Abstract
Whereas vertebrate achaete–scute complex (as-c) and atonal (ato) homologs are required for neurogenesis, their neuronal determination activities in the central nervous system (CNS) are not yet supported by loss-of-function studies, probably because of genetic redundancy. Here, to address this problem, we generated mice double mutant for the as-c homolog Mash1 and the ato homolog Math3. Whereas in Mash1 or Math3 single mutants neurogenesis is only weakly affected, in the double mutants tectal neurons, two longitudinal columns of hindbrain neurons and retinal bipolar cells were missing and, instead, those cells that normally differentiate into neurons adopted the glial fate. These results indicated that Mash1 and Math3 direct neuronal versus glial fate determination in the CNS and raised the possibility that downregulation of these bHLH genes is one of the mechanisms to initiate gliogenesis.
Keywords: bHLH/gliogenesis/Mash1/Math3/neurogenesis
Introduction
Neurons and glial cells differentiate from common precursor cells, and development of each cell type is regulated by many transcription factors. Among them, basic helix–loop–helix (bHLH) factors have been shown to play an important role in neuronal determination and differentiation of both invertebrates and vertebrates (Jan and Jan, 1994; Guillemot, 1995; Anderson and Jan, 1997; Kageyama and Nakanishi, 1997; Lee, 1997; Anderson, 1999).
In Drosophila, there are at least two classes of proneural bHLH genes, the achaete–scute complex (as-c) and atonal (ato), which not only determine the neural fate but also specify the identities of the external sensory organ and the chordotonal organ, respectively (Jan and Jan, 1994; Anderson and Jan, 1997). Thus, neural fate determination seems to be coupled to specification of the neuronal subtype identities, and these two steps are regulated by proneural bHLH genes. Similarly, in vertebrates, the as-c homolog Mash1 and the distant ato homologs ngns are expressed in a complementary manner and involved in generation of different subsets of neurons (Guillemot and Joyner, 1993; Gradwohl et al., 1996; Sommer et al., 1996; Ma et al., 1997). In the central nervous system (CNS), Mash1 and ngns are involved in specification of distinct subtypes of neurons such as ventral and dorsal telencephalic neurons (Blader et al., 1997; Casarosa et al., 1999; Torii et al., 1999; Fode et al., 2000). In the peripheral nervous system (PNS), where neural crest cells give rise to autonomic and sensory neurons, Mash1 is involved in generation of the former neurons while ngns are involved in generation of the latter (Guillemot et al., 1993; Sommer et al., 1995; Blaugrund et al., 1996; Hirsch et al., 1998; Ma et al., 1999; Perron et al., 1999). Misexpression studies demonstrated that Mash1 and ngns can induce both pan-neuronal properties and subtype-specific identities in precursor cells (Lo et al., 1998; Perez et al., 1999). Moreover, transient expression of proneural bHLH genes is sufficient to convert P19 embryonal carcinoma cells into neurons (Farah et al., 2000). These gain-of-function studies indicate that vertebrate bHLH genes are involved in both neuronal commitment and specification of particular subtypes such as Drosophila proneural genes. However, in Mash1- or ngn-deficient mice, although subsets of neurons are missing, it is not clear whether the block of generation of neurons occurs at the neuronal versus glial fate determination or just after commitment to the neuronal fate (Sommer et al., 1995; Fode et al., 1998; Hirsch et al., 1998; Ma et al., 1998, 1999). Particularly, in the CNS of Mash1- or ngn-deficient mice there are still many neurons generated (Casarosa et al., 1999; Fode et al., 2000) and thus the neuronal commitment activity suggested by a gain-of-function study (Lo et al., 1998; Perez et al., 1999; Farah et al., 2000) is not yet fully supported by a loss-of-function study in the CNS, probably because of genetic redundancy.
The loss-of-function evidence that a bHLH gene is important for neuronal versus glial fate choice is shown in the retina. In mice mutant for Mash1 or the distant ato homolog NeuroD, retinal neurons decrease whereas glial cells increase, suggesting that bHLH genes are involved in the process of neuronal versus glial fate determination (Tomita et al., 1996b; Morrow et al., 1999). However, the defects observed in the mutant retina are only partial and still all the neuronal types are generated, suggesting that the bHLH genes could be redundant or not be a key molecule but only influence the differentiation or survival of committed cells. In addition, even if genetic redundancy is the case, Mash1 and ngns are expressed in a complementary manner while other bHLH genes such as NeuroD function at later stages of development (Lee et al., 1995; Miyata et al., 1999; Liu et al., 2000; Schwab et al., 2000) and, therefore, it is not clear which bHLH genes are actually redundant at the neuronal commitment process.
Here, to address the question about the determination activity of vertebrate bHLH genes in the CNS, we generated mice double mutant for two classes of bHLH genes, Mash1 and Math3, another distant ato homolog belonging to a different subclass from ngns (Takebayashi et al., 1997). Unlike other ato homologs, Math3 is coexpressed with Mash1 in various regions of the developing CNS and therefore it is possible that these two genes could be functionally redundant. Although the previous misexpression study indicated that the Xenopus counterpart of Math3 has a neuronal determination activity (Takebayashi et al., 1997), we found that Math3(–/–) mice exhibited only a partial loss of cerebellar granule neurons due to apoptosis of their committed precursors, suggesting that Math3 is not essential for neuronal commitment. However, in Math3(–/–)-Mash1(–/–) double-homozygous mice, generation of neurons is blocked at the neural precursor stage in the regions where the two genes are coexpressed. Strikingly, the cells that normally differentiate into neurons adopted the glial fate, indicating a fate switch from neurons to glial cells. These loss-of-function data demonstrated that the vertebrate as-c and ato homologs are key molecules in neuronal versus glial fate determination.
Results
Spatio-temporal expression patterns of Math3
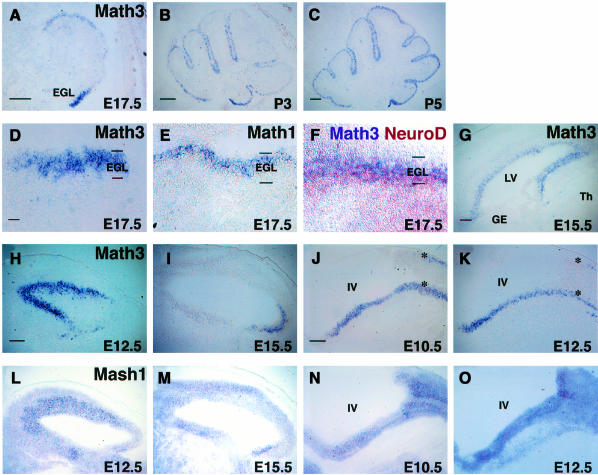
The spatio-temporal expression patterns of Math3 were determined by in situ hybridization. In the cerebellum, Math3 expression was observed in the external granular layer (EGL) at embryonic day 17.5 (E17.5) (Figure 1A). The EGL contains dividing neuronal precursors, which are derived from the rhombic lip and committed to the granule cell fate. The EGL cells then stop cell division, migrate inwardly through the Purkinje cell layer and differentiate into mature granule cells, which form the internal granular layer (IGL). Those dividing precursors and postmitotic premigratory cells are present in the outer and inner regions of the EGL, respectively. Math3 was expressed mainly in the outer region of the EGL (Figure 1D and F), and this expression pattern is very similar to the ato homolog Math1 (Akazawa et al., 1995) (Figure 1E). The Math3 expression region is different from but slightly overlapped with that of another ato homolog, NeuroD, which is expressed mainly in the inner region (Miyata et al., 1999) (Figure 1F). Math3 expression continued postnatally (Figure 1B and C) but was not detectable in the adult (data not shown). In the forebrain, Math3 expression was restricted to the ventricular zone, which contains neural precursor cells (Figure 1G). Math3 was expressed mainly in the dorsal forebrain but not in the ventral forebrain (Figure 1G). This expression pattern is very similar to ngns but different from Mash1 (Guillemot and Joyner, 1993; Gradwohl et al., 1996; Sommer et al., 1996; Ma et al., 1997). Thus, in the forebrain and cerebellum Math3 is coexpressed with other ato homologs (Math1 and ngns) but not with Mash1.
