Abstract
The RNA-binding motif protein 3 (RBM3) was initially discovered as a putative cancer biomarker based on its differential expression in various cancer forms in the Human Protein Atlas (HPA). We previously reported an association between high expression of RBM3 and prolonged survival in breast and epithelial ovarian cancer (EOC). Because the function of RBM3 has not been fully elucidated, the aim of this study was to use gene set enrichment analysis to identify the underlying biologic processes associated with RBM3 expression in a previously analyzed EOC cohort (cohort 1, n = 267). This revealed an association between RBM3 expression and several cellular processes involved in the maintenance of DNA integrity. RBM3-regulated genes were subsequently screened in the HPA to select for putative prognostic markers, and candidate proteins were analyzed in the ovarian cancer cell line A2780, whereby an up-regulation of Chk1, Chk2, and MCM3 was demonstrated in siRBM3-treated cells compared to controls. The prognostic value of these markers was assessed at the messenger RNA level in cohort 1 and the protein level in an independent EOC cohort (cohort 2, n = 154). High expression levels of Chk1, Chk2, and MCM3 were associated with a significantly shorter survival in both cohorts, and phosphorylated Chk2 was an adverse prognostic marker in cohort 2. These results uncover a putative role for RBM3 in DNA damage response, which might, in part, explain its cisplatin-sensitizing properties and good prognostic value in EOC. Furthermore, it is demonstrated that Chk1, Chk2, and MCM3 are poor prognostic markers in EOC.
Introduction
Epithelial ovarian cancer (EOC) is the fifth most common cause of cancer-related death in women and carries the highest mortality rate of gynecological malignancies in the western world. In 2008, it was estimated that 21,650 new ovarian cancer cases would be diagnosed in the United States and that 15,520 would die of the disease [1]. The poor ratio of survival to incidence in EOC is related to the high percentage of cases that are diagnosed at an advance stage and the lack of effective therapies for advanced refractory disease. Adjuvant systemic chemotherapy for ovarian cancer is empiric and initial treatment involves paclitaxel-platinum-based regimens, which continue to show improved outcomes compared with other cytotoxic agents such as gemcitabine, topotecan, and liposomal doxorubicin [2]. Despite aggressive surgery and chemotherapy, most patients relapse within 3 to 5 years, and the median time to relapse is 15 months after diagnosis [3]. Such poor statistics indicate the urgent need for the development of new diagnostic, prognostic, and predictive biomarkers, which are essential for the development of personalized therapeutic regimens for ovarian cancer patients [4].
RNA-binding proteins with RNA-binding motifs (RBM) are involved in many aspects of RNA processing and regulation of gene transcription [5,6]. The RNA-binding motif protein 3 (RBM3) protein has been shown to bind to both DNA and RNA [7]. We initially described RBM3 as a putative cancer biomarker based on its differential expression in various cancer forms in the Human Protein Atlas (HPA) (www.proteinatlas.org) [8,9]. Within this context, we described RBM3 as a prognostic biomarker in breast cancer, which is associated with an improved survival, particularly in estrogen receptor-positive tumors [10]. We subsequently reported an association between RBM3 messenger RNA (mRNA) and protein expression and good prognosis in two independent EOC cohorts and demonstrated that RBM3 expression conferred sensitivity to cisplatin in vitro [11].
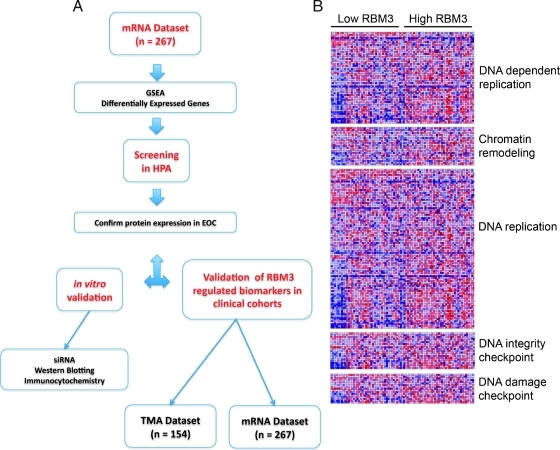
These data suggest that RBM3 may play a key role in both breast and ovarian tumorigenesis and progression; however, its exact function is still to be fully elucidated. The aim of this study was to identify the underlying biologic processes associated with RBM3 expression in EOC and use this approach to identify new prognostic and predictive biomarkers. Our secondary objective was to improve understanding of the molecular mechanisms underlying the prognostic value of RBM3 in EOC. This approach involved the integration of transcriptomic and antibody-based proteomic data whereby gene set enrichment analysis (GSEA) was performed in a cohort of 267 EOC cases from a publicly available data set [12], in which we have previously demonstrated that high RBM3 expression levels independently predict a prolonged survival [11]. The HPA was then screened to select promising EOC biomarker candidates identified from the aforementioned GSEA. These biomarkers were subsequently validated in vitro and in an independent EOC tissue microarray (TMA). This method, schematically described in Figure 1A, highlights a novel approach to biomarker discovery whereby transcriptomic and proteomic data can be integrated to identify new biomarkers.
Figure 1.
Identification of cellular processes associated with RBM3 expression in EOC. (A) Flowchart illustrating a novel approach to biomarker discovery whereby transcriptomic and proteomic data can be integrated to identify new biomarkers. (B) GSEA demonstrated that increased RBM3 expression was associated with a number of processes including DNA-dependent replication P < .01, chromatin remodeling P < .05, DNA replication P < .01, DNA integrity checkpoint P < .05, and DNA damage checkpoint P < .05.
