Abstract
Objective:
Mutations in mitofusin 2 (MFN2) are the most common cause of axonal Charcot-Marie-Tooth disease (CMT2). Over 50 mutations have been reported, mainly causing autosomal dominant disease, though families with homozygous or compound heterozygous mutations have been described. We present 3 families with early-onset CMT2 associated with compound heterozygous MFN2 mutations. Transcriptional analysis was performed to investigate the effects of the mutations.
Methods:
Patients were examined clinically and electrophysiologically; parents were also examined where available. Genetic investigations included MFN2 DNA sequencing and dosage analysis by multiplex ligation-dependent probe amplification. MFN2 mRNA transcripts from blood lymphocytes were analyzed in 2 families.
Results:
Compound heterozygosity for MFN2 mutations was associated with early-onset CMT2 of varying severity between pedigrees. Parents, where examined, were unaffected and were heterozygous for the expected mutations. Four novel mutations were detected (one missense, one nonsense, an intragenic deletion of exons 7 + 8, and a 3–base pair deletion), as well as 2 previously reported missense mutations. Transcriptional analysis demonstrated aberrant splicing of the exonic deletion and indicated nonsense-mediated decay of mutant alleles with premature truncating mutations.
Conclusions:
Our findings confirm that MFN2 mutations can cause early-onset CMT2 with apparent recessive inheritance. Novel genetic findings include an intragenic MFN2 deletion and nonsense-mediated decay. Carrier parents were asymptomatic, suggesting that MFN2 null alleles can be nonpathogenic unless coinherited with another mutation.
Mitofusin 2 (MFN2) mutations are the most prevalent cause of axonal Charcot-Marie-Tooth disease (CMT).1–3 Over 50 mutations have been reported in MFN2,4 the majority causing autosomal dominant CMT2; however, homozygous and compound heterozygous MFN2 mutations have also been reported in early-onset CMT2.2,5–7 In some reported cases, the phenotype of patients with 2 MFN2 mutations in trans have severe early onset axonal neuropathy (SEOAN),8 though patients with mild and moderate phenotypes have been reported with either homozygous or compound heterozygous mutations.6,7 We describe 3 families with compound heterozygous MFN2 mutations causing early-onset CMT2. The severity of the disease varied between pedigrees.
METHODS
Patients.
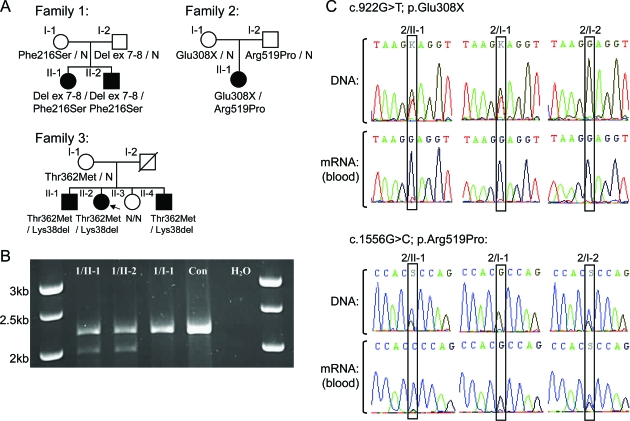
We examined 5 affected patients belonging to 2 British families (families 1 and 2) and one Italian family (family 3) (figure, A). The parents were reviewed, when available, clinically and neurophysiologically, and tested for MFN2 mutations.
