Abstract
RNA polymerase I (Pol I) is dedicated to transcription of the large ribosomal DNA (rDNA). The mechanism of Pol I recruitment onto rDNA promoters is poorly understood. Here we present evidence that subunit A43 of Pol I interacts with transcription factor Rrn3: conditional mutations in A43 were found to disrupt the transcriptionally competent Pol I–Rrn3 complex, the two proteins formed a stable complex when co-expressed in Escherichia coli, overexpression of Rrn3 suppressed the mutant phenotype, and A43 and Rrn3 mutants showed synthetic lethality. Consistently, immunoelectron microscopy data showed that A43 and Rrn3 co-localize within the Pol I–Rrn3 complex. Rrn3 has several protein partners: a two-hybrid screen identified the C-terminus of subunit Rrn6 of the core factor as a Rrn3 contact, an interaction supported in vitro by affinity chromatography. Our results suggest that Rrn3 plays a central role in Pol I recruitment to rDNA promoters by bridging the enzyme to the core factor. The existence of mammalian orthologues of A43 and Rrn3 suggests evolutionary conservation of the molecular mechanisms underlying rDNA transcription in eukaryotes.
Keywords: A43 subunit/RNA polymerase I/Rrn3/Saccharomyces cerevisiae/transcription initiation
Introduction
In eukaryotes the large ribosomal RNAs (rRNAs) are synthesized by RNA polymerase I (Pol I) (Warner, 1989; Nomura et al., 1993). Pol I transcription accounts for ∼60% of the total transcriptional activity of exponentially growing yeast cells and the rRNA synthesis rate is tightly linked to the cellular growth rate (Warner, 1999). Pol I is the ultimate sensor of a complex series of molecular interactions that regulate rRNA gene activity.
Purified yeast Pol I contains 14 distinct subunits whose structural genes have been cloned (for a review see Carles and Riva, 1998). Like all eukaryotic forms of RNA polymerase, Pol I contains two large subunits, A190 and A135, homologous to the eubacterial β and β′ subunits, respectively, which harbour the active site of the enzyme taken in a broad sense (Riva et al., 1987). Among the Pol I subunits, four polypeptides (A49, A43, A34.5 and A14) are specific to the enzyme as they have no counterparts in Pol II or Pol III. While A49, A34.5 and A14 are non-essential (Liljelund et al., 1992; Smid et al., 1995; Gadal et al., 1997), A43 is strictly required for cell viability and rRNA synthesis in vivo (Thuriaux et al., 1995). Until now no information has been available on the nature of the essential function of A43.
Yeast Pol I requires four general transcription factors for initiation of rDNA transcription (Nomura, 1998). Two multimeric complexes (UAF and CF) and the TATA-binding protein (TBP) are assembled onto the rDNA promoter to form the pre-initiation complex. The core factor (CF) and the upstream activating factor (UAF) bind to the core element and to the upstream element, respectively. The CF comprises three subunits (Rrn6, Rrn7 and Rrn11), which are all essential in vivo (Keys et al., 1994). UAF contains histones H3 and H4 and three non-essential subunits, Rrn5, Rrn9 and Rrn10 (Keener et al., 1997). In vitro, CF is strictly required for basal rDNA transcription whereas UAF is not, but facilitates the recruitment of CF in conjunction with TBP (Keys et al., 1996). TBP, which interacts with both CF (via Rrn6) and UAF (via Rrn9) (Lin et al., 1996; Steffan et al., 1996), appears to be necessary only for the UAF-dependent recruitment of CF (Steffan et al., 1996, 1998; Keener et al., 1998). A schematic model of assembly of the pre-initiation complex, based on known interactions between TBP and the subunits of UAF and CF, was proposed by Nomura and colleagues (Steffan et al., 1996).
In addition to CF, UAF and TBP, initiation of transcription by Pol I requires the transcription factor Rrn3 (Yamamoto et al., 1996; Keener et al., 1998; Milkereit and Tschochner, 1998), which is functionally conserved between yeast and human (Moorefield et al., 2000). Two populations of Rrn3 were found in yeast extracts: a major form of free factor and a minor one associated with Pol I, which is the initiation-competent form of the enzyme in vitro (Milkereit et al., 1997). The Pol I subunits responsible for Rrn3 binding are unknown. Interestingly, Rrn3 dissociates from Pol I during the transcription cycle and cannot subsequently reassociate with the enzyme in vitro (Milkereit and Tschochner, 1998).
