compartments, a property consistent with its potential function as a transport receptor.
Identification of the ubiquitin-conjugating enzyme, UbcM2, as a candidate importin-11 transport cargo
We performed yeast dihybrid screens of a random-primed murine library created from a whole 10 day embryo, using full-length importin-11 as the bait for the first screen and importin-11(78–975) as bait in a second screen. These screens identified 12 positives, of which one was Ran and one corresponded to a putative nucleoporin, called Npap60L (Fan et al., 1997). The appearance of Ran among the positives provided a validation of the specificity of the screen. Several other clones were derived from a gene that encodes UbcM2, a murine, E2-type ubiquitin-conjugating enzyme (Ubc) (Pestov et al., 1998). UbcM2 belongs to a family of E2 Ubcs that possess N-terminal extensions of unknown function (Matuschewski et al., 1996) (Figure 4A). The human homolog, UBE2E3, has 100% identity to its mouse counterpart (Ito et al., 1999). Among the yeast ubiquitin-conjugating enzymes, UbcM2 is most closely related to Ubc4 and Ubc5.
Both the full-length and N-terminal deletion mutant of importin-11 interacted with UbcM2, whereas a different transport receptor (CAS) did not interact with this enzyme, demonstrating that the interaction with importin-11 is specific (Figure 4B). Both forms of importin-11 also interacted with an N-terminally truncated protein, UbcM2(53–206), which lacks most of the N-terminal extension. No clones of UbcM2 were identified from the dihybrid screen that lacked a complete catalytic domain.
Surprisingly, importin-11(78–975) appeared to interact more robustly with truncated UbcM2 than did wild-type importin-11, as indicated by the lack of growth of yeast expressing importin-11 and UbcM2(53–206) on plates containing the histidine synthesis inhibitor, 3-AT (Figure 4B). However, the relative levels of expression of importin-11 and importin-11(78–975) were not normalized, so the significance of this observation remains to be determined.
Importin-11 and UbcM2 interact directly
To test for direct binding between importin-11 and UbcM2 we produced the UbcM2 in bacteria as a C-terminal fusion to glutathione S-transferase (GST) (GST–UbcM2). This protein was soluble, although there was some proteolysis during preparation (Figure 4C). Recombinant H6S-Imp-11 was bound to S–agarose beads in the presence of the GST–UbcM2 or GST. Proteins associating with the beads were detected by immunoblotting. The results of this experiment (Figure 4C) reveal that importin-11 and UbcM2 can interact directly. Furthermore, they suggest that the catalytic domain of UbcM2 must be intact for this interaction to take place, as none of the C-terminal, GST–UbcM2 degradation products were precipitated. Similar results were obtained using a recombinant N-terminal fusion of UbcM2 to GFP-GFP-His6 (Ubc-GGH6) (data not shown).
Interaction specificity was tested using importin-β as an alternative possible binding partner for UbcM2. In this experiment 35S-labeled importin-11 and importin-β were each expressed in a reticulocyte lysate by in vitro transcription–translation. Recombinant Ubc-GGH6, GFP-His6 and His6-tagged importin-α were attached to Ni2+–agarose beads and incubated with the lysates, then washed. Importin-11 bound to Ubc-GGH6 but not to GFP or importin-α, whereas the importin-β bound only to importin-α (Figure 4D). These data demonstrate that the association of importin-11 with Ubc-GGH6 is highly specific.
Importin-11–UbcM2 complex is dissociated by Ran:GTP
The prevailing model of receptor-mediated nuclear protein import proposes that transport receptor–cargo complexes form in the cytoplasm and are dissociated in the nucleus by Ran:GTP (Görlich and Kutay, 1999). We therefore wished to test the susceptibility of the importin-11–UbcM2 complex to disruption by Ran:GTP in vitro. This assay is also a stringent test for the specificity of the interaction between the UbcM2 cargo and importin-11, because dissociation is induced by a conformational change in the importin triggered by association with the Ran:GTP (Chook and Blobel, 1999).
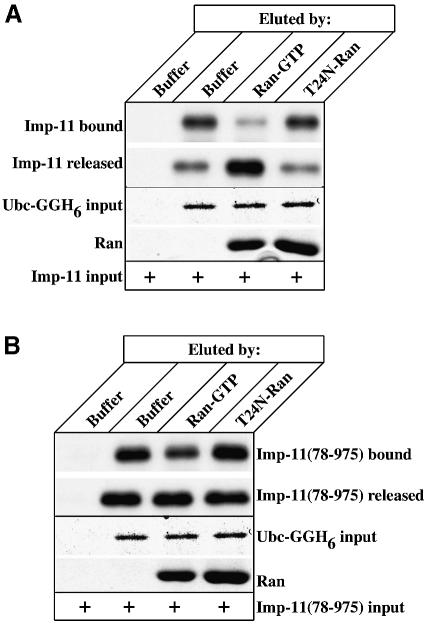
35S-labeled importin-11 and importin-11(78–975) were incubated with recombinant Ubc-GGH6 and the protein complexes were bound to Ni2+–agarose beads as above. After washing to remove free proteins, the beads were incubated with wild-type Ran that had been loaded with GTP, or with a mutant Ran(T24N), which is defective in GTP binding, or with buffer alone. Proteins that remained bound to the beads, as well as those eluted, were then assessed by SDS–PAGE and fluorography. Ran:GTP efficiently disrupted the importin-11–Ubc-GGH6 interaction (Figure 5A). Notably, dissociation of the importin-11(78–975) from Ubc-GGH6 occurred even in the control, with buffer alone, and was only marginally increased by the presence of Ran:GTP, consistent with our observation that importin-11(78–975) is defective in its ability to bind Ran (Figure 5B). The increased release from Ubc-GGH6 in the control sample suggests that cargo binding to importin-11 may be stabilized by the N-terminal domain of the karyopherin.