Fig. 1. Spatio-temporal expression pattern of Math3 and other bHLH genes. Distribution of bHLH genes on parasagittal sections was examined by in situ hybridization. In all sections, anterior is to the left and dorsal is up. (A–C) Math3 expression in the developing cerebellum at E17.5 (A), P3 (B) and P5 (C). Math3 is expressed in the EGL. (D) At E17.5, Math3 is expressed at the outer region of the EGL, which contains dividing precursors of cerebellar granule cells. (E) Math1 is also expressed in the outer region of the EGL. (F) NeuroD (brown) is expressed mainly in the inner region of the EGL, which contains postmitotic premigratory cells, whereas Math3 (purple) is expressed mainly in the outer region. (G) At E15.5, Math3 is expressed in the ventricular zone of the dorsal telencephalon but not of the ventral telencephalon. (H) At E12.5, Math3 is expressed at a high level in the ventricular zone of the anterior two thirds of the midbrain. (I) At E15.5, Math3 expression is shifted to the ventricular zone of the posterior midbrain. (J) At E10.5, Math3 is expressed in two longitudinal columns of the hindbrain (asterisks). (K) At E12.5, Math3 expression is observed in two longitudinal columns of the hindbrain (asterisks). (L) At E12.5, Mash1 is expressed in the midbrain. The expression level is higher in the ventricular zone of the anterior two thirds of the midbrain. (M) At E15.5, Mash1 is expressed in the developing midbrain. (N and O) At E10.5 (N) and E12.5 (O), Mash1 is expressed in the ventricular zone of the hindbrain. GE, ganglionic eminence; IV, the fourth ventricle; LV, lateral ventricle; Th, thalamus. Scale bar, 300 μm (A–C and G–O); 30 μm (D–F).
In the midbrain, Math3 was expressed at a high level in the ventricular zone of the anterior two thirds and at a low level in the posterior region at E12.5 (Figure 1H). However, Math3 was expressed at a high level in the posterior part of the midbrain at E15.5 (Figure 1I). In the hindbrain, Math3 was expressed in two longitudinal columns at E10.5 and E12.5 (Figure 1J and K, asterisks). Interestingly, in these midbrain and hindbrain regions Mash1 was also expressed (Figure 1L–O), suggesting that Math3 and Mash1 may cooperatively regulate neural development in these regions.
Generation and characterization of Math3 knockout mice
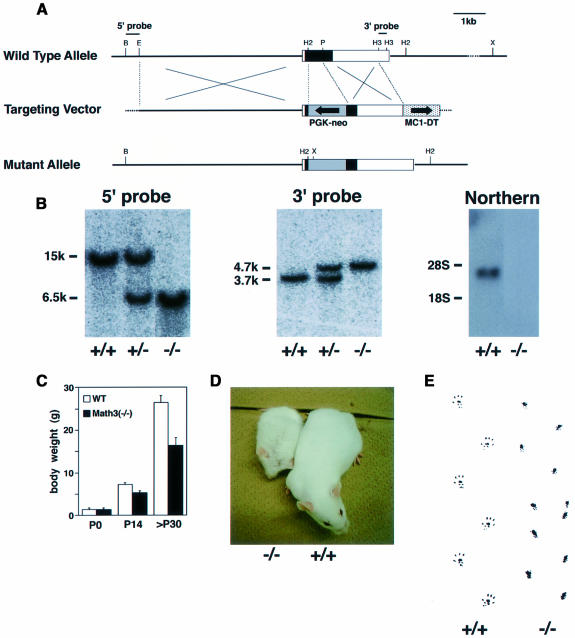
The Math3 function in neural development was next determined by generating knockout mice. Math3 gene consists of two exons, and the whole protein-coding region is present in the second exon (Tsuda et al., 1998). In ES cells, the majority of the coding region of the Math3 gene was replaced with the neo gene by homologous recombination (Figure 2A and B). Heterozygous mice generated from the mutant ES cells were fertile and looked normal. Homozygous mice, which lost Math3 expression (Figure 2B), were born according to the Mendelian rule and initially the body size was normal (Figure 2C). However, their growth was progressively retarded (Figure 2C and D) and, by 3 weeks after birth, >40% of them died partly because they did not drink milk well. In addition, the body weight of those that survived was still ∼40% smaller than the wild type (Figure 2C). Approximately 50% of the homozygous mutants survived beyond several months after birth but most of them died within 1 year.
Fig. 2. Generation of Math3-deficient mice. (A) Strategy for homologous recombination of Math3 gene by a targeting vector. Schematic representation of the wild-type Math3 gene (top), Math3 targeting vector (middle) and mutant allele (bottom). The neo gene was inserted into the second exon. The closed and open boxes represent the coding and non-coding regions, respectively, of the second exon of the Math3 gene. (B) Southern and northern blot analyses. Genotypes were determined by Southern blot analyses (left and middle). 5′-external probe detected 15 kb wild-type and 6.5 kb mutant bands of XhoI–BamHI-digested genomic DNA. 3′-external probe detected 3.7 kb wild-type and 4.7 kb mutant bands of HincII-digested genomic DNA. Northern blot analysis (right) showed that the functional Math3 mRNA is not present in Math3 homozygous mutant mice. (C) The body weight of wild-type and Math3(–/–) mice. The average with a standard error (n = 13 at P0, 53 at P14 and 26 at >P30) is shown. The growth of Math3(–/–) mice was progressively retarded. (D) Seven-week-old wild-type and Math3(–/–) mice. (E) Hind footprint pattern of wild-type and Math3(–/–) mice. The mutant mice showed ataxic gait.
Strikingly, Math3-deficient mice became ataxic by 1 week after birth. Adult mutant mice also exhibited ataxic gait (Figure 2E) and, when put on a rod (Ø = 8 mm), they stayed only for 9.8 ± 0.7 s (n = 5), whereas the wild-type mice remained balanced for 33.9 ± 2.8 s (n = 5), indicating that Math3 mutation causes severe motor defects.
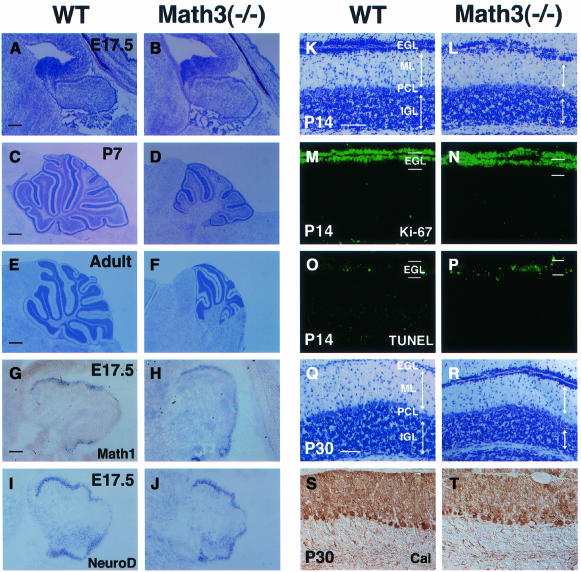
At E17.5, the gross morphology of the nervous system of Math3-deficient mice appeared normal. Histological analysis of the homozygous mutants showed that the cerebellar anlage had the EGL and was normal in size at E17.5 (Figure 3A and B). Thus, Math3 may not be involved in the initial specification of granule cell fate in the rhombic lip. However, at postnatal day 7 (P7) and in adults, the mutant cerebellum was smaller in size than the wild type (Figure 3C–F). The lobule formation was poor and some of them were missing (Figure 3F). Both the IGL and the molecular layer, which contains parallel fibers of granule cells, were thinner in Math3-deficient mice than in the wild type (Figure 3K, L, Q and R), suggesting that the granule cell number in the IGL was reduced in Math3-deficient mice. In contrast, the Purkinje cell layer (calbindin+) looked normal (Figure 3S and T). The observed reduction of the granule cell number in the IGL could be due to a decrease in dividing cells and/or an increase in apoptosis. We thus monitored cell proliferation by Ki-67, a nuclear antigen expressed by proliferating cells, and cell death by the TUNEL assay. Whereas a normal number of Ki-67+ proliferating cells were present in the EGL (Figure 3M and N), TUNEL+ dying cells were increased in the EGL of Math3(–/–) mutants (Figure 3O and P), suggesting that Math3 is critical for survival but not proliferation of the EGL cells. These cerebellar defects may be a primary cause of ataxia.
