Abstract
Cutaneous malignant melanoma is a highly aggressive and frequently chemoresistant cancer, whose incidence continues to rise. Epidemiological studies reveal that the major etiological melanoma risk factor is ultraviolet (UV) solar radiation, with the highest risk associated with intermittent burning doses, especially during childhood1,2. We have experimentally validated these epidemiological findings using the hepatocyte growth factor/scatter factor (HGF/SF) transgenic mouse model, which develops lesions in stages highly reminiscent of human melanoma with respect to biological, genetic and etiologic criteria, but only when irradiated as neonatal pups with UVB, not UVA3,4. However, mechanisms underlying UVB-initiated, neonatal-specific melanomagenesis remain largely unknown. Here we introduce a mouse model permitting fluorescence-aided melanocyte imaging and isolation following in vivo UV irradiation. We use expression profiling to show that activated neonatal skin melanocytes isolated following a melanomagenic UVB dose bear a distinct, persistent interferon response signature, including genes associated with immunoevasion. UVB-induced melanocyte activation, characterized by aberrant growth and migration, was abolished by antibody-mediated systemic blockade of interferon-γ (IFN-γ), but not type-I interferons. IFN-γ was produced by macrophages recruited to neonatal skin by UVB-induced ligands to the chemokine receptor Ccr2. Admixed recruited skin macrophages enhanced transplanted melanoma growth by inhibiting apoptosis; notably, IFN-γ blockade abolished macrophage-enhanced melanoma growth and survival. IFN-γ-producing macrophages were also identified in 70% of human melanomas examined. Our data reveal an unanticipated role for IFN-γ in promoting melanocytic cell survival/immunoevasion, and suggest that IFN-γ-R signaling represents a novel therapeutic melanoma target.
Mechanisms associated with UV-mediated alterations to melanocytes and their microenvironment have been inscrutable because they cannot be adequately studied in cultured cells. Moreover, melanocytes represent only ~1% of skin cells, and bear few specific cell surface markers permitting efficient isolation. To enable detailed study of melanocyte biology in vivo, we generated a mouse model in which expression of the reverse tetracycline-activated transactivator rtTA2s-M2, characterized by minimal leakiness and background, was regulated by the melanocyte-specific dopachrome tautomerase (Dct) gene promoter (Supplementary Fig. 1a). Dct-rtTA mice bred with transgenic mice bearing a histone H2B-GFP fusion construct controlled by the tetracycline response element (TRE) created Dct-rtTA/TRE-H2BGFP bi-transgenic mice (hereafter iDct-GFP) (Supplementary Fig. 1b). iDct-GFP mice exhibited an inducible GFP profile from embryonic through adult stages consistent with known Dct expression patterns. GFP expression was observed in embryonic neural crest, retinal pigment epithelium and telencephalon, as expected (Fig. 1a; Supplementary Fig. 2). Neonatal and adult skin GFP+ cells were strictly localized to hair follicles, where most GFP+ cells were in bulb regions, with smaller numbers in the outer root sheath and bulge regions, harboring melanocyte precursors5 (Fig. 1b). Co-localization of GFP and anti-Dct antibody by immunohistochemistry (IHC) unequivocally identified GFP+ cells as melanocytes (Fig. 1c). No background GFP expression was detectable without doxycycline. Full GFP induction was achieved within 12–18 hours of a single intraperitoneal injection of a non-toxic doxycycline dose in neonatal or adult mice (Supplementary Fig. 1c).
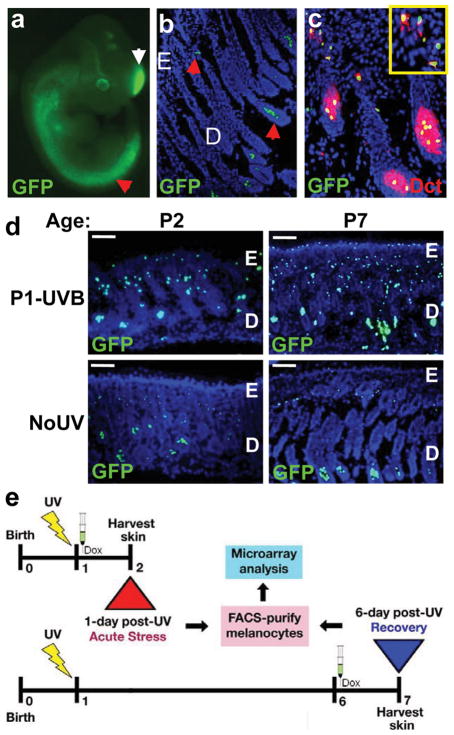
Figure 1. Melanocyte-specific GFP expression reveals UVB-induced activation.
a, E11.5 iDct-GFP embryo showing GFP+ cells in neural crest (red arrow) and telencephalon (white arrow). b, In 7-day old pup skin GFP+ cells are located in the bulb (lower arrow) and bulge (upper arrow) regions of hair follicles. Blue = DAPI; E, epidermis; D, dermis. c, Immunohistochemistry with anti-Dct antibody shows co-localization with GFP in iDct-GFP skin. d, UVB-induced activation of melanocytes, characterized by proliferation and migration towards epidermis. Dorsal skins were examined at 1 day (at age P2) and 6 days (P7) post-irradiation. Scale bars = 40 μm. e, Schematic of the regime for isolating GFP+ melanocytes by FACS. Pups are irradiated at P1, and dorsal skins harvested at either P2 (24 h post-UV) or P7 (6 d post-UV). Doxycycline injections are always given after irradiation, 24 h prior to skin harvest.
Reasoning that novel clues to molecular mechanism(s) underlying UV-induced melanomagenesis would be found within the genomic response of melanocytes to UV, we used the iDct-GFP mouse to examine responses to UVB vs. UVA of melanocytes residing in situ, within their natural morphologic and physiologic microenvironment. Precisely defined wideband wavelengths and physiologically relevant doses4 (see Methods) of UV (Supplementary Fig. 3) were used to irradiate postnatal day-1 (P1) iDct-GFP mice and skins were harvested at time points post-irradiation. To avoid potential toxicity from chronic expression and interference with UV absorption, GFP was doxycycline-induced after UV irradiation, 24 hours prior to skin harvest. GFP+ melanocytes were phenotypically activated, characterized by elevated melanocyte numbers and migration towards the epidermis6, 24 to 48 hours after exposure to UVB, but not UVA, peaking at 3 days and lasting at least 10 days post-irradiation (Fig. 1d; Supplementary Fig. 4a). UVB-induced melanocyte activation was specific to neonatal irradiation; adult mice irradiated at P29 did not exhibit this response (Supplementary Fig. 4b).