Fig. 5. The importin-11–UbcM2 complex is dissociated specifically by Ran:GTP. (A) 35S-labeled importin-11, expressed by in vitro transcription–translation, was combined with either 1 µg of recombinant Ubc-GGH6 immobilized on Ni2+–agarose beads or with beads alone. Bead-associated proteins were eluted by buffer, or by Ran loaded with GTP (Ran:GTP) or a mutant of Ran that cannot bind nucleotide (T24N-Ran). Proteins remaining associated with the beads (Imp-11 bound and Ubc-GGH6 input) and aliquots of those eluted (Imp-11 released and Ran) were resolved by SDS–PAGE and detected by staining with Coomassie Brilliant Blue or fluorography. (B) As for (A), but using the N-terminal mutant, Imp-11(78–975).
UbcM2 is a predominantly nuclear, shuttling protein, and its localization depends on importin-11 function
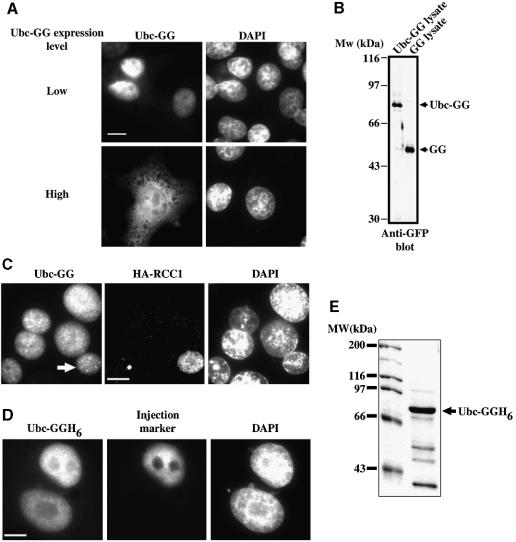
To examine the functional significance of the importin-11–UbcM2 interaction, we first determined the steady-state intracellular distribution of UbcM2 in intact cells and asked whether it constitutively shuttles between the nuclear and cytoplasmic compartments. Because the size of the UbcM2 protein is below the diffusion limit for the nuclear pores, we expressed a fusion protein of UbcM2 bearing a C-terminal GFP-GFP tag (Ubc-GG), with a mol. wt of ∼80 kDa. Interestingly, it localized exclusively within the nuclei of those transfected cells expressing relatively low amounts of the fusion protein, but was distributed in both the cytoplasm and nuclei of cells that expressed higher levels of the fusion protein (Figure 6A). An immunoblot demonstrated that the expressed protein was not significantly degraded into small, freely diffusible fragments when expressed in the BHK cells (Figure 6B). These data indicate that the nuclear import and/or retention of Ubc-GG is a saturable process, and that the import of Ubc-GG is unlikely to occur via passive diffusion.
Fig. 6. UbcM2 is a constitutively shuttling, nuclear protein. (A) BHK cells were transfected with plasmid encoding Ubc-GG then fixed, permeabilized and analyzed by fluorescence microscopy as described in Materials and methods. Cells expressing relatively low amounts of protein were imaged at 500 ms exposures (Low) and those expressing larger amounts at 150 ms (High). (B) Cells were transfected with plasmids encoding Ubc-GG or a GFP-GFP fusion (GG). Protein extracts were immunoblotted for GFP. Full-length forms of each fusion protein are denoted by arrows. (C) Fusion assays were carried out on donor BHK cells expressing Ubc-GG, and acceptor tsBN2/RCC1 cells that express HA-tagged RCC1. After fusion, cells were processed for immunofluorescence using 12CA5 antibody and a Texas Red-conjugated secondary. Left panel shows the GFP fluorescence (Ubc-GG); middle panel shows Texas Red fluorescence (HA-RCC1); right panel shows DNA staining (DAPI). The white arrow in the left panel denotes the acceptor nucleus within the fused cell (n = 20). (D) Binucleated BHK cells were micro-injected into one nucleus with a mixture of Ubc-GGH6 (3 µM) and TRITC-labeled dextran (Injection marker) and incubated for 30 min before fixation (n = 40). Bars, 10 µm. (E) Coomassie Blue-stained gel of the Ubc-GGH6 protein used for all micro-injection experiments. Full-length Ubc-GGH6 is denoted with an arrow.