Fig. 3. The cerebellar defects of Math3-deficient mice. (A–F) The wild-type (A, C and E) and Math3(–/–) (B, D and F) cerebellum at E17.5 (A and B), P7 (C and D) and adult (E and F). The Math3(–/–) cerebellar anlage has the EGL and appears normal at E17.5. However, at P7 and adult the Math3(–/–) cerebellum is smaller and the lobule formation is poor. The posterior region is more severely affected in Math3(–/–) cerebellum. (G and H) In situ hybridization of Math1 in E17.5 wild-type (G) and Math3(–/–) (H) cerebellum. Math1 expression appears normal in Math3(–/–) cerebellum. (I and J) In situ hybridization of NeuroD in E17.5 wild-type (I) and Math3(–/–) (J) cerebellum. NeuroD expression appears normal in Math3(–/–) cerebellum. (K and L) Histology of wild-type (K) and Math3(–/–) (L) cerebellum at P14. The molecular layer (ML) and IGL of Math3(–/–) cerebellum are smaller, suggesting that granule cell number is reduced. The Purkinje cell layer (PCL) appears normal in Math3(–/–) cerebellum. (M and N) Staining with anti-Ki-67 antibody. Cells in the outer region of the EGL are mitotic in both wild-type (M) and Math3(–/–) (N) cerebellum at P14. (O and P) TUNEL assay. Only some cells are TUNEL+ in the wild-type EGL (O) whereas many cells are TUNEL+ in the Math3(–/–) EGL (P), indicating that many Math3(–/–) precursors are dying in the EGL. (Q and R) Histology of wild-type (Q) and Math3(–/–) (R) cerebellum at P30. The ML and IGL of Math3(–/–) cerebellum are still smaller, suggesting that granule cell number is reduced. (S and T) Staining with anti-calbindin antibody of wild-type (S) and Math3(–/–) (T) cerebellum at P30. The Purkinje cell layer appears normal in Math3(–/–) cerebellum. In all sections, anterior is to the left and dorsal is up. Scale bar, 200 μm (A and B); 500 μm (C and D); 800 μm (E and F); 100 μm (G–J); 50 μm (K–T).
It has been shown that cerebellar granule cell development is regulated by two sequentially expressed ato homologs, Math1 and NeuroD (Akazawa et al., 1995; Ben-Arie et al., 1997; Miyata et al., 1999). Math1 is essential for the initial specification of granule cells while NeuroD is required for development of postmitotic cells. In Math3(–/–) cerebellum, both Math1 and NeuroD expression was not affected (Figure 3G–J), indicating that Math3 is required for granule cell development even though Math1 and NeuroD are properly expressed.
Besides the cerebellum, the histology of the postnatal Math3(–/–) nervous system appeared normal (data not shown). Thus, from examination of the mutant mice, Math3 was shown to be important for the survival of differentiating cerebellar neurons, but there was no direct evidence that Math3 is involved in neuronal fate determination. We assumed that these rather weak phenotypes of Math3-deficient mice may be due to compensation by other bHLH genes. Since Math3 is coexpressed with Mash1 in various regions (Figure 1), we next generated Math3(–/–)-Mash1(–/–) double-mutant mice to explore the possible neuronal determination function. By crossing Math3(–/–)-Mash1(+/–) male and Math3(+/–)-Mash1(+/–) female mice, we obtained 15 Math3(–/–)-Mash1(–/–) embryos out of 123 embryos (12.2%) at E15.5. Since this ratio is almost the same as expected (12.5%), most double mutants survived until E15.5. However, we currently failed to obtain double mutants at E17.5, suggesting that they died between E15.5 and E17.5. The double mutants exhibited more severe defects in the midbrain, hindbrain and retina than Math3(–/–) or Mash1(–/–) single mutants, as shown below, suggesting that the two genes cooperatively regulate neural development.
Block of neurogenesis and ectopic gliogenesis in Math3(–/–)-Mash1(–/–) midbrain
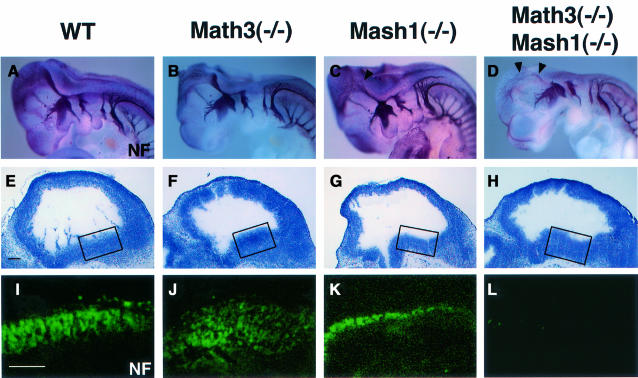
Staining in whole mount with antibody to neurofila ment (NF) revealed that both cranial and dorsal root ganglia looked normal in Math3(–/–), Mash1(–/–) and Math3(–/–)-Mash1(–/–) embryos at E10.5 (Figure 4A–D). However, compared with the wild-type and Math3(–/–) embryos, neurite extension was slightly reduced in the midbrain and hindbrain of Mash1(–/–) embryos (Figure 4C, arrowhead) and more severely reduced in those of Math3(–/–)-Mash1(–/–) embryos (Figure 4D, arrowheads), suggesting that development of the double-mutant brain was more severely affected. Histological analysis indicated that whereas many NF+ neurons differentiated in the midbrain of the wild-type and Math3(–/–) embryos (Figure 4I and J), there were fewer neurons in Mash1(–/–) (Figure 4K) and virtually no neurons in the Math3(–/–)-Mash1(–/–) mutants at E11.5 (Figure 4L), suggesting that neurogenesis is blocked in the double-mutant midbrain.
Fig. 4. The midbrain defects of Math3-Mash1 mutant mice. (A–D) Whole-mount immunostaining with anti-neurofilament (NF) antibody of E10.5 embryos of wild type (A), Math3(–/–) (B), Mash1(–/–) (C) and Math3(–/–)-Mash1(–/–) (D). Neurite extension is slightly reduced in Mash1(–/–) (C, arrowhead) and more severely reduced in Math3(–/–)-Mash1(–/–) (D, arrowheads). (E–H) Histology of E11.5 midbrain of wild type (E), Math3(–/–) (F), Mash1(–/–) (G) and Math3(–/–)-Mash1(–/–) (H). (I–L) Immunohistochemical staining with anti-NF antibody of the boxed regions in (E–H). Neurons are normally generated in Math3(–/–) (J). However, there are fewer neurons in Mash1(–/–) (K) and virtually no neurons in Math3(–/–)-Mash1(–/–) (L). Thus, neuronal differentiation is blocked in Math3(–/–)-Mash1(–/–) midbrain. In all sections, anterior is to the left and dorsal is up. Scale bar, 150 μm (E–L).
