Abstract
N-acetylglucosaminyltransferase I (GnT I) serves as the gateway from oligomannose to hybrid and complex N-glycans and plays a critical role in mammalian development and possibly all metazoans. We have determined the X-ray crystal structure of the catalytic fragment of GnT I in the absence and presence of bound UDP-GlcNAc/Mn2+ at 1.5 and 1.8 Å resolution, respectively. The structures identify residues critical for substrate binding and catalysis and provide evidence for similarity, at the mechanistic level, to the deglycosylation step of retaining β-glycosidases. The structuring of a 13 residue loop, resulting from UDP-GlcNAc/Mn2+ binding, provides an explanation for the ordered sequential ‘Bi Bi’ kinetics shown by GnT I. Analysis reveals a domain shared with Bacillus subtilis glycosyltransferase SpsA, bovine β-1,4-galactosyl transferase 1 and Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase. The low sequence identity, conserved fold and related functional features shown by this domain define a superfamily whose members probably share a common ancestor. Sequence analysis and protein threading show that the domain is represented in proteins from several glycosyltransferase families.
Keywords: evolution/glycosylation/glycosyltransferases/N-acetylglucosaminyltransferase I/X-ray crystal structure
Introduction
Eukaryotic cell surface and secreted proteins are post-translationally modified by O- (Ser/Thr-linked) and N-linked (Asn-linked) oligosaccharide chains. In many cases, these oligosaccharides serve as ligands for processes mediated by specific protein–carbohydrate interaction, such as targeting, adhesion and signalling (Varki, 1993). The N-linked glycans can be subdivided into three types: oligomannose, hybrid and complex. Oligomannose structures are found in all eukaryotes, whereas hybrid and complex N-glycans are characteristic of multicellular organisms. It has been proposed, therefore, that the appearance of these structures correlates with the evolution of multicellular organisms and the need for structures capable of mediating cell–cell interactions. It has been suggested that the enzymes required for the production of these information-bearing oligomannose structures were grafted onto a primordial pathway that was designed to produce oligomannose core saccharides for purely structural roles (Kornfeld and Kornfeld, 1980; Drickamer and Taylor, 1998).
The enzyme at the gateway from oligomannose structures to hybrid and complex N-glycans is UDP-N-acetylglucosamine:α-3-d-mannoside β-1,2-N-acetyl glucosaminyltransferase I (GnT I, EC 2.4.1.101) (Stanley et al., 1975; Narasimhan et al., 1977; Harpaz and Schachter, 1980). It transfers the first N-acetyl glucosamine (GlcNAc) residue onto the oligomannose core; all other enzymes in the hybrid and complex pathway depend on its prior action (Schachter, 1986, 1991). GnT I plays a fundamental role in mammalian development, as shown by studies in mice with a null mutation in the GnT I gene (Ioffe and Stanley, 1994; Metzler et al., 1994). Moreover, mutation or misregulation of several of the enzymes dependent on GnT I action is associated with human disease and metastasis (Jaeken et al., 1993, 1994; Charuk et al., 1995; Tan et al., 1996; Granovsky et al., 2000).
The known glycosyltransferases have been classified into 47 different families, based on both sequence similarity and substrate/product stereochemistry (inverting or retaining) (Campbell et al., 1997, 1998; Coutinho and Henrissat, 1999). GnT I (family 13) is an inverting glycosyltransferase: the α-linked GlcNAc moiety from the UDP-α-GlcNAc donor is transferred to the Man-α- 1,3-Man-β-R arm (3-arm) of the Man5GlcNAc2 acceptor, creating the β-linked GlcNAc-β-1,2-Man-α-1,3-Man-β-R product (Vella et al., 1984; Möller et al., 1992; Reck et al., 1995).
Mechanistically, glycosyltransferases are believed to proceed through an oxocarbenium-ion-like transition state similar to that proposed for glycosidases (Kim et al., 1988; Qiao et al., 1996; Murray et al., 1997; Davies et al., 1998a). The reaction is thought to involve general base catalysis and in many cases has been shown to depend on Mn2+ ions (Nishikawa et al., 1988; Murray et al., 1996). To date, the X-ray crystal structures of four glycosyltransferases have been determined: the unclassified inverting bacteriophage T4 β-glucosyltransferase (BGT) (Vrielink et al., 1994; Moréra et al., 1999), the inverting Bacillus subtilis nucleoside–diphospho-sugar glycosyltransferase SpsA (family 2) (Charnock and Davies, 1999), the inverting bovine β-1,4-galactosyltransferase 1 (β4Gal-T1, family 7) (Gastinel et al., 1999) and the inverting Escherichia coli N-acetylglucosaminyl transferase MurG (family 28) (Ha et al., 2000). BGT and glycogen phosphorylase have been proposed to share a common ancestor (Artymiuk et al., 1995; Holm and Sander, 1995), a relationship that may now extend to MurG and family 1 UDP-glucuronosyltransferases (Ha et al., 2000). A clear picture of the structural basis for catalysis has not yet emerged, however, in part because the complete donor nucleotide–sugar was not observed in any of these structures.
Here we present the X-ray crystal structure of GnT I, both in its uncomplexed form and in complex with UDP-GlcNAc and a Mn2+ ion. The structure provides insight into the reaction mechanism of inverting glycosyltransferases, as well as similarity to that found for the deglycosylation step of retaining β-glycosidases. More over, the structure shows that GnT I possesses a domain that is structurally very similar to one found in SpsA (the SGC domain, SpsA GnT I core domain). This domain is also found, in a somewhat modified form, in β4Gal-T1 and E.coli N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) (Brown et al., 1999). These proteins, with their low sequence identity, common fold and related functional features, define the SGC superfamily. In conjunction with sequence analysis and protein threading, we identify several other glycosyltransferase families containing the SGC domain.
Results and discussion
Overall structure
The catalytic fragment of rabbit GnT I (residues 106–447) (Sarkar et al., 1998) was crystallized in the presence of UDP-GlcNAc and Mn2+ and solved by the multi-wavelength anomalous diffraction (MAD) phasing method using a methylmercury chloride derivative and data collected at two wavelengths (Table I). In total, the structure was refined against data sets from three different crystals. Of these, no bound nucleotide sugar or Mn2+ ion was observed in the mercury ‘derivative’ (refined at 1.4 Å resolution) nor in the structure refined against the data set that we have termed ‘native’ (1.5 Å resolution). Since the native and derivative data sets were collected from crystals that had been grown (at room temperature) 4 weeks before the X-ray data were collected, we assume that in these cases the UDP-GlcNAc had been hydrolysed. In contrast, both components were observed in the ‘complex’ (1.8 Å resolution) where samples were 5 days old.