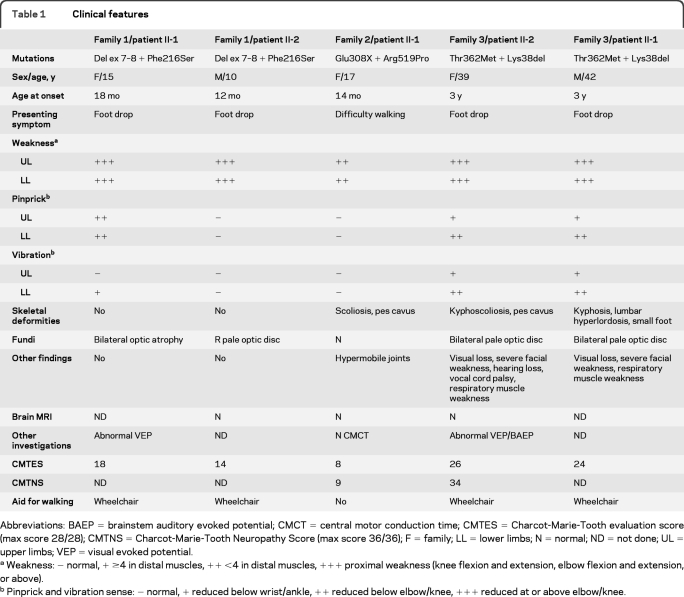
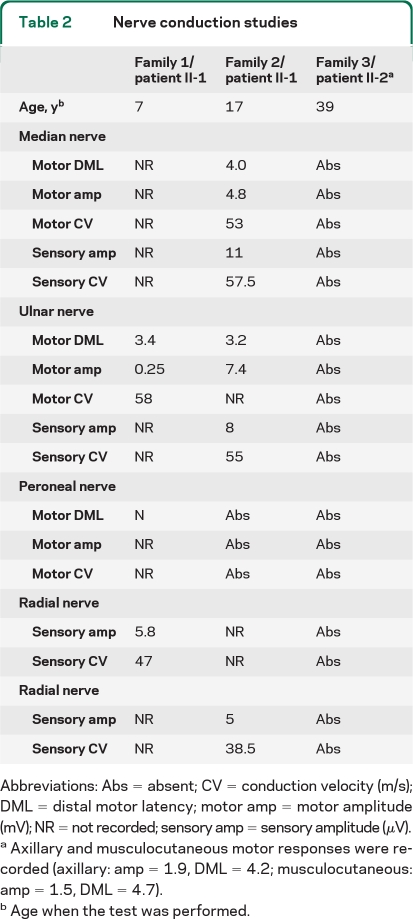
Figure. Pedigrees and transcript analysis.
(A) Pedigrees. Arrow = index case; N = normal genotype. (B) Family 1 MFN2 mRNA PCR from exons 4 to 19. All bands were sequenced. Two transcripts were separated by agarose gel electrophoresis in 1/II-1 and 1/II-2; the upper band encodes the maternal c.647T>C mutation, the lower paternal band, encodes a shortened transcript in which exon 6 is spliced to exon 9, confirmed by sequencing. 1/I-1 was confirmed to heterozygously express c.647T>C. Con = normal control PCR; H2O = water control PCR. (C) Family 2 MFN2 DNA and mRNA sequencing. The thymine allele at base 922 was not evident in 2/II-1 and 2/I-1 in the mRNA sequencing. This allele is likely subjected to nonsense-mediated decay. Because of the nonsense-mediated decay of her maternal allele, 2/II-1 only expresses a cytosine at base c.1556, while it is heterozygously expressed in 2/I-2.
Ethical approval and patient consent.
The study was approved by the ethics committees of The National Hospital for Neurology and Neurosurgery and the Institutional Review Board of the Carlo Besta Neurological Institute IRCCS Foundation and all patients or guardians of patients provided written consent.
Genetic analysis.
MFN2 sequencing was performed with the BigDye Terminator kit v1.1 and a 3730XL genetic analyzer (Applied Biosystems). A total of 350 British and 200 Italian control chromosomes were screened for the novel MFN2 sequence mutations. A multiplex ligation probe amplification (MLPA) assay (MRC Holland kit P143) was used for MFN2 copy-number analysis in families 1 and 2. For transcriptional analysis, blood was collected and extracted using the Qiagen/PreAnalytix blood RNA system. cDNA was synthesized using the Applied Biosystems high capacity cDNA reverse transcription kit.
RESULTS
Clinical and neurophysiologic findings.