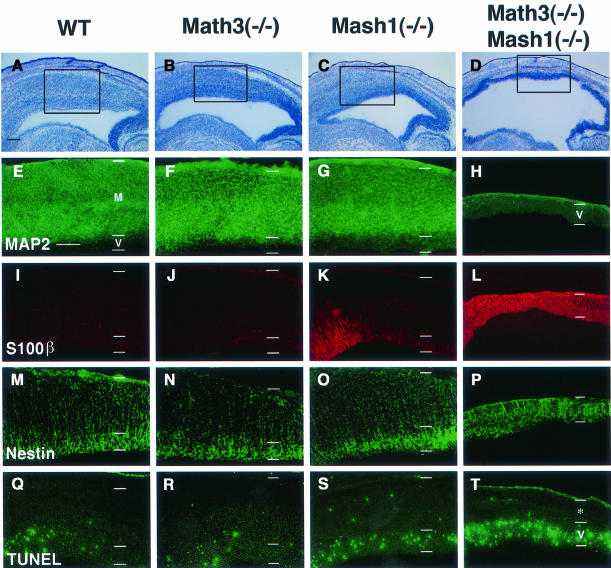
At E15.5, the tectum region of wild-type, Math3(–/–) and Mash1(–/–) midbrain consisted of two layers, the ventricular zone and the mantle layer (Figure 5A–C). Although the size of the Math3(–/–) and Mash1(–/–) tectum was slightly reduced compared with the wild type (Figure 5A–C), their mantle layer contained massive neurons (MAP2+) (Figure 5E–G). In contrast, the double-mutant tectum was much thinner and neurons were still completely missing (Figure 5D and H). The double-mutant tectum region consisted of only the ventricular zone (Nestin+) (Figure 5P). Surprisingly, the majority of the cells in the double-mutant ventricular zone expressed S100β, an early astrocyte-specific marker (Figure 5L), whereas the wild-type and Math3(–/–) cells did not (Figure 5I and J). These results indicate that the ventricular cells that normally differentiate into neurons at E15.5 were blocked from neuronal differentiation and instead adopted the glial fate in the absence of Math3 and Mash1. Interestingly, in Mash1(–/–) tectum, subsets of cells also ectopically expressed S100β (Figure 5K), suggesting that gliogenesis is slightly enhanced in the absence of Mash1. A later glial marker, glial fibrillary acidic protein (GFAP), was not expressed in the mutant tectum (data not shown), suggesting that the ectopic glial cells were still immature. The TUNEL assay showed that apoptosis was significantly enhanced in the Math3(–/–)-Mash1(–/–) tectum (Figure 5T) whereas it was normal in Math3(–/–) and slightly enhanced in the Mash1(–/–) tectum (Figure 5Q–S). Since there was no difference in the number of TUNEL+ cells at E11.5 (data not shown), when neurogenesis is already blocked (Figure 4L), the cell death observed at E15.5 in the double mutants is likely to be a secondary effect due to the block of neurogenesis.
Fig. 5. The defects of the tectum of Math3-Mash1 mutant mice. (A–D) Histology of E15.5 tectum. The tectum is slightly thinner in Math3(–/–) (B) and Mash1(–/–) (C) and much thinner in Math3(–/–)-Mash1(–/–) (D) than in the wild type (A). (E–H) Immunohistochemistry with anti-MAP2 antibody of the boxed regions in (A–D). The wild-type (E), Math3(–/–) (F) and Mash1(–/–) tectum (G) consist of the ventricular zone (V) and the mantle layer (M), which contains massive MAP2+ neurons. In contrast, Math3(–/–)-Mash1(–/–) tectum consists of only the ventricular zone and lacks neurons (H). (I–L) Immunohistochemistry with anti-S100β antibody. There are no S100β+ cells in the wild-type (I) and Math3(–/–) tectum (J) whereas some cells expressed S100β in the Mash1(–/–) tectum (K). Strikingly, in Math3(–/–)-Mash1(–/–) tectum (L) the majority of the cells expressed S100β, indicating that gliogenesis is significantly enhanced in the double mutants. (M–P) Immunohistochemistry with anti-Nestin antibody. The double-mutant tectum (P) consists only of the ventricular zone (Nestin+). (Q–T) TUNEL assay. Only some cells are TUNEL+ in the wild-type (Q) and Math3(–/–) (R) tectum. In contrast, there are slightly more TUNEL+ cells in Mash1(–/–) (S) and many more TUNEL+ cells in Math3(–/–)-Mash1(–/–) (T). Thus, double-mutant neural precursor cells are blocked from neuronal differentiation and result in extensive apoptosis. The asterisk in (T) indicates non-neural tissues including the skull. In all sections, anterior is to the left and dorsal is up. Scale bar, 300 μm.
Block of neurogenesis and ectopic gliogenesis in the Math3(–/–)-Mash1(–/–) hindbrain
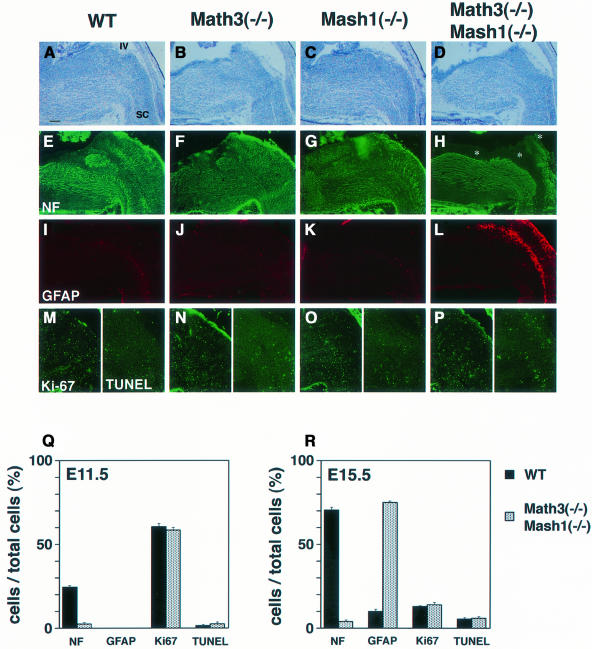
We next examined defects of the hindbrain, which normally expresses both Math3 and Mash1 in two longitudinal columns. In the wild-type, Math3(–/–) and Mash1(–/–) hindbrain, many MAP2+ or NF+ neurons differentiated at E11.5 but only a few neurons were generated in the Math3(–/–)-Mash1(–/–) hindbrain (Figure 6Q), suggesting that neurogenesis is blocked. At E15.5, neuronal differentiation was almost completed in the wild-type, Math3(–/–) and Mash1(–/–) hindbrain (Figure 6E–G). However, in the Math3(–/–)-Mash1(–/–) hindbrain, there were still very few neurons in the two longitudinal columns (Figure 6H, asterisks, and R) whereas the other neurons were generated. Strikingly, in the adjacent sections, the regions devoid of neurons contained ectopic glial cells (GFAP+) (Figure 6L and R). The majority (>70%) of the cells expressed GFAP in the double-mutant longitudinal columns, in contrast to the wild type, Math3(–/–) and Mash1(–/–) where only ∼10% of the cells expressed GFAP at this stage (Figure 6I–K and R). This defect was likely a fate switch from neurons to glial cells, but it was also possible that neurons died while glial cells proliferated. To distinguish between these possibilities, cell proliferation and apoptosis were examined by Ki-67 staining and TUNEL assay. No difference in Ki-67 and TUNEL staining was detected at E11.5 and E15.5 (Figure 6M–R), indicating that cell proliferation and death are not involved in these defects. These results indicate that the cells that normally differentiate into neurons adopted the glial fate in the double mutants.