Materials and Methods
Patients
Cohort 1. Cohort 1 is composed of 285 cases of serous and endometrioid carcinoma of the ovary, fallopian tube, and peritoneum. The cohort has been described previously [12]. Most patients underwent laparotomy for staging and debulking and, subsequently, received first-line platinum/taxane-based chemotherapy. In most cases, tumor tissue was excised at the time of primary surgery, before the administration of chemotherapy. Eighteen patients who received neoadjuvant platinum-based chemotherapy were excluded from this study; hence, the total number or patients examined was 267. Optimal debulking was defined as less than 1 cm (diameter) residual disease, and suboptimal debulking was more than 1 cm (diameter) residual disease. Recurrence-free survival (RFS) was defined as the time interval between the date of diagnosis and the first confirmed sign of disease recurrence based on GCIG definitions. Overall survival (OS) was defined as the time interval between the date of histological diagnosis and the date of death from any cause. Median follow-up was 29 months (range = 0–214 months).
RNA was extracted from tumors and hybridized to Affymetrix U133 Plus 2 arrays as previously described [12]. Complete expression data were downloaded from GEO (www.ncbi.nlm.nih.gov/geo) (Accession GSE9899). R package “Affy” (www.bioconductor.org) was used to normalize the CEL files using the Robust Multichip Average (RMA) method [13]. For RBM3 analysis, normalized gene expression values were extracted from the data set and used without modification. Tumor samples were classified using a previously published method [14].
Cohort 2. This cohort is a merge of all incident cases of EOCs in the large, population-based prospective cohort studies Malmö Diet and Cancer Study [18] (n = 101) and Malmö Preventive Medicine Study [19] (n = 108) until December 31, 2008, and has been described previously [11]. Thirty-five patients participated in both studies, and archival tumor tissue could be retrieved from 154 of the total number of 174 cases. After a median follow-up of 2.65 years (range = 0–21 years), 105 patients (68.2%) were dead and 49 (31.8%) were alive. All tumors were reevaluated regarding histological subtype and histological grade. Information regarding clinical stage was obtained from the medical charts, following the standardized classification of tumor staging of the International Federation of Gynecology and Obstetrics. Information on residual disease after surgery was not available. Standard adjuvant therapy was platinum-based chemotherapy from the 1990s, given in combination with paclitaxel.
Ethical permission was obtained from the ethics committee at Lund University (reference no. 447-07 and 35/08), whereby informed consent was deemed not to be required other than by the opt-out method.
Human Protein Atlas TMAs
Tissue microarrays containing triplicate 1-mm cores of 48 different types of normal tissue, duplicate 1-mm cores of 216 different cancer tissues, and a cell microarray including 47 different cell lines and 12 patient cell samples were generated as previously described [15,16].
GSEA and Selection of Interesting Genes
The microarray data set was downloaded from the GEO Web site (http:/www.ncbi.nlm.nih.gov/geo). Data were analyzed using Bioconductor 1.9 (http://bioconductor.org) running on R 2.6.0 [17]. Probe set expression measures were calculated using the Affymetrix package's RMA default method [18]. The function GeneSetTest from the limma package [19] was used to assess whether each sample had a tendency to be associated with an up-regulation or down-regulation of RBM3. All samples were ranked on this enrichment, from the most significant to the least significant. The top and bottom 50 samples was extracted from the data set and given the names of “high-RBM3” and “low-RBM3,” respectively. Differential gene expression was assessed using the signal-to-noise ratio. Gene set enrichment was performed using GSEA software (http://www.broadinstitute.org/gsea/index.jsp) as previously described [20,21]. Heat maps were drawn using expression data showing the probes that mapped to the biologic processes of DNA dependent DNA replication, chromatin remodeling, DNA replication, DNA integrity checkpoint, and DNA damage checkpoint.
Cell Lines and Reagents
The human ovarian cancer cell line A2780 (received as a gift from Prof R. Brown, Imperial College, London) was maintained in RPMI-1640 supplemented with glutamine, 10% fetal bovine serum, and 1% penicillin/streptomycin in a humidified incubator of 5% CO2 at 37°C.
Real-time Quantitative Polymerase Chain Reaction and Western Blot Analysis
Total RNA isolation (RNeasy; QIAgen, Hilden, Germany), complementary DNA synthesis (Reverse Transcriptase Kit; Applied Biosystems, Warrington, United Kingdom), and real-time quantitative polymerase chain reaction (QPCR) analysis with SYBR Green PCR master mix (Applied Biosystems) were performed as previously described [22,23]. Quantification of expression levels was done using the comparative Ct method, normalization according to housekeeping genes HMBS, YWHAZ, and UBC. Primer sequences are given in Table W1. All primers were designed using Primer Express (Applied Biosystems).
For immunoblot analysis, cells were lysed in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1% Triton X-100, 50 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride) and supplemented with protease inhibitor cocktail Complete Mini (Roche, Basel, Switzerland). For Western blot analysis, 20 to 50 µg of protein was separated on 15% SDS-PAGE gels and transferred onto nitrocellulose membranes (Hybond ECL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). The membranes were probed with primary antibodies followed by horseradish peroxidase. conjugated secondary antibodies (Amersham Life Science, Alesbury, United Kingdom) and visualized using the enhanced chemiluminescence detection system (ECL) and ECL films (Amersham Pharmacia Biotech). RBM3 was detected by the mouse monoclonal anti-RBM3 antibody (AAb030038; Atlas Antibodies AB, Stockholm, Sweden) diluted 1:500 in blocking solution (5% bovine serum albumin, 1x PBS, 0.1% Tween 20). Dilutions of the investigative antibodies are given in Table W2. Membranes were stripped and reprobed with an anti-β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:1000, to provide a loading control.
