Abstract
Genome-wide association studies have broadened our understanding of the genetic architecture of cancer to include common variants, in addition to the rare variants previously identified by linkage analysis. We review current knowledge on the genetic architecture of four cancers—breast, lung, prostate and colorectal—for which the balance of common and rare alleles identified ranges from fewer common alleles (lung cancer) to more common alleles (prostate cancer). Although most variants are cancer specific, pleiotropy has been observed for several variants, for example, variants at the 8q24 locus and breast, ovarian and prostate cancers or variants in KITLG in relation to hair color and testicular cancer. Although few studies have been adequately powered to investigate heterogeneity among ancestry groups, effect sizes associated with common variants have been reported to be fairly homogenous among ethnic groups. Some associations appear to be ancestry specific, such as HNF1B, which is associated with prostate cancer in European Americans and Latinos but not in African-Americans. Studies of cancer and other complex diseases suggest that a simple dichotomy between rare and common allelic architectures may be too simplistic and that future research is needed to characterize a fuller spectrum of allele frequency (common (>5%), uncommon (1–5%) and rare (<<1%) alleles) and effect size. In addition, a broadening of the concept of genetic architecture to encompass both population architecture, which reflects differences in exposures, genetic factors and population level risk among diverse groups of people, and genomic architecture, which includes structural, epigenomic and somatic variation, is envisioned.
Introduction
Studying the genetic predisposition to developing cancer has been a vast and productive undertaking since early linkage studies of high-risk families identified rare and highly penetrant loci (those with large effect size) such as BRCA1 and BRCA2 for breast cancer and APC for colorectal cancer (1,2). However, linkage studies tended to identify variants of very low frequency (<1/1000) in the general population. Subsequent candidate gene studies were designed to identify common (allele frequency ≥5%) low-penetrance susceptibility alleles that could be easily studied in groups of unrelated cases and controls, choosing one or at most a few variants that were hypothesized to associate with disease and relying heavily on biological assumptions or where available, data from small clinical collections. As a whole, candidate gene studies in cancer, as in other complex diseases, were limited by inconsistency across studies (3,4), leading many to call for replication as a necessary standard (5). As the common disease–common variant hypothesis gained traction and studies such as the HapMap project generated comprehensive data on the common patterns of DNA sequence variation in the human genome, it became practical to design studies that would capture a substantial proportion of common human genome variation with a minimum subset (although still hundreds of thousands) of single nucleotide polymorphisms (SNPs). Concerns about population stratification, or confounding by race or ethnicity, could be addressed by utilizing the large number of SNPs to be genotyped to empirically assess and control for ancestry (6) or by appropriate case–control matching (7). These genome-wide association (GWA) studies no longer relied on correctly specified candidate genes and would go on to discover hundreds of variants, many in intergenic regions or genes not known to be associated with disease (Figure 1) (8). In contrast to the rare high-penetrance loci described above [often with minor allele frequency < 0.001% and odds ratios (ORs) > 10], these common variants generally have modest effect sizes (OR < 2).
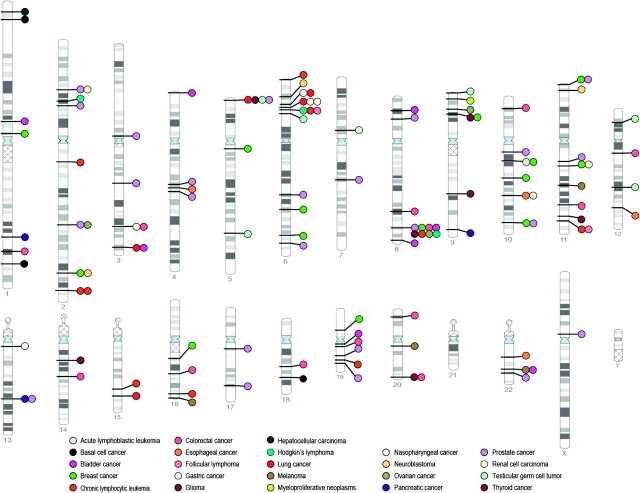
Fig. 1.
Cancer-associated genetic variants identified through GWA studies. Genetic variants were identified from the NHGRI Genome-wide Association Study catalog (www.genome.gov/gwastudies) and include all cancer associations at P < 5 × 10−8 through 2010.
Only a small proportion of cancer heritability can be explained by as yet identified genetic variants from linkage studies, candidate gene studies and GWA studies (1). However, the ever-growing catalog of genetic variants predisposing to cancer will inform us about the genetic architecture of various cancers in terms of the various types of risk alleles, their frequencies and effect sizes, what proportion of risk or heritability they explain and how they interact with environmental factors (9). The goal of this review is to examine the genetic architecture of several key cancers in comparison with the genetic architecture of other complex diseases, in order to develop strategies for future studies to identify relevant variants. We describe the concept of population architecture of a genetic variant (similar to the genetic architecture of a complex trait), which includes defining population-based estimates of relative and absolute risk; assessing generalizeability of genotype–phenotype disease groups across groups defined by ethnicity, gender and environmental factors and characterizing the spectrum of traits influenced by a genetic variant. Finally, we anticipate and describe future research endeavors that will further extend characterization of the genetic architecture of cancer.