Detailed clinical features and nerve conduction studies are summarized in tables 1 and 2. The affected individuals in families 1 and 3 had SEOAN; the proband in family 2 had early onset moderate axonal neuropathy. The parents of families 1 and 2 were unaffected on clinical examination. Only the mother in family 2 had neurophysiology that was normal. The mother in family 3 was unavailable but reportedly unaffected; the deceased father was also thought to be unaffected.
Table 1.
Clinical features
Abbreviations: BAEP = brainstem auditory evoked potential; CMCT = central motor conduction time; CMTES = Charcot-Marie-Tooth evaluation score (max score 28/28); CMTNS = Charcot-Marie-Tooth Neuropathy Score (max score 36/36); F = family; LL = lower limbs; N = normal; ND = not done; UL = upper limbs; VEP = visual evoked potential.
Weakness: − normal, + ≥4 in distal muscles, ++ <4 in distal muscles, +++ proximal weakness (knee flexion and extension, elbow flexion and extension, or above).
Pinprick and vibration sense: − normal, + reduced below wrist/ankle, ++ reduced below elbow/knee, +++ reduced at or above elbow/knee.
Table 2.
Nerve conduction studies
Abbreviations: Abs = absent; CV = conduction velocity (m/s); DML = distal motor latency; motor amp = motor amplitude (mV);NRbnot recorded; sensory amp = sensory amplitude (μV).
Axillary and musculocutaneous motor responses were recorded (axillary: amp = 1.9, DML = 4.2; musculocutaneous: amp = 1.5, DML = 4.7).
Age when the test was performed.
Genetic results.
Family 1.
MFN2 sequencing identified a maternally inherited missense mutation, c.647T>C; p.Phe216Ser in exon 7 in both children. MLPA analysis detected a deletion of MFN2 exons 7 and 8 in the children and their father, confirmed by long PCR (data not shown). MLPA on 306 further patients with CMT2 (with and without MFN2 sequence mutations) detected no other rearrangements. The MFN2 transcript was amplified in the children and the mother (figure, B). Translation of the mutant paternal transcript would result in a truncated protein of 199 in-frame amino acids, followed by 61 out-of-frame amino acids before a premature stop codon.
Family 2.
MFN2 sequencing identified 2 novel mutations in the proband, a maternally inherited nonsense mutation in exon 9 (c.922G>T; p.Glu308X), and a paternally inherited missense mutation in exon 15 (c.1556G>C; p.Arg519Pro). Neither mutation was detected in 350 British control chromosomes. MLPA analysis of the proband detected no MFN2 rearrangements. Sequencing of MFN2 transcripts demonstrated nonsense-mediated decay of the maternal allele (figure, C).
Family 3.
Two MFN2 mutations were detected in the proband (3/II-2), c.1085C>T; p.Thr362Met and c.113_115delAGA; p.Lys38del, and her 2 affected brothers; neither were detected in her unaffected sister. c.1085C>T was maternally inherited; no DNA was available from the deceased father. The novel c.113_115delAGA; p.Lys38del mutation was not detected in 200 Italian and 350 British control chromosomes.