TMA Construction
Before TMA construction, all cases were histopathologically reevaluated on hematoxylin and eosin-stained slides. Areas representative of cancer were then marked, and TMAs were constructed as previously described [24]. In brief, two to four 1.0-mm cores were taken from each tumor and mounted in a new recipient block using a semiautomated arraying device (TMArrayer; Pathology Devices, Inc, Westminster, MD).
Immunohistochemistry and Analysis of Staining
For immunohistochemical analysis, 4-µm TMA sections were automatically pretreated using the PT-link system (DAKO, Copenhagen, Denmark) and then stained in a Techmate 500 (DAKO) with the mouse monoclonal anti-RBM3 antibody (AAb030038) diluted 1:5000 and MCM3 (HPA 004789) diluted 1:1000 from Atlas Antibodies. The following antibodies were purchased from Cell Signaling Technologies (Danvers, MA): Chk1 (mouse monoclonal, clone 2G1D5, no. 2360) diluted 1:100, Chk2 (mouse, monoclonal, clone 1C12, no. 3440,) diluted 1:2000, pSer345-Chk1 (rabbit monoclonal, no. 2348) diluted 1:150, and pT68-Chk2 (rabbit polyclonal no. 2661) diluted 1:50.
Chk1, Chk2, MCM3, and phosphorylated Chk1 and Chk2 were mainly expressed in the nuclei, and both the fraction of positive cells and staining intensity were taken into account using a semiquantitative scoring system as described previously for the assessment of RBM3 staining [14]. Nuclear fraction (NF) was categorized into four groups, namely 0 (0%–1%), 1 (2%–25%), 2 (26%–75%), and 3 (>75%) and nuclear staining intensity (NI) denoted as 0 to 2, whereby 0 = negative, 1 = intermediate, and 2 = moderate to strong intensity. A combined nuclear score (NS) of NFxNI, which had a range of 0 to 6, was then constructed.
Cell Pellet Arrays
Cell lines were fixed in 4% formalin and processed in gradient alcohols. Cell pellets were cleared in xylene and washed multiple times in molten paraffin. Once processed, cell lines were arrayed in duplicate 1.0-mm cores using a manual tissue arrayer (Beecher, Inc, Sun Prairie, WI), and immunohistochemistry was performed on 5-µm sections using the same antibodies as for immunohistochemistry with the following dilutions: RBM3, 1:1000; Chk1, 1:100; Chk2 and MCM3, 1:2000; pSer345-Chk1 and pT68-Chk2, 1:50.
Small Interfering RNA-Mediated Knockdown of RBM3 Gene Expression
Transfection with small Interfering RNA (siRNA) against RBM3 (Applied Biosystems, Carlsbad, CA) or control siRNA (Applied Biosystems) was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with a final concentration of 50 nM siRNA. All siRNA experiments were performed using three independent RNA oligonucleotides (nos. 58, 59, and 60) targeting RBM3.
Statistics
Spearman ρ tests were used for comparison of Chk1, Chk2, and MCM3 expression with RBM3 expression and relevant clinicopathological characteristics. Kaplan-Meier analysis, using mean expression value to dichotomize data, and log-rank test were used to illustrate differences in RFS and overall survival (OS) according to CHK1, CHK2, and MCM3 gene expression and OS according to Chk1, Chk2, and MCM3 protein level. Cox regression proportional hazards models were used to estimate the impact of RBM3 expression on RFS and OS in both univariate and multivariate analyses, adjusted for stage and differentiation grade (both cohorts) and volume of residual tumor (0 vs >0) in cohort 1. Patients who had received neoadjuvant chemotherapy in cohort 1 (n = 18) were excluded from the survival analyses. All calculations were performed using SPSS version 15.0 (SPSS, Inc, Chicago, IL). All statistical tests were two-sided, and P < .05 was considered statistically significant. Experimental data are expressed as mean ± SEM of at least three independent experiments. Statistical significance of differences between means was determined by Student's t test.
Results
Identification of Cellular Processes Associated with RBM3 Expression in EOC
In an attempt to profile the effect of RBM3 expression in EOC, we used gene expression data from a previously described cohort [12] to compare the gene profiles of treatment-naive tumors with high RBM3 mRNA levels to those tumors showing no or low RBM3 expression. Comparison of the 50 tumors expressing the highest levels of RBM3 mRNA to the 50 tumors expressing the lowest levels of RBM3 mRNA using GSEA demonstrated that increased RBM3 expression was associated with a number of processes including DNA-dependent replication, DNA replication, chromatin remodeling, and DNA integrity checkpoint (Figure 1B). Low RBM3 mRNA expression was associated with a variety of different processes including cAMP G protein signaling, transcription factor activity, and the protein kinase cascade.
Validation of Selected Candidates by Western Blot Analysis and Real-time QPCR in siRBM3-Treated A2780 Ovarian Cancer Cells
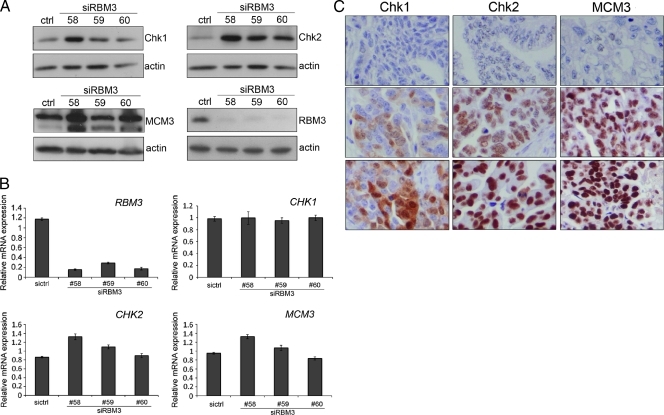
The HPA platform was then used to select the most promising biomarkers identified from the GSEA for further validation. As increased RBM3 was associated with an improved prognosis and cisplatin sensitivity, we concentrated on cellular processes associated with high RBM3 mRNA expression. From the list of differentially expressed genes associated with DNA-dependent replication, DNA replication, chromatin remodeling, and DNA integrity checkpoint, we selected corresponding proteins with a differential expression pattern in EOC in the HPA, with available validated antibodies. In total, 28 genes were selected for an initial validation in the human ovarian cancer cell line A2780 (Table W2). Of these 28 genes, 3 were chosen for further validation; the DNA damage checkpoint kinases (CHK1 and CHK2) and minichromosome maintenance protein 3 (MCM3). These markers were chosen because they play a role in DNA integrity, and we have previously shown that RBM3 sensitizes A2780 cells to the DNA-damaging agent cisplatin.