Fig. 6. The defects of the hindbrain of Math3-Mash1 mutant mice. (A–D) Histology of wild-type (A), Math3(–/–) (B), Mash1(–/–) (C) and Math3(–/–)-Mash1(–/–) hindbrain (D) at E15.5. (E–H) Immunohistochemistry with anti-NF antibody. Neurons are generated in the whole region of wild-type (E), Math3(–/–) (F) and Mash1(–/–) hindbrain (G). In contrast, neurons are missing in the two longitudinal columns of Math3(–/–)-Mash1(–/–) hindbrain (H, asterisks). (I–L) Immunohistochemistry with anti-GFAP antibody of adjacent sections of (E–H). Whereas only a few glial cells are present in wild-type (I), Math3(–/–) (J) and Mash1(–/–) (K) hindbrain, many glial cells are generated in the regions that lack neurons in Math3(–/–)-Mash1(–/–) hindbrain (L). (M–P) Ki-67 (left) and TUNEL staining (right). Each panel shows the right half of (A–D). No significant difference in Ki-67 and TUNEL staining is observed in the wild-type and mutant hindbrain. In all sections, anterior is to the left and dorsal is up. IV, the fourth ventricle; SC, spinal cord. Scale bar, 250 μm. (Q and R) Quantification of the cells in the longitudinal columns of the hindbrain. The cell number of the two longitudinal columns was counted in a section (16 μm thick) of at least three independent samples of the wild-type and Math3(–/–)-Mash1(–/–) embryos at E11.5 (Q) and E15.5 (R). The ratios of NF+, GFAP+, Ki-67+ and TUNEL+ cells per the total cells in the columns are shown with bars to represent the standard error.
Lack of bipolar cells and ectopic generation of glial cells in the Math3(–/–)-Mash1(–/–) retina
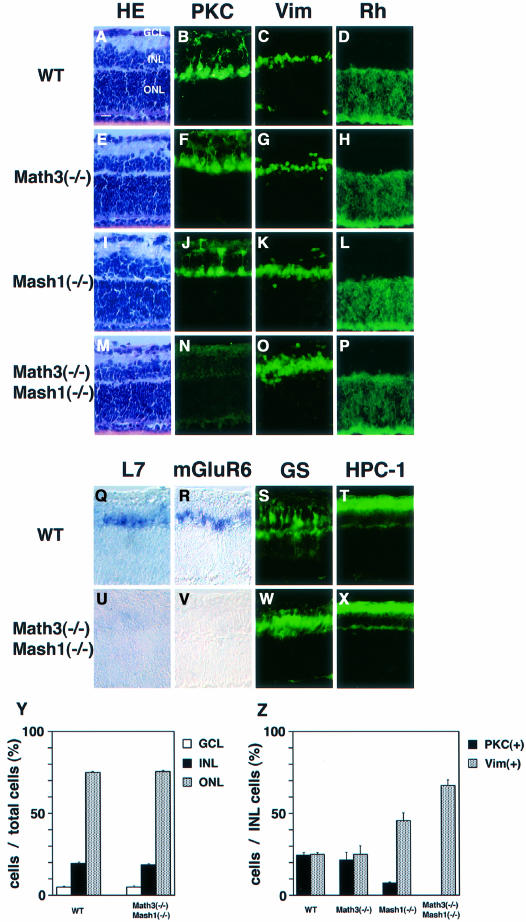
To examine further the possible fate switch in the double mutants, we next examined the retina. The retina is an ideal system to analyze the cell fate because it has only six types of neuron and one type of glial cells, which can be clearly identified by position, cell morphology and specific markers (Cepko et al., 1996). The mature retina consists of three cellular layers, the ganglion cell layer (GCL), the inner nuclear layer (INL) and the outer nuclear layer (ONL). The INL contains three types of interneuron (amacrine, bipolar and horizontal cells) and one type of glia (Müller glial cells). It has been shown that both Math3 and Mash1 are expressed by differentiating bipolar cells (Jasoni and Reh, 1996; Roztocil et al., 1997; Takebayashi et al., 1997). Because most retinal cells including glial cells differentiate postnatally, we used the retinal explant culture, which mimics in vivo development well. Retinal explants were prepared from E15.5 embryos and cultured for 2 weeks, during which period the majority of retinal cells finish differentiation. By this method, it was possible to monitor the later stage of cell differentiation well after the mutant hosts died. After 2 weeks of culture, the wild-type and mutant retina consisted of three cellular layers (Figure 7A, E, I and M). Hematoxylin–eosin (HE) staining indicated that the cell number of each layer was normal in Math3(–/–), Mash1(–/–) and Math3(–/–)-Mash1(–/–) retina (Figure 7A, E, I, M and Y). However, whereas bipolar cells (PKC+, mGluR6+, L7+) were normally present in Math3(–/–) (Figure 7F and Z), they were reduced in number in Mash1(–/–) retina (Figure 7J and Z) and completely missing in the Math3(–/–)-Mash1(–/–) retina (Figure 7N, U, V and Z). Instead, Müller glial cells (vimentin+, glutamine synthetase+) were slightly increased in Mash1(–/–) (Figure 7K and Z) and significantly increased in the double-mutant retina (Figure 7O, W and Z). The other cell types such as rods (rhodopsin+) and amacrine cells (HPC-1+) were not affected in the double-mutant retina (Figure 7P, X and Y). To determine whether any changes of birth date, which is important for cell type specification, are involved in the defects, the retinal explants were examined at days 5, 7 and 10 of culture. At day 5, no glial cells were detectable whereas at days 7 and 10 there were differentiating glial cells in both the wild type and the double mutants (data not shown), indicating that the time course of gliogenesis is not affected in the double mutants. In addition, there were already more glial cells in the double mutants at day 7, suggesting that more cells initially adopt the glial fate. In contrast, bipolar cells were not detectable in the double mutants at any time points, excluding the possibility that once borne bipolar cells die in the double mutants. Furthermore, there was no significant difference in the Ki-67 staining and TUNEL assay between the wild type and double mutants (data not shown). These results indicated that cell proliferation and apoptosis are not involved in the loss of bipolar cells and concomitant increase of Müller glial cells, supporting the idea that there is a fate switch from neurons to glial cells in the absence of Math3 and Mash1.
Fig. 7. The retinal explant cultures from mutant embryos. Retinal explants prepared from E15.5 embryos of wild-type (A–D and Q–T), Math3(–/–) (E–H), Mash1(–/–) (I–L) and Math3(–/–)-Mash1(–/–) (M–P and U–X) were cultured for 2 weeks and subjected to immunohistochemistry and in situ hybridization. HE staining (A, E, I and M) indicates that the cell number of each layer is not affected in all the mutants. However, whereas there are the normal number of bipolar cells (PKC+) in Math3(–/–) (F), there are fewer in Mash1(–/–) (J) and virtually no bipolar cells in Math3(–/–)-Mash1(–/–) (N). In contrast, the number of Müller glial cells (Vim+) is slightly increased in Mash1(–/–) (K) and significantly increased in Math3(–/–)-Mash1(–/–) (O). The number of rods (Rh+) (D, H, L and P) appears normal in all the mutants. (Q–X) In situ hybridization of L7 (Q and U) and mGluR6 (R and V) (bipolar cell markers) and immunohistochemistry with anti-glutamine synthetase (S and W) (a Müller glial marker) and anti-HPC-1 antibodies (T and X) (an amacrine cell marker). Both L7 and mGluR6 expression is lost whereas GS+ cells are increased in the double mutants. Amacrine cells (HPC-1+) are not affected (X). Scale bar, 25 μm. (Y and Z) Quantification of retinal cells. The cell number was counted in a section (16 μm thick, 500 μm wide) of the central region of at least three independent samples from each genotype, and the relative ratios of retinal cells were determined. The percentage of GCL, INL and ONL cells per the total retinal cells (Y) and that of PKC+ and Vim+ cells per the total INL cells (Z) are shown with bars to represent the standard error.