To determine whether UbcM2 is a shuttling protein, two types of experiments were performed. First, a heterokaryon fusion assay was performed between BHK cells expressing Ubc-GG and a cell line (tsBN2) that stably expresses hemagglutinin (HA)-tagged RCC1. We demonstrated previously that RCC1 does not shuttle (Nemergut and Macara, 2000), and this line therefore both provides acceptor nuclei and functions as a negative control for shuttling. As shown in Figure 6C, the Ubc-GG equilibrates between the donor and acceptor nuclei, while the HA-RCC1 does not, demonstrating that the Ubc-GG can shuttle between the nuclear and cytoplasmic compartments. As a second approach, we micro-injected recombinant Ubc-GGH6 into one nucleus of a multinucleated BHK cell (Figure 6D). This protein, which was predominantly full length (Figure 6E), rapidly equilibrated into the uninjected nucleus, confirming the conclusion from the heterokaryon assay that UbcM2 is a shuttling protein.
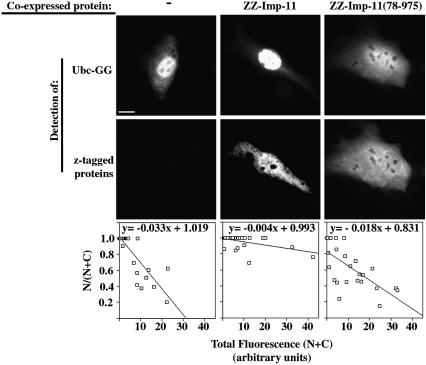
To test whether increasing the amount of importin-11 could alter the distribution of UbcM2, we co-expressed Ubc-GG with zz-importin-11 in BHK cells and measured the ratio of nuclear GFP fluorescence (N) to total cell fluorescence (N+C). These data were compared with those obtained from cells expressing only Ubc-GG and from those co-expressing Ubc-GG and importin-11(78–975) or importin-β. When plotted as the fraction of the Ubc-GG in the nucleus, N/(N+C), versus the expression level in the cell (N+C), for a range of cells, the slopes of the graphs provide a measure of the ability of the importin-11 to facilitate nuclear accumulation of the UbcM2 (Figure 7, bottom panels).
Fig. 7. Nuclear accumulation of UbcM2 is dependent on functional importin-11. BHK cells ectopically expressing Ubc-GG alone (–) or together with zz-tagged importin-11 (ZZ-Imp-11), the mutant ZZ-Imp-11(78–975) or importin-β were processed for immunofluorescence as described for Figure 3. GFP was imaged under conditions such that no pixels were saturated, and quantitated to obtain total fluorescence (N+C) and nuclear fluorescence (N) for each cell. Graphs show fractional nuclear GFP fluorescence for a range of expression levels of the Ubc-GG, in the presence or absence of the co-expressed importins. The data in the graphs were compiled from 10–20 cells in each condition for two separate experiments. Bar, 10 µm. (See Supplementary data for method of image analysis and importin-β graph.)
The results (Figure 7) show that: (i) as the level of Ubc-GG expression increases, the ratio of nuclear/total cell fluorescence decreases, as seen previously (Figure 6A) and (ii) this effect of expression level on intracellular distribution is overcome by co-expression of importin-11 but not by importin-11(78–975) or importin-β (see Supplementary data, available at The EMBO Journal Online). The behavior of importin-11(78–975) is consistent with our observation that this mutant does not efficiently release cargo in response to high Ran:GTP concentrations. Therefore, these data indicate that the limiting factor in Ubc-GG nuclear accumulation in this assay is the concentration of endogenous importin-11, and that importin-11 is a bona fide import receptor for UbcM2.
Nuclear accumulation of UbcM2 requires Ran function
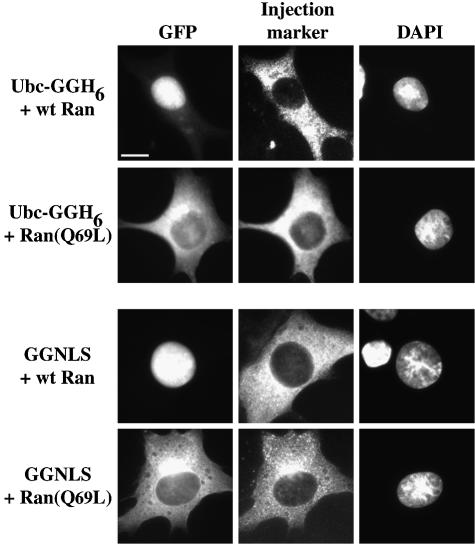
One important criterion of karyopherin-mediated nuclear import is that it is inhibitable by constitutively active mutants of Ran. Ran (Q69L) is resistant to RanGAP, and so remains GTP bound within the cell. When present in the cytosol, the Ran (Q69L) will bind the importin and inappropriately trigger cargo release.