To demonstrate an association between RBM3 and the selected candidates, A2780 cells were transfected with RBM3-specific siRNA followed by Western blot analysis. siRNA-mediated knockdown of RBM3 resulted in an increase in Chk1, Chk2, and MCM3 protein expression (Figure 2A). Real-time QPCR demonstrated that siRNA-mediated knockdown of RBM3 did not alter transcription of the CHK1, CHK2, and MCM3 genes, suggesting that RBM3 may silence translation of these proteins (Figure 2B). Evaluation of Chk1, Chk2, and MCM3 protein expression in EOC tumor tissue demonstrated nuclear expression of all three proteins, with Chk1 also expressed occasionally in the cytoplasm (Figure 2C).
Figure 2.
Downregulation of RBM3 affects the expression of MCM3, Chk1, and Chk2. The expression of MCM3, Chk1, Chk2, and RBM3 were examined by (A) Western blot analysis and (B) reverse transcription-PCR in A2780 cells 48 hours after transfection of cells with three different siRNAs targeting RBM3 (nos. 58, 59, and 60). Data shown are mean ± SEM of four, for siRBM3 nos. 58 and 59, and three for siRBM3 no. 60, independent experiments performed in triplicate. (C) Immunohistochemical staining of Chk1, Chk2, and MCM3 in EOC tumors denoted as negative, intermediate, and strong.
Survival Analysis of RBM3-Regulated Biomarkers in EOC
Cohort 1 (n = 267) was used to examine the prognostic value of CHK1, CHK2, and MCM3 at the mRNA level, and immunohistochemistry was performed on a TMA consisting of 154 prospectively collected EOC cases (cohort 2) using antibodies against the corresponding proteins. As visualized in Table 1, the relationship between RBM3 and the candidate biomarkers demonstrated a negative correlation between RBM3 and MCM3, CHK1, and CHK2 at the mRNA level in cohort 1 but not at the protein level in cohort 2. MCM3, CHK1, and CHK2 correlated significantly with each other and with a lower differentiation grade in both cohorts. MCM3 expression was associated with a more advanced clinical stage in both cohorts, and the same was seen for CHK1 in cohort 1, whereas CHK2 mRNA or protein levels were not significantly associated with clinical stage.
Table 1.
Associations between Chk1, Chk2, and MCM3 and Patient and Tumor Characteristics in Cohorts 1 and 2.
| Cohort | 1 | 2 | ||||
| Variable | CHK1 | CHK2 | MCM3 | Chk1 | Chk2 | MCM3 |
| Age | ||||||
| R | 0.102 | 0.069 | 0.168 | -0.009 | 0.038 | 0.126 |
| P | .1 | .265 | .006 | .914 | .652 | .139 |
| n | 263 | 263 | 263 | 141 | 145 | 140 |
| Differentiation grade | ||||||
| R | 0.329 | 0.186 | 0.323 | 0.328 | 0.270 | 0.286 |
| P | <.001 | .003 | <.001 | <.001 | .001 | .001 |
| n | 263 | 260 | 260 | 141 | 145 | 140 |
| Clinical stage | ||||||
| R | 0.13 | 0.108 | 0.141 | 0.129 | 0.116 | 0.181 |
| P | .035 | .080 | .022 | .141 | .185 | .04 |
| n | 263 | 263 | 263 | 131 | 133 | 130 |
| RBM3 | ||||||
| R | -0.247 | -0.192 | -0.253 | 0.126 | 0.074 | 0.074 |
| P | <.001 | .002 | <.001 | .138 | .38 | .386 |
| n | 263 | 263 | 263 | 140 | 143 | 140 |
| CHK1/Chk1 | ||||||
| R | 0.440 | 0.599 | 0.451 | 0.462 | ||
| P | <.001 | <.001 | <.001 | <.001 | ||
| n | 263 | 263 | 139 | 137 | ||
| CHK2/Chk2 | ||||||
| R | 0.440 | 0.412 | 0.451 | 0.509 | ||
| P | <.001 | <.001 | <.001 | <.001 | ||
| n | 263 | 263 | 139 | 139 | ||
n indicates number of tumor samples; R, Spearman correlations coefficient.
P < .005 in bold.