We performed an expression microarray study on melanocytes isolated from dorsal skin of iDct-GFP pups irradiated at P1 with UVB or UVA (Fig. 1e). Doxycycline-induced GFP-labeled melanocytes were isolated via fluorescence-activated cell sorting (FACS) at 1-day and 6-days post-irradiation (ages P2 and P7, respectively); arrays from 1-day post-UV would reflect the acute UV stress response of in vivo melanocytes, while the 6-day post-UV time point should uncover responses persisting after the acute stress response subsides. FACS isolation consistently yielded >95% melanocyte enrichment (Supplementary Fig. 5). Gene expression profiling produced robust data with good reproducibility among biological triplicates (Fig. 2a), and confirmed the absence of detectable levels of contaminating skin cell types, including keratinocytes, fibroblasts and adipocytes (Supplementary Fig. 6).
Figure 2. UVB-induced melanocyte activation is mediated by interferon-γ.
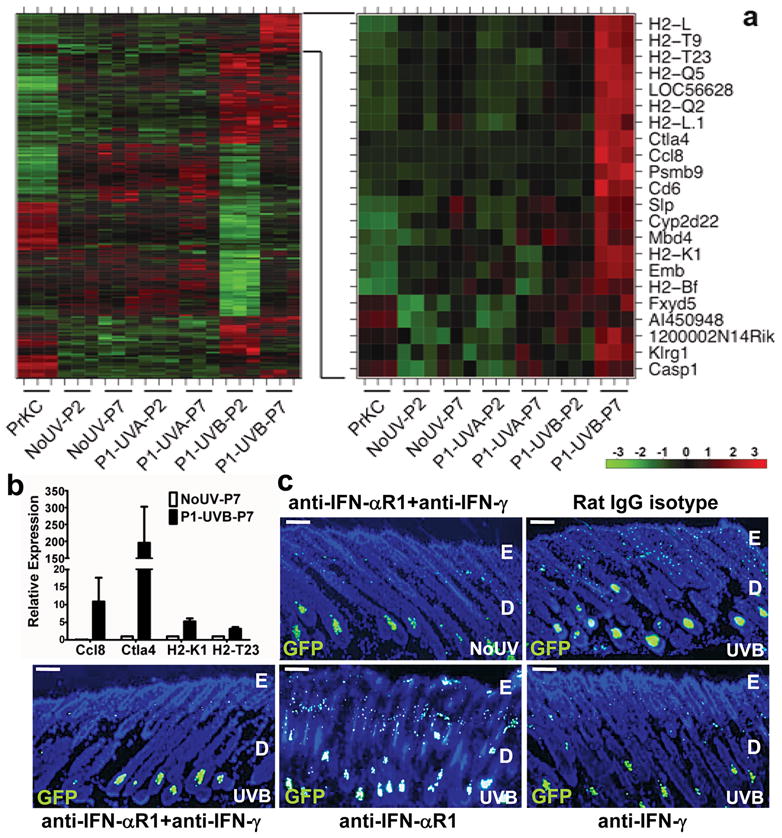
a, Unsupervised clustering of cDNA microarray analysis of gene expression in FACS-sorted melanocytes from 1 day (P2) or 6 days (P7) following UVB or UVA irradiation, and respective unirradiated controls. The expanded heatmap (right) shows the delayed induced gene subset, which includes multiple genes known to be induced by IFN-γ. Primary mouse keratinocytes (PrKC) were included as controls. All groups included biological triplicates. b, qRT-PCR validation of expression of 4 genes (n=3 samples each) from IFN signature (error bars = s.e.m.). c, Antibody-mediated blockade of interferons by treating pups with intraperitoneal injections of anti-IFN-αR1, anti-IFN-γ, or both in combination, 1 h prior to and 3 days after UVB irradiation at P1. The dorsal skins were harvested (n=3 each group) and analyzed for melanocyte activation. Representative images are shown. E, epidermis; D, dermis. Scale bars = 40 μm.
UVB elicited a potent, transient, acute stress response in melanocytes, including increased expression of p53 target genes (e.g., p21Waf1/Cip1, Cyclin G1, and Reprimo), while UVA-associated changes were subtle (Fig. 2a; Supplementary Fig. 7). Intriguingly, a small subset of genes exhibited a delayed response evident at 6-day post-UVB (Fig. 2a), including a putative interferon (IFN) responsive gene signature7 (Fig. 2a; Supplementary Table 1). Four upregulated genes (Ccl8, Ctla4, H2-K1 and H2-T23) from this group were validated by qRT-PCR (Fig. 2b). The response was neonate specific (Supplementary Fig. 8), and included genes implicated in conferring immunoevasiveness (i.e., Ctla4, H2-T23, H2-M3, H2-Bf, and Slp).
To determine whether IFN signaling plays a biologically significant role in UVB-induced melanocyte activation, we blocked both type-I (IFN-α and IFN-β) and type-II (IFN-γ) interferons by neonatal administration of anti-IFN-αR1 and anti-IFN-γ antibodies, respectively. Melanocytes from 6-day post-UVB skin were activated in the presence of isotype control antibody, while the anti-IFN-αR1+anti-IFN-γ antibody combination completely abolished this response (Fig. 2c). Moreover, while antibody-mediated blockade of type-I IFN-α/β signaling alone failed to overtly affect UVB-mediated activation, blockade of IFN-γ alone dramatically inhibited this response (Fig. 2c). These results were corroborated by flow cytometric quantification of GFP+ skin cells from each antibody-treated group (Supplementary Fig. 9). We next isolated GFP+ melanocytes from UV-irradiated neonates blocked with either anti-IFN-γ or anti-IFN-αR1, and compared their expression patterns to control. Anti-IFN-γ and anti-IFN-αR1 antibodies repressed expression of a common gene set associated with IFN response (Supplementary Fig. 10). However, IFN-γ inhibition also more potently repressed expression of several non-classical major histocompatibility complex (MHC) class Ib antigens (e.g., H2-T23 and H2-Q2), as well as Psmb9, Gbp2, Icam, Irf1 and Fosb; members of this gene subset should be responsible for the observed melanocytic phenotypes. Notably, IFN-γ blockade exclusively and potently inhibited expression of chemokine Ccl8/MCP-2.