Genetic architecture of cancer—knowledge to date
What we know about genetic architecture has been shaped by prevailing hypotheses about disease biology and genetic influences on disease susceptibility as well as the available studies and technologies to test those hypotheses. Different study designs have been utilized for identifying common versus rare variants. Linkage studies and family-based studies of cancer, for example, have identified rare variants with frequency much <1% and with high penetrance. These studies were consistent with a polygenic model of inheritance in which no one variant is the main culprit (1,10). Candidate gene and GWA studies have identified common variants, generally present at 5% allele frequency or higher. There is probably an intermediate class of variants, with ∼1–5% frequency, which currently is not well characterized but is becoming more amenable to discovery as genotyping and sequencing costs decrease. These variants are not likely to have effects large enough to have been detected through linkage nor frequencies high enough to discover through GWA studies. Some have posited that combinations of rare alleles might explain some of this missing heritability or that common variants associated with complex disease may in fact reflect the contributions of undiscovered rare alleles which, by chance, track with the GWA study-identified allele (11). How many such variants exist and how large their effect sizes are will greatly influence our understanding of genetic architecture. Recent data from the 1000 Genomes Project (http://www.1000genomes.org) confirm that existing genome-wide association study (GWAS) platforms assay a substantial proportion of variants with allele frequencies >4% and less coverage at frequencies of 1–2%. These data support the idea that extending the allele frequency range to include rare variants may explain an increasing proportion of phenotypic variation (12). As genotyping arrays become larger in scope, to be superseded eventually by sequencing, the relative dichotomy of ‘common’ and ‘rare’ variation will resolve to more of a spectrum of variation (9,10). It is also worth noting that few studies have examined gene environment interactions, which will be important in delineating the specific environmental contexts or exposures in which the genetic association is manifested.
Published GWAs in cancer are summarized in supplementary Table 1A, available at Carcinogenesis Online. Over 150 associations at P < 5 × 10−8 have been identified with a variety of cancer phenotypes, however, the functional implications of these variants are often unclear. As seen for other complex diseases, the vast majority of cancer-associated variants identified in GWA studies are either intronic (39%) or intergenic (52%). Effect sizes associated with these common variants are modest (median OR 1.25, 75% with OR < 1.5) (8), consistent with the idea that variants with large effect sizes could be subject to selection pressure (13), though selection pressures are thought to have changed dramatically over the course of human evolution. Associations with OR >2 were observed for SNPs associated with prostate cancer, testicular cancer and myeloproliferative neoplasms.
Most variants identified to date are cancer type and, in some cases, subtype specific, though important exceptions include TERT, CLPTM1L, TP53 and the 8q24 region. In addition to true differences in the genetic architecture of various cancers, the tumor-specific productivity of the GWA approach may reflect the ability to assemble large sample sizes with enough statistical power to identify variants of small effect; the genetic heterogeneity of some tumor types relative to others; the relative contribution of genetic and environmental factors or the differing selection pressures on cancers showing major differences in incidence or course by age or sex. Few studies have followed-up variants from GWA studies in families, either to study unexplained disease or as genetic modifiers of known variants. The overwhelming majority of studies have included cancer risk as the primary GWAS endpoint. A minority of studies have looked at survival, prognosis or treatment response in individuals with cancer, with limitations due to small sample size or lack of replication (14–20).
To date, the gaps in genetic architecture reflect the areas where genotyping technology is still being developed and/or applied. Thus, the contribution of rare variants other than those segregating in affected families has yet to be defined but may well be clarified as very high density SNP arrays (2 million or more SNPs), exome and whole genome sequencing become feasible in population-based studies. For reasons that are largely unknown, epidemiologic studies of some cancer types have been more successful at identifying less common, high penetrance variants and others at identifying high frequency susceptibility alleles of smaller effect. Linkage scans of prostate and testicular cancer, for example, have identified only a limited number of highly penetrant variants (21), although GWA studies of prostate cancer have identified the largest number of variants to date (1). Studies of breast and colorectal cancer have identified high-risk mutations in BRCA1, BRCA2 and APC, MLH1 and MSH2, respectively, and fewer susceptibility alleles overall. Below, we highlight specific results from the four most common cancers—breast, colorectal, prostate and lung. We discuss findings from both candidate gene studies and GWAS, recognizing that many GWAS findings may still be regarded as preliminary until functional consequences are identified.
Breast cancer
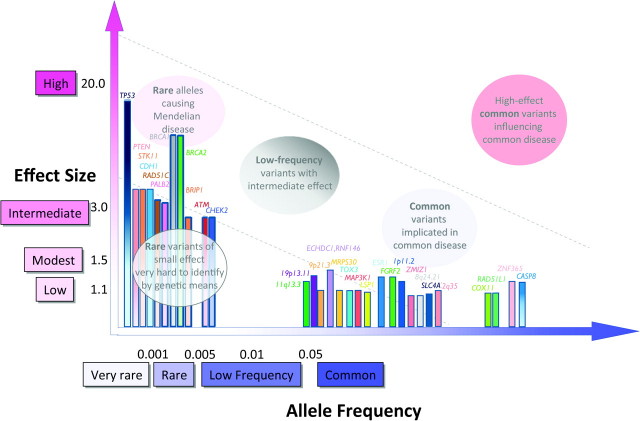
Over two dozen loci and variants have been implicated in susceptibility to breast cancer (Figure 2). Fewer than 10% of breast cancer cases are attributed to rare, high-penetrance genes, with a similar estimate attributed to SNPs identified through GWA studies (supplementary Table 1B is available at Carcinogenesis Online) (1,2). BRCA1 and BRCA2, the two most well-characterized genes, are associated with greater than a 10-fold increase of breast cancer risk relative to the general population (22). The other rare and highly penetrant loci are reported in rare hereditary cancer syndromes such as Li-Fraumeni syndrome and Cowden’s syndrome. An intermediate risk group of breast cancer variants—in ATM, BRIP1, CHEK2, PALB2 and RAD51C generally confer a 2- to 3-fold increased risk of breast cancer. Minor allele frequencies for these variants are typically <0.1%, although there are some notable exceptions (for example, CHEK2 has a frequency of ∼1% in the Ashkenazi Jewish population) (2). GWA-identified variants to date have shown ORs <1.5 and are frequent (most with risk allele frequencies of >20%). In contrast to the rare and intermediate effect variants, where functional significance is often recognized (for example, in tumor suppressor or loss-of-function mutations), the GWA loci harbor susceptibility variants whose functions are largely unknown. In fact, some, such as the variants in 8q24.21, are in regions of the genome that are devoid of known genes.