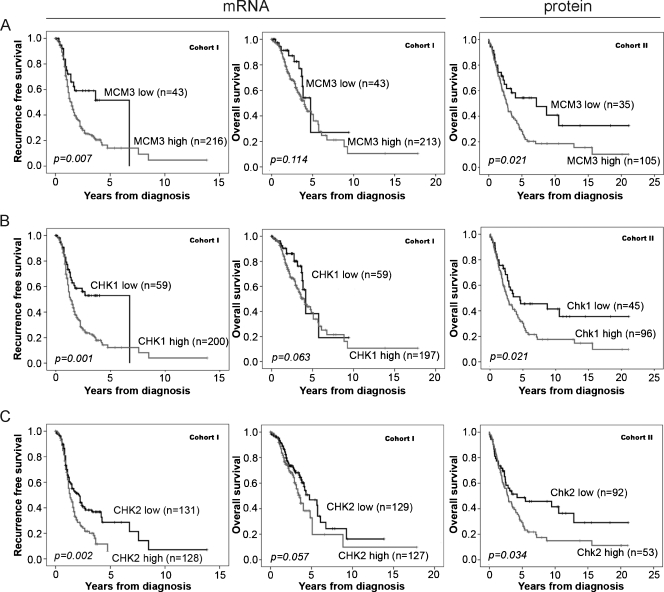
Kaplan-Meier analysis demonstrated an association between high MCM3 mRNA and protein expression and a significantly reduced RFS, but not OS, in cohort 1 and a reduced OS in cohort 2 (Figure 3A). Cox univariate analysis confirmed the association between increased MCM3 mRNA expression and decreased RFS (hazard ratio [HR] = 1.98, 95% confidence interval [CI] = 1.18–3.31, P = .010) in cohort 1 and increased MCM3 protein expression (NS > 3) and poor OS in cohort 2 (HR = 1.82, 95% CI = 1.09–3.04, P = .022). However, multivariate Cox regression analysis did not confirm MCM3 as an independent prognostic marker in either cohort (Table 2).
Figure 3.
Increased mRNA (cohort 1) and protein expression (cohort 2) of MCM3, Chk1, and Chk2 are associated with an impaired survival. Kaplan-Meier analysis of RFS and OS according to (A) MCM3, (B) CHK1, and (C) CHK2 mRNA levels in cohort 1. Kaplan-Meier analysis of OS according to immunohistochemical (A) MCM3, (B) Chk1, and (C) Chk2 staining in cohort 2.
Table 2.
Cox Univariate and Multivariate Analyses of RFS of Chk1, Chk2, and MCM3 According to mRNA Expression (Cohort 1) and Protein Expression (Cohort 2).
| Cohort 1 (mRNA) | Cohort 2 (Protein) | ||||
| HR (95% CI) | P | HR (95% CI) | P | ||
| MCM3 | MCM3 | ||||
| Univariate | Univariate | ||||
| Low | 1.00 | Low | 1.00 | ||
| High | 1.98 (1.18–3.31) | .01 | High | 1.82 (1.09–3.04) | .022 |
| Multivariate | Multivariate | ||||
| Low | 1.00 | Low | 1.00 | ||
| High | 2.39 (1.30-0.72) | .383 | High | 1.05 (0.60–1.83) | .871 |
| Chk1 | Chk1 | ||||
| Univariate | Univariate | ||||
| Low | 1.00 | Low | 1.00 | ||
| High | 2.05 (1.33–3.15) | .001 | High | 1.70 (1.08–2.68) | .023 |
| Multivariate | Multivariate | ||||
| Low | 1.00 | Low | 1.00 | ||
| High | 1.37 (0.84–2.24) | .203 | High | 1.22 (0.71–2.10 | .47 |
| Chk2 | Chk2 | ||||
| Univariate | Univariate | ||||
| Low | 1.00 | Low | 1.00 | ||
| High | 1.61 (1.19–2.19) | .002 | High | 1.59 (1.03–2.46) | .036 |
| Multivariate | Multivariate | ||||
| Low | 1.00 | Low | 1.00 | ||
| High | 1.52 (1.08–2.13) | .015 | High | 1.21 (0.74–1.97) | .448 |
Multivariate analysis included adjustment for age (continuous), stage (I–II vs III–IV), grade (1–2 vs 3), and residual disease (none vs any, only available for cohort 1).
Kaplan-Meier analysis revealed an association between high CHK1 mRNA levels and a reduced RFS in cohort 1 (Figure 3B), which was further confirmed by Cox univariate analysis (HR = 2.05, 95% CI = 1.33–3.15, P = .001) (Table 2). Cox multivariate analysis demonstrated that CHK1 was not an independent predictor of RFS (HR = 1.37, 95% CI = 0.84–2.24, P = .203) in cohort 1 (Table 2), and CHK1 mRNA expression was not associated with OS (Figure 3B). In cohort 2, Chk1 protein expression (NS > 0) was associated with a reduced OS (Figure 3B) confirmed by Cox univariate analysis (HR = 1.70, 95% CI = 1.08–2.68, P = .023). However, multivariate analysis did not confirm Chk1 protein expression as an independent predictor of OS in cohort 2 (HR = 1.23, 95% CI = 0.71–2.10, P = .47) (Table 2).
High levels of Chk2, both mRNA and protein levels, were associated with an impaired survival in EOC (Figure 3C). Cox univariate analysis confirmed the association between increased CHK2 mRNA expression and RFS in cohort 1 (HR = 1.61, 95% CI = 1.19–2.19, P = .002) and Chk2 protein expression (NS > 0) and OS in cohort 2 (HR = 1.59, 95% CI = 1.03–2.47, P = .036). Multivariate Cox regression analysis confirmed the association between high CHK2 mRNA expression and poor outcome in cohort 1 (HR = 1.52, 95% CI = 1.08–2.13, P = .015); however, this was not replicated at the protein level in cohort 2 (HR = 1.21, 95% CI = 9.74-1.97, P = .448) (Table 2).