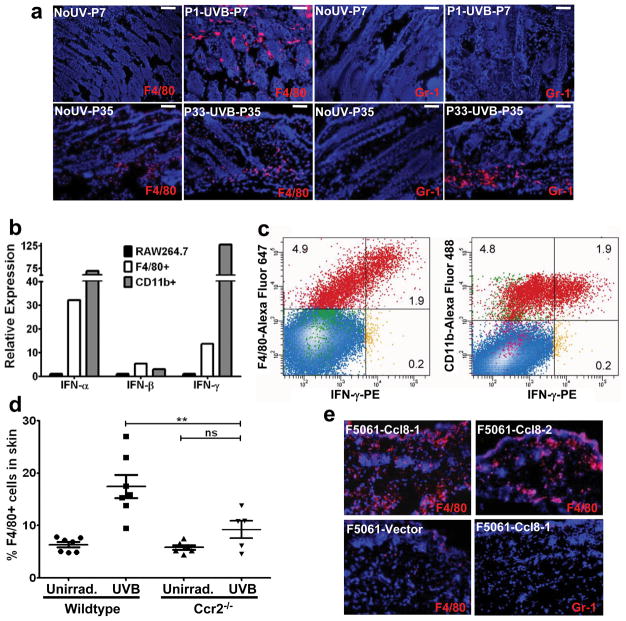
We determined the IFN-γ source by interrogating 6-day post-UVB-irradiated skin for immune cell infiltration. IHC failed to detect T-cells, B-cells, dendritic cells, NK, or NK-T cells; however, CD11b+ cells of myeloid origin were evident (Supplementary Fig. 11). Anti-F4/80 and anti-Gr-1 antibodies identified these as macrophages (F4/80+Gr-1−), not neutrophils (Fig. 3a, upper panel). Nearly 90% of CD11b+ cells were also F4/80+ (Supplementary Fig. 12). Notably, the adult response was distinct; dorsal skins from P35 mice 2-days post-UVB showed predominant Gr-1+ cell infiltration, and minimal F4/80+ cells (Fig. 3a, lower panel), as reported8.
Figure 3. UVB induces chemoattraction of IFN-γ-producing macrophages into neonatal skin.
a, IHC with anti-F4/80 and anti-Gr-1 antibodies in dorsal skins of UVB-irradiated and unirradiated neonatal (upper panel) and adult (lower panel) mice. Scale bars = 40 μm. b, qRT-PCR for IFN-α, IFN-β, and IFN-γ expression in F4/80+ and CD11b+ cells FACS isolated from P1-UVB-P7 neonatal dorsal skins, compared with non-activated RAW264.7 macrophages. c, Flow cytometric analysis of P1-UVB-P7 skin cell suspensions identified IFN-γ+ macrophages. d, Flow cytometric analysis of macrophage (F4/80+) infiltration into skin, 2 days post UVB irradiation in Ccr2-deficient pups (irradiated at P1), as compared to wildtype pups. **p<0.01; One-Way ANOVA test with post-hoc Tukey analysis. e, F5061 melanoma cells ectopically expressing Ccl8 chemoattract F4/80+ macrophages (red), but not Gr-1+ cells, to sites of subcutaneous inoculation in syngeneic FVB/N mice. One vector-transfected and two Ccl8-transfected clone cells were used. Blue = DAPI.
To determine if infiltrating macrophages expressed IFN-γ, as has been suggested9, type-I and type-II interferon mRNAs were quantified from FACS-purified CD11b+ and F4/80+ cells. Both had upregulated IFN-α and IFN-γ expression, and to a lesser extent IFN-β (Fig. 3b). Flow cytometry demonstrated that 28% of both CD11b+ and F4/80+ cells expressed IFN-γ (Fig. 3c). Our data support the notion that UVB-recruited, infiltrating macrophages secrete IFN-γ, inducing the IFN signature detected in activated melanocytes. Although we were unable to detect NK cell markers in FACS-isolated CD11b+ cells by qRT-PCR (Supplementary Fig. 13), we cannot rule out possible contribution by undetectably low number of NK or other inflammatory cells.
That UVB-induced chemokines were responsible for neonatal macrophage recruitment, particularly Ccr2/Ccr5 ligands, was suggested by our melanocyte microarray data and confirmed by qRT-PCR (Supplementary Fig. 14a, b). These results were further corroborated through treatment of cultured melan-c normal immortalized melanocyte cell line with non-cytocidal UV (Supplementary Fig. 14c). In contrast, upregulation of these chemokines was not detected in UV-irradiated whole skin (Supplementary Fig. 15) or isolated keratinocytes10. Ccr2 and Ccr5 were highly expressed in skin-infiltrating macrophages, but not in non-activated RAW264.7 macrophages (Supplementary Fig. 16). Finally, mice deficient in Ccr2 were significantly inhibited in their ability to recruit F4/80+ macrophages into neonatal skin (Fig. 3d); in contrast, Ccr5-deficient mice showed no significant difference (data not shown).
The arrival of IFN-γ-expressing macrophages coincided with a >100-fold upregulation in melanocyte expression of the Ccr2 ligand Ccl8, a known IFN-γ-response gene11, while expression of other Ccr2 ligands had returned to baseline. We propose that recruited IFN-γ+ macrophages enhance melanocyte Ccl8 expression, reinforcing macrophage-melanocyte interactions and fueling an inflammatory positive feedback loop. To confirm its ability to potently chemoattract macrophages Ccl8 was ectopically expressed in F5061 cells, established from a UV-induced melanoma from an immunocompetent HGF/SF-transgenic mouse. F5061-Ccl8 cells were subcutaneously inoculated into syngeneic FVB/N mice, markedly enhancing macrophage infiltration into the transplantation site (Fig. 3e). Conditioned media from F5061-Ccl8 cells also significantly elevated trans-membrane migration of RAW264.7 macrophages (Supplementary Fig. 17).