Murine rDNA transcription has also been extensively studied (Grummt, 1998). Efficient transcription initiation requires Pol I and four transcription factors: TIF-IA, TIF-IB, TIF-IC and UBF. TIF-IB (corresponding to the human factor SL1) contains TBP (Eberhard et al., 1993) and functions analogously to the yeast CF (Clos et al., 1986; Schnapp et al., 1990a; Eberhard et al., 1993): TIF-IB/SL1 is essential for transcription initiation in vitro, binds to the core promoter element and recruits Pol I. UBF stimulates transcription initiation and therefore appears to be functionally homologous to yeast UAF, even if these proteins are structurally unrelated (Bell et al., 1988; Jantzen et al., 1992; Kuhn and Grummt, 1992; Hempel et al., 1996). UBF may also participate in Pol I recruitment by contacting the polymerase-associated factor PAF53, a murine homologue of the yeast A49 subunit (Muramatsu et al., 1998). Efficient initiation of transcription by murine Pol I requires two additional factors (TIF-IA and TIF-IC), both associated with Pol I (Schnapp et al., 1990b, 1994).
Interestingly, yeast Rrn3 and murine TIF-IA share several properties. They are required for transcription initiation by Pol I (Schnapp et al., 1993; Yamamoto et al., 1996), are associated with Pol I and dissociate from the enzyme during transcription (Schnapp et al., 1993; Brun et al., 1994; Milkereit and Tschochner, 1998) and are growth regulated (Grummt, 1998; Milkereit and Tschochner, 1998). Finally, Rrn3 and TIF-IA are monomeric proteins of ∼75 kDa. Recent data indeed demonstrate that TIF-IA is the mouse homologue of yeast Rrn3 (Bodem et al., 2000).
While extensive studies have focused on the characterization and formation of the pre-initiation complex, the Pol I recruitment step itself remains unclear. An important question is to identify the factors within the pre-initiation complex that recruit Pol I and the enzyme subunit(s) involved.
In this work we show that the Pol I subunit A43 is a major determinant for the formation and/or stabilization of the Pol I–Rrn3 complex. Subunit A43 and Rrn3 were found to co-localize on the RNA polymerase. Thus, A43 is the first Pol I subunit shown to interact directly with a class I general transcription factor. We also present evidence in favour of an interaction between Rrn3 and Rrn6, a subunit of the CF. We propose that Rrn3 plays a central role in Pol I recruitment by bridging Pol I to the CF. The conservation of A43 and Rrn3 in higher eukaryotes suggests that the molecular mechanisms of Pol I recruitment on the rDNA promoter have been evolutionarily conserved from yeast to human.
Results
Mutational analysis of RPA43
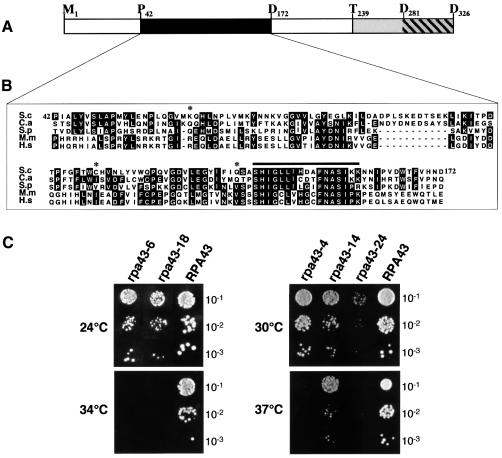
A43 (encoded by the RPA43 gene) is the only Pol I-specific subunit that is essential for cell viability. While no structural homologues of RPA43 were found in the genomes of eubacteria or archaebacteria, putative orthologues of A43 are present in Schizosaccharomyces pombe, Candida albicans and higher eukaryotes, suggesting a conserved role for this subunit in the mechanism of rDNA transcription. A region of strong homology was restricted to the central part of the Saccharomyces cerevisiae protein (residues P42–D172; see Figure 1A and B). Within this region, 23% identity and 58% similarity were found between the S.cerevisiae and the human sequences. A highly conserved motif of 15 residues was present in the five sequences analysed (Figure 1B). To characterize the role of A43, we generated mutants by random mutagenesis. Five rpa43 alleles that conferred variable thermosensitive growth phenotypes were isolated (Figure 1C). Interestingly, the three point mutations of the rpa43-6 allele (K63E, C118R and Q140R) were localized in the conserved domain. The rpa43-14 allele encoded a truncated polypeptide lacking the 79 C-terminal residues, which were replaced by a divergent amino acid sequence of nine residues, indicating that the highly acidic C-terminus of A43 was not required for its essential function.