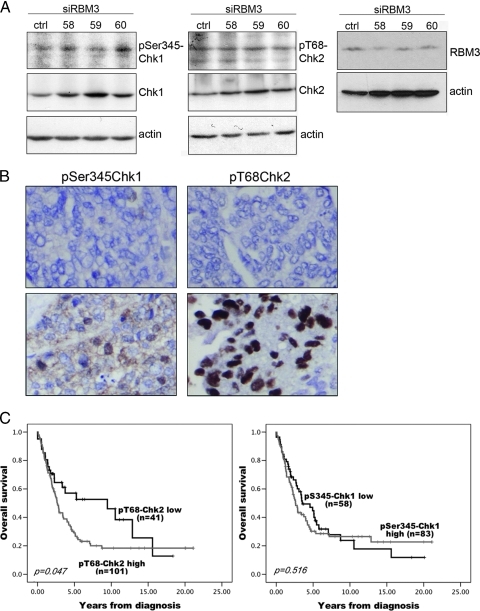
Down-regulation of RBM3 Generates an Increase in Phosphorylation of Chk1
The protein kinases Chk1 and Chk2 are crucial checkpoint proteins activated in response to DNA damage by signals from ATM and ATR leading to cell cycle arrest and DNA repair through activation of a complex signaling network [25–27]. Taken in the context of a previous study by Sureban et al. [28], who demonstrated that down-regulation of RBM3 in the human colon adenocarcinoma cell line HCT116 resulted in activation of DNA damage response by phosphorylation of the checkpoint proteins Chk1 and Chk2 [28], we hypothesized that RBM3 activates Chk1 and Chk2 in EOC. To address this issue, we examined the phosphorylation of Chk1 at Ser-345 and Chk2 at Thr-68 by Western blot analysis in A2780 cells transfected with siRNA targeting RBM3. A slightly higher level of pSer345-Chk1, but not pThr68-Chk2, was observed in the siRBM3 transfected A2780 cells (Figure 4A), indicating activation of the checkpoint proteins by down-regulation of RBM3 in the absence of DNA damage. This suggests that RBM3 may restrain a checkpoint response in the absence of DNA damage by regulating the protein levels of Chk1 and Chk2 to maintain a relative low cellular level of the phosphorylated and total proteins in absence of a DNA damage stimulus.
Figure 4.
The impact of RBM3 on phosphorylation of Chk1 and Chk2 and the association of pSer345-Chk1 and pT68-Chk2 with OS in EOC. The impact of RBM3 on checkpoint response was examined in siRBM3-transfected cells by Western blot analysis using antibodies against (A) pSer345-Chk1 and pT68-Chk2. (B) Immunohistochemical staining of pSer345-Chk1 and pT68-Chk2 in EOC tumors. (C) Kaplan Meier analysis of OS according to immunohistochemical staining of pS345-Chk1 and pT68-Chk2 in cohort 2 in strata defined as high versus low expression.
Phosphorylated Chk2 on Thr-68 Is Associated with an Impaired Survival
Having demonstrated that RBM3 regulates Chk1 and Chk2 protein expression in vitro and that both Chk1 and Chk2 are associated with an impaired survival in EOC, we next sought to examine the relationship between phosphorylated Chk1 and Chk2 and prognosis. pSer345-Chk1 and pThr68-Chk2 were thus assessed using immunohistochemistry in cohort 2 (Figure 4B).
RBM3 protein expression did not correlate with phosphorylated Chk1 or Chk2; however, there was a significant correlation between pSer345-Chk1 and pThr68-Chk2 (R = 0.298, P < .001). Neither pSer345-Chk1 nor pThr68-Chk2 was associated with any clinico-pathological parameters (data not shown). Kaplan-Meier analysis demonstrated no prognostic significance of pSer345-Chk1; however, pThr68-Chk2 positivity (NS > 0) was associated with a reduced OS (P = .047; Figure 4C). Cox univariate analysis confirmed the association between pThr68-Chk2 and a reduced OS (HR = 1.62, 95% CI = 1.00–2.63, P = .049); however, this did not remain significant in multivariate analysis.
Discussion
We previously reported an association between RBM3 and a prolonged survival in breast cancer and EOC [10,11]. In the present study, we identified differentially expressed genes in EOC tumors with high versus low RBM3 expression, aiming to discover novel prognostic EOC biomarkers and to gain a deeper understanding of the function of RBM3. GSEA revealed an association between RBM3 expression and a number of cellular processes involved in the maintenance of DNA integrity including regulation of DNA replication, DNA replication, chromatin remodeling, and DNA integrity checkpoint. In the light of previous findings demonstrating a relationship between RBM3 and cisplatin sensitivity [11], these results suggest that RBM3 may be involved in the cellular response to DNA damage. Further investigations are, however, required to prove this hypothesis and to determine the exact function of RBM3 in this context.
The unearthing of RBM3 as a putative prognostic tissue biomarker in EOC was the result of an antibody-based approach, using the HPA as a discovery tool [9], followed by further validation in clinically well-annotated tumor samples from two independent EOC cohorts [11]. In this study, we used an integrated transcriptomic and proteomic approach, based on tumor samples from the same clinical cohorts, to identify novel putative EOC biomarkers among RBM3-associated genes and their corresponding proteins. Our results provide, to our knowledge, the first description of an association between high expression of Chk1, Chk2, and MCM3 and poor prognosis in EOC patients.
Chk1 has previously been associated with tumor grade and cell proliferation in breast cancer [29], and CHK2 mutations have been frequently studied in the context of hereditary breast cancer [30]. The negative correlation demonstrated between RBM3 and DNA damage checkpoint proteins Chk1 and Chk2 in vitro suggests that RBM3 might be involved in DNA damage response. These serine/threonine protein kinases play crucial roles in maintaining genomic stability by mediating the signaling cascade initiated by the checkpoint proteins ATM and ATR in response to DNA damage leading to DNA repair, cell cycle arrest, or apoptosis [27]. Chk1 and Chk2 are phosphorylated by ATM and ATR in response to DNA damage, and once activated, they can phosphorylate downstream targets and control cell cycle progression by regulating the activities of Cdc25 phosphatases [31–33], p53 [34], and DNA repair factors [35]. Traditionally, the signaling network has been divided into two major protein kinase pathways: ATM activating Chk2 in response to double-stranded breaks and ATR operating together with Chk1 in response to single-stranded breaks and stalled replication forks during the S phase. The negative correlation between RBM3 and Chk1/Chk2 further emphasizes that RBM3 expression may predict response to platinum-based chemotherapy by silencing these important regulators of cellular DNA damage response. Inhibition of Chk1 and ATR was recently shown to generate the greatest impact on cisplatin response in ovarian cancer cell lines as illustrated in an RNAi screen [36], which might, in part, explain the cisplatin-sensitizing effect of RBM3 that we have previously described [11]. Inhibition of Chk1 has been reported to sensitize tumor cells to chemotherapy in various cell lines [37–39], and several Chk1 inhibitors have been developed and evaluated in clinical trials [40].