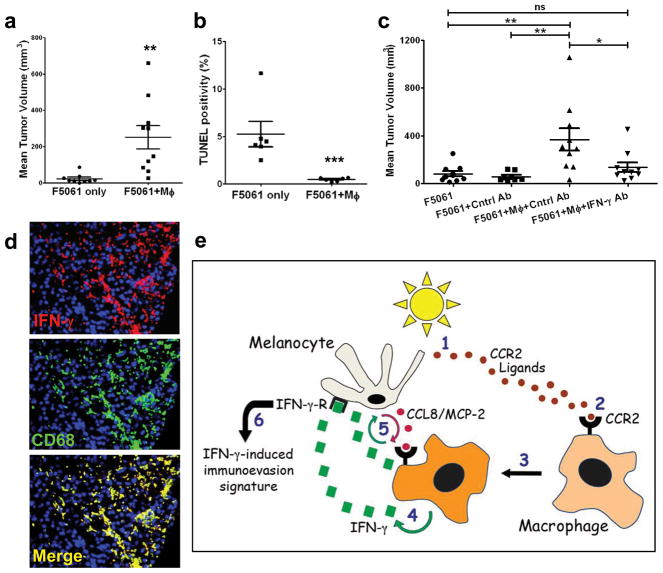
Macrophages exhibit either anti- or pro-tumourigenic properties12. F4/80+ macrophages were isolated from 6-day post-UVB neonatal skin, admixed 1:5 with F5061 melanoma cells and transplanted subcutaneously into FVB/N mice. Admixed transplants exhibited significantly increased growth relative to controls (Fig. 4a; Supplementary Fig. 18), indicating that these activated macrophages were pro-tumourigenic. In contrast, macrophages isolated from spleens of unirradiated control pups did not affect tumour growth (Supplementary Fig. 19). Ki-67 IHC showed no difference in proliferation between admixed tumors vs. controls (Supplementary Fig. 20); however, TUNEL assays revealed significantly less apoptosis in admixed tumors (Fig. 4b; Supplementary Fig. 21), demonstrating that UVB-recruited macrophages promote melanoma cell survival. IHC confirmed that a subset of tumour-associated macrophages maintained IFN-γ expression (Supplementary Fig. 22). Moreover, macrophage presence strongly correlated with enhanced CTLA4 expression in F5061 melanoma cells (Supplementary Fig. 23), recapitulating the functional consequence of macrophage infiltration observed in neonatal skin melanocytes (Fig. 2a, b).
Figure 4. IFN-γ mediates pro-tumourigenic effects of UVB-recruited skin macrophages.
a, Mean volumes (±s.e.m.) of F5061 melanomas admixed with skin macrophages isolated from P1-UVB-P7 pups (F5061+MΦ, n=10), vs. F5061 only controls (n=9), following subcutaneous transplantation in syngeneic FVB/N mice; **p<0.01. b, Percentages of TUNEL+ cells (±s.e.m.) in admixed and control tumours (n=6 each); ***p<0.001. c, Antibody-mediated blockade of IFN-γ significantly inhibits pro-tumourigenic effects of macrophages; *p<0.05; **p<0.01; ns = not significant. One-Way ANOVA test with post-hoc Tukey analysis. d, Dual IHC with anti-IFN-γ (red) and anti-CD68 (green) antibodies on a human melanoma tissue microarray (TMA) exhibits IFN-γ-expressing macrophages (yellow). Representative tumour is shown. e, Schematic representation of the UVB-induced inflammatory cascade leading to IFN-γ-mediated immunoevasion and survival of melanocytes during sunburn-associated remodeling. UV induces release of CCR2 ligands (1) that activate CCR2+ macrophages (2), which are recruited to neonatal skin (3). Macrophages secrete IFN-γ (4), which activates melanocytes, inducing expression of genes that include CCL8/MCP-2, fueling inflammation (5) and immunoevasion (6).
Despite its well-documented anti-tumourigenic activity13, IFN-γ has also been implicated as a pro-tumourigenic factor14,15. To determine if macrophage-secreted IFN-γ was responsible for the enhanced melanoma growth observed in admixed tumours, we included intraperitoneal administration of either anti-IFN-γ or control antibodies. While admixed melanomas in the control group showed the expected enhanced growth, those in mice given anti-IFN-γ antibody exhibited significantly reduced growth (Fig. 4c). Immunophenotyping of tissue microarrays (TMA) containing UVB-induced mouse melanomas showed most (66%) were macrophage-rich, with fewer having T-cells (59%) and B-cells (32%) (Supplementary Fig. 24). To determine if human melanoma-associated macrophages produce IFN-γ, we performed dual IHC for CD68 and IFN-γ using human melanoma TMA. We discovered that 19 of 27 (70%) melanomas examined contained abundant macrophages (CD68+), of which all 19 demonstrated CD68+IFN-γ+ dual positivity (Fig. 4d; Supplementary Fig. 25).
In this report we show that UV incites melanomagenesis not only through DNA mutagenesis, but also by altering interactions between melanocytes and their microenvironment to regulate remodeling of UV-damaged skin. Based on our results, a model implicating a neonatal-specific UV-induced pro-melanomagenic inflammatory cascade emerges (Fig. 4e). In accord with their relative tumourigenicity in albino HGF/SF-transgenic mice4 UVB, not UVA, induces melanocytic expression of multiple chemoattracting Ccr2 ligands (Ccl2/MCP-1, Ccl7/MCP-3, Ccl8/MCP-2, Ccl12/MCP-5), recruiting Ccr2+ macrophages into neonatal skin. IFN-γ from recruited macrophages stimulates melanocyte proliferation and migration, and expression of genes implicated in immunoevasion/survival.
Erythemal neonatal UVB causes robust macrophage infiltration and is melanomagenic; adult skin responds with a rapid, short-lived neutrophil influx, but no melanoma. We propose that mechanisms underlying neonatal UVB-induced melanomagenesis operate within the immunoediting paradigm13: UVB-activated mutant neonatal melanocytes, particularly progenitors16, exposed to inflammation evade immune-mediated elimination, persisting through an extended equilibrium phase before evolving into clinically-significant melanoma. Macrophage-induced melanocyte proliferation would more efficiently fix UV-induced mutations in prospective melanoma cells, while enhanced melanocyte migration could facilitate UVB-associated long-term tolerance to melanocytic antigens17 by promoting aberrant melanocyte-immune cell interactions. Moreover, enduring inflammation-associated epigenetic alterations occur in transformed cells18, and perhaps in long-lived macrophage subpopulations19, indefinitely extending their biological effects.