To test whether UbcM2 import is sensitive to Ran (Q69L), recombinant mutant Ran was co-injected into BHK cells with Ubc-GGH6. As a control, we also used a fusion protein of GST–GFP-NLS (GGNLS), which contains the SV-40 large-T-antigen NLS. GGNLS is imported by the classical importin-α/β system (Welch et al., 1999). As shown in Figure 8, wild-type Ran had no effect on the nuclear localization of the Ubc-GGH6 or of GGNLS, while the Ran (Q69L) mutant dramatically inhibited import of both fusion proteins. This result confirms that the import of the UbcM2 fusion protein does not occur by passive diffusion, and suggests that the import mechanism is mediated by a Ran-dependent process.
Fig. 8. Nuclear import of UbcM2 is Ran dependent. BHK cells were micro-injected into the cytoplasm with mixtures of Ubc-GGH6 (2.5 µM), TRITC-labeled dextran (1 mg/ml) plus either wild-type Ran (60 µM) or mutant Ran(Q69L) (50 µM). Cells were incubated for 30 min at 37°C before fixation. Control injections were done with GGNLS (19 µM). Left panel shows GFP fluorescence (GFP); middle panel shows the TRITC fluorescence (Injection marker); right panel shows the DNA staining (DAPI). Bar, 10 µm; 30–40 cells were injected, with similar results.
Importin-11 imports UbcM2 into the nuclei of permeabilized cells
To test more directly the hypothesis that importin-11 is the transport receptor for UbcM2, we carried out in vitro nuclear import assays in digitonin-permeabilized HeLa cells. The import substrate for these experiments was Ubc-GGH6. Because recombinant importin-11 precipitated out of solution when transferred into the import assay buffer it could not be used as a source of transport factor. Instead, we used a cytosolic extract prepared from human embryonic kidney (HEK) cells transiently transfected with an HA3-importin-11 expression plasmid. This plasmid expresses importin-11 with an N-terminal, triple HA tag. Control extracts were prepared from cells transiently transfected with either empty vector or an HA3-importin-β expression plasmid (Figure 9A).
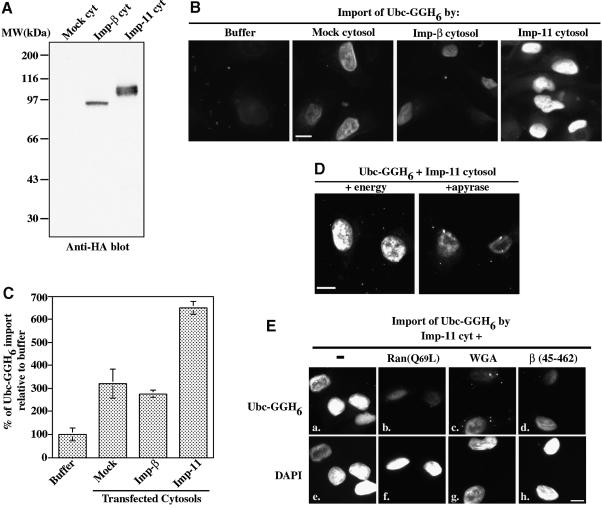
Fig. 9. Importin-11 can mediate the import of UbcM2 in vitro. (A) Aliquots of the cytosolic extracts used in (B), (D) and (E) were analyzed by immunoblotting with 12CA5 anti-HA antibody. Importin-11 migrates as a doublet. (B) Representative images of permeabilized HeLa cells overlayed with mixtures of Ubc-GGH6 (1.2 µM) and assay buffer + 4.5 µM Ran (Buffer), or cytosolic extracts containing no importin-11 (Mock cytosol), HA3-tagged importin-β (Imp-β cytosol) or HA3-tagged importin-11 (Imp-11 cytosol). (C) Relative levels of Ubc-GGH6 nuclear accumulation. Data were collected and pooled from two experiments and represent between 100 and 200 cells/condition. (D) Photomicrographs of cells incubated with Ubc-GGH6, Imp-11 cytosol, and either an energy regenerating system (+energy) or the inhibitor apyrase (+apyrase). (E) Digitonin-permeabilized HeLa cells were overlayed with mixtures of recombinant Ubc-GGH6, Imp-11 cytosol, energy and either no inhibitor (–) (a), mutant Ran:GTP [Ran(Q69L)] (6 µM) (b), wheatgerm agglutinin (0.28 mg/ml) (c) or importin-β (45–462) (2 µM) (d). The corresponding DAPI-stained nuclei for each sample are also shown (e–h). Bars, 10 µm.