Table I. Crystallographic data and refinement statistics.
| Derivative (MeHgCl) |
Native | Complex with UDP-GlcNAc and Mn2+ | ||
|---|---|---|---|---|
| Edge | Peak | |||
| Crystal parameters | ||||
| space group | P212121 | P212121 | P212121 | |
| a (Å) | 40.4 | 40.5 | 40.5 | |
| b (Å) | 82.4 | 82.4 | 82.2 | |
| c (Å) | 102.5 | 102.5 | 102.0 | |
| Diffraction statistics | ||||
| wavelength (Å) | 1.0093 | 1.0075 | 0.9914 | 1.0713 |
| resolution range (Å) | 31.7–1.4 | 31.7–1.4 | 38.2–1.5 | 34.3–1.8 |
| measured reflections (n) | 348 028 | 325 287 | 401 605 | 64 537 |
| unique reflections (n) | 102 627 | 102 233 | 99 934 | 42 919 |
| completeness (%) | 78.7 | 78.4 | 94.2 | 70.3 |
| Rsyma | 0.047 | 0.053 | 0.065 | 0.092 |
| sites (n) | 1 | 1 | – | – |
| phasing powerb: | ||||
| dispersive | – | 1.64 | – | – |
| anomalous | 2.26 | 2.65 | – | – |
| figure of merit, before solvent flattening | 0.581 | – | – | – |
| Refinement statistics | ||||
| Rcryst | 0.167 | – | 0.166 | 0.185 |
| Rfree | 0.189 | – | 0.194 | 0.229 |
| total atoms (n) | 3204 | – | 3167 | 3138 |
| protein | 2710 | – | 2710 | 2811 |
| substrate | 0 | – | 0 | 40 |
| water | 492 | – | 457 | 275 |
| r.m.s.d.c bond length (Å) | 0.011 | – | 0.013 | 0.010 |
| r.m.s.d. bond angle (°) | 1.5 | – | 1.6 | 1.5 |
| mean B value (Å2) | 14.2 | – | 14.4 | 16.2 |
| protein | 12.3 | – | 12.3 | 16.0 |
| domain 1 (106–317) | 11.5 | – | 11.3 | 14.2 |
| loop (318–330) | – | – | – | 28.3 |
| linker (331–353) | 12.1 | – | 12.1 | 15.4 |
| domain 2 (354–447) | 14.1 | – | 14.6 | 18.7 |
| substrates | – | – | – | 23.0 |
| water | 26.6 | – | 27.9 | 25.9 |
aRsym = |I – <I>|/I, where I is the observed intensity and <I> is the average intensity obtained from multiple observations of symmetry-related reflections.
bPhasing power, root mean square (r.m.s.) FH/r.m.s. ε, where ε is lack of closure and FH is the calculated heavy atom structure factor.
cr.m.s.d., root mean square deviation.
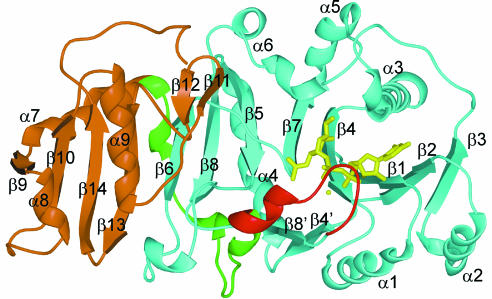
GnT I is a two-domain protein, with overall dimensions of ∼65 × 40 × 50 Å (Figure 1). The N-terminal domain (domain 1: residues 106–317) is an eight-stranded mixed β-sheet (β1–β8), flanked by six α-helices (α1–α6) and a small two-stranded antiparallel β-sheet (β4′ and β8′). It contains two disulfide bonds, the first (C115–C145) connecting β-strands β1 and β2, and the second (C239– C305) connecting β-strands β5 and β8. The smaller C-terminal domain (domain 2: residues 354–447) is a four-stranded mixed β-sheet (β9, β10, β13 and β14), flanked by three α-helices (α7–α9) and a short β-finger (β11 and β12). The two domains are connected by a linker region (residues 331–353), which wraps halfway around domain 1 before starting the first helix of domain 2. The ∼1050 Å2 interface between domains 1 and 2 is quite hydrophilic and contains 20 bridging water molecules. The residues buried in the interface on domain 1 are 53% polar, while those in domain 2 are 36% polar.
Fig. 1. GnT I ribbon diagram. Domain 1 is shown in cyan, the loop structured upon UDP-GlcNAc binding in red, the linker connecting domains 1 and 2 in green, domain 2 in brown and UDP-GlcNAc and Mn2+ ion in yellow. All molecular images were prepared using SPOCK (Christopher, 1998) and rendered using Raster3D (Bacon and Anderson, 1988; Merritt and Murphy, 1994).
The α-helices α3, α5 and α6 sit on ‘top’ of the central β-sheet and create a pocket for the nucleotide–sugar and oligosaccharide acceptor. Electrostatic potential analysis shows that this pocket is largely acidic, in contrast to the rest of the protein surface, which is primarily positively charged. The nucleotide sugar itself sits between helices α3 and α6 and β-strands β1, β2 and β4. The topology and structure of β-strands β1–β4, and helices α1–α3, are similar to those of the corresponding elements in domains possessing the Rossmann fold (Rossmann et al., 1974); however, the orientation of the nucleotide with respect to these elements differs from that seen here.
In the native and derivative structures, where UDP-GlcNAc and Mn2+ were not observed, there is also no electron density for a 13 residue loop (residues 318–330) adjacent to the nucleotide–sugar binding site. The ‘missing loop’ is presumed to be disordered in these crystals, as SDS–PAGE analysis of washed crystals showed the protein to be intact. In the complex, the loop is structured and found to form a flap that partially covers the UDP-GlcNAc moiety. Although structured by UDP-GlcNAc binding, only the tip of the loop makes direct interactions with it. Approximately 50 Å2 is buried between the tip of the loop and the UDP-GlcNAc phosphates. Structuring the loop also buries ∼600 Å2 of protein surface next to the nucleotide–sugar binding site. In these crystals, the active site and the loop itself are exposed to a large solvent channel and are not involved in crystal contacts. Aside from structuring the loop, there is no major conformational change associated with UDP-GlcNAc binding. The native and complex structures show a root mean square deviation (r.m.s.d.) of 0.28 Å, based on the α-carbon atoms of residues 106–317 and 331–447.
Interestingly, two GnT I loss-of-function mutations in cultured cell lines have been identified. The first is a mutation in G320, one of the loop residues structured upon UDP-GlcNAc binding (Opat et al., 1998), and the second, C123, is the site of mercury binding in our derivative structure (Puthalakath et al., 1996). The former presumably prevents loop structuring, while the latter probably destabilizes the protein as C123 points into the protein core.
The nucleotide sugar and metal-binding sites
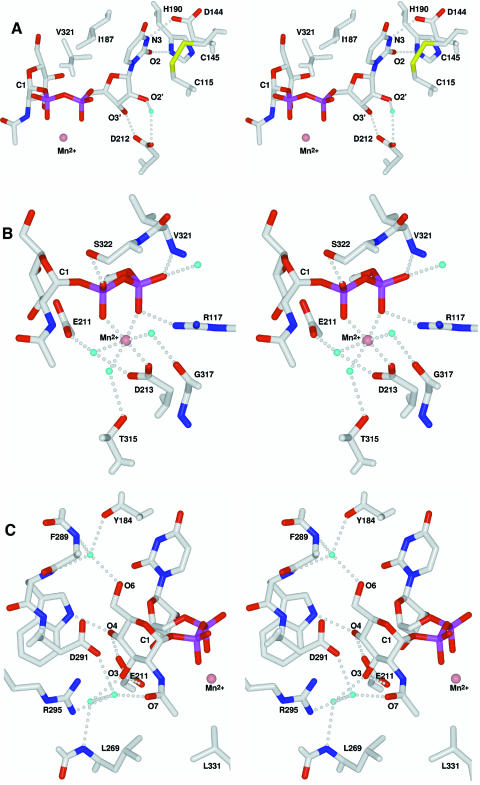
As shown by the complex, GnT I binds UDP-GlcNAc with the uracil and ribose moieties in the anti conformation. The uracil ring is sandwiched between I187 and the C115–C145 cysteine bridge, and its N3 and O2 atoms make key hydrogen bond interactions with D144 and H190, respectively (Table II; Figure 2A). Moreover, its C5 atom is in van der Waals contact with V321, part of the loop structured by UDP-GlcNAc binding. The ribose O2′ and O3′ atoms make a water-mediated and a direct hydrogen bond, respectively, with the carboxyl side chain of D212.