Discussion
Math3 and Mash1 are neuronal determination genes
In this study, we examined the neuronal fate determination activities of mammalian as-c and ato homologs. Whereas the previous misexpression studies indicated that vertebrate proneural genes can convert uncommitted cells into the neuronal fate (Ma et al., 1996; Blader et al., 1997; Takebayashi et al., 1997; Lo et al., 1998; Perez et al., 1999), disruption of each gene alone failed to demonstrate that they function at the neuronal versus glial fate determination (Guillemot et al., 1993; Sommer et al., 1995; Hirsch et al., 1998). Or rather, some of these loss-of-function studies indicated that vertebrate proneural genes only regulate the late stage development of committed neuronal cells. This apparent discrepancy between the gain- and loss-of-function studies may be due to genetic redundancy. Here, we showed that Math3 and Mash1 are coexpressed in various regions of the CNS and that mice double mutant for the two bHLH genes exhibit the block of the neuronal versus glial fate choice. These results support the conclusion that the vertebrate as-c and ato homologs function as neuronal determination genes in the CNS.
Fate switch from neurons to glia
The double mutants displayed loss of neurons and concomitant gliogenesis in the tectum, hindbrain and retina. Since neuronal apoptosis and glial proliferation cannot account for these defects, it is most likely that the cells that normally differentiate into neurons adopted the glial fate in the absence of Math3 and Mash1, indicating that Math3 and Mash1 not only specify the neuronal fate but also prevent ectopic gliogenesis. These data suggest that, whereas most neural precursor cells initially differentiate into neurons and only later into glial cells, they already have the potential to differentiate into glial cells from the early stages of development. Thus, the apparent sequence of neurons first and glial cells afterwards during development may not be the change of competence but the result of early expression of the proneural bHLH genes, which direct neurogenesis and inhibit gliogenesis. These data also raise the possibility that at least one mechanism to initiate gliogenesis is downregulation of neuronal determination genes. Recently, we and others have found that misexpression of Notch or its effectors, Hes1 and Hes5, which can functionally antagonize proneural bHLH genes (Kageyama and Nakanishi, 1997; Ohtsuka et al., 1999), induces gliogenesis at the expense of the neuronal fate (Furukawa et al., 2000; Gaiano et al., 2000; Hojo et al., 2000; Morrison et al., 2000). The mechanism for Notch-induced gliogenesis is not yet understood, but it is likely that Notch signaling suppresses proneural bHLH genes such as Math3 and Mash1, thereby promoting gliogenesis, although it remains to be determined whether this is the only mechanism for Notch-induced gliogenesis.
Is gliogenesis a default pathway after inactivation of proneural bHLH genes? Recent studies indicate that oligodendrocyte development is promoted by two related bHLH genes, Olig1 and Olig2 (Lu et al., 2000; Zhou et al., 2000). These results suggest that, like neurogenesis, gliogenesis in general is positively regulated by bHLH genes. Thus, glial development may not be a simple default pathway but may require glia-specific bHLH genes. It is likely that proneural bHLH genes inhibit gliogenesis by suppressing such gliogenic genes, although the precise mechanism for the possible suppression by proneural bHLH genes remains to be determined.
A similar fate switch was reported in the inner ear of mice mutant for Math1, a positive regulator of hair cell development (Bermingham et al., 1999; Zheng and Gao, 2000). In the inner ear, common precursors give rise to hair cells (sensory neurons) and support cells. Null mutation of Math1 leads to agenesis of hair cells and an increase in support cells, suggesting a fate switch from hair cells to support cells (Bermingham et al., 1999). Thus, vertebrate as-c and ato homologs specify the neuronal versus non-neuronal cell fate in various regions. Interestingly, the opposite effect is observed in mutations for the Notch ligands, which exhibit extra hair cells at the expense of support cells (Haddon et al., 1998; Lanford et al., 1999). Since the Notch ligand Jagged2 is expressed by hair cells in mice, it is likely that Jagged2 suppresses the neighboring cells from adopting the hair cell fate by activation of Notch signaling.
Redundant functions of mammalian as-c and ato homologs
We showed that mice double mutant for Math3 and Mash1 exhibited severe defects in the regions that normally express both genes. This redundant function of as-c and ato homologs in various parts of the CNS is interesting since, in Drosophila, the two groups of proneural bHLH genes as-c and ato direct generation of different subsets of neurons and thus their redundant functions are not known. These two classes of proneural genes seem to have intrinsically different functions since ato mutation cannot be rescued by scute (Chien et al., 1996). Domain-swapping analysis indicated that the basic region of the proneural proteins encodes the neuronal subtype-specific information (Chien et al., 1996). It has been shown that vertebrate bHLH genes also have subtype-specific activities (Lo et al., 1998; Perez et al., 1999; Fode et al., 2000). Although our experiments suggest redundant functions of Math3 and Mash1, they do not formally prove whether Math3 and Mash1 compensate each other in the same precursor cells or whether Math3-dependent precursors substitute Mash1-dependent precursors and vice versa in single mutants. Since no significant change of cell proliferation or death was observed in the hindbrain and retina of single mutants, we prefer the idea that Math3 and Mash1 function in the same precursors and that, only when neither is expressed do precursor cells choose the glial fate. The subtype specificity of the as-c homolog Mash1 and the ato homolog Math3 may not be so strict and the two bHLH genes could be functionally interchangeable to some extent.
Cooperative functions of bHLH and homeobox genes
Retinal explant assays demonstrated that Math3 and Mash1 are essential for bipolar cell development. It has been shown that the homeobox gene Chx10 is also essential for bipolar cell development: Chx10-null mice lack bipolar cells (Burmeister et al., 1996). Thus, at least three genes, Math3, Mash1 and Chx10, are involved in bipolar cell development although it is not clear at which stage Chx10 functions. Interestingly, in contrast to Math3(–/–)-Mash1(–/–) double-mutant retina, Müller glial cells are not increased and therefore a fate switch does not occur in Chx10-null mice, suggesting that the cells that normally differentiate into bipolar cells are likely to result in apoptosis in the absence of Chx10 rather than differentiate into glial cells. These results strongly suggest that the bHLH genes and Chx10 have distinct functions in bipolar cell development. Chx10 may not only confer the cell type-specific identities, as suggested by other homeobox genes (Tanabe and Jessell, 1996), but also regulate cell survival. In contrast, the main function of Math3 and Mash1 is determination of the neuronal fate but not cell survival. It remains to be analyzed whether the two bHLH genes can also directly confer some of the bipolar cell-specific identities. Further characterization of bHLH and homeobox genes will be necessary to decipher the roles of each factor in specification of particular neuronal subtypes.
Materials and methods
Generation of Math3 mutant mice
TT2 ES cells (1 × 107) (Gibco) were electroporated with 30 µg of the linearized targeting vector in 0.8 ml of phosphate-buffered saline (PBS) and selected with 250 µg/ml G418 (Gibco). ES cell lines with Math3 mutation were identified by Southern blot analysis using 5′ and 3′ probes shown in Figure 2. Chimeric mice were generated by aggregating the Math3-mutant ES cells with CD1 morula and then by implanting them into the uteri of pseudopregnant foster mothers. The resulting chimeric males were bred with CD1 females. Homozygous mice generated from two independent mutated ES cell lines showed identical phenotypes.