HeLa cells were permeabilized with digitonin and overlaid with mixtures of Ubc-GGH6 and each of the cytosolic extracts. Ubc-GGH6 nuclear accumulation was increased 2- to 3-fold in the presence of the HA3-importin-11 cytosol, as compared with the levels achieved with either the mock or importin-β control extracts, and 6.5-fold as compared with buffer supplemented with Ran (control for transport by residual factors associated with permeabilized cells) (Figure 9B and C). The intermediate levels of import obtained with the control cytosols establish the import competency of each extract and are likely to be due to endogenous importin-11. To demonstrate further that the nuclear accumulation of Ubc-GGH6 occurs via an active, receptor-mediated translocation through the nuclear pores, rather than, for example, through binding to chromatin in over-permeabilized nuclei, we asked whether the process was sensitive to inhibition by energy depletion, or by the dominant interfering mutants Ran(Q69L) and importin-β(45–462), or by wheatgerm agglutinin. As shown in Figure 9D, energy depletion by addition of apyrase substantially reduced Ubc-GGH6 accumulation in the nuclei. The transport inhibitors also blocked nuclear accumulation of Ubc-GGH6 (Figure 9E) as effectively as they inhibited the importin-α/β pathway (data not shown). Together, these data support the hypothesis that importin-11 is a bona fide nuclear transport receptor for the UbcM2 ubiquitin-conjugating enzyme.
Discussion
It is remarkable that although most of the nuclear proteins that have been studied to date contain a ‘classical’, basic NLS, which interacts with the importin-α/importin-β receptor for translocation into the nucleus, there also exists a large family of related transport receptors that recognize other, distinct NLS sequences. In the budding yeast, S.cerevisiae, there are at least eight importins, and in mammalian cells there are many more. Why such variety? An answer to this question requires that we identify cargo proteins for each. With these cargoes in hand, it should be possible to determine whether specific regulatory processes control their import, and to what extent cross-talk occurs between different import pathways.
Among the mammalian importins, several have been shown to transport ribosomal proteins into the nucleus; transportin carries hnRNPA1, and transportin-SR imports proteins that contain SR domains. Importin-β can import proteins that contain Arg-rich sequences, and collaborate with a variety of adaptors to import classical NLS cargo (with importin-α), m7G-capped Usn RNAs (with snurportin), RPA (with XRIPα) and histone H1 (with importin-7) (for review see Görlich and Kutay, 1999). Clearly, these cases represent only the tip of the iceberg. To seek new mammalian importins and cargoes, we searched the databases for sequences related to an uncharacterized member of the S.cerevisiae karyopherin family, Lph2p. An EST encoding a C-terminal protein fragment related to Lph2p was identified and used to obtain a full-length open reading frame encoding a novel member of the karyopherin family, which we termed importin-11. Importin-11 is more similar to the yeast Lph2p protein than to other members of the karyopherin family. Using a yeast dihybrid approach, we identified a ubiquitin-conjugating enzyme, UbcM2, as a transport cargo for this importin. In addition, the screen identified Ran and a putative nucleoporin, Npap60L, as binding partners. Importin-11 and the UbcM2 interact specifically and directly. Ran:GTP can also bind directly to importin-11, and triggers the dissociation of the UbcM2 cargo. Interestingly, the N-terminally deleted mutant of importin-11, which is defective in Ran binding, appears to possess a slightly lower affinity for the UbcM2 cargo, suggesting that one function of the N-terminal domain may be to stabilize cargo binding in the absence of Ran:GTP.
When ectopically expressed, UbcM2 is predominantly nuclear, and co-expression of importin-11 accentuates the nuclear accumulation. When added to permeabilized cells, a cell extract containing importin-11 drives nuclear import of a recombinant UbcM2-GFP-GFP fusion protein. This import requires energy and is inhibited by dominant interfering mutants of Ran and importin-β, and by wheatgerm agglutinin. Technical problems in obtaining sufficient soluble importin-11 from a bacterial expression system for a reconstituted in vitro import assay precluded a test of whether importin-11 requires other soluble cofactors for the import of UbcM2. However, the permeabilized cells inevitably contain residual factors that could function catalytically in nuclear import, so this criticism also applies to prior studies of other nuclear transporters. The fact that importin-11 can bind cargo in a Ran-dependent fashion, can interact with a nucleoporin, and can shuttle and stimulate the import of its cargo, together fulfill the accepted criteria for a nuclear transport receptor.
The size of UbcM2 is such that the protein could in principle diffuse passively through the nuclear pores. Nevertheless, like histone H1, ribosomal proteins and Ran itself, which are also well below the diffusion limit of the pores, the cell uses specific, facilitated mechanisms of import, perhaps to allow the accumulation of a high concentration of these proteins within the nuclear compartment. This accumulation may help drive the assembly of ribosomes or chromatin in the case of the ribosomal proteins and histones, and is essential to prevent rapid depletion of the Ran:GTP gradient. In the case of UbcM2, import could be required to prevent its accumulation in the cytoplasm. However, this hypothesis implies that the protein leaks out of the nucleus by diffusion and our data suggest that this is not the case. Ubc-GGH6, which is above the diffusion limit, rapidly equilibrates between the nuclei of a heterokaryon, indicating that export may also be a facilitated process. The receptor for UbcM2 export remains to be identified, but it is not sensitive to the Crm1 inhibitor, leptomycin B (our unpublished data).