Table II. The UDP-GlcNAc binding site; distances between the UDP-GlcNAc, the Mn2+, bound waters and the protein atoms involved in their binding are listed.
| Interacting atoms |
|
Distance (Å) |
Interacting atoms |
|
Distance (Å) |
| Uracil N3 | D144 Oδ2 | 2.8 | GlcNAc O6 | H2O 4 | 2.7 |
| Uracil O2 | H190 Nδ1 | 2.7 | Mn2+ | D213 Oδ2 | 2.1 |
| Ribose O2′ | D212 Oδ1 | 3.2 | Mn2+ | H2O 38 | 2.4 |
| Ribose O2′ | H2O 40 | 2.9 | Mn2+ | H2O 87 | 2.4 |
| Ribose O3′ | D212 Oδ1 | 2.9 | Mn2+ | H2O 116 | 2.1 |
| α-phosphate O1α | V321 N | 2.7 | H2O 4 | Y184 O | 2.9 |
| α-phosphate O1α | H2O 72 | 2.8 | H2O 4 | F289 N | 2.8 |
| α-phosphate O2α | R117 NH2 | 2.8 | H2O 4 | W290 N | 3.2 |
| α-phosphate O2α | Mn2+ | 2.1 | H2O 27 | L269 N | 3.0 |
| β-phosphate O1β | S322 Oγ | 2.5 | H2O 38 | E211 Oε1 | 2.4 |
| β-phosphate O2β | Mn2+ | 2.1 | H2O 38 | D213 Oδ1 | 2.8 |
| β-phosphate O2β | GlcNAc N2 | 2.9 | H2O 40 | D212 Oδ2 | 3.0 |
| GlcNAc O7 | H2O 263 | 2.8 | H2O 87 | T315 Oγ1 | 3.0 |
| GlcNAc O3 | E211 Oε1 | 2.7 | H2O 116 | G317 O | 2.6 |
| GlcNAc O3 | H2O 27 | 2.6 | H2O 263 | D291 Oδ1 | 2.9 |
| GlcNAc O4 | E211 Oε2 | 2.6 | H2O 263 | R295 NH2 | 3.0 |
| GlcNAc O4 | W290 Nε1 | 2.8 |
Fig. 2. Stereo view of the UDP-GlcNAc/Mn2+ binding site. Carbon, oxygen, nitrogen, sulfur and phosphorus are coloured white, red, blue, yellow and purple, respectively; water molecules are cyan and the Mn2+ ion is salmon. Hydrogen bonds are shown as dotted lines. The C1 of the N-acetylglucos amine moiety is labelled for reference. (A) Uracil and ribose interactions; (B) Mn2+ and phosphate interactions; (C) N-acetylglucosamine interactions.
The Mn2+ ion shows an octahedral geometry coordinated by six inner sphere oxygen atoms (Table II; Figure 2B). The α- and β-phosphates of UDP-GlcNAc each contribute a coordinating oxygen atom, as do three water molecules. These water molecules are, in turn, hydrogen bonded to outer sphere protein residues E211, D213, T315 and G317. The remaining inner sphere metal ligand is provided by the carboxyl group of D213, the only direct interaction with the protein. As such, it seems that GnT I does not have an independent metal-binding site capable of binding Mn2+ in the absence of UDP-GlcNAc. In addition to coordinating the Mn2+ ion in a conformation typical of bidentate divalent metal-bound nucleotides (Black et al., 1994; Klevickis and Grisham, 1996), the phosphates make direct interactions with the protein. The α-phosphate makes a salt bridge with R117 and a hydrogen bond to the amide nitrogen of V321, and the β-phosphate hydrogen bonds to the hydroxyl group of S322. These interactions with V321 and S322 are an important component of the UDP-GlcNAc-dependent structuring of the loop.
Finally, the GlcNAc moiety itself makes several interactions with the protein (Table II; Figure 2C). The vicinal O3 and O4 hydroxyls are hydrogen bonded to the carboxyl group of E211 and the O4 hydroxyl also makes a hydrogen bond with the Nε1 atom of W290. The O6 hydroxyl is hydrogen bonded to a tightly bound water molecule seen in both the uncomplexed and complex structures. Van der Waals interactions are also important, most notably between the N-acetyl methyl group and the side chains of L269 and L331. The GlcNAc N2 makes a hydrogen bond with one of the oxygen atoms of the β-phosphate (Table II; Figure 2B and C). This intramolecular hydrogen bond may be an important component of catalysis, as will be discussed below.
The glycosyltransferase DxD motif
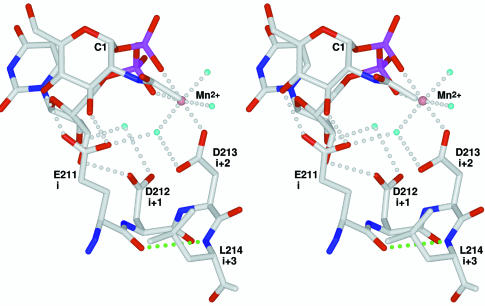
The so-called DxD motif has been identified in many glycosyltransferase families (Breton et al., 1998; Shibayama et al., 1998; Wiggins and Munro, 1998; Breton and Imberty, 1999) and is thought to be involved in Mn2+ ion binding and catalysis (Busch et al., 1998; Wiggins and Munro, 1998; Shibayama et al., 1999; Hodson et al., 2000). Although the canonical motif contains two aspartic acid residues, the first (in the first position of the motif) is relatively variable while the second (in the third position of the motif) is quite well conserved. The DxD motif is flanked by apolar residues (hhhhDxDxh) and site-directed mutagenesis has shown that both acidic residues are required for activity in yeast α-1,3-mannosyltransferase (Wiggins and Munro, 1998). In GnT I, the motif is present in the form 211EDD213 and with L214 forms residues i to i+3 of a type I β-turn connecting β-strands β4 and β4′ (Figure 3). As such, the first and third positions of the motif are directed towards the same face of the turn. The fact that β4 runs through the core of the protein is consistent with the observed presence of several apolar residues on the N-terminal side of the motif.
Fig. 3. The DxD motif. Atom colours and labels are as in Figure 2. ‘i’, ‘i+1’, ‘i+2’ and ‘i+3’ correspond to the residues of the type I β-turn. The hydrogen bond characteristic of this turn type is shown in green.
The interactions with UDP-GlcNAc and the Mn2+ ion illustrate the importance of the motif. As discussed above, the third position (D213) makes the only direct interaction with the bound Mn2+ ion. In addition, it makes a hydrogen bond with one of the metal coordinating water molecules, which itself is hydrogen bonded to the first position of the motif (E211). These residues are further constrained by the well defined octahedral geometry characteristic of Mn2+ ion coordination. Since the phosphates of the nucleotide– sugar also coordinate the Mn2+ ion, the relative orientation of the nucleotide–sugar and the conserved acidic residues is well defined. In GnT I, this arrangement positions the GlcNAc moiety of the donor sugar for interaction with the first position of the motif. Owing to this geometry, this position would also be expected to play a ‘carbohydrate binding’ role in other sugar–nucleoside diphosphate/Mn2+-dependent glycosyltransferases, regardless of the nucleo tide–sugar type.