Fig. 2.
Allele frequency and effect sizes for genetic variants associated with breast cancer. Allele frequencies and ORs are taken from published literature where available and are not depicted to scale. Associations identified through GWA or GWA follow-up studies are shown with solid colored bars; all others are shaded from dark (top) to light (bottom).
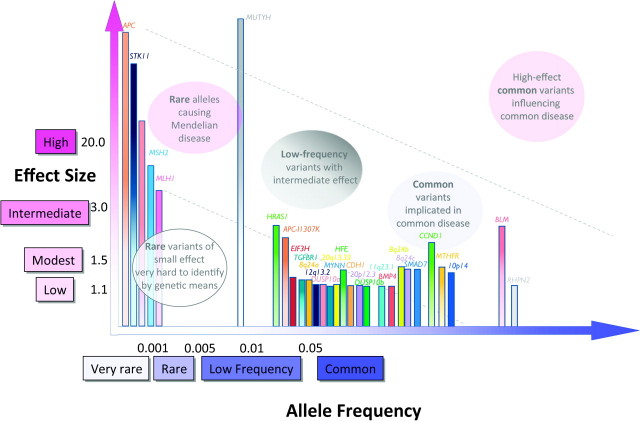
Colorectal cancer
As in breast cancer, several key loci in colorectal cancer were discovered in familial or syndromic cases (including APC, MUTYH and mismatch repair genes MLH1, MSH2, MSH6 and PMS2; Figure 3, supplementary Table 1C is available at Carcinogenesis Online). Together, these loci account for ∼20% of the genetic predisposition to colorectal cancer (23). A handful of low to moderate penetrance variants, including APC-I1307K, BLM, HRAS1, TGFβR1, HFE, CCND1 and MTHFR, have also been identified in candidate gene association studies (23). GWA studies have identified a number of common variants (those at BMP4, CDH1, EIF3H, RHPN2, SMAD7, 8q24, 10p14, 11q23.1 and 20p12.3), which are associated with small increases in relative risk and which cumulatively account for ∼6% of the excess familial risk (24).
Fig. 3.
Allele frequency and effect sizes for genetic variants associated with colorectal cancer. Allele frequencies and ORs are taken from published literature where available and are not depicted to scale. Associations identified through GWA or GWA follow-up studies are shown with solid colored bars; all others are shaded from dark (top) to light (bottom).
Prostate cancer
Prostate cancer demonstrates strong familial clustering (25) but high penetrance genes have yet to be identified and replicated. The most likely candidate so far is BRCA2, which is associated with a 20-fold increased risk relative to the general population. Linkage studies show a number of susceptibility loci with modest LOD scores; however, most were not replicated across studies (26).The GWA study approach has been fruitful in identifying many replicated variants, all with OR <2 and most with OR <1.3 (Figure 4; supplementary Table 1D is available at Carcinogenesis Online). In addition to variants in the EHBP1, IGF1/IGF2 (11p15), ITGA6, KLK3, LMTK2, MSMB, NKX3.1, NUDT10/NUDT11, PDLIM5, SLC22A3, TCF2, THADA and TET2 genes, many of these variants are in intergenic regions at 3p12, 3q21, 8q24, 11q13, 17q24, 19q13 and 22q13.2. Furthermore, the cumulative effects of these loci explain at least 20% of the familial risk (27). Interestingly, several of these variants localize to distinct linkage disequilibrium blocks on 8q24 (28).
Fig. 4.
Allele frequency and effect sizes for genetic variants associated with prostate cancer. Allele frequencies and ORs are taken from published literature where available and are not depicted to scale. Associations identified through GWA or GWA follow-up studies are shown with solid colored bars; all others are shaded from dark (top) to light (bottom).
GWA studies have identified more signals for prostate cancer than for any other cancer type. This may reflect the true underlying genetic architecture of prostate cancer (reasonably common variants with modest effect sizes) and/or the ways in which prostate cancer has been particularly amenable to the GWA approach, including reduced influence of environmental factors, limited disease subtype heterogeneity and long survival after diagnosis. Notably, the variants identified thus far do not distinguish between less and more aggressive tumor types (29), suggesting that they may be more involved in the initiation of cancer than its severity or course. Finally, that prostate cancer presents largely in older age suggests that alleles predisposing to prostate cancer may be under less selective pressure than those that predispose to cancers at younger ages (27) or that age-related senescence plays a key role in etiology. However, the overall picture is probably more complicated, as genotypes are selected based on the phenotypes they produce and focusing on a variant’s influence on a single cancer ignores possible pleiotropic effects (30).