What is the function of nuclear UbcM2? The related proteins in yeast, Ubc4/5, are known to play an important role in the degradation of the transcription factor, MATα2 (Chen et al., 1993), and UbcM2 can partially complement a Ubc4/5 double mutant (Matuschewski et al., 1996). In mammalian cells, the ectopic expression of UbcM2 can inhibit cell proliferation (Pestov et al., 1998). Vertebrate Ubc4 homologs can mediate ubiquitylation of p53 and cyclins in vitro (Hatakeyama et al., 1997). Another member of the ubiquitin-conjugating enzyme family, UbcH5b/c, has been implicated in the polyubiquitylation of nuclear, TGF-β-activated Smad2 (Lo and Massague, 1999). Thus, it is likely that there exists a large number of nuclear targets for regulated ubiquitylation, and that the localization of specific Ubc isoforms plays a critical role in the control of this process.
Materials and methods
Cloning and recombinant protein expression
pK-UbcM2-GFP-GFP(Ubc-GG) was generated by PCR amplification of UbcM2 and ligation of the PCR product into pK-GFP-GFP, a derivative of the mammalian expression plasmid, pRK7 (Lounsbury and Macara, 1997). The bacterial vector pQE60-Ubc-GGH6 was constructed by PCR amplification, using pK-UbcM2-GFP-GFP as the template, and ligation of the PCR product into pQE60 (Qiagen).
5′ RACE was performed to obtain the complete open reading frame of importin-11. RACE was carried out using a human brain Matchmaker cDNA library (Clontech) as template, and a sense primer (pACT2 5′ primer) to the pACT2 cDNA vector, plus two nested primers complementary to nucleotides 461–491 (primer #1) and 395–425 (primer #2) of the importin-11 cDNA sequence obtained from ESTAA082435. A product of the expected size was TA-cloned and sequenced. Sequences were aligned using CLUSTAL, and phylogenetic analysis was performed using the PAUP algorithm.
The full-length importin-11 open reading frame was assembled by ligating a 270 bp, PCR-amplified 5′ RACE product to the 5′ end of ESTAA082435. The importin-11 cDNA was then moved into pK-zz, pKH3, pGBT10 and pET30a (Novagen). pK-zz expresses fusion proteins bearing two N-terminal z domains, the region of protein A that binds IgG. pKH3 expresses fusion proteins bearing an N-terminal, triple HA tag. pGBT10 was derived from the yeast vector, pGBT9 (Clontech), which expresses proteins bearing the GAL4 DNA-binding domain; and pET30a (Novagen) is a bacterial vector that expresses proteins bearing N-terminal His6 and S-peptide tags. The protein fragment encoded by ESTAA082435 is referred to as importin-11(78–975).
Ubc-GGH6, GST-UbcM2 and H6S-Imp-11 were expressed in Escherichia coli BL21(DE3), by induction with 2 mM isopropyl-β-d-thiogalactopyranoside at room temperature for 4 h in Terrific Broth. Ubc-GGH6 was purified using Talon beads (Clontech), GST–UbcM2 was purified over glutathione–Sepharose beads (Pharmacia) and H6S-Imp-11 was purified using Ni2+-NTA–agarose beads (Qiagen). The expression and purification of wild-type Ran, Ran Q69L and Ran T24N have been described previously (Lounsbury and Macara, 1997; Nemergut and Macara, 2000).
Northern blot analysis
A human poly(A)+ RNA multiple tissue northern blot (Clontech) was hybridized according to the manufacturer’s instructions. [α-32P]dCTP-labeled probes were generated with Random Primer DNA labeling system kit (Gibco-BRL), according to the manufacturer’s instructions, and the ESTAA082435 DNA or full-length human Ran was used as template.
Yeast dihybrid and conjugation assays
Full-length importin-11 and importin-11(78–975) were used separately as baits for yeast dihybrid screens of a random-primed, murine library created from a whole, 10 day embryo. From 1.2 × 106 transformants in the first screen, five positive clones were identified, which included two UbcM2 clones. From 5.4 × 106 transformants in the second screen, seven positive clones were identified of which one corresponded to UbcM2.
Mating assays were performed as described previously (Neudauer et al., 1998). Importin-11, importin-11(78–975) and CAS were all expressed in the S.cerevisiae HF7c (MATa) strain as C-terminal fusions to the DBD of the GAL4 protein. UbcM2, UbcM2(53–207) and Ran were expressed in the W303α (MATα) strain as C-terminal fusions to the TA domain of VP16.
Transient transfections
BHK cells and HEK cells were seeded 20 h before transfection and transfected by a calcium phosphate precipitation procedure (Plafker and Macara, 2000).
Immunofluorescence
Cells to be processed for fluorescence microscopy were seeded onto poly-lysine-coated coverslips and processed as described (Plafker and Macara, 2000). zz-tagged proteins were detected with Texas Red-conjugated, rabbit anti-goat antibodies (1:20 000). HA3-tagged RCC1 was detected with mouse monoclonal anti-HA antibody 12CA5 (1:500), followed by 4× 5 min phosphate-buffered saline (PBS) washes, and incubation with Texas Red-conjugated, anti-mouse secondary antibodies (1:1500). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.01 µg/ml in PBS). Cells were examined using a Nikon inverted microscope equipped with a 60× water immersion objective lens, and images were captured with a Hamamatsu charge-coupled device camera and Openlab software (Improvision).