Reaction mechanism
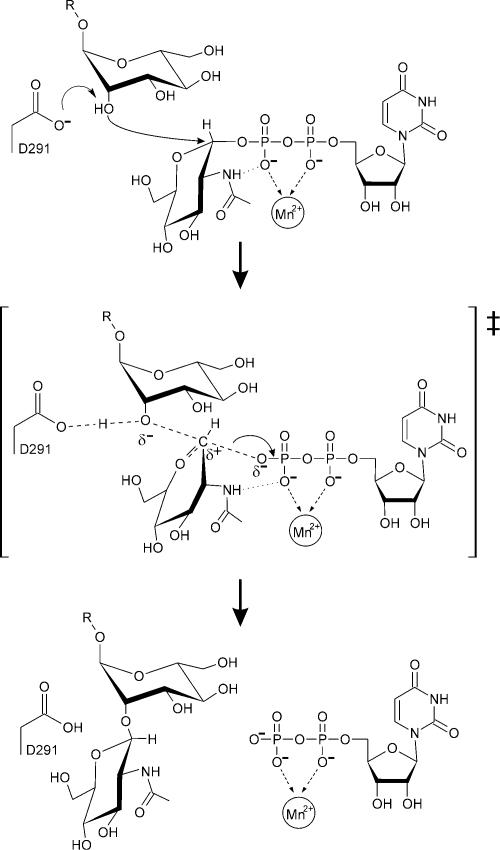
Catalysis by inverting glycosyltransferases is believed to involve a general base, which serves to assist in the deprotonation of the nucleophilic hydroxyl of the acceptor (Figure 4). In GnT I, the only residue capable of playing this role is D291, 4.7 Å away from the GlcNAc C1 (Figure 2C). The structure shows that the acceptor will be able to approach the UDP-GlcNAc donor, so as to permit nucleophilic attack and inversion of stereochemistry at the GlcNAc C1. Many glycosyltransferases also require a metal ion cofactor and, as shown in Figure 2C, the Mn2+ ion is disposed to neutralize the negative charge that develops on the β-phosphate of the UDP leaving group. As with other metal-dependent enzyme-mediated reactions, the ability to do so is maintained by limiting the number of negatively charged inner sphere ligands (in this case, three positions are filled by neutral water molecules) (Cowan, 1998; Dudev et al., 1999).
Fig. 4. Schematic representation of the GnT I reaction mechanism. The oxocarbenium-ion-like transition state is enclosed in large square brackets.
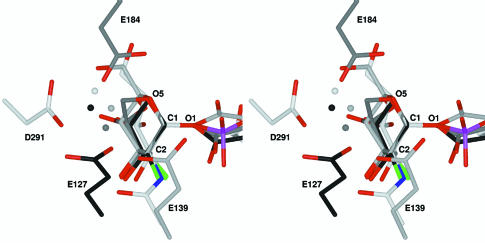
Mechanistically, the glycosyltransferase reaction is thought to involve an oxocarbenium-ion-like transition state similar to that proposed for glycosidases. In retaining glycosidases, two such transition states and a covalent glycosyl-enzyme intermediate are required; hydrolysis of the intermediate can be likened to the reaction performed by an inverting glycosyltransferase. To examine this parallel, we superimposed the covalently bound α-linked sugar moiety from three ‘trapped’ retaining β-glycosidases (Davies et al., 1998b; Notenboom et al., 1998; Sabini et al., 1999) on to the α-linked GlcNAc moiety of the GnT I– UDP-GlcNAc complex (Figure 5). In this overlay, the sugar C1 position is found to be ringed by the catalytic bases, each of which is hydrogen bonded to a water molecule (3.5–4.0 Å from C1) positioned for nucleophilic attack (in GnT I, the water molecule will be displaced by the O2 hydroxyl group of the incoming acceptor). The conserved base and tight clustering of these water molecules strongly supports the suggestion of a shared mechanism among glycosidases and glycosyltransferases.
Fig. 5. Stereo diagram of an overlay of the GnT I active site with three glycosyl-enzyme glycosidase intermediates. Carbon atoms are coloured white in GnT I (D291), light grey in Bacillus agaradhaerens Cel5A [E139, Protein Data Bank (PDB) entry code 6A3H] (Davies et al., 1998b), dark grey in B.agaradhaerens Xyl11 (E184, PDB code 1QH6) (Sabini et al., 1999) and black in Cellulomonas fimi Cex (E127, PDB code 2XYL) (Notenboom et al., 1998). Water molecules are shown as spheres in the same colour as the carbons of the protein to which they are bound. Oxygen (except for waters), nitrogen, fluorine and phosphorus atoms are coloured in red, blue, green and purple, respectively. The structures were overlaid using the positions of the carbohydrate atoms C1, O1, C2 and O5. In all cases the water molecules are positioned for nucleophilic attack at the donor sugar C1; in GnT I the water molecule will be replaced by the attacking 2-hydroxyl of the 3-arm mannose of the Man5GlcNAc2 acceptor. The structures of the glycosyl-enzyme intermediates of the glycosidases Streptomyces lividans CelB2 (PDB code 2NLR) (Sulzenbacher et al., 1999) and Bacillus circulans BCX (PDB code 1BVV) (Sidhu et al., 1999) can be overlaid with GnT I in the same way and yield a very similar picture.
Although the oxocarbenium-ion-like transition state requires a flattening of the sugar ring at C1, in only one of these glycosyl–enzyme glycosidase intermediates (Sabini et al., 1999) is the ring found to be distorted (see Figure 5). Since these intermediates have been trapped by 2-fluoro substitution and interactions with the 2-hydroxyl have been shown to be important for transition state stabilization, the presence or absence of distortion in these complexes is difficult to interpret (Zechel and Withers, 2000). As shown in Figure 2B and C, the intramolecular hydrogen bond between the N2 of the GlcNAc moiety and the β-phosphate suggests that interactions with position 2 might also play a role in stabilizing the glycosyltransferase transition state.
In GnT I, the first example of a glycosyltransferase showing the nucleotide–sugar, the donor sugar is also found in the standard 4C1 conformation. Although enzyme-induced ring distortion might be expected in the case of glycosidases, the low energy conformation seen here in GnT I is perhaps more consistent with the glycosyltransferase reaction. Since a distorted nucleotide– sugar would be susceptible to unwanted hydrolysis, we propose that in the case of an inverting glycosyltransferase, acceptor binding is likely to be an additional requirement for driving the reaction toward the transition state (a similar requirement presumably exists for retaining glycosyltransferases). By using acceptor binding energy in this way, glycosyltransferases can confer acceptor specificity on their reactions.
Loop structuring and the acceptor-binding pocket
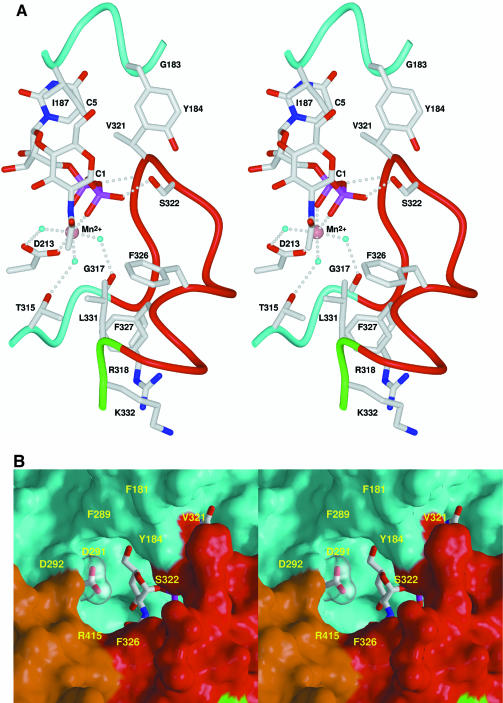
Comparison of the uncomplexed and complex structures shows that UDP-GlcNAc binding structures the 318–330 loop, forming a flap that partly covers the UDP-GlcNAc (Figure 6A). Central to this structuring is V321, at the tip of the loop, whose side chain sits in an apolar pocket formed by residues G183, Y184 and I187 and the C5 atom of the uracil moiety. In addition, the backbone amide nitrogen of V321 and side-chain hydroxyl of S322 make hydrogen bonds to the α- and β-phosphates of the UDP-GlcNAc. Residues 320–323 form a type IV β-turn, while residues 324–330 make one complete turn of an α-helix. The loop folds upon itself, burying residue F327 against R318 and the non-loop residues T315, L331 and K332. The only conformational changes other than structuring the loop itself are a peptide flip (F316–G317) and a reorientation of the T315 side chain. These changes are critical as the G317 carbonyl and the T315 hydroxyl are repositioned to make hydrogen bonds with two of the Mn2+ ion coordinating water molecules (see Figures 6A and 2B).