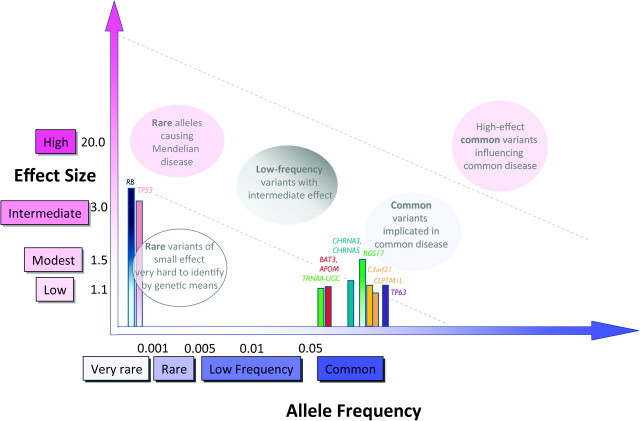
Lung cancer
The etiology of lung cancer is recognized to be strongly environmental, although there is increasing evidence that genetic factors may also play a role. Family and linkage studies have identified high-penetrance variants in TP53, RB1 and 6q23–25 (31–33). More recently, GWA studies have consistently identified three loci at genome-wide significance (Figure 5; supplementary Table 1E is available at Carcinogenesis Online)—variants at 5p15.33 (TERT-CLPTM1L), 6p21(BAT3, APOM)/6p22.1 (TRNAA-UGC) and 15q25.1 (CHRNA3, CHRNA4 and CHRNA5), which explain ∼7% of the familial risk of lung cancer (27). Interestingly, 15q25.1 variants are also associated with smoking behavior, a risk factor for lung cancer. Part of the association of 15q25.1 variants with lung cancer can probably be explained by associations with smoking behavior; however, not all studies agree whether they have some degree of independent effects (34–36). Specific variants have also been consistently identified for strong lung cancer risk factors, such as smoking quantity, smoking initiation or cessation (37,38) but have not met strict definitions of genome-wide significance.
Fig. 5.
Allele frequency and effect sizes for genetic variants associated with lung cancer. Allele frequencies and ORs are taken from published literature where available and are not depicted to scale. Associations identified through GWA or GWA follow-up studies are shown with solid colored bars; all others are shaded from dark (top) to light (bottom).
Complexities in genetic architecture
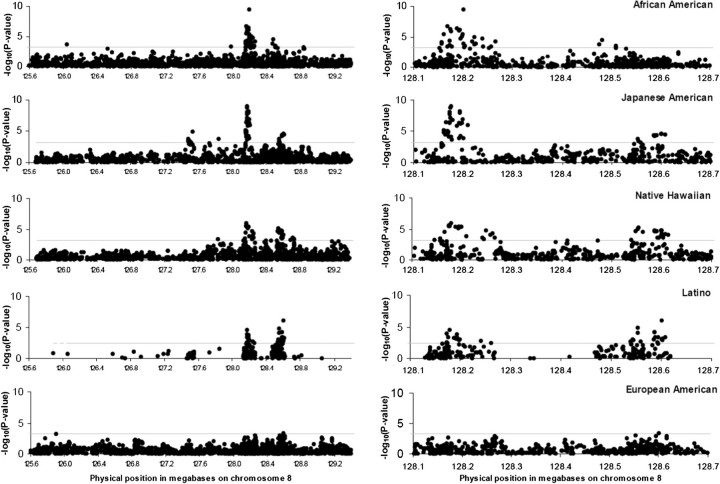
The emerging picture of the genetic architecture of cancer is complex and varies somewhat by cancer type, as discussed above. Adding to this complexity are several additional dimensions, including allelic heterogeneity, phenotypic heterogeneity and pleiotropy. Allelic heterogeneity, or the association of a single trait with multiple variants within the same gene or locus, has been described for several cancers. For example, within the 8q24 region, five loci are independently associated with prostate cancer susceptibility (28,39).Additionally, these loci may be population specific, reflecting differences in linkage disequilibrium (Figure 6) as well as disease risks conferred (39). Allelic heterogeneity is evident across the spectrum of rare and common variation, as for colorectal cancer and variants in the APC gene (23). In the presence of allelic heterogeneity, the value of any one approach, whether GWA-, family- or candidate gene-based, is limited; results from all of these study designs (and probably more) are needed to identify the multiplicity of alleles that underlie genetic architecture.
Fig. 6.
Transethnic linkage disequilibrium plots for 8q24 locus and prostate cancer. Results for scanning across the admixture peak for each population separately. Data for the entire admixture peak (125.6–129.4 Mb) is on the left, with a blow up of the region of highest interest on the right (128.1–128.7 Mb). Results for African-Americans are restricted to cases age <72 (the group that initially gave the admixture signal in Freedman et al. 2006). The number of variants tested is: 2111 for African-Americans, 1565 for Japanese Americans, 1565 for Native Hawaiians, 275 for Latinos and 2056 for European Americans. Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics (39), (2007).
Cancer is not only genomically but also phenotypically complex. Studies of common variant discovery, particularly GWA studies, have typically been designed to minimize phenotypic and ancestral heterogeneity and maximize sample sizes. Cancer cases are generally aggregated across cancer stages, presenting future opportunities to refine existing studies by studying disease progression, subtypes or intermediate endpoints. Cancers with short survival times after diagnosis, such as pancreatic cancer, acute leukemia and small cell lung cancer, may be limited in recruiting enough participants in a case–control study design. Further exploration is needed but the emerging data in breast and lung cancer suggest that some variants are associated with tumor subtype; integration of knowledge of somatic and germ line variants is also needed. Associations with several rare and common loci, including FGFR2, MAP3K1, 8q24.21, 2q35 and 5q11.2 are stronger in estrogen receptor-positive breast cancer (41), BRCA1 variants are associated with estrogen receptor-negative breast cancer (42) and the TERT-CLPTM1L variant at 5p13.33 is associated with adenocarcinoma of the lung but not other subtypes of lung cancer (43). Although a limited number of common variants have been associated with intermediate phenotypes relevant to cancers, such as mammographic density as it relates to breast cancer, it would be interesting to extend these studies to the GWA study setting.