MCM proteins are key components of the DNA replication licensing system essential for maintenance of precise chromosome duplication [41,42]. Disruption of genetic stability has been reported to be a consequence of deregulation of the MCM complexes in yeast, and abnormal expression of MCM proteins has been observed in human cancers. A high expression of MCM3 protein has been reported to be associated with an impaired survival in malignant glioma [43], medulloblastoma [44], and malignant melanoma [45].
The negative association between RBM3 and CHK1, CHK2, and MCM3 genes seen in cohort 1 was not replicated at the protein level in cohort 2, which could potentially be explained by the smaller number of patients in the latter. However, in vitro experiments showed a clear inverse association between RBM3 and Chk1, Chk2 and MCM3 at the protein level, whereby siRNA-mediated down-regulation of RBM3 in A2780 cells resulted in an obvious increase in Chk1, Chk2, and MCM3 48 hours after transfection, in contrast to a nonsignificant alteration at the mRNA level. Considering the fact that RBM3 is a RNA-binding protein [46], it could be speculated that RBM3 binds and destabilizes the transcripts of CHK1, CHK2, and MCM3 in RBM3 high tumors, hence the inverse relationship observed in vivo. In addition, a reason to why we did not detect a significant increase in the mRNA levels of Chk1, Chk2, and MCM3 in response to a downregulation of RBM3 in vitro might be that RBM3 primarily acts at the translational rather than the transcriptional level. RBM3 has indeed been reported to be involved in translation contributing to an enhanced rather than suppressed global translation [47–49]. Another hypothesis is that RBM3 indirectly contributes to low levels of some checkpoint proteins by enhanced translation of proteins involved in the turnover of these proteins. Additional, more detailed investigations are required to gain further mechanistic insight into how RBM3 affects the levels of Chk1, Chk2, and MCM3. A limitation to this study is that the in vitro experiments have been performed on only one cell line, and future studies should include additional cell models.
Although the functional role of RBM3 in DNA damage requires further investigation, our data indicate a possible suppressive role of RBM3 on the checkpoint response in the absence of DNA damage, illustrated by the observed increased phosphorylation of Chk1 on silencing of RBM3 in the A2780 cells. In line with this observation, down-regulation of RBM3 in colorectal cancer cell lines led to activation of both Chk1 and Chk2 [28]. Immunohistochemical analysis revealed a negative prognostic value for pT68-Chk2 but not for pS345-Chk1 in cohort 2. The negative prognostic value observed for pT68-Chk2-expressing tumors could be because these tumors have an activated checkpoint response and are thus undergoing pressure for selection of a mutated, more aggressive, phenotype [50,51]. Along this line, it could be speculated that an attenuated DNA damage response imposed by RBM3 could explain the association with a good prognosis observed in RBM3 high breast cancers, irrespective of adjuvant chemotherapy [10].
In conclusion, we have, for the first time, revealed a link between RBM3 in DNA damage response. In addition, three novel potential biomarkers in EOC have been identified: MCM3, Chk1, and Chk2. The negative correlation between RBM3 and Chk1 and Chk2 protein levels in vitro might, in part, explain the positive effect of RBM3 on cisplatin response observed in ovarian cancer cell lines. Further investigations are required to understand the mechanisms behind the observed findings and to explain the function of RBM3, particularly its association with a good prognosis in EOC and other cancer forms.
Supplementary Material
Acknowledgments
The authors thank Elise Nilsson for excellent technical assistance.