In situ hybridization
In situ hybridization on frozen sections was performed, as described previously (Akazawa et al., 1992). The digoxigenin- or FITC-labeled probes used in this study were as follows: Math3 (Takebayashi et al., 1997), Mash1 (Guillemot and Joyner, 1993), Math1 (Akazawa et al., 1995), NeuroD (Lee et al., 1995), L7 (Oberdick et al., 1990) and mGluR6 (Nakajima et al., 1993).
Whole-mount immunohistochemistry
Whole-mount immunohistochemistry was performed as described previously (Mark et al., 1993). In brief, E10.5 embryos were fixed in 4% paraformaldehyde in PBS at 4°C for 3 h and were bleached with 0.1% H2O2 overnight at 4°C. Then the embryos were incubated with anti-NF antibody (2H3, Hybridoma Bank) for 3 days at 4°C and next with peroxidase-conjugated secondary antibody (Chemicon) overnight at 4°C. The peroxidase deposits were visualized by 4-chloro-1-naphthol.
Histological analysis
For examination of adult brain, mice were deeply anesthetized with pentobarbital and perfused transcardially with 4% paraformaldehyde in 0.1 M sodium phosphate buffer (PB; pH 7.3). The brain was dissected rapidly, incubated in 25% sucrose in PB overnight at 4°C and frozen sections were made.
For immunohistochemistry, sections were fixed with 4% paraformaldehyde in PBS for 10 min, preincubated in PBS containing 5% normal goat serum, 1% bovine serum albumin, 0.1% Triton X-100 and 0.02% sodium azide for 30 min and then incubated with the following antibodies: monoclonal antibody against Ki-67 (Pharmingen), calbindin-D (Sigma), 160 kDa NF (Amersham), MAP2 (Sigma), PKC (Amersham), Nestin (Pharmingen), glutamine synthetase (Chemicon), HPC-1 (Sigma), S100β (Sigma) and vimentin (Histofine), and rabbit polyclonal antibody against GFAP (DAKO) and rhodopsin (LSL). As a secondary antibody, biotinylated goat antibodies against mouse and rabbit IgG (Vector) were used. The antibody complex was visualized by avidin-labeled fluorescein or ABC kit (Vector). TUNEL assay was performed as described previously (Tomita et al., 1996a).
Retinal explant culture
The explant of neural retina was placed onto a Millicell chamber filter (Millipore) with the ganglion cell layer upward. The chamber was transferred to a six-well culture plate. Explants were cultured with 50% MEM with HEPES, 25% Hanks’ solution, 25% heat-inactivated horse serum, 200 µM l-glutamine and 6.75 mg/ml glucose at 34°C in 5% CO2 (Tomita et al., 1996a).
Acknowledgments
Acknowledgements
We thank Drs S.Takada, D.Watanabe and J.Kitano for technical advice and Dr H.Ohkubo for NeuroD cDNA. Monoclonal antibody 2H3 was obtained from the Developmental Studies Hybridoma Bank. This work was supported by Special Coordination Funds for Promoting Science and Technology and research grants from the Ministry of Education, Science, Sports, and Culture of Japan and Japan Society for the Promotion of Science.
References
- Akazawa C., Sasai,Y., Nakanishi,S. and Kageyama,R. (1992) Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J. Biol. Chem., 267, 21879–21885. [PubMed] [Google Scholar]
- Akazawa C., Ishibashi,M., Shimizu,C., Nakanishi,S. and Kageyama,R. (1995) A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem., 270, 8730–8738. [DOI] [PubMed] [Google Scholar]
- Anderson D.J. (1999) Lineages and transcription factors in the specification of vertebrate primary sensory neurons. Curr. Opin. Neurobiol., 9, 517–524. [DOI] [PubMed] [Google Scholar]
- Anderson D.J. and Jan,Y.N. (1997) The determination of the neuronal phenotype. In Cowan,W.M. (ed.), Molecular and Cellular Approaches to Neural Development. Oxford University Press, New York, NY, pp. 26–63. [Google Scholar]
- Ben-Arie N., Bellen,H.J, Armstrong,D.L., McCall,A.E., Gordadze,P.R., Guo,Q., Matzuk,M.M. and Zoghbi,H.Y. (1997) Math1 is essential for genesis of cerebellar granule neurons. Nature, 390, 169–172. [DOI] [PubMed] [Google Scholar]
- Bermingham N.A., Hassan,B.A., Price,S.D., Vollrath,M.A., Ben-Arie,N., Eatock,R.A., Bellen,H.J., Lysakowski,A. and Zoghbi,H.Y. (1999) Math1: an essential gene for the generation of inner ear hair cells. Science, 284, 1837–1841. [DOI] [PubMed] [Google Scholar]
- Blader P., Fischer,N., Gradwohl,G., Guillemot,F. and Strähle,U. (1997) The activity of Neurogenin1 is controlled by local cues in the zebrafish embryo. Development, 124, 4557–4569. [DOI] [PubMed] [Google Scholar]
- Blaugrund E., Pham,T.D., Tennyson,V.M., Lo,L., Sommer,L., Anderson,D.J. and Gershon,M.D. (1996) Distinct subpopulations of enteric neuronal progenitors defined by time of development, sympatho adrenal lineage markers and Mash-1-dependence. Development, 122, 309–320. [DOI] [PubMed] [Google Scholar]
- Burmeister M. et al. (1996) Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nature Genet., 12, 376–384. [DOI] [PubMed] [Google Scholar]
- Casarosa S., Fode,C. and Guillemot,F. (1999) Mash1 regulates neurogenesis in the ventral telencephalon. Development, 126, 525–534. [DOI] [PubMed] [Google Scholar]
- Cepko C.L., Austin,C.P., Yang,X., Alexiades,M. and Ezzeddin,D. (1996) Cell fate determination in the vertebrate retina. Proc. Natl Acad. Sci. USA, 93, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C., Hsiao,C.-D., Jan,L.Y. and Jan,Y.N. (1996) Neuronal type information encoded in the basic helix-loop-helix domain of proneural genes. Proc. Natl Acad. Sci. USA, 93, 13239–13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M.H., Olson,J.M., Sucic,H.B., Hume,R.I., Tapscott,S.J. and Turner,D.L. (2000) Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development, 127, 693–702. [DOI] [PubMed] [Google Scholar]
- Fode C., Gradwohl,G., Morin,X., Dierich,A., LeMeur,M., Goridis,C. and Guillemot,F. (1998) The bHLH protein NEUROGENIN2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron, 20, 483–494. [DOI] [PubMed] [Google Scholar]
- Fode C., Ma,Q., Casarosa,S., Ang,S.-L., Anderson,D.J. and Guillemot,F. (2000) A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev., 14, 67–80. [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Mukherjee,S., Bao,Z.-Z., Morrow,E.M. and Cepko,C.L. (2000) rax, Hes1, and notch1 promote the formation of Müller glia by postnatal retinal progenitor cells. Neuron, 26, 383–394. [DOI] [PubMed] [Google Scholar]
- Gaiano N., Nye,J.S. and Fishell,G. (2000) Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron, 26, 395–404. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Fode,C. and Guillemot,F. (1996) Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol., 180, 227–241. [DOI] [PubMed] [Google Scholar]
- Guillemot F. (1995) Analysis of the role of basic-helix-loop-helix transcription factors in the development of neural lineages in the mouse. Biol. Cell, 84, 3–6. [DOI] [PubMed] [Google Scholar]
- Guillemot F. and Joyner,A.L. (1993) Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech. Dev., 42, 171–185. [DOI] [PubMed] [Google Scholar]
- Guillemot F., Lo,L.-C., Johnson,J.E., Auerbach,A., Anderson,D.J. and Joyner,A.L. (1993) Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell, 75, 463–476. [DOI] [PubMed] [Google Scholar]
- Haddon C., Jiang,Y.-J., Smithers,L., and Lewis,J. (1998) Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development, 125, 4637–4644. [DOI] [PubMed] [Google Scholar]