DISCUSSION
CMT2 caused by MFN2 mutations is typically an autosomal dominant disease, with the assumption that mutations act via a dominant-negative or toxic gain-of-function mechanism.9 There are previous reports of patients with CMT2 with 2 MFN2 mutations in trans 2,5–7; in most cases the inheritance was recessive, though semidominant inheritance has also been described, where the parents were slightly affected.5 MFN2 is not unique among CMT-related genes in causing disease via multiple inheritance patterns. In most cases recessive forms are more severe, though exceptions are known.10,11
In our series, SEOAN was associated with compound heterozygosity for a deletion of MFN2 exons 7 and 8 and the missense mutation c.647T>C; p.Phe216Ser in family 1. The mechanism of the pathogenic effect of the exon 7 + 8 deletion, carried by the unaffected father, is uncertain. This transcript is likely to be subject to nonsense-mediated decay, as would be expected for any stop codon located >55 bp from a downstream intron/exon boundary.12 This is suggested by the reduced intensity of the lower band in the figure, C. Although the PCR performed here was not a strictly quantitative assay, preferential amplification of a shorter template would be expected to generate a more intense shorter band if template concentrations were equal. If translated, the deletion transcript would result in a truncated, out-of-frame protein lacking nearly 50% of the GTPase domain, both coiled-coil domains and the transmembrane domain. Depending on the amount of protein synthesized and its activity, this may therefore be a null mutation or have a dominant-negative or toxic gain-of-function effect with reduced penetrance (unless coexpressed with a second mutation). Interestingly, homozygosity for the Phe216Ser mutation caused early-onset mild CMT2 in another patient.7
We detected nonsense-mediated decay of the maternal transcript carrying the Glu308X mutation in family 2, who has a milder phenotype. As a result, the proband in family 2 expresses only her unaffected father's Arg519Pro allele. Despite only expressing a single MFN2 allele the mother remains unaffected. This suggests that MFN2 is not sensitive to haploinsufficiency, confirming that the pathogenic dominant missense mutations described to date are likely to have a dominant-negative or a toxic gain-of-function effect and highlighting that nonsense mutations in MFN2 should not necessarily be assumed to be pathogenic without considering the likelihood of nonsense-mediated decay and the presence of a mutation on the other allele. This is supported by experiments showing that dominant MFN2 mutations do not affect the stability or targeting of the protein.9 In addition, heterozygous MFN2-knockout mice do not develop peripheral neuropathy.13 Thus the pathogenic mechanism in family 2 may be hemizygous expression of a missense mutation that is not sufficient to cause disease when coexpressed with normal protein.
The inheritance patterns in families 1 and 2 appear recessive though one or more of the parents (age range 36–46) may yet develop symptoms.
In family 3, coinheritance of c.1085C>T; p.Thr362Met and c.113_115delAGA; p.Lys38del caused SEOAN. The mode of inheritance is unknown; neither parent was known to be affected. The disease severity might be caused by the presence of 2 abnormal MFN2 proteins which in the heterozygous state can be compensated by normal MFN2. Given that the c.1085C>T; p.Thr362Met mutation has been reported in autosomal dominant3 and semidominant5 pedigrees, it is likely to have a dominant-negative effect. The pathogenic effect of the c.113_115delAGA; p.Lys38del mutation is unknown at present.
We have presented 3 pedigrees supporting the hypothesis that coinheritance of 2 non- or minimally pathogenic MFN2 mutations causes early-onset CMT2. Similar to earlier reports, the disease was very severe in 2 families, but moderate in one (family 2). The lack of rearrangements in 306 additional patients with CMT2 indicates that exonic rearrangements are rare in MFN2 but should be considered, particularly in cases of early-onset neuropathy where one mutation has been identified by sequencing. Thorough genetic analysis is absolutely required for accurate prognosis and prediction of recurrence risk. Further work is required to understand the underlying molecular mechanisms of MFN2 mutations in dominant and apparently recessive families; long-term follow-up of carrier parents will help to clarify the inheritance patterns involved.
Footnotes
- CMT
- Charcot-Marie-Tooth disease
- MLPA
- multiplex ligation probe amplification
- SEOAN
- severe early onset axonal neuropathy
AUTHOR CONTRIBUTIONS
Dr. Polke: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, study supervision. Dr. Laurá: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, study supervision. Dr. Pareyson: drafting/revising the manuscript, analysis or interpretation of data, acquisition of data. Dr. Taroni: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, contribution of vital reagents/tools/patients, acquisition of data, obtaining funding. M. Milani: analysis or interpretation of data, acquisition of data. G. Bergamin: analysis or interpretation of data, acquisition of data. Dr. Gibbons: analysis or interpretation of data, acquisition of data. Prof. Houlden: drafting/revising the manuscript, analysis or interpretation of data, acquisition of data. S.C. Chamley: analysis or interpretation of data, acquisition of data. Dr. Blake: drafting/revising the manuscript, analysis or interpretation of data, acquisition of data. Dr. DeVile: drafting/revising the manuscript, acquisition of data. Dr. Sandford: drafting/revising the manuscript, acquisition of data. M.G. Sweeney: drafting/revising the manuscript, analysis or interpretation of data, acquisition of data, study supervision. Dr. Davis: drafting/revising the manuscript, analysis or interpretation of data, study supervision. Prof. Reilly: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, study supervision.