Fig. 6. Stereo diagram of the structured loop and the acceptor-binding pocket. Atom colours and labels are as in Figure 2. Backbone tubes and molecular surfaces are colour-coded as follows: red, structured loop; green, linker region; cyan, domain 1; brown, domain 2. (A) Structured loop and UDP-GlcNAc/Mn2+ interactions; (B) Surface representation of the acceptor binding pocket. The side chain of the catalytic base (D291) and the N-acetylglucosamine moiety of the UDP-GlcNAc are seen at the base of the pocket.
As shown in Figure 6B, loop structuring creates one side of a deep pocket terminating over the proposed catalytic base (D291) and the GlcNAc moiety. Two loop residues (S322 and F326), fully conserved among active GnT I sequences, are presented to the pocket (Figure 6B). To explore the potential roles played by these and other residues in the binding pocket, we modelled the 3-arm mannose of the acceptor into the site. With the attacking O2 hydroxyl group positioned between the D291 Oδ2 and the UDP-GlcNAc C1 (at the conserved water position shown in Figure 5), only one general orientation leads to reasonable steric and chemical interactions with the protein. In this orientation, the exocyclic C6 hydroxymethyl group of the mannose interacts with S322 and F326, while the O3 and O4 point toward D291, R295 and R415. The importance of the mannose O3, O4 and O6 predicted by this model is consistent with substrate studies using synthetic analogues of the trimannose core of the acceptor (Möller et al., 1992; Reck et al., 1995).
Enzyme kinetics
GnT I proceeds through an ordered sequential ‘Bi Bi’ kinetic mechanism (Nishikawa et al., 1988). The enzyme binds first Mn2+/UDP-GlcNAc and then the Man5GlcNAc2 acceptor; the oligosaccharide product is then released, followed by UDP. The GnT I structures provide a model for these observations. Since UDP-GlcNAc binding is required to structure the loop and create the acceptor-binding site, it is clear that the nucleotide–sugar must bind before the acceptor. An explanation for the observed ordered product release is also provided by the structure of the loop and the central role played by V321. This residue, anchored in the apolar pocket, is positioned adjacent to the scissile bond and splits the loop into two parts: the N-terminal portion leading to the apolar pocket and the C-terminal end forming part of the acceptor/product-binding pocket. Once the bond between the GlcNAc C1 and the β-phosphate is broken, the now terminal β-phosphate acquires an additional negative charge and greater mobility. The hydrogen bond to S322 is likely to be broken, leading to disruption of the C-terminal end of the loop and oligosaccharide product release. V321 may be unaffected, owing to its interactions with the apolar pocket and the α-phosphate. Since the uracil C5 forms part of the apolar pocket, V321 would continue to hold the UDP in place. Once the oligosaccharide product has been removed, bulk solvation of the UDP/Mn2+ complex presumably completes loop disruption and finally UDP/Mn2+ release, a process aided by the fact that only a single direct interaction is made between the Mn2+ ion and the protein surface (through D213).
The SGC domain
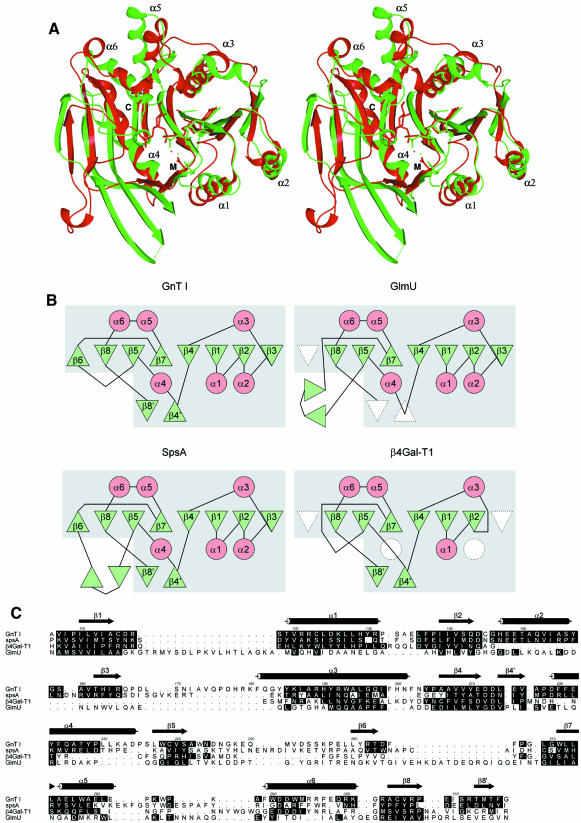
Structural alignment shows that domain 1 of GnT I is very similar to a corresponding domain in B.subtilis glycosyltransferase SpsA (residues 2–217) (Charnock and Davies, 1999). It has identical topology and all of the major secondary structural elements characterizing the domain are found in both structures (Figure 7A). The domain is also found, with some modification in secondary structure (the topology remains the same), in β4Gal-T1 (residues 180–346) (Gastinel et al., 1999) and GlmU (residues 4–227, Figure 7B) (Brown et al., 1999). Structural align ment of GnT I domain 1 and the domains in SpsA, β4Gal-T1 and GlmU using the program DALI (Holm and Sander, 1993) yields Z-scores of 15.7, 10.6 and 9.8, respectively. The very strong structural similarity between GnT I domain 1 and SpsA suggests the existence of a canonical core domain, the SGC domain, represented, to date, in these four structures.
Fig. 7. The SGC domain. (A) Stereo ribbon overlay of the SGC domains of GnT I (red) and SpsA (green). For clarity, only the α-helices are labelled. UDP (SpsA) and UDP-GlcNAc (GnT I) are shown in stick representation. M and C label the side chains of the metal binding and catalytic aspartic acid residues, respectively, which are also shown in stick representation. (B) Topology diagrams of the SGC domains of GnT I, SpsA, GlmU and β4Gal-T1. β-strands are shown as green triangles and α-helices as red circles, with missing elements shown in white. The secondary structural elements are labelled as in GnT I. The boxed grey region corresponds to the SGC domain. (C) A structure-based sequence alignment of GnT I, SpsA, β4Gal-T1 and GlmU, produced by STAMP (Russell and Barton, 1992) and modified slightly by manual intervention. Shown are residue numbers and secondary structure for GnT I. Residues with their α-carbon atoms <2.5 Å from the structurally equivalent GnT I α-carbon atom are shaded in black (as is the GnT I residue itself). The alignment was rendered using ALSCRIPT (Barton, 1993).