Heterokaryon shuttling assays
Transfected BHK cells were mixed with an equal number of GSN2 cells or stably transformed BHK cells that express HA3-RCC1. The mixed cell populations were seeded onto poly-lysine-coated coverslips, grown overnight at 37°C, and fused with 50% polyethylene glycol 8000 (PEG; Sigma) in Hanks’ buffered saline solution for 2 min at 23°C. The fusogen was rinsed off and the coverslips were placed in growth medium supplemented with 35 µM cycloheximide. After 2 h at 37°C, the cells were processed for indirect immunofluorescence. Ubc-GG shuttling assays were carried out with BHK cells stably transformed with HA-tagged RCC1 (Nemergut and Macara, 2000). Importin-11 shuttling was tested using GSN2 cells (kind gift of Bryce Paschal, University of Virginia, VA) (Black et al., 1999).
Microinjections
Microinjections were performed as described previously (Plafker and Macara, 2000). Ubc-GGH6 was injected at 5 µM, Ran at 40–50 µM and GGNLS at 19 µM. TRITC-labeled dextran (1 mg/ml) was included in all mixtures as an injection site marker. Following injections, samples were incubated for 30 min at 37°C before being fixed, permeabilized and stained with DAPI.
Preparation of cytosolic extracts
HEK cells were transfected as described above and cytosolic extracts were prepared 2 days post-transfection as described previously (Holaska and Paschal, 1998). The extract was clarified by centrifugation for 30 min in a 4°C microcentrifuge, and the resulting supernatant was filtered through a pre-filter disc (0.45 µm pores) then centrifuged at 100 000 g for 10 min in an airfuge (Beckman). The protein concentration of the cytosolic extracts was determined by Bradford assay. HA3-tagged protein in each sample was immunoblotted with a horseradish peroxidase (HRP)-conjugated, 12CA5 antibody (1:5000).
Binding assays
All binding assays were performed at 4°C. 35S-labeled proteins were synthesized by in vitro transcription–translation from pKH3 vectors, according to the manufacturer’s instructions (Promega). Ubc-GGH6, GFP-His6 and importin-α H6 (all at 1 µg/sample) were incubated in 100 µl reaction mixtures containing the 35S proteins, plus 20 µl of Ni2+-NTA–agarose beads, and transport buffer (TB; 20 mM HEPES–KOH pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate and 0.5 mM EGTA) plus 0.1% v/v Tween-20 and 3% w/v bovine serum albumin (BSA) for 60 min, shaken at 1400 r.p.m. Following two 500 µl rinses with TB–Tween and one with PBS, bound proteins were resolved by SDS–PAGE, stained and visualized by fluorography. For the Ran-elution experiments, the bead-bound complexes were rinsed twice, then combined with 20 µM Ran pre-loaded with GTP, 20 µM Ran(T24N) or TB. After 30 min incubation, 20% of the supernatant from each reaction was solubilized for SDS–PAGE and fluorography. The remaining bead-bound complexes were rinsed with 500 µl of PBS, then analyzed as described above.
For direct binding, GST–Ubc (1.3 µg) was combined with either 20 µl of peptide-S–agarose beads alone or beads plus recombinant His-S-Imp-11 (1.3 µg) in RanBP binding buffer (Lounsbury and Macara, 1997) and shaken for 60 min. GST (1 µg) was used as a negative control. After 3× 500 µl washes, the bound fractions were resolved by SDS–PAGE and detected by immunoblotting. GST–Ubc and GST were detected with an anti-GST antibody (1:4000), an anti-mouse-HRP secondary antibody (1:20 000) and enhanced chemiluminescence (ECL). His-S-Imp-11 was detected with an S-protein–HRP conjugate (Novagen; 1:5000) and ECL.
In vitro nuclear import assays
HeLa cells were plated on poly-lysine-coated coverslips 20–24 h before being permeabilized on ice with 0.005% digitonin (Calbiochem-Novabiochem Corp). Import assays were performed as described previously (Plafker and Macara, 2000). All import cocktails contained 1 µg/µl nucleoplasmin core, and either an energy-regenerating system or apyrase. Ubc-GGH6 was used at a final concentration of 1.8 µM and GGNLS at 18 µM. Transfected cytosols were used at final concentrations ranging from 0.6 to 0.9 µg/µl. Import inhibitors were at the following concentrations: Ran(Q69L) 6 µM, WGA 0.28 mg/ml and Imp-β(45–462) 2 µM. (See Supplementary data for quantitation methods.)
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank members of the Macara laboratory for helpful discussions, Dirk Görlich for the nucleoplasmin core expression vector and Michael Black (ITS, University of Virginia, VA) for help with the PAUP analysis. Lester Lau kindly provided a mouse UbcM2 clone. This work was supported by a grant from the National Institutes of Health, DHHS to I.G.M. (GM50526). S.M.P. is a recipient of a postdoctoral NRSA fellowship from the National Institutes of Health (F32 GM20478).