Notably, our systemic antibody blockade experiments demonstrating the importance of physiologically relevant IFN-γ in UVB-induced melanocyte activation and melanoma cell survival strongly support the notion that IFN-γ can be pro-tumourigenic as well as anti-tumourigenic, depending on the context, intensity and durability of the IFN-γ signal14,15. In fact, serum IFN-γ has been implicated as an independent prognostic indicator for melanoma recurrence20. We propose that IFN-γ-associated survival mechanisms operational in neonatal melanocytes are recapitulated in melanoma, contributing to selection of more aggressive, therapeutically resistant phenotypes. Relevance to human melanoma is supported by detection of macrophage-associated IFN-γ expression in most patient samples examined, and a clinical trial showing that IFN-γ may have adverse effects regarding melanoma patient relapse and mortality21. We provide the first evidence that IFN-γ-R signaling can facilitate melanoma progression, a remarkable discovery considering that high-dose IFN-α is used to treat melanoma, albeit with limited success22. Non-overlapping functions of type-I and type-II interferons are well described13, and strongly supported by our data.
The IFN “survival signature” associated with UVB-activated mouse melanocytes contains genes involved in human melanoma immunoevasion, including non-classical MHC class Ib antigens (mouse H2-M3/human HLA-G; mouse H2-T23/Qa-1/human HLA-E)23,24. HLA-E suppresses NK and cytotoxic T-lymphocytes25,26. This IFN signature also features complement isoforms C4a and C4b, implicated in systemic autoimmunity suppression27, and CTLA4, a potent immune evasion facilitator. CTLA4 is also highly upregulated in mouse melanoma cells admixed with neonatal macrophages, and expressed on human melanoma cells, where it may be involved in immune escape28,29.
We here identify novel cellular/molecular inflammatory mechanisms centered on IFN-γ signaling that may underlie the initiation, survival and/or outgrowth of UVB-induced melanoma cells. We propose that such mechanisms are highly relevant to strategies employed by melanoma cells to evade immunosurveillance in patients. In what could prove to be a paradigm shift, our data strongly suggest that IFN-γ/IFN-γ-R or its downstream pathway members represent promising prognostic markers and/or efficacious therapeutic targets in an appropriate subset of melanoma patients.
Methods Summary
All mouse studies were performed under the strict guidelines of Animal Study Protocols approved by the Animal Care and Use Committees at NCI/NIH and GWUMC. The Dct-rtTA transgenic mice express the rtTA2s-M2 under the control of the Dct (Trp2) gene promoter. Thirty-one founder transgenic mice were generated by standard microinjection techniques, individually crossed with the TRE-H2BGFP transgenic mice, and screened for a line that produced no leaky background expression of GFP without doxycycline and quickly responded to a single intraperitoneal doxycycline injection. One such line was selected to make a double homozygous Dct-rtTA/TRE-H2BGFP transgenic mouse line in albino FVB/N background, and males were crossed with wildtype FVB/N females to produce litters that were 100% double heterozygous (iDct-GFP). One day after birth, the pups were irradiated with UV radiation (UVB, UVA, or sham)4 (Supplementary Fig. 3). Dorsal skins were harvested at 24 hours and 6 days post irradiation (or sham irradiation). Twenty-four hours prior to sacrifice of mice and harvest of dorsal skins, neonates were injected intraperitoneally with doxycycline at 80 μg/g body weight to activate GFP expression. Skins from 6–8 pups of a litter were pooled to prepare single cell suspensions using a published protocol30, followed by isolation of GFP+ melanocytes via FACS. RNA samples isolated from these melanocytes were subjected to cDNA microarray on Illumina Murine Beadchips v 2.0 (Illumina, Inc.) qRT-PCR validation was performed using primers listed in Supplementary Table 2. Immunohistochemical and flow cytometric analyses were performed using antibodies listed in Supplementary Table 3.
Methods
Generation of Dct-rtTA and iDct-GFP mice
DNA fragments from the following plasmids were used to construct the plasmid pDct-rtTA-βGlo, which harbors the transgene fragment used to make the Dct-rtTA transgenic mice: 1. phβGlobin - The vector backbone is pBluescript II KS+ and the insert is a BamHI/PstI genomic fragment from human beta globin gene, which is comprised of partial exon 2, full intron 2, full exon 3 and poly A signal. 2. pPDct - This plasmid contains the 3.4 kb BamHI/Eco47III fragment from the dopachrome tautomerase (Dct) gene promoter31. 3. pBS/IRES-M2 - This plasmid contains the rtTA2s-M2 fragment. It is a variant of pUHrT62-132, and was obtained from Drs. Carlo Toniatti and Hermann Bujard. The Dct-rtTA transgenic mice were generated using standard microinjection techniques in FVB/N background strain. Dct-rtTA mice were bred with the TRE-H2BGFP transgenic mice33 to generate the bi-transgenic iDct-GFP mice.
Mice
Wildtype FVB/N female breeders, Ccr2−/− mice (strain B6.129S4-Ccr2tm1Ifc/J), and Ccr5−/− mice (strain B6.129P2-Ccr5tm1Kuz/J) were obtained from The Jackson Laboratories (Bar Harbor, ME, USA) and housed under strict guidelines of the IACUC-approved protocols. Transportation of mice between NCI-Frederick and GWUMC animal facilities was performed by special animal courier service approved by IACUC.
In Vivo UV irradiation
The Standard Erythemal Dose (SED) is used to compare the sunburning effectiveness of different ultraviolet (UV) emitting sources. These sources include UV emitting lamps in suntanning beds, welder’s arcs and sunlight among others. By determining the SED one is able to compare how efficient the UV emitting sources are at inducing sunburn or reddening in skin relative to each other. In experimental UV studies, many laboratories use different types of UV sources for a variety of experiments. Thus, determining the SED allows one to produce equivalent amounts of “sunburning” radiation regardless of the UV spectral output of the different sources used. The SED is produced by multiplying the spectral output or irradiance (Watts/m2 of the UV emitting source) with the CIE Standard Erythemal Action Spectrum (note that 1 Watt = 1 Joule (J) per second.) The CIE Standard Erythemal Action Spectrum is thus used to “weight” the incoming UV radiation on an erythemal wavelength basis. The product curve produced from this convolution will give the erythemal “effective” irradiance [(Watts erythemal effective)/m2] upon integration of the area under this curve. The “erythemal effective” irradiance is used to calculate the erythemal or sunburning dose by the equation: Dose (erythemal-effective) = Irradiance (erythemal-effective) × time (seconds). By definition 1 SED = 100 J/m2.