DISCLOSURE
Dr. Polke, Dr. Laurá, and Dr. Pareyson report no disclosures. Dr. Taroni serves as an Associate Editor for Neurological Sciences and receives research support from ApoPharma, AIFA (Italian Drug Administration), the Italian Ministry of Health, and Telethon-Italia. M. Milani, G. Bergamin, and Dr. Gibbons report no disclosures. Prof. Houlden receives research support from the MRC UK and the Wellcome Trust. S.C. Chamley, Dr. Blake, and Dr. DeVile report no disclosures. Dr. Sandford serves as an Associate Editor for Nephron-Experimental Nephrology and receives research support from the Wellcome Trust, the MRC UK, Kidney Research UK, NIHR, Cambridge Biomedical Research Centre, and the PBC Foundation. M.G. Sweeney reports no disclosures. Dr. Davis reports no disclosures. Prof. Reilly serves on the editorial advisory board of Brain, on the editorial boards of Neuromuscular Disorders and Journal of Neurology, Neurosurgery & Psychiatry.
REFERENCES
- 1. Züchner S, Mersiyanova IV, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 2004;36:449–451 [DOI] [PubMed] [Google Scholar]
- 2. Verhoeven K, Claeys KG, Züchner S, et al. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain 2006;129:2093–2102 [DOI] [PubMed] [Google Scholar]
- 3. Chung KW, Kim SB, Park KD, et al. Early onset severe and late-onset mild Charcot-Marie-Tooth disease with mitofusin 2 (MFN2) mutations. Brain 2006;129:2103–2118 [DOI] [PubMed] [Google Scholar]
- 4. Cartoni R, Martinou JC. Role of mitofusin 2 mutations in the physiopathology of Charcot-Marie-Tooth disease type 2A. Exp Neurol 2009;218:268–273 [DOI] [PubMed] [Google Scholar]
- 5. Nicholson GA, Magdelaine C, Zhu D, et al. Severe early-onset axonal neuropathy with homozygous and compound heterozygous MFN2 mutations. Neurology 2008;70:1678–1681 [DOI] [PubMed] [Google Scholar]
- 6. Calvo J, Funalot B, Ouvrier RA, et al. Genotype-phenotype correlations in Charcot-Marie-Tooth disease type 2 caused by mitofusin 2 mutations. Arch Neurol 2009;66:1511–1516 [DOI] [PubMed] [Google Scholar]
- 7. Vallat JM, Ouvrier RA, Pollard JD, et al. Histopathological findings in hereditary motor and sensory neuropathy of axonal type with onset in early childhood associated with mitofusin2 mutations. J Neuropathol Exp Neurol 2008;67:1097–1102 [DOI] [PubMed] [Google Scholar]
- 8. Ouvrier RA, McLeod JG, Morgan GJ, Wise GA, Conchin TE. Hereditary motor and sensory neuropathy of neuronal type with onset in early childhood. J Neurol Sci 1981;51:181–187 [DOI] [PubMed] [Google Scholar]
- 9. Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci 2007;27:422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marques W, Jr, Sweeney MG, Wood NW. Thr(118)Met amino acid substitution in the peripheral myelin protein 22 does not influence the clinical phenotype of Charcot-Marie-Tooth disease type 1A due to the 17p11.2-p12 duplication. Braz J Med Biol Res 2003;36:1403–1407 [DOI] [PubMed] [Google Scholar]
- 11. Abe A, Numakura C, Saito K, et al. Neurofilament light chain polypeptide gene mutations in Charcot-Marie-Tooth disease: nonsense mutation probably causes a recessive phenotype. J Hum Genet 2009;54:94–97 [DOI] [PubMed] [Google Scholar]
- 12. Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998;23:198–199 [DOI] [PubMed] [Google Scholar]
- 13. Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 2003;160:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]