- Hirsch M.-R., Tiveron,M.-C., Guillemot,F., Brunet,J.-F. and Goridis,C. (1998) Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development, 125, 599–608. [DOI] [PubMed] [Google Scholar]
- Hojo M., Ohtsuka,T., Hashimoto,N., Gradwohl,G., Guillemot,F. and Kageyama,R. (2000) Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development, 127, 2515–2522. [DOI] [PubMed] [Google Scholar]
- Jan Y.N. and Jan,L.Y. (1994) Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu. Rev. Genet., 28, 373–393. [DOI] [PubMed] [Google Scholar]
- Jasoni C.L. and Reh,T.A. (1996) Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J. Comp. Neurol., 369, 319–327. [DOI] [PubMed] [Google Scholar]
- Kageyama R. and Nakanishi,S. (1997) Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr. Opin. Genet. Dev., 7, 659–665. [DOI] [PubMed] [Google Scholar]
- Lanford P.J., Lan,Y., Jiang,R., Lindsell,C., Weinmaster,G., Gridley,T. and Kelley,M.W. (1999) Notch signalling pathway mediates hair cell development in mammalian cochlea. Nature Genet., 21, 289–292. [DOI] [PubMed] [Google Scholar]
- Lee J.E. (1997) Basic helix-loop-helix genes in neural development. Curr. Opin. Neurobiol., 7, 13–20. [DOI] [PubMed] [Google Scholar]
- Lee J.E., Hollenberg,S.M., Snider,L., Turner,D.L., Lipnick,N. and Weintraub,H. (1995) Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science, 268, 836–844. [DOI] [PubMed] [Google Scholar]
- Liu M., Pleasure,S.J., Collins,A.E., Noebels,J.L., Naya,F.J., Tsai,M.-J. and Lowenstein,D.H. (2000) Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc. Natl Acad. Sci. USA, 97, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L., Tiveron,M.-C. and Anderson,D.J. (1998) MASH1 activates expression of the paired homeodomain transcription factor Phox2a, and couples pan-neuronal and subtype-specific components of autonomic neuronal identity. Development, 125, 609–620. [DOI] [PubMed] [Google Scholar]
- Lu Q.R., Yuk,D., Alberta,J.A., Zhu,Z., Pawlitzky,I., Chan,J., McMahon,A.P., Stiles,C.D. and Rowitch,D.H. (2000) Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron, 25, 317–329. [DOI] [PubMed] [Google Scholar]
- Ma Q., Kintner,C. and Anderson,D.J. (1996) Identification of neurogenin, a vertebrate neuronal determination gene. Cell, 87, 43–52. [DOI] [PubMed] [Google Scholar]
- Ma Q., Sommer,L., Cserjesi,P. and Anderson,D.J. (1997) Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing Notch ligands. J. Neurosci., 17, 3644–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Chen,Z., Barrantes,I.B., de la Pompa,J.L. and Anderson,D.J. (1998) neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron, 20, 469–482. [DOI] [PubMed] [Google Scholar]
- Ma Q., Fode,C., Guillemot,F. and Anderson,D.J. (1999) NEUROGENIN1 and NEUROGENIN2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev., 13, 1717–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M., Lufkin,T., Vonesch,J.-L., Ruberte,E., Olivo,J.-C., Dollé,P., Gorry,P., Lumsden,A. and Chambon,P. (1993) Two rhombomeres are altered in Hoxa-1 mutant mice. Development, 119, 319–338. [DOI] [PubMed] [Google Scholar]
- Miyata T., Maeda,T. and Lee,J.E. (1999) NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev., 13, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Perez,S.E., Qiao,Z., Verdi,J.M., Hicks,C., Weinmaster,G. and Anderson,D.J. (2000) Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest cells. Cell, 101, 499–510. [DOI] [PubMed] [Google Scholar]
- Morrow E.M., Furukawa,T., Lee,J.E. and Cepko,C.L. (1999) NeuroD regulates multiple functions in the developing neural retina in rodent. Development, 126, 23–36. [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Iwakabe,H., Akazawa,C., Nawa,H., Shigemoto,R., Mizuno,N. and Nakanishi,S. (1993) Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for l-2-amino-4-phosphonobutyrate. J. Biol. Chem., 268, 11868–11873. [PubMed] [Google Scholar]
- Oberdick J., Smeyne,R.J., Mann,J.R., Zackson,S. and Morgan,J.I. (1990) A promoter that drives transgene expression in cerebellar Purkinje and retinal bipolar neurons. Science, 248, 223–226. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Ishibashi,M., Gradwohl,G., Nakanishi,S., Guillemot,F. and Kageyama,R. (1999) Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J., 18, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez S.E., Rebelo,S. and Anderson,D.J. (1999) Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development, 126, 1715–1728. [DOI] [PubMed] [Google Scholar]
- Perron M., Opdecamp,K., Butler,K., Harris,W.A. and Bellefroid,E.J. (1999) X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl Acad. Sci. USA, 96, 14996–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roztocil T., Matter-Sadzinski,L., Alliod,C., Ballivet,M. and Matter,J.-M. (1997) NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development, 124, 3263–3272. [DOI] [PubMed] [Google Scholar]
- Schwab M.H. et al. (2000) Neuronal basic helix-loop-helix proteins (NEX and BETA2/NeuroD) regulate terminal granule cell differentiation in the hippocampus. J. Neurosci., 20, 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L., Shah,N., Rao,M. and Anderson,D.J. (1995) The cellular function of MASH1 in autonomic neurogenesis. Neuron, 15, 1245–1258. [DOI] [PubMed] [Google Scholar]
- Sommer L., Ma,Q. and Anderson,D.J. (1996) neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci., 8, 221–241. [DOI] [PubMed] [Google Scholar]
- Takebayashi K., Takahashi,S., Yokota,C., Tsuda,H., Nakanishi,S., Asashima,M. and Kageyama,R. (1997) Conversion of ectoderm into a neural fate by ATH-3, a vertebrate basic helix-loop-helix gene homologous to Drosophila proneural gene atonal. EMBO J., 16, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y. and Jessell,T.M. (1996) Diversity and pattern in the developing spinal cord. Science, 274, 1115–1123. [DOI] [PubMed] [Google Scholar]
- Tomita K., Ishibashi,M., Nakahara,K., Ang,S.-L., Nakanishi,S., Guillemot,F. and Kageyama,R. (1996a) Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron, 16, 723–734. [DOI] [PubMed] [Google Scholar]
- Tomita K., Nakanishi,S., Guillemot,F. and Kageyama,R. (1996b) Mash1 promotes neuronal differentiation in the retina. Genes Cells, 1, 765–774. [DOI] [PubMed] [Google Scholar]
- Torii M., Matsuzaki,F., Osumi,N., Kaibuchi,K., Nakamura,S., Casarosa,S., Guillemot,F. and Nakafuku,M. (1999) Transcription factors Mash-1 and Prox-1 delineate early steps in differentiation of neural stem cells in the developing central nervous system. Development, 126, 443–456. [DOI] [PubMed] [Google Scholar]
- Tsuda H., Takebayashi,K., Nakanishi,S. and Kageyama,R. (1998) Structure and promoter analysis of Math3 gene, a mouse homolog of Drosophila proneural gene atonal. J. Biol. Chem., 273, 6327–6333. [DOI] [PubMed] [Google Scholar]
- Zheng J.L. and Gao,W.-Q. (2000) Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nature Neurosci., 3, 580–586. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wang,S. and Anderson,D.J. (2000) Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron, 25, 331–343. [DOI] [PubMed] [Google Scholar]