Thus, our UVB source, which consists of a UVB bandpass interference filter with a half-band width of ± 5 nm (dimensions: 2″ × ″) blocked to 10−3 to 10−4 outside the main bandpass (280–320 nm). This custom-made filter when coupled to our 2.5kW Xenon arc allows for very clean wavelength resolution. The 100 cm2 exposure area is large enough to accommodate up to 12 neonates (1 to 3 days-old), which are situated in a 3-chambered animal holder covered with a quartz lid that allows for UV transmission. Holes are drilled into the sides to allow for air exchange. This unit sits atop a turntable rotating at approximately 3 rpm to average out beam uniformity. The irradiance is measured with a spectroradiometer (StellarNet) and under standard conditions regularly produces an irradiance of approximately 0.30 CIE-effective Watts/m2. The exposure time is constant at 90 minutes. These conditions regularly produce a CIE effective dose of approximately 14 to 16 SED. This level of CIE-effective irradiance agrees well with our previous calculations using a Radiation Transfer algorithm over a global latitude/longitude grid at Northern or Southern summer mid latitudes34. Direct terrestrial measurements at these latitudes indicate that under summer sunlight conditions on clear days in Northern or Southern Hemisphere between 11 AM and 3 PM one can measure SED’s in the range of 10 to 20 depending on exposure conditions. As an example and as part of a project to estimate the number of SED’s one might receive during the day using, in this case, cyclists training in Spain during summer and winter, a recent paper by Serrano et al. shows just such levels in good agreement with our measurements35.
Fluorescence activated cell sorting (FACS)
FACS was performed on a FACSAria flow cytometer (BD Biosciences, San Jose, CA) equipped with a fixed-alignment cuvette flow cell and a Coherent Sapphire solid state laser providing 13 mW of 488 nm excitation wavelength. Processing rates averaged about 5,000 events per second at 20 psi. Cells of interest were gated by a combination of forward and orthogonal light scatter, and GFP fluorescence was captured in the detector with a 530/30 bandpass filter. GFP-positive cells suspended in phosphate-buffered saline plus 1% bovine serum albumin (BSA) were sorted as bulk populations into 12×75 mm polypropylene tubes. Dorsal skins from a combined litter of 6–8 pups gave a disaggregated cell suspension containing about 30–50×106 cells. FACS resulted in collection of 1–3×105 GFP-positive melanocytes per litter. Following FACS collection of cells into PBS+1% BSA solution, the cells were centrifuged at 4°C, and the pelleted cells were lysed in Trizol Reagent (Invitrogen, Inc., Carlsbad, CA, USA) for isolation of RNA. Total RNA was isolated from cells by Trizol organic extraction followed by RNeasy Micro Kit (Qiagen Sciences, MD, USA) procedure. Skin macrophages were isolated by immunostaining with anti-F4/80-Alexa Fluor 647 antibody (BioLegend, Inc., San Diego, CA.)
Microarray analysis of gene expression
Three biological replicates for each group were used for cDNA microarray analyses. RNA was quantitated fluorimetrically and assayed for integrity using the Agilent Bioanalyzer (Agilent Technologies, CA, USA). 100 ng RNA was converted to biotinylated cRNA using one round of amplification with the Illumina Labeling Kit (Illumina, Inc., San Diego, CA, USA) and one round of T7 polymerase amplification and hybridized to Illumina Murine Beadchips v. 2.0. After hybridization and staining, the arrays were scanned in an Illumina Bead Station, and the images processed using Illumina Bead Studio software.
Statistical analysis of microarray data
The raw microarray data were extracted from the BeadStudio software and imported into the R statistical programming environment (R Development Core Team)36 and Bioconductor37. A variance stabilization transformation38 followed by quantile normalization and quality assessment were performed using the lumi package38. Differential expression between groups was calculated using a linear model and empirical Bayes-moderated false discovery rates (FDR) for each treated versus untreated line were calculated using the limma package39. A heatmap including probes showing differential expression between UVB-treated and untreated with a false discovery rate (FDR) of 0.05 was generated (Fig. 2a and Supplementary Fig. 7). Hierarchical clustering of genes was performed using single linkage and Pearson correlation distance metric. Samples were simply ordered naturally.
Quantitative real-time RT-PCR
The most highly significant genes identified through the statistical analysis of microarray data were validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Reverse transcription (RT) was performed on 0.1–1.0 μg RNA with Superscript III RT system (Invitrogen) following manufacturer’s protocol. Real-time PCR was performed with Quantitect SYBR Green PCR system (Qiagen Sciences, MD, USA) on a 7900HT Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA). 18S rRNA was used as the normalizer. The primers used in qRT-PCR are listed in Supplementary Table 2.
Histology and immunohistochemistry
Skin and tumor samples were preserved in OCT compound and stored frozen at −80°C. 5μm sections were cut using a cryostat (Leica Microsystems, IL, USA) and were observed under a fluorescence microscope (Nikon Instruments, Inc., NY, USA) with DAPI (Vectashield, Vector Laboratories, CA, USA) counterstaining. The antibodies for fluorescence immunohistochemistry were obtained as listed in Supplementary Table 3.
In vivo antibody-mediated blockade
For IFN blockade experiment in pups: the anti-murine IFN-γ neutralizing antibody was the rat IgG clone XMG-6 (3.19 mg/ml)40. The anti-murine IFN-αR1 antibody was the mouse IgG1 clone MARI-5A3 (purified Mabs, 4 mg/ml)41. The control antibodies were rat IgG1 clone GL113 (1.52 mg/ml) ascites. The in vivo applications of these antibodies have been described42–43. 0.1–0.2 mg antibody was injected per pup intraperitoneally, 1–2 hours prior to and 3 days after UVB irradiation. For IFN-γ blockade in tumourigenesis experiment: anti-IFN-γ antibody was XMG-6. The control antibody was rat IgG1 against horseradish peroxidase (HRPN) (BioXcell, West Lebanon, NH, USA). Antibodies were administered intraperitoneally as follows: 1 mg per mouse on days −1, 0, 1, 3, 6; and 0.5 mg per mouse on days 9, 12, and 15 after inoculation. Tumours were harvested at day 18.
Immunofluorescence flow cytometry and FACS
The immunofluorescence-based flow cytometry and FACS sorting were done using antibodies listed in Supplementary Table 3, using standard procedures.
In Vitro UV irradiation
The melan-c melanocyte cell line (gift from Dr. Vincent Hearing) was irradiated with the Junggust Box, a hand-made enclosed module that houses two F20 sunlamps outputting 60% UVB and 38% UVA wavebands. A total UV dose of 175 J/m2 was given at a dose rate of 2.0 W/m2. The cells were irradiated in 10 cm culture dishes at 80% confluence overlaid with 3 ml PBS and the dish covered with plastic saran wrap to filter out any UVC. Following irradiation, the cells were incubated for the desired time in normal media (RPMI1640, 10% FBS, 100,000 U/L penicillin, 100 mg/L streptomycin sulphate, 2 mM glutamine, and 200 nM TPA).