Fig. 1. Analysis of RPA43 mutants. (A) Schematic representation of the A43 subunit of S.cerevisiae. The black box represents the domain conserved in higher eukaryotes (see B). The grey box corresponds to the non-essential C-terminal part absent in the mutant protein encoded by the rpa43-14 allele (see C) and the hatched box to the hyperacidic C-terminal domain. (B) Sequence alignment of the conserved domain between putative homologues of the A43 subunit. S.c., Saccharomyces cerevisiae; C.a., Candida albicans; S.p., Schizosaccharomyces pombe; M.m., Mus musculus; H.s., Homo sapiens. Black boxes indicate the residues identical in at least three sequences. The black line localizes a 15 residue motif highly conserved from yeast to human. The asterisks indicate the three residues mutated in the protein encoded by the rpa43-6 allele. (C) Five rpa43 mutant alleles (rpa43-4, 6, 14, 18 and 24) borne on a centromeric plasmid were used to complement a chromosomal disruption of RPA43. Growth on YPD medium at different temperatures of serial dilutions of the resulting strains was analysed.
Pol I containing a mutated A43 subunit is impaired in specific transcription
The availability of conditional mutants prompted us to correlate their phenotype with a transcriptional defect in vitro. The rpa43-6 mutant was chosen for this biochemical investigation because growth of this strain was affected at all temperatures tested and the three point mutations found in A43-6 were all located in the conserved domain of the protein.
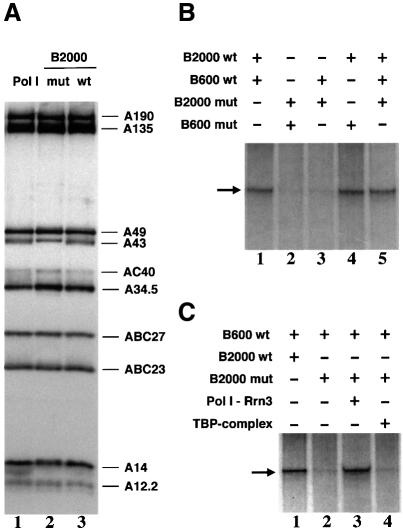
Since A43 is not required for core Pol I activity (Lanzendörfer et al., 1997), we investigated whether this subunit was required for in vitro specific transcription of rDNA. The B2000 fraction, which contains the initiation-competent form of Pol I required for this assay (Milkereit and Tschochner, 1998), was purified from the rpa43-6 mutant (B2000mut) and from wild-type isogenic cells (B2000wt) grown at 24°C. The subunit composition of Pol I present in the B2000mut and B2000wt fractions was determined by western blot analysis using anti-Pol I antibodies and was found to be indistinguishable (Figure 2A, lanes 2 and 3). Activity of Pol I present in the B2000mut and B2000wt fractions was found to be identical in a non-specific in vitro transcription assay (data not shown).
Fig. 2. A43-6 containing Pol I is inactive in specific transcription in vitro. (A) Subunit composition of Pol I contained in the B2000 fraction prepared from the rpa43-6 mutant (mut) and from a wild-type (wt) strain was analysed by western blotting using anti-Pol I antibodies. Purified RNA Pol I was used as a positive control. (B) Specific in vitro transcription of a mini 26S transcription unit was carried out with the B600 and B2000 fractions purified from the rpa43-6 mutant (mut) or a wild-type (wt) strain. B2000 fractions were normalized by their activity in a non-specific in vitro transcription assay. RNA was analysed by urea–PAGE followed by autoradiography. (C) Pol I–Rrn3 and TBP-containing complexes, purified from a wild-type strain, were added to the B2000mut fraction and specific in vitro transcription was performed as in (B).
We next investigated whether the B2000mut fraction was competent for in vitro specific transcription of an rDNA gene. In addition to the B2000 fraction, specific in vitro transcription of rDNA requires the B600 fraction (Milkereit and Tschochner, 1998). Whereas the B600wt and B2000wt fractions governed accurate transcription of a mini rDNA gene (Figure 2B, lane 1), no specific transcript was synthesized using the B600mut and B2000mut fractions (Figure 2B, lane