Despite the structural similarity shown by these enzymes, they do not show significant amino acid sequence identity. Even with the structure-based sequence alignment program STAMP (Russell and Barton, 1992), the structurally equivalent residues yield identities of only 13, 18 and 8% between the SGC domain of GnT I and that of SpsA, β4Gal-T1 and GlmU, respectively (Figure 7C). These levels of identity make it difficult, if not impossible, to establish whether or not these enzymes stem from a common ancestor. Analysis of residues critical for function may, however, shed light on this question. The position of the UDP moiety in the GnT I complex is virtually identical to that found in the SpsA complex (Figure 7A) and very similar to that seen in the β4Gal-T1 and GlmU complexes. The DxD motif is present in all four of these proteins and forms a superimposable type I β-turn in each case. Finally, at position D291, the proposed catalytic base in GnT I, both SpsA (D191) and β4Gal-T1 (D318) also possess aspartic acid residues (GlmU does not, but it is not a glycosyltransferase). Not only are these key residues and functional features identical in these structures, but also they are found at the same position on the structural/topological framework. Taken together, the low sequence identity, common fold and related functional features shown by these enzymes define a protein superfamily whose members are likely to share a common evolutionary origin (Murzin et al., 1995).
The SGC superfamily
The lack of sequence identity between glycosyltransferases with different specificities has led to a classification that now includes 47 different families (Coutinho and Henrissat, 1999). GnT I, for example, is in a family of its own, and a position-specific iterated BLAST (PSI-BLAST) search, using the GnT I sequence, identifies no other related glycosyltransferases. Based on the knowledge that the GnT I SGC domain is structurally similar to the corresponding domain in SpsA, we decided to make a new attempt at finding sequence similarity between these and other glycosyltransferases. We reasoned that the SpsA sequence, coming from a much larger glycosyltransferase family, would provide a more robust profile and used it to seed a PSI-BLAST search (Altschul et al., 1997). Searched in this way, similarity between SpsA (family 2) and rabbit GnT I (family 13) was identified. Similarity was also found between SpsA and the β-1,4-GalNAc transferases (family 12), the ceramide glucosyltransferases (family 21) and the polypeptide GalNAc transferases (family 27); neither β4Gal-T1 (family 7) nor GlmU appeared in the searches.
To explore further the possible relationships among the glycosyltransferase families, we used protein threading to determine the compatibility of representative glycosyltransferase sequences with the SGC domain. Using the program THREADER 2 (Jones et al., 1992), a single, arbitrarily selected sequence from each of the 27 glycosyltransferase families described by Campbell et al. (1997, 1998) was run against a database of 1900 structures, which included the SGC domain of GnT I, SpsA, β4Gal-T1 and GlmU. In both the normal and randomized test scores, the selected sequence from family 2, family 7 and family 13 ranked first or second against the SGC domain of SpsA, β4Gal-T1 and GnT I, respectively, as would be expected. The sequence from family 3, family 6, family 16 and family 26 also ranked first or second in the two tests, and sequences from several other families also received high scores. These results, and those based on PSI-BLAST searching, suggest that the SGC domain is widely represented among glycosyltransferase families, including both inverting and retaining enzymes.
Conclusion
The structure of the catalytic domain of GnT I has provided the basis for its Mn2+/UDP-GlcNAc-binding properties, as well as insight into its catalytic and kinetic mechanisms. A structural basis for the suggested parallel with glycosidase mechanism has also been shown. The structure of the DxD motif shows that the first conserved residue plays a role in binding the donor sugar, while the second coordinates the essential Mn2+ ion. These roles are likely to be conserved in other DxD-containing glycosyltransferases, regardless of donor specificity. In addition, structural analysis has defined the SGC domain, seen in GnT I, SpsA, β4Gal-T1 and GlmU. Sequence analysis and protein threading show that the SGC domain is contained in enzymes from several of the existing inverting and retaining glycosyltransferase families. Among these are enzymes involved in mammalian N- and O-linked oligosaccharide biosynthesis, bacterial cell wall production and the synthesis of glycogen, chitin and cellulose. Together, they constitute the SGC superfamily.
Materials and methods
Expression and purification
The catalytic fragment of rabbit (Oryctolagus cuniculus) GnT I (residues 106–447) was expressed in the baculovirus/insect cell system (Sarkar et al., 1998). Sf9 cells were grown in a BioFlo 3000 bioreactor (New Brunswick Scientific) using ESF921 serum-free media (Expression Systems, Woodland, CA). Media were harvested on day 3 after baculovirus infection (multiplicity of infection of 1.0). After clarification by centrifugation, the supernatants (∼3.0 l) were treated with 10 g/l polyethyleneimine–cellulose (Sigma–Aldrich) and recentrifuged. The supernatant was dialysed against 2 mM MES pH 6.5, 50 mM NaCl and 0.02% NaN3 and loaded onto a 15.0 ml carboxymethyl (CM) HyperD F column (BioSepra) at a flow rate of 5.0 ml/min. The sample was eluted with a linear NaCl gradient (50 mM to 1.0 M) and assayed for enzyme activity (Sarkar, 1994). The GnT I-containing fractions (which eluted at ∼250 mM NaCl) were pooled and loaded directly onto a 10.0 ml Ni2+–NTA SuperFlow column (Qiagen) at 2.0 ml/min and eluted with a step gradient containing 10 mM Tris pH 8.5, 100 mM NaCl, 5% glycerol and 100 mM imidazole. Enterokinase was used to remove the N-terminal His6 tag and the protein was repurified over the CM column as described above, except that NaCl was replaced by KCl and 2 mM MnCl2 was added. The protein was then concentrated to 10 mg/ml, in a solution containing 10 mM MES buffer pH 5.5, 270 mM KCl, 2 mM MnCl2 and 10 mM UDP-GlcNAc.
Crystallographic methods
Crystals were grown using the hanging drop vapour diffusion method, by mixing 1.5 µl of protein solution (10 mg/ml GnT I catalytic fragment, 10 mM MES buffer pH 5.5, 270 mM KCl, 2 mM MnCl2 and 10 mM UDP-GlcNAc) with 1.5 µl of well solution [15–25% polyethylene glycol (PEG) 8000, 100 mM Tris buffer pH 7.9 and 0–5% glycerol] and equilibrating against 1 ml of well solution. A mercury derivative was obtained by soaking a crystal in well solution containing 20 mM MeHgCl. All data were collected using Quantum 4 charge-coupled device detectors on the F2 beamline of the Cornell High Energy Synchrotron Source. Crystals were flash-frozen in a 100K N2 stream after a short soak in a cryoprotectant solution (21–25% PEG 8000, 100 mM Tris buffer pH 7.9 and 15% glycerol) (Table I). Data were integrated, scaled and reduced with DENZO and SCALEPACK (Otwinowski and Minor, 1997). The mercury position was identified with SOLVE (Terwilliger and Berendzen, 1999) and refined using SHARP (La Fortelle and Bricogne, 1997). Solvent flattening and histogram matching were performed using DM (Cowtan, 1994). The resultant experimental map was traced using the program O (Jones et al., 1991) and the model was refined with multiple rounds of manual rebuilding using O and OOPS2 (Kleywegt and Jones, 1996), alternated with simulated annealing and positional and B-factor refinement using CNS (Brünger et al., 1998). The initial model was also refined against the ‘native’ and ‘complex’ data in a similar fashion.
Acknowledgments
Acknowledgements
We would like to thank Y.Bourne, G.Davies and L.Gastinel for access to the coordinates of GlmU, SpsA and β4Gal-T1 prior to public release, as well as J.R.Brisson for coordinates of the trimannose core. This work was funded by the Protein Engineering Network of Centres of Excellence and GLYCODesign Inc., and by a University of Toronto Open Doctoral Fellowship to U.Ü. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR-9311772, using the Macromolecular Diffraction at the CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health. The atomic coordinates and structure factors of the three structures have been deposited in the Protein Data Bank (codes 1FO8, 1FO9 and 1FOA).