Chemotactic assays for Ccl8
The cDNA for mouse Ccl8 was amplified by RT-PCR and cloned into pcDNA3.1 expression vector (Clontech). F5061 cells were transfected using FuGene HD (Roche). Empty pcDNA3.1 vector was transfected as control. In vitro chemotactic assay was performed on RAW264.7 macrophage cell line in Transwell system with 8 μm pore size (Corning). 1×105 cells were seeded in the top well, and the migrant cells were counted following hematoxylin staining. The experiment was done in duplicate. Five random microscopic fields from each replicate were counted for the number of migrant cells. For the in vivo chemotactic assay, 2.5×105 F5061-Ccl8 or F5061-vector cells were inoculated subcutaneously in syngeneic FVB/N mice. Skin samples from the area of the inoculation were harvested two days post inoculation, preserved in OCT compound, cryosectioned, and immunohistochemistry with anti-F4/80 antibody was performed to assess macrophage infiltration.
Tumourigenicity assay
F4/80+ macrophages were FACS-isolated from dorsal skins of pups 6 days post UVB irradiation at P1. Macrophages were admixed with F5061 melanoma cells in 1:5 ratio (0.5×105 macrophages : 2.5×105 F5061) and inoculated subcutaneously in syngeneic FVB/N mice (n=10). Control mice were inoculated with 2.5×105 F5061 cells without macrophages (n=10). Tumours were harvested after three weeks and were measured in three dimensions. Tumor volumes were calculated by the formula: V=length*width*depth*π*1/6. For the tumourigenicity experiment with IFN-γ blockade: for each of the two groups with macrophages, 4 mice had 0.5×105 macrophages but other 6 mice from each group had 0.42×105 macrophages mixed with 2.5×105 F5061 cells. No differences in tumour growth were detected between the mice with these differing numbers of macrophages. All four groups had 10 mice, but two mice from F5061+Cntrl Ab group died prematurely and were taken out of the analysis.
TUNEL assay
The TUNEL assay was performed using TACS TdT Fluorescein kit (R&D Systems, Minneapolis, MN, USA) following the manufacturer’s protocol.
Human and Mouse melanoma tissue microarrays
The human melanoma tissue microarray was obtained from Dr. Stephen Hewitt at NCI-Frederick, National Institutes of Health, Frederick, MD, USA. Antigen retrieval was performed by microwave boiling in citrate buffer (pH 6). Immunohistochemistry with anti-CD68 and anti-IFN-γ antibodies was performed serially with appropriate fluorescence-tagged secondary antibodies. The mouse melanoma tissue microarray has been generated by our laboratory and contains melanoma tissues from our HGF/SF transgenic mouse model, induced by a single neonatal UV radiation dose. Anti-F4/80 (macrophages), anti-CD3 (pan T-cells) and anti-B220 (B-cells) antibodies were used in conjunction with the Vectastain Elite ABC system (Vector Laboratories, Burlingame, CA, USA) following the manufacturer’s protocol.
Statistical analyses
All statistical analyses were performed using Graphpad Prism software. Two-tailed Student’s t-test and ANOVA (Analysis of Variance) with post-hoc Tukey analyses were performed as indicated to validate significant differences. Means ± s.e.m. are indicated for all statistical analyses. A probability (P) value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We would like to thank the following individuals for their support: Stuart Yuspa for primary keratinocytes; Carlo Toniatti and Herman Bujard for the rtTA2sM2 construct; Vince Hearing for melan-c cell line; Stephen Hewitt for the human melanoma tissue microarray; Miriam Anver for immunohistochemical staining and production/analysis of mouse melanoma TMA; Kevin Blas and Eleazar Vega-Valle for technical help; Nick Restifo and Andy Hurwitz for suggestions and discussions. M.R.Z. was supported by NCI Director’s Innovation Career Development Award. E.C.D. and F.P.N. were supported by grants from National Institutes of Health, USA (awards CA53765 and CA92258), and the Melanoma Research Foundation, Hillsborough, NJ, USA.
Footnotes
Supplementary Information accompanies the online version of the paper at www.nature.com/nature.
Competing Interests. Authors have no competing interests.
Authors’ Contributions. M.R.Z. designed and performed experiments, interpreted data and wrote manuscript. S.D. performed statistical analysis of microarray data and generated heatmaps. F.P.N. interpreted data and reviewed manuscript. C.G-C. managed mouse colonies. T.S.H. performed flow cytometry and FACS. R.L.W. performed cDNA microarrays. L.F. produced Dct-rtTA transgenic mice. E.F. provided TRE-H2BGFP mice. L.L. helped design interferon blockade experiments. H.A.Y. interpreted data and reviewed manuscript. T.J.H. evaluated GFP expression in skin and reviewed manuscript. H.A. evaluated embryonic expression of GFP and reviewed manuscript. G.T. designed interferon blockade experiments, provided antibodies, and reviewed manuscript. P.S.M. designed and performed analysis of microarray data and reviewed manuscript. E.C.D. designed, measured and performed UV irradiation experiments, supervised project, and reviewed manuscript. G.M. designed experiments, interpreted data, supervised project and wrote manuscript.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database www.ncbi.nlm.nih.gov/geo under accession number XXXXXXX.