One of the benefits of agnostic, genome-wide approaches is the identification of genetic variants associated with multiple outcomes (supplementary Table 1F is available at Carcinogenesis Online). These examples of pleiotropy may be illustrative of etiological pathways in common or biological interactions underlying seemingly unrelated outcomes. For example, an early GWA study of prostate cancer unexpectedly identified a variant in HNF1B/TCF2, a gene known to be involved in diabetes (44). Interestingly, an inverse relationship was observed: the variant was associated with a reduced risk of prostate cancer and an increased risk of diabetes, corroborating findings from previous observational studies that individuals with diabetes seemed to be at lower risk of developing prostate cancer (45). Notably, this association does not seem to extend to other cancer types, even those that have been positively associated with diabetes (40). Other examples of pleiotropy include the JAZF1 locus, associated with height (46), prostate cancer (47) and type 2 diabetes (48); the KITLG gene with testicular cancer (49,50) and hair color (51); the CDKN2A-CDKN2B (9p21.3) locus with type 2 diabetes (48), myocardial infarction (52), glioma (53,54), melanoma (55) and nevus density (56), and the TERT-CLPML1 (5p15.33) with basal cell carcinoma, glioma, bladder, prostate and lung cancer (40,57–59). Interestingly, the 8q24 locus harbors a number of distinct susceptibility loci, some of which demonstrate pleiotropic effects in different cancer types. Five distinct 8q24 subregions have emerged, displaying patterns of association that appear to be specific for breast, ovarian, colorectal and prostate cancer (60). Three regions were associated with prostate cancer only, one was associated with breast cancer only and one was associated with breast, ovarian and prostate cancer.
Genetic architecture of other complex diseases
For the vast majority of complex disease, variants with a range of allele frequencies and associated risks have been identified, their discoveries reflecting the use of the methods that are best powered to detect them (9,13). Nonetheless, there is a wide range in the number of variants identified to date and the proportion of heritability explained. For example, in age-related macular degeneration, five alleles, ranging in allele frequency from 5 to 57%, account for approximately half of the familial risk (61). For Crohn’s disease, however, only 10% of the variance in risk can be explained by 32 variants identified to date with frequencies from 2 to 93% (62). For the more common outcomes of type 2 diabetes and height, 18 variants explain 6% of the familial risk and 40 variants explain 5% of the trait variance, respectively (63,64). It is clear, however, that much of the heritability underlying cancer and other common diseases remains undiscovered (65). An interesting observation from several complex traits, such as type 2 diabetes, body mass index and QT interval, is that GWAS variants are found in genes known also to be involved in Mendelian disease. Such loci have not yet been identified for cancer but are under active investigation. Possible explanations include a different etiologic model for cancer whereby genes involved in the Mendelian two-hit model of cancer do not generalize to non-familial cancers, and possibly, smaller sample sizes for cancer studies relative to those of complex diseases.
As rare variant SNP genotyping, whole exome or genome sequencing becomes increasingly feasible in large-scale population studies, the catalog of rare variation is likely to expand. How these yet-to-be-identified signals overlap with the known variants and whether their associations will be independent of them will be of great interest. To the extent that rare disease-predisposing alleles exist and are spread evenly across the genome, as opposed to concentrated in regions of the genome where GWA signals have been found, genome-wide resequencing may be necessary to improve our understanding of the genetic architecture of complex disease (9,66). Replication will probably remain the gold standard for validating associations, necessitating large sample sizes. Lack of replication may also be informative, however, suggesting the need to investigate populations where genetic background or environmental exposures may differ (67). Cancer variants spanning the full spectrum of genetic variation, from very rare to common variants, is probably relevant and should be catalogued.
Population architecture of cancer
Describing the population architecture of genetic variants in relation to cancer necessitates expanding genetic studies to include diverse populations and person-level factors, and accounting for gene-by-environment interactions (68). Although effect sizes associated with common variants in candidate genes have been reported to be fairly homogenous among ethnic groups (69), there are some notable exceptions. For example, only 7 of 23 prostate cancer-associated variants identified from GWAS in European descent, African descent or diverse populations replicated in Japanese populations (70). As genetic architecture of cancer may vary according to ancestral group, it is important to perform studies in multiple ancestral groups having different linkage disequilibrium patterns, allele frequencies and environmental exposures. So far, heterogeneity in associations by ancestry has been evaluated in a limited number of cancer types. In one prostate cancer study, most associations with several variants and prostate cancer were consistently and positively associated with disease risk among African-Americans, European Americans, Latinos, Japanese Americans and Native Hawaiians. However, ethnic-specific heterogeneity was observed for four variants (71). In a more recent study of prostate cancer susceptibility loci identified primarily in men of European descent, all regions at chromosome 8q24 were validated in men of African descent, but most associations outside of this region were not validated (72). An earlier fine mapping study of the 8q24 locus and prostate cancer in multiple ethnic groups (39) demonstrated that some variants were unique to certain populations, others conferred varying magnitudes of risk, and even variants that were similar in effect sizes across populations were found at different allele frequencies among ethnic groups (Table I). To this last point, seven variants in the 8q24 locus may explain substantially more of the disease burden in African-Americans compared with European Americans, although population attributable risk, the measure used, can be problematic to interpret (73). Thus, even when effect sizes of associated variants are similar among subpopulations, large differences in allele frequency may imply that some groups may be disproportionately affected in ways that are relevant to health disparities. That the emerging knowledge from GWA studies to date largely reflects studies in European descent populations (7,74) highlights the need to include more diverse populations, to understand genomic as well as population architecture in populations with disproportionate disease burdens. Our gap in knowledge regarding population architecture is not limited to race or ethnic considerations. Whether and how the genotype-cancer associations differ by person-level dimensions such as age, gender and presence or absence of other risk factors (genetic or environmental) remain largely to be seen.
Table I.