References
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk P.J., Rice,D.W., Poirrette,A.R. and Willett,P. (1995) β-Glucosyltransferase and phosphorylase reveal their common theme. Nature Struct. Biol., 2, 117–120. [DOI] [PubMed] [Google Scholar]
- Bacon D.J. and Anderson,W.F. (1988) A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph., 6, 219–220. [Google Scholar]
- Barton G.J. (1993) ALSCRIPT a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Black C.B., Huang,H.-W. and Cowan,J.A. (1994) Biological coordination chemistry of magnesium, sodium, and potassium ions. Protein and nucleotide binding sites. Coord. Chem. Rev., 135/136, 165–202. [Google Scholar]
- Breton C. and Imberty,A. (1999) Structure–function studies of glycosyltransferases. Curr. Opin. Struct. Biol., 9, 563–571. [DOI] [PubMed] [Google Scholar]
- Breton C., Bettler,E., Joziasse,D.H., Geremia,R.A. and Imberty,A. (1998) Sequence–function relationships of prokaryotic and eukaryotic galactosyltransferases. J. Biochem., 123, 1000–1009. [DOI] [PubMed] [Google Scholar]
- Brown K., Pompeo,F., Dixon,S., Mengin-Lecreulx,D., Cambillau,C. and Bourne,Y. (1999) Crystal structure of the bifunctional N-acetyl glucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J., 18, 4096–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Busch C., Hofmann,F., Selzer,J., Munro,S., Jeckel,D. and Aktories,K. (1998) A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem., 273, 19566–19572. [DOI] [PubMed] [Google Scholar]
- Campbell J.A., Davies,G.J., Bulone,V. and Henrissat,B. (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J., 326, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.A., Davies,G.J., Bulone,V. and Henrissat,B. (1998) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J., 329, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock S.J. and Davies,G.J. (1999) Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry, 38, 6380–6385. [DOI] [PubMed] [Google Scholar]
- Charuk J.H., Tan,J., Bernardini,M., Haddad,S., Reithmeier,R.A., Jaeken,J. and Schachter,H. (1995) Carbohydrate-deficient glyco protein syndrome type II. An autosomal recessive N-acetylglucos aminyltransferase II deficiency different from typical hereditary erythroblastic multinuclearity, with a positive acidified-serum lysis test (HEMPAS). Eur. J. Biochem., 230, 797–805. [DOI] [PubMed] [Google Scholar]
- Christopher J.A. (1998) SPOCK: The Structural Properties Observation and Calculation Kit Program Manual. The Center for Macromolecular Design, Texas A&M University, College Station, TX. [Google Scholar]
- Coutinho P.M. and Henrissat,B. (1999) Carbohydrate-active enzymes server. http://afmb.cnrs-mrs.fr/~pedro/cazy/db.html
- Cowan J.A. (1998) Magnesium activation of nuclease enzymes—the importance of water. Inorg. Chim. Acta, 275–276, 24–27. [Google Scholar]
- Cowtan K. (1994) ‘dm’: an automated procedure for phase improvement by density modification. CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Davies G., Sinnott,M.L. and Withers,S.G. (1998a) Glycosyl transfer. In Sinnott,M.L. (ed.), Comprehensive Biological Catalysis: a Mechanistic Reference, Vol. I. Academic Press, London, UK, pp. 119–209. [Google Scholar]
- Davies G.J., Mackenzie,L., Varrot,A., Dauter,M., Brzozowski,A.M., Schülein,M. and Withers,S.G. (1998b) Snapshots along an enzymatic reaction coordinate: analysis of a retaining β-glycoside hydrolase. Biochemistry, 37, 11707–11713. [DOI] [PubMed] [Google Scholar]
- Drickamer K. and Taylor,M.E. (1998) Evolving views of protein glycosylation. Trends Biochem. Sci., 23, 321–324. [DOI] [PubMed] [Google Scholar]
- Dudev T., Cowan,J.A. and Lim,C. (1999) Competitive binding in magnesium coordination chemistry: water versus ligands of biological interest. J. Am. Chem. Soc., 121, 7665–7673. [Google Scholar]
- Gastinel L.N., Cambillau,C. and Bourne,Y. (1999) Crystal structures of the bovine β4-galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J., 18, 3546–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovsky M., Fata,J., Pawling,J., Muller,W.J., Khokha,R. and Dennis,J.W. (2000) Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nature Med., 6, 306–312. [DOI] [PubMed] [Google Scholar]
- Ha S., Walker,D., Shi,Y. and Walker,S. (2000) The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci., 9, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz N. and Schachter,H. (1980) Control of glycoprotein synthesis. Bovine colostrum UDP-N-acetylglucosamine:α-d-mannoside β2-N- acetylglucosaminyltransferase I. Separation from UDP-N-acetyl glucosamine:α-d-mannoside β2-N-acetylglucosaminyltransferase II, partial purification, and substrate specificity. J. Biol. Chem., 255, 4885–4893. [PubMed] [Google Scholar]
- Hodson N., Griffiths,G., Cook,N., Pourhossein,M., Gottfridson,E., Lind,T., Lidholt,K. and Roberts,I.S. (2000) Identification that KfiA, a protein essential for the biosynthesis of the Escherichia coli K5 capsular polysaccharide, is an α-UDP-GlcNAc glycosyltransferase: the formation of a membrane associated K5 biosynthetic complex requires KfiA, KfiB and KfiC. J. Biol. Chem., 275, 27311–27315. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1995) Evolutionary link between glycogen phosphorylase and a DNA modifying enzyme. EMBO J., 14, 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioffe E. and Stanley,P. (1994) Mice lacking N-acetylglucos aminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl Acad. Sci. USA, 91, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeken J., Carchon,H. and Stibler,H. (1993) The carbohydrate-deficient glycoprotein syndromes: pre-Golgi and Golgi disorders? Glycobiology, 3, 423–428. [DOI] [PubMed] [Google Scholar]
- Jaeken J., Schachter,H., Carchon,H., De Cock,P., Coddeville,B. and Spik,G. (1994) Carbohydrate deficient glycoprotein syndrome type II: a deficiency in Golgi localised N-acetyl-glucosaminyltransferase II. Arch. Dis. Child., 71, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) A new approach to protein fold recognition. Nature, 358, 86–89. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim S.C., Singh,A.N. and Raushel,F.M. (1988) Analysis of the galactosyltransferase reaction by positional isotope exchange and secondary deuterium isotope effects. Arch. Biochem. Biophys., 267, 54–59. [DOI] [PubMed] [Google Scholar]
- Klevickis C. and Grisham,C.M. (1996) Phosphate–metal ion interactions of nucleotides and polynucleotides. In Sigel,A. and Sigel,H. (eds), Metal Ions in Biological Systems, Vol. 32. Marcel Dekker, New York, NY, pp. 1–26. [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1996) Efficient rebuilding of protein structures. Acta Crystallogr. D, 52, 829–832. [DOI] [PubMed] [Google Scholar]
- Kornfeld R. and Kornfeld,S. (1980) Structure of glycoproteins and their oligosaccharide units. In Lennarz,W.J. (ed.), The Biochemistry of Glycoproteins and Proteoglycans. Plenum Press, New York, NY, pp. 1–34. [Google Scholar]