Footnotes
This study was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Cancer Society, Gunnar Nilsson's Cancer Foundation, and the Research funds of Malmö University Hospital. The UCD Conway Institute is funded by the Programme for Third-Level Institutions, as administered by the Higher Education Authority of Ireland. The authors have no conflicts of interest to declare.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and are available online at www.transonc.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Guarneri V, Piacentini F, Barbieri E, Conte PF. Achievements and unmet needs in the management of advanced ovarian cancer. Gynecol Oncol. 2010;117:152–158. doi: 10.1016/j.ygyno.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet. 2009;374:1371–1382. doi: 10.1016/S0140-6736(09)61338-6. [DOI] [PubMed] [Google Scholar]
- 4.Brennan DJ, O'Connor DP, Rexhepaj E, Ponten F, Gallagher WM. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat Rev Cancer. 2010;10:605–617. doi: 10.1038/nrc2902. [DOI] [PubMed] [Google Scholar]
- 5.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94:5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 7.Wright CF, Oswald BW, Dellis S. Vaccinia virus late transcription is activated in vitro by cellular heterogeneous nuclear ribonucleoproteins. J Biol Chem. 2001;276:40680–40686. doi: 10.1074/jbc.M102399200. [DOI] [PubMed] [Google Scholar]
- 8.Bjorling E, Lindskog C, Oksvold P, Linne J, Kampf C, Hober S, Uhlen M, Ponten F. A web-based tool for in silico biomarker discovery based on tissue-specific protein profiles in normal and cancer tissues. Mol Cell Proteomics. 2008;7:825–844. doi: 10.1074/mcp.M700411-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas—a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 10.Jogi A, Brennan DJ, Ryden L, Magnusson K, Ferno M, Stal O, Borgquist S, Uhlen M, Landberg G, Pahlman S, et al. Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod Pathol. 2009;22:1564–1574. doi: 10.1038/modpathol.2009.124. [DOI] [PubMed] [Google Scholar]
- 11.Ehlen A, Brennan DJ, Nodin B, O'Connor DP, Eberhard J, Alvarado-Kristensson M, Jeffrey IB, Manjer J, Brandstedt J, Uhlen M, et al. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J Transl Med. 2010;8:78. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 13.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 14.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Andersson AC, Stromberg S, Backvall H, Kampf C, Uhlen M, Wester K, Ponten F. Analysis of protein expression in cell microarrays: a tool for antibody-based proteomics. J Histochem Cytochem. 2006;54:1413–1423. doi: 10.1369/jhc.6A7001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005;4:384–393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Team RDC. A language and environment for statistical computing. R Foundation for Statistical Computing. 2008 [Google Scholar]
- 18.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 19.Smyth G. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005:397–420. [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lofstedt T, Jogi A, Sigvardsson M, Gradin K, Poellinger L, Pahlman S, Axelson H. Induction of ID2 expression by hypoxia-inducible factor-1: a role in dedifferentiation of hypoxic neuroblastoma cells. J Biol Chem. 2004;279:39223–39231. doi: 10.1074/jbc.M402904200. [DOI] [PubMed] [Google Scholar]
- 23.Wellmann S, Buhrer C, Moderegger E, Zelmer A, Kirschner R, Koehne P, Fujita J, Seeger K. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 24.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 25.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 27.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 28.Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, Morrison AR, Dieckgraefe BK, Brackett DJ, Postier RG, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundgren K, Holm K, Nordenskjold B, Borg A, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10:R81. doi: 10.1186/bcr2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narod SA. Testing for CHEK2 in the cancer genetics clinic: ready for prime time? Clin Genet. 2010;78:1–7. doi: 10.1111/j.1399-0004.2010.01402.x. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko YS, Watanabe N, Morisaki H, Akita H, Fujimoto A, Tominaga K, Terasawa M, Tachibana A, Ikeda K, Nakanishi M. Cell-cycle-dependent and ATM-independent expression of human Chk1 kinase. Oncogene. 1999;18:3673–3681. doi: 10.1038/sj.onc.1202706. [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of theChk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 34.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 35.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 36.Arora S, Bisanz KM, Peralta LA, Basu GD, Choudhary A, Tibes R, Azorsa DO. RNAi screening of the kinome identifies modulators of cisplatin response in ovarian cancer cells. Gynecol Oncol. 2010;118:220–227. doi: 10.1016/j.ygyno.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Ganzinelli M, Carrassa L, Crippa F, Tavecchio M, Broggini M, Damia G. Checkpoint kinase 1 down-regulation by an inducible small interfering RNA expression system sensitized in vivo tumors to treatment with 5-fluorouracil. Clin Cancer Res. 2008;14:5131–5141. doi: 10.1158/1078-0432.CCR-08-0304. [DOI] [PubMed] [Google Scholar]
- 38.Paparatto D, Fletcher D, Piwowar K, Baldino K, Morel C, Dunaway S. The Schizosaccharomyces pombe checkpoint kinases Chk1 and Cds1 are important for cell survival in response to cisplatin. PLoS One. 2009;4:e6181. doi: 10.1371/journal.pone.0006181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- 40.Bucher N, Britten CD. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer. 2008;98:523–528. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailis JM, Forsburg SL. MCM proteins: DNA damage, mutagenesis and repair. Curr Opin Genet Dev. 2004;14:17–21. doi: 10.1016/j.gde.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Lei M. The MCM complex: its role in DNA replication and implications for cancer therapy. Curr Cancer Drug Targets. 2005;5:365–380. doi: 10.2174/1568009054629654. [DOI] [PubMed] [Google Scholar]
- 43.Soling A, Sackewitz M, Volkmar M, Schaarschmidt D, Jacob R, Holzhausen HJ, Rainov NG. Minichromosome maintenance protein 3 elicits a cancer-restricted immune response in patients with brain malignancies and is a strong independent predictor of survival in patients with anaplastic astrocytoma. Clin Cancer Res. 2005;11:249–258. [PubMed] [Google Scholar]
- 44.Lau KM, Chan QK, Pang JC, Li KK, Yeung WW, Chung NY, Lui PC, Tam YS, Li HM, Zhou L, et al. Minichromosome maintenance proteins 2, 3 and 7 in medulloblastoma: overexpression and involvement in regulation of cell migration and invasion. Oncogene. 2010;29:5475–5489. doi: 10.1038/onc.2010.287. [DOI] [PubMed] [Google Scholar]
- 45.Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 46.Derry JM, Kerns JA, Francke U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum Mol Genet. 1995;4:2307–2311. doi: 10.1093/hmg/4.12.2307. [DOI] [PubMed] [Google Scholar]
- 47.Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci USA. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smart F, Aschrafi A, Atkins A, Owens GC, Pilotte J, Cunningham BA, Vanderklish PW. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J Neurochem. 2007;101:1367–1379. doi: 10.1111/j.1471-4159.2007.04521.x. [DOI] [PubMed] [Google Scholar]
- 49.Wellmann S, Truss M, Bruder E, Tornillo L, Zelmer A, Seeger K, Buhrer C. The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr Res. 2010;67:35–41. doi: 10.1203/PDR.0b013e3181c13326. [DOI] [PubMed] [Google Scholar]
- 50.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 51.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.