Ethnic-specific patterns of association of genetic variants at 8q24 with prostate cancer.
| Marker, region and position | African-Americansab (1614/837) | Japanese Americans (722/728) | Native Hawaiiansa(111/112) | Latinosa (637/633) | European Americansb (1182/942) | Phetc | Pooled OR (95% CI)d (unadjusted for other markers) | Pooled OR (95% CI)e (adjusted for other markers) |
| rs13254738, region 2 and 128173525 | 1.24 (1.09–1.42) 58% | 1.57 (1.33–1.83) 62% | 1.46 (1.00–2.12) 50% | 1.25 (1.07–1.46) 49% | 1.11 (0.97–1.26) 33% | 0.02 | 1.26 (1.18–1.36) | 1.18 (1.09–1.27) |
| rs6983561, region 2 and 128176062 | 1.34 (1.18–1.53) 40% | 1.78 (1.47–2.15) 16% | 3.17 (1.87–5.36) 12% | 1.99 (1.34–2.96) 3% | 1.16 (0.86–1.58) 4% | 0.001 | 1.51 (1.37–1.67) | 1.42 (1.28–1.58) |
| Broad11934905f, region 2 and 128200991 | 2.45 (1.65–3.62) 2% | — (—) <1% | — (—) 0% | — (—) <1% | — (—) 0% | — | 2.45 (1.65–3.62) | 2.24 (1.43–3.21) |
| rs6983267, region 3 and 128482487 | 1.43 (1.17–1.75) 84% | 1.22 (1.05–1.42) 32% | 1.29 (0.88–1.89) 28% | 1.05 (0.89–1.24) 62% | 1.13 (0.99–1.28) 51% | 0.17 | 1.18 (1.09–1.27) | 1.14 (1.03–1.26) |
| rs7000448g, region 3 and 128510352 | 1.33 (1.12–1.58) 61% | 1.23 (1.04–1.46) 24% | 1.38 (0.89–2.14) 22% | 1.29 (1.07–1.56) 29% | 1.14 (0.93–1.40) 37% | 0.83 | 1.26 (1.15–1.38) | 1.19 (1.08–1.32) |
| DG8S737–8g, region 1 and 128545681 | 1.25 (1.06–1.49) 16% | 1.48 (1.16–1.88) 16% | 2.55 (1.33–4.89) 15% | 1.46 (1.05–2.02) 6% | 1.45 (0.96–2.19) 5% | 0.27 | 1.39 (1.23–1.57) | 1.23 (1.08–1.40) |
| rs10090154, region 1 and 128601319 | 1.11 (0.94–1.32) 16% | 1.49 (1.23–1.81) 15% | 2.54 (1.61–4.02) 17% | 1.98 (1.49–2.61) 7% | 1.44 (1.17–1.76) 9% | 0.0005 | 1.43 (1.30–1.58) | 1.32 (1.17–1.50) |
Each cell of the table gives ORs (and 95% confidence intervals) for allele dosage effects along with the risk allele frequency in controls. ORs in this table do not correct for local ancestry estimates in African Africans, Latinos and Native Hawaiians, as we know local ancestry is correlated to some of these alleles.
Adjusted for genome-wide European ancestry.
OR adjusted for study.
P value testing for heterogeneity of allelic effects across all populations.
OR adjusted for population, study and genome wide European ancestry (African Africans, Latinos and Native Hawaiians).
OR adjusted for population, study and genome-wide European ancestry (African Africans, Latinos and Native Hawaiians) and all other markers in the same region (i.e. region 1, 2 or 3). Within a region, individuals missing data for any marker were excluded from analysis.
Analysis of Broad11934905 is presented for African-Americans only, as this is the only population in which the risk variant has an appreciable frequency.
A smaller number of subjects were genotyped for rs7000448 (2422 affected individuals and 2311 controls) and the microsatellite (3036 cases and 2208 controls).
Differences between populations in the distribution of risk factors for disease, health behaviors and lifestyle choices are associated with differences in disease etiology and prevalence and can also modify the association between genetic variants and disease (75). The identification of gene–environment interactions may improve understanding of biological mechanisms or pathways. Additionally, since environmental factors are more easily modified than genetic factors, identifying strata of individuals who may stand to benefit most from changing their environments or behaviors may help to target preventative or treatment measures. Few GWAS to date have included analyses of gene–environment interactions, which require at least tens of thousands of participants to detect modest interactions (interaction OR < 1.5) (76). The need for massive sample sizes requires more stringent efforts to harmonize environmental studies among multiple studies within a consortium, a non-trivial effort. Studies that take advantage of already collected environmental measures in extant populations, particularly longitudinal studies, and efforts to harmonize measures across studies will facilitate the large meta-analyses that will be needed for adequately powered analyses (77). Additionally, the complexity of genome-wide interaction analyses emerging from thousands or hundreds of thousands of possible SNP candidates and possible higher order interactions necessitates refinement of existing analytic and bioinformatic approaches and development of new tools where necessary (78).
Ongoing resources and efforts
The implementation of large-scale and high-throughput genotyping and analysis, an approach first pioneered in linkage studies and continuing today in GWA and sequencing studies, has now been applied to a vast range of common complex diseases. Resolution of the genetic architecture of each disease, then, has proceeded within the resources of each research community. However, efforts are underway to facilitate approaches and resources that can be easily shared and adopted across disease communities. For example, the National Cancer Institute’s Cancer Post-GWAS Initiative [http://epi.grants.cancer.gov/pgwas/ (4 December 2011, date last accessed)] supports highly collaborative transdisciplinary research teams designed to accelerate post-GWA research towards the identification of causal variants. Additionally, the investigators supported by the initiative will leverage existing GWA studies of cancer for novel discoveries: meta or pooled analyses of previously generated ‘initial scan’ GWA study data will allow for the detection of common variants with smaller genetic effects or less frequent risk alleles. Critically, the increased sample size of these pooled analyses will also mean that clinically significant subgroup analyses may be performed and additional phenotypes such as risk of relapse or survival could be considered using existing data. Finally, each collaborative team was challenged to consider more sophisticated models of susceptibility (pathways, gene gene and gene environment interactions) as well as to extend GWA study findings to diverse populations, which ideally would add to our understanding of racial and ethnic disparities in cancer. The National Human Genome Research Institute’s (NHGRI) Population Architecture using Genomics and Epidemiology (PAGE) program [www.pagestudy.org (4 December 2011, date last accessed)], a consortium of population-based studies, is further characterizing promising variants identified through GWA studies in ∼80 000–100 000 participants of European, African, Asian, Native American and Hispanic descent, with the specific aim of further clarifying the population architecture of these variants (79). High-priority efforts within PAGE include assessing whether associations identified through GWA studies generalize to diverse ethnic groups or are modified by known risk factors or environmental measures. Additionally, investigators are using the breadth and depth of the phenotype data to investigate in a hypothesis-free way whether these genetic variants are associated with phenotypes beyond the initially targeted phenotype (80).