References
- Black B.E., Levesque,L., Holaska,J.M., Wood,T.C. and Paschal,B.M. (1999) Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol. Cell. Biol., 19, 8616–8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer R.A., Lehner,C.F., Eppenberger,H.M. and Nigg,E.A. (1989) Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell, 56, 379–390. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson,P., Sommer,T., Jentsch,S. and Hochstrasser,M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell, 74, 357–369. [DOI] [PubMed] [Google Scholar]
- Chi N.C. and Adam,S.A. (1997) Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol. Biol. Cell, 8, 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y.M. and Blobel,G. (1999) Structure of the nuclear transport complex karyopherin-2-Ran.GppNHp. Nature, 399, 230–237. [DOI] [PubMed] [Google Scholar]
- Cole C.N. and Hammell,C.M. (1998) Nucleocytoplasmic transport: driving and directing transport. Curr. Biol., 8, R368–R372. [DOI] [PubMed] [Google Scholar]
- Corbett A.H. and Silver,P.A. (1997) Nucleocytoplasmic transport of macromolecules. Microbiol. Mol. Biol. Rev., 61, 193–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M. and Silver,P.A. (2000) Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell, 5, 133–140. [DOI] [PubMed] [Google Scholar]
- Davis L.I. (1995) The nuclear pore complex. Annu. Rev. Biochem., 64, 865–896. [DOI] [PubMed] [Google Scholar]
- Fan F., Liu,C.P., Korobova,O., Heyting,C., Offenberg,H.H., Trump,G. and Arnheim,N. (1997) cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics, 40, 444–453. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Jensen,J.P. and Weissman,A.M. (1997) Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J. Biol. Chem., 272, 15085–15092. [DOI] [PubMed] [Google Scholar]
- Holaska J.M. and Paschal,B.M. (1998) A cytosolic activity distinct from Crm1 mediates nuclear export of protein kinase inhibitor in permeabilized cells. Proc. Natl Acad. Sci. USA, 95, 14739–14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Kato,S., Matsuda,Y., Kimura,M. and Okano,Y. (1999) cDNA cloning, characterization and chromosome mapping of UBE2E3 (alias UbcH9), encoding an N-terminally extended human ubiquitin-conjugating enzyme. Cytogenet. Cell Genet., 84, 99–104. [DOI] [PubMed] [Google Scholar]
- Jakel S. and Görlich,D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts,B.L., Richardson,W.D. and Smith,A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Kunzler M. and Hurt,E.C. (1998) Cse1p functions as the nuclear export receptor for importin α in yeast. FEBS Lett., 433, 185–190. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997a) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde,E., Bischoff,F.R., Mattaj,I.W. and Görlich,D. (1997b) Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J., 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R.S. and Massague,J. (1999) Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nature Cell Biol., 1, 472–478. [DOI] [PubMed] [Google Scholar]
- Lounsbury K.M. and Macara,I.G. (1997) Ran-binding protein 1 (RanBP1) forms a ternary complex with ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J. Biol. Chem., 272, 551–555. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Matuschewski K., Hauser,H.P., Treier,M. and Jentsch,S. (1996) Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J. Biol. Chem., 271, 2789–2794. [DOI] [PubMed] [Google Scholar]
- Melchior F. and Gerace,L. (1998) Two-way trafficking with Ran. Trends Cell Biol., 8, 175–179. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nemergut M.E. and Macara,I.G. (2000) Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol., 149, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudauer C.L., Joberty,G., Tatsis,N. and Macara,I.G. (1998) Distinct cellular effects and interactions of the Rho-family GTPase TC10. Curr. Biol., 8, 1151–1160. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum,J.S. and Blobel,G. (1999) Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol., 145, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov D.G., Grzeszkiewicz,T.M. and Lau,L.F. (1998) Isolation of growth suppressors from a cDNA expression library. Oncogene, 17, 3187–3197. [DOI] [PubMed] [Google Scholar]
- Plafker K. and Macara,I.G. (2000) Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol. Cell. Biol., 20, 3510–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Dobbelstein,M., Freedman,D.A., Shenk,T. and Levine,A.J. (1998) Nucleo-cytoplasmic shuttling of the mdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J., 17, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D., Fahrenkrog,B. and Aebi,U. (1999) The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol., 11, 391–401. [DOI] [PubMed] [Google Scholar]
- Tao W. and Levine,A.J. (1999) P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl Acad. Sci. USA, 96, 6937–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (1997) The ubiquitin system. Trends Biochem. Sci., 22, 383–387. [DOI] [PubMed] [Google Scholar]
- Welch K., Franke,J., Kohler,M. and Macara,I.G. (1999) RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-α3. Mol. Cell. Biol., 19, 8400–8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R. (2000) Gatekeepers of the nucleus. Science, 288, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Wozniak R.W., Rout,M.P. and Aitchison,J.D. (1998) Karyopherins and kissing cousins. Trends Cell Biol., 8, 184–188. [DOI] [PubMed] [Google Scholar]