Full Methods and associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Garibyan L, Fisher DE. How sunlight causes melanoma. Curr Oncol Rep. 2010;12:319–326. doi: 10.1007/s11912-010-0119-y. [DOI] [PubMed] [Google Scholar]
- 2.Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control. 2001;12:69–82. doi: 10.1023/a:1008980919928. [DOI] [PubMed] [Google Scholar]
- 3.Noonan FP, et al. Neonatal sunburn and melanoma in mice. Nature. 2001;413:271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- 4.De Fabo EC, Noonan FP, Fears T, Merlino G. Ultraviolet B but not ultraviolet A radiation initiates melanoma. Cancer Res. 2004;64:6372–6376. doi: 10.1158/0008-5472.CAN-04-1454. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura EK, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 6.Walker GJ, et al. Murine neonatal melanocytes exhibit a heightened proliferative response to ultraviolet radiation and migrate to the epidermal basal layer. J Invest Dermatol. 2009;129:184–193. doi: 10.1038/jid.2008.210. [DOI] [PubMed] [Google Scholar]
- 7.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 8.Wolnicka-Glubisz A, et al. Deficient inflammatory response to UV radiation in neonatal mice. J Leukoc Biol. 2007;81:1352–1361. doi: 10.1189/jlb.1206729. [DOI] [PubMed] [Google Scholar]
- 9.Darwich L, et al. Secretion of interferon-gamma by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126:386–393. doi: 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, et al. Rays and arrays: the transcriptional program in the response of human epidermal keratinocytes to UVB illumination. Faseb J. 2001;15:2533–2535. doi: 10.1096/fj.01-0172fje. [DOI] [PubMed] [Google Scholar]
- 11.Proost P, Wuyts A, Van Damme J. Human monocyte chemotactic proteins-2 and -3: structural and functional comparison with MCP-1. J Leukoc Biol. 1996;59:67–74. doi: 10.1002/jlb.59.1.67. [DOI] [PubMed] [Google Scholar]
- 12.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 14.He YF, et al. Sustained low-level expression of interferon-gamma promotes tumor development: potential insights in tumor prevention and tumor immunotherapy. Cancer Immunol Immunother. 2005;54:891–897. doi: 10.1007/s00262-004-0654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beatty GL, Paterson Y. IFN-gamma can promote tumor evasion of the immune system in vivo by down-regulating cellular levels of an endogenous tumor antigen. J Immunol. 2000;165:5502–5508. doi: 10.4049/jimmunol.165.10.5502. [DOI] [PubMed] [Google Scholar]
- 16.Hirobe T. Histochemical survey of the distribution of the epidermal melanoblasts and melanocytes in the mouse during fetal and postnatal periods. Anat Rec. 1984;208:589–594. doi: 10.1002/ar.1092080414. [DOI] [PubMed] [Google Scholar]
- 17.Wolnicka-Glubisz A, Noonan FP. Neonatal susceptibility to UV induced cutaneous malignant melanoma in a mouse model. Photochem Photobiol Sci. 2006;5:254–260. doi: 10.1039/b506974b. [DOI] [PubMed] [Google Scholar]
- 18.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy J, Summer R, Wilson AA, Kotton DN, Fine A. The prolonged lifespan of alveolar macrophages. Am J Respir Cell Mol Biol. 2008;38:380–385. doi: 10.1165/rcmb.2007-0224RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porter GA, et al. Significance of plasma cytokine levels in melanoma patients with histologically negative sentinel lymph nodes. Ann Surg Oncol. 2001;8:116–122. doi: 10.1007/s10434-001-0116-3. [DOI] [PubMed] [Google Scholar]
- 21.Meyskens FL, Jr, et al. J Natl Cancer Inst. 1995;87:1710–1713. doi: 10.1093/jnci/87.22.1710. [DOI] [PubMed] [Google Scholar]
- 22.Ascierto PA, Kirkwood JM. Adjuvant therapy of melanoma with interferon: lessons of the past decade. J Transl Med. 2008;6:62. doi: 10.1186/1479-5876-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebmann V, Wagner S, Grosse-Wilde H. HLA-G expression in malignant melanoma. Semin Cancer Biol. 2007;17:422–429. doi: 10.1016/j.semcancer.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Derre L, et al. Expression and release of HLA-E by melanoma cells and melanocytes: potential impact on the response of cytotoxic effector cells. J Immunol. 2006;177:3100–3107. doi: 10.4049/jimmunol.177.5.3100. [DOI] [PubMed] [Google Scholar]
- 25.Lee N, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U S A. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wischhusen J, Waschbisch A, Wiendl H. Immune-refractory cancers and their little helpers--an extended role for immunetolerogenic MHC molecules HLA-G and HLA-E? Semin Cancer Biol. 2007;17:459–468. doi: 10.1016/j.semcancer.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Koralov SB, Kelsoe G. Complement C4 inhibits systemic autoimmunity through a mechanism independent of complement receptors CR1 and CR2. J Exp Med. 2000;192:1339–1352. doi: 10.1084/jem.192.9.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah KV, Chien AJ, Yee C, Moon RT. CTLA-4 is a direct target of Wnt/beta-catenin signaling and is expressed in human melanoma tumors. J Invest Dermatol. 2008;128:2870–2879. doi: 10.1038/jid.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolnicka-Glubisz A, King W, Noonan FP. SCA-1+ cells with an adipocyte phenotype in neonatal mouse skin. J Invest Dermatol. 2005;125:383–385. doi: 10.1111/j.0022-202X.2005.23781.x. [DOI] [PubMed] [Google Scholar]
- 31.Budd PS, Jackson IJ. Structure of the mouse tyrosinase-related protein-2/dopachrome tautomerase (Tyrp2/Dct) gene and sequence of two novel slaty alleles. Genomics. 1995;29:35–43. doi: 10.1006/geno.1995.1212. [DOI] [PubMed] [Google Scholar]
- 32.Urlinger S, et al. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Fabo EC, Noonan FP, Frederick JE. Biologically effective doses of sunlight for immune suppression at various latitudes and their relationship to changes in stratospheric ozone. Photochem Photobiol. 1990;52:811–817. doi: 10.1111/j.1751-1097.1990.tb08686.x. [DOI] [PubMed] [Google Scholar]
- 35.Serrano MA, Canada J, Moreno JC. Erythemal ultraviolet exposure of cyclists in Valencia, Spain. Photochem Photobiol. 2010;86:716–721. doi: 10.1111/j.1751-1097.2009.00693.x. [DOI] [PubMed] [Google Scholar]
- 36.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2008. [Google Scholar]
- 37.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 39.Gentleman R. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. [Google Scholar]
- 40.Cherwinski HM, Schumacher JH, Brown KD, Mosmann TR. Two types of mouse helper T cell clone. III. Further differences in lymphokine synthesis between Th1 and Th2 clones revealed by RNA hybridization, functionally monospecific bioassays, and monoclonal antibodies. J Exp Med. 1987;166:1229–1244. doi: 10.1084/jem.166.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehan KC, et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 42.Goldszmid RS, et al. TAP-1 indirectly regulates CD4+ T cell priming in Toxoplasma gondii infection by controlling NK cell IFN-gamma production. J Exp Med. 2007;204:2591–2602. doi: 10.1084/jem.20070634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gramzinski RA, et al. Interleukin-12- and gamma interferon-dependent protection against malaria conferred by CpG oligodeoxynucleotide in mice. Infect Immun. 2001;69:1643–1649. doi: 10.1128/IAI.69.3.1643-1649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.