Considering genetic architecture across the full spectrum of allelic frequencies necessitates the availability of a public, easily accessible knowledge base where data on both common and rare variation, in relation to cancer and other complex traits, are available. Although such a comprehensive resource has yet to be developed for a broadly inclusive range of common diseases, examples of compendia where association data are available include the NHGRI Genome-wide Association Study Catalog [www.genome.gov/gwastudies (4 December 2011, date last accessed)], the Center for Disease Control and Prevention’s HuGENavigator resource [http://hugenavigator.net/ (4 December 2011, date last accessed)], the Genetic Association Database [http://geneticassociationdb.nih.gov/ (4 December 2011, date last accessed)] and the Cancer Genome Atlas [http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp. (4 December 2011, date last accessed)].
Future directions
Although the research following up on GWA findings is still in its relative infancy, clinical and population-based studies of cancer remain forward looking in refining approaches to study design and statistical methods, incorporating novel genetic and genomic technologies and maximizing existing resources. Expanding the catalog of genomic cancer variation will open up opportunities for additional GWA studies, meta-analyses of existing family and population-based data, sequencing studies, epigenomic studies and gene expression studies (10). In Europeans, the increase in genomic coverage and accumulation of large numbers of densely genotyped samples for many common diseases will reach a point where only variants with very small effects or very low allele frequencies remain to be discovered. In addition to pursuing dense genotyping and sequencing in these populations, an important opportunity to pursue now is to extend discovery efforts to underrepresented populations. European ancestry populations have less genetic diversity than African ancestry populations and patterns of linkage disequilibrium are known to vary by population. Additional studies within cancer subtypes may help to clarify not only the biological impact of known variants but the differing pathogenesis and course of these diseases. The goal of fully characterizing the genetic architecture of cancer extends beyond understanding etiology; it is a gateway to improvements in prevention, therapeutics and clinical care. In this regard, following up pleiotropic variants involved with more than one cancer may provide insights into common cancer mechanisms. Additionally, risk prediction models can be developed, and where variants with large effect sizes are known, gene-based development of diagnostics and therapeutics may be facilitated. Beyond targeting rare and common genetic variants, studies are moving toward understanding the genomic architecture of cancer, going beyond inherited DNA sequence variation to include epigenomics, structural and somatic variation and pathway interactions. The interplay between inherited and acquired variation has yet to unfold, although early efforts have uncovered overlap among genes identified through tumor sequencing and GWA studies [for example, in the 9p21 region and glioma or lung cancer (81, 82) or the TERT gene and lung cancer (82)]. Additionally, examining the contributions of variants and genes across biological pathways that link diseases will be an important part of characterizing the overlap in genomic architecture across related diseases (83). Finally, future efforts to further our understanding of the population architecture of cancer will put the fruits of all human genetic studies—from family-based to population-based, from rare variant- to common variant-focused—into clinical and public health context. This is an exciting time in research related to the genetic susceptibility to cancer, one in which we look forward to the convergence of knowledge of genetic and environmental causes of cancer to improve human health.
Supplementary Material
Acknowledgments
We thank Darryl Leja, Lin Gyi, Camilla Benedicto, Kelly Bennett, Peggy Hall and Jaya Mehta for assistance with figures and literature searches.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- GWA
genome-wide association
- NHGRI
National Human Genome Research Institute
- OR
odds ratio
References
- 1.Fletcher O, et al. Architecture of inherited susceptibility to common cancer. Nat. Rev. Cancer. 2010;10:353–361. doi: 10.1038/nrc2840. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD. Inherited susceptibility to common cancers. N. Engl. J. Med. 2008;359:2143–2153. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 3.Hirschhorn JN, et al. A comprehensive review of genetic association studies. Genet. Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Todd JA. Statistical false positive or true disease pathway? Nat. Genet. 2006;38:731–733. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 5.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 6.Tian C, et al. Accounting for ancestry: population substructure and genome-wide association studies. Hum. Mol. Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg NA, et al. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy MI. Exploring the unknown: assumptions about allelic architecture and strategies for susceptibility variant discovery. Genome Med. 2009;1:66. doi: 10.1186/gm66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cazier JB, et al. General lessons from large-scale studies to identify human cancer predisposition genes. J. Pathol. 2010;220:255–262. doi: 10.1002/path.2650. [DOI] [PubMed] [Google Scholar]
- 11.Dickson SP, et al. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard JK, et al. The allelic architecture of human disease genes: common disease-common variant.or not? Hum. Mol. Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 14.Bolton KL, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horinouchi M, et al. Association of genetic polymorphisms with hepatotoxicity in patients with childhood acute lymphoblastic leukemia or lymphoma. Pediatr. Hematol. Oncol. 2010;27:344–354. doi: 10.3109/08880011003739422. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70:9721–9729. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- 17.Penney KL, et al. Genome-wide association study of prostate cancer mortality. Cancer Epidemiol. Biomarkers Prev. 2010;19:2869–2876. doi: 10.1158/1055-9965.EPI-10-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato Y, et al. Genome-wide association study on overall survival of advanced non-small cell lung cancer patients treated with carboplatin and paclitaxel. J. Thorac. Oncol. 2010;6:132–138. doi: 10.1097/JTO.0b013e318200f415. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, et al. Genome-wide examination of genetic variants associated with response to platinum-based chemotherapy in patients with small-cell lung cancer. Pharmacogenet. Genomics. 2010;20:389–395. doi: 10.1097/FPC.0b013e32833a6890. [DOI] [PubMed] [Google Scholar]