- La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Metzler M., Gertz,A., Sarkar,M., Schachter,H., Schrader,J.W. and Marth,J.D. (1994) Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J., 13, 2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller G., Reck,F., Paulsen,H., Kaur,K.J., Sarkar,M., Schachter,H. and Brockhausen,I. (1992) Control of glycoprotein synthesis: substrate specificity of rat liver UDP-GlcNAc:Manα3R β2-N-acetylglucos aminyltransferase I using synthetic substrate analogues. Glycoconj. J., 9, 180–190. [DOI] [PubMed] [Google Scholar]
- Moréra S., Imberty,A., Aschke-Sonnenborn,U., Rüger,W. and Freemont,P.S. (1999) T4 phage β-glucosyltransferase: substrate binding and proposed catalytic mechanism. J. Mol. Biol., 292, 717–730. [DOI] [PubMed] [Google Scholar]
- Murray B.W., Takayama,S., Schultz,J. and Wong,C.H. (1996) Mechanism and specificity of human α-1,3-fucosyltransferase V. Biochemistry, 35, 11183–11195. [DOI] [PubMed] [Google Scholar]
- Murray B.W., Wittmann,V., Burkart,M.D., Hung,S.C. and Wong,C.H. (1997) Mechanism of human α-1,3-fucosyltransferase V: glycosidic cleavage occurs prior to nucleophilic attack. Biochemistry, 36, 823–831. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Narasimhan S., Stanley,P. and Schachter,H. (1977) Control of glycoprotein synthesis. Lectin-resistant mutant containing only one of two distinct N-acetylglucosaminyltransferase activities present in wild type Chinese hamster ovary cells. J. Biol. Chem., 252, 3926–3933. [PubMed] [Google Scholar]
- Nishikawa Y., Pegg,W., Paulsen,H. and Schachter,H. (1988) Control of glycoprotein synthesis. Purification and characterization of rabbit liver UDP-N-acetylglucosamine:α-3-d-mannoside β-1,2-N-acetylglucosaminyltransferase I. J. Biol. Chem., 263, 8270–8281. [PubMed] [Google Scholar]
- Notenboom V., Birsan,C., Warren,R.A.J., Withers,S.G. and Rose,D.R. (1998) Exploring the cellulose/xylan specificity of the β-1,4-glycanase Cex from Cellulomonas fimi through crystallography and mutation. Biochemistry, 37, 4751–4758. [DOI] [PubMed] [Google Scholar]
- Opat A.S., Puthalakath,H., Burke,J. and Gleeson,P.A. (1998) Genetic defect in N-acetylglucosaminyltransferase I gene of a ricin-resistant baby hamster kidney mutant. Biochem. J., 336, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Puthalakath H., Burke,J. and Gleeson,P.A. (1996) Glycosylation defect in Lec1 Chinese hamster ovary mutant is due to a point mutation in N-acetylglucosaminyltransferase I gene. J. Biol. Chem., 271, 27818–27822. [DOI] [PubMed] [Google Scholar]
- Qiao L., Murray,B.W., Shimazaki,M., Schultz,J. and Wong,C.H. (1996) Synergistic inhibition of human α-1,3-fucosyltransferase V. J. Am. Chem. Soc., 118, 7653–7662. [Google Scholar]
- Reck F., Springer,M., Meinjohanns,E., Paulsen,H., Brockhausen,I. and Schachter,H. (1995) Synthetic substrate analogues for UDP-GlcNAc:Manα1-3R β1-2-N-acetylglucosaminyltransferase I. Sub strate specificity and inhibitors for the enzyme. Glycoconj. J., 12, 747–754. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Moras,D. and Olsen,K.W. (1974) Chemical and biological evolution of nucleotide-binding protein. Nature, 250, 194–199. [DOI] [PubMed] [Google Scholar]
- Russell R.B. and Barton,G.J. (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins, 14, 309–323. [DOI] [PubMed] [Google Scholar]
- Sabini E., Sulzenbacher,G., Dauter,M., Dauter,Z., Jørgensen,P.L., Schülein,M., Dupont,C., Davies,G.J. and Wilson,K.S. (1999) Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase. Chem. Biol., 6, 483–492. [DOI] [PubMed] [Google Scholar]
- Sarkar M. (1994) Expression of recombinant rabbit UDP-GlcNAc: α3-d-mannoside β-1,2-N-acetylglucosaminyltransferase I catalytic domain in Sf9 insect cells. Glycoconj. J., 11, 204–209. [DOI] [PubMed] [Google Scholar]
- Sarkar M., Pagny,S., Ünligil,U., Joziasse,D., Mucha,J., Glössl,J. and Schachter,H. (1998) Removal of 106 amino acids from the N-terminus of UDP-GlcNAc: α-3-d-mannoside β-1,2-N-acetylglucosaminyl transferase I does not inactivate the enzyme. Glycoconj. J., 15, 193–197. [DOI] [PubMed] [Google Scholar]
- Schachter H. (1986) Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem. Cell Biol., 64, 163–181. [DOI] [PubMed] [Google Scholar]
- Schachter H. (1991) The ‘yellow brick road’ to branched complex N-glycans. Glycobiology, 1, 453–461. [DOI] [PubMed] [Google Scholar]
- Shibayama K., Ohsuka,S., Tanaka,T., Arakawa,Y. and Ohta,M. (1998) Conserved structural regions involved in the catalytic mechanism of Escherichia coli K-12 WaaO (RfaI). J. Bacteriol., 180, 5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama K., Ohsuka,S., Sato,K., Yokoyama,K., Horii,T. and Ohta,M. (1999) Four critical aspartic acid residues potentially involved in the catalytic mechanism of Escherichia coli K-12 WaaR. FEMS Microbiol. Lett., 174, 105–109. [DOI] [PubMed] [Google Scholar]
- Sidhu G., Withers,S.G., Nguyen,N.T., McIntosh,L.P., Ziser,L. and Brayer,G.D. (1999) Sugar ring distortion in the glycosyl-enzyme intermediate of a family G/11 xylanase. Biochemistry, 38, 5346–5354. [DOI] [PubMed] [Google Scholar]
- Stanley P., Narasimhan,S., Siminovitch,L. and Schachter,H. (1975) Chinese hamster ovary cells selected for resistance to the cytotoxicity of phytohemagglutinin are deficient in a UDP-N-acetylglucosamine–glycoprotein N-acetylglucosaminyltransferase activity. Proc. Natl Acad. Sci. USA, 72, 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzenbacher G., Mackenzie,L.F., Wilson,K.S., Withers,S.G., Dupont,C. and Davies,G.J. (1999) The crystal structure of a 2-fluorocellotriosyl complex of the Streptomyces lividans endoglucanase CelB2 at 1.2 Å resolution. Biochemistry, 38, 4826–4833. [DOI] [PubMed] [Google Scholar]
- Tan J., Dunn,J., Jaeken,J. and Schachter,H. (1996) Mutations in the MGAT2 gene controlling complex N-glycan synthesis cause carbohydrate-deficient glycoprotein syndrome type II, an autosomal recessive disease with defective brain development. Am. J. Hum. Genet., 59, 810–817. [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology, 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella G.J., Paulsen,H. and Schachter,H. (1984) Control of glycoprotein synthesis. IX. A terminal Man αl-3Manβ1-sequence in the substrate is the minimum requirement for UDP-N-acetyl-d-glucosamine: α-d-mannoside (GlcNAc to Manα1-3) β2-N-acetylglucosaminyl transferase I. Can. J. Biochem. Cell Biol., 62, 409–417. [DOI] [PubMed] [Google Scholar]
- Vrielink A., Rüger,W., Driessen,H.P. and Freemont,P.S. (1994) Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J., 13, 3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins C.A. and Munro,S. (1998) Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl Acad. Sci. USA, 95, 7945–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel D.L. and Withers,S.G. (2000) Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res., 33, 11–18. [DOI] [PubMed] [Google Scholar]