- 20.Azzato EM, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol. Biomarkers Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpten J, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 22.Antoniou A. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat. Rev. Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 24.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemminki K, et al. Genetic epidemiology of multistage carcinogenesis. Mutat. Res. 2001;473:11–21. doi: 10.1016/s0027-5107(00)00162-7. [DOI] [PubMed] [Google Scholar]
- 26.Easton DF, et al. Where are the prostate cancer genes?–A summary of eight genome wide searches. Prostate. 2003;57:261–269. doi: 10.1002/pros.10300. [DOI] [PubMed] [Google Scholar]
- 27.Varghese JS, et al. Genome-wide association studies in common cancers–what have we learnt? Curr. Opin. Genet. Dev. 2010;20:201–209. doi: 10.1016/j.gde.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Al Olama AA, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 29.Witte JS. Prostate cancer genomics: towards a new understanding. Nat. Rev. Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller MC, et al. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav. Brain Sci. 2006;29:385–404. doi: 10.1017/S0140525X06009095. discussion, 405–452. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher O, et al. Lifetime risks of common cancers among retinoblastoma survivors. J. Natl Cancer Inst. 2004;96:357–363. doi: 10.1093/jnci/djh058. [DOI] [PubMed] [Google Scholar]
- 32.Hwang SJ, et al. Lung cancer risk in germline p53 mutation carriers: association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Hum. Genet. 2003;113:238–243. doi: 10.1007/s00439-003-0968-7. [DOI] [PubMed] [Google Scholar]
- 33.You M. Fine mapping of chromosome 6q23-25 region in familial lung cancer families reveals RGS17 as a likely candidate gene. Clin. Cancer Res. 2009;15:2666–2674. doi: 10.1158/1078-0432.CCR-08-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung RJ, et al. Inherited predisposition of lung cancer: a hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol. Biomarkers Prev. 2007;16:2736–2744. doi: 10.1158/1055-9965.EPI-07-0494. [DOI] [PubMed] [Google Scholar]
- 35.Amos CI. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chanock SJ, et al. Genomics: when the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 37.Caporaso N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott KS. Evaluation of association of HNF1B variants with diverse cancers: collaborative analysis of data from 19 genome-wide association studies. PLoS One. 2010;5:e10858. doi: 10.1371/journal.pone.0010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Closas M, et al. Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin. Cancer Res. 2008;14:8000–8009. doi: 10.1158/1078-0432.CCR-08-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner NC, et al. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 43.Landi MT. A genome-wide association study of lung cancer identifies a region of chromosome 5p.15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 45.Kasper JS, et al. A meta-analysis of diabetes mellitus and the risk of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:2056–2062. doi: 10.1158/1055-9965.EPI-06-0410. [DOI] [PubMed] [Google Scholar]
- 46.Soranzo N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eeles RA. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voight BF. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanetsky PA, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat. Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rapley EA, et al. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 52.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 53.Shete S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wrensch M, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falchi M, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stacey SN, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baird DM. Variation at the TERT locus and predisposition for cancer. Expert. Rev. Mol. Med. 2010;12:e16. doi: 10.1017/S146239941000147X. [DOI] [PubMed] [Google Scholar]
- 60.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maller J, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 62.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visscher PM. Sizing up human height variation. Nat. Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 65.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iles MM. What can genome-wide association studies tell us about the genetics of common disease? PLoS Genet. 2008;4:e33. doi: 10.1371/journal.pgen.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greene CS, et al. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4:e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haiman CA, et al. Exploring genetic susceptibility to cancer in diverse populations. Curr. Opin. Genet. Dev. 2010;20:330–335. doi: 10.1016/j.gde.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ioannidis JP, et al. 'Racial' differences in genetic effects for complex diseases. Nat. Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 70.Ioannidis JP. Population-wide generalizability of genome-wide discovered associations. J. Natl Cancer Inst. 2009;101:1297–1299. doi: 10.1093/jnci/djp298. [DOI] [PubMed] [Google Scholar]
- 71.Waters KM, et al. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol. Biomarkers Prev. 2009;18:1285–1289. doi: 10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang BL. Validation of genome-wide prostate cancer associations in men of African descent. Cancer Epidemiol. Biomarkers Prev. 2010;20:23–32. doi: 10.1158/1055-9965.EPI-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rockhill B, et al. Use and misuse of population attributable fractions. Am. J. Public Health. 1998;88:15–19. doi: 10.2105/ajph.88.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Need AC, et al. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 75.Hunter DJ. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 76.Dempfle A, et al. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur. J. Hum. Genet. 2008;16:1164–1172. doi: 10.1038/ejhg.2008.106. [DOI] [PubMed] [Google Scholar]
- 77.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat. Rev. Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moore JH, et al. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445–455. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matise TC, et al. The next PAGE in understanding complex traits: study design for analysis of Population Architecture using Genetics and Epidemiology. Am. J. of Epidemiol. doi: 10.1093/aje/kwr160. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pendergrass SA, et al. Phenome-Wide Association Study (PheWAS) for Exploration of Novel Genotype-Phenotype Relationships and Pleiotropy Discovery. Genet. Epidemiol. doi: 10.1002/gepi.20589. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verhaak RG. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weir BA. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barrenas F, et al. Network properties of complex human disease genes identified through genome-wide association studies. PLoS One. 2009;4:e8090. doi: 10.1371/journal.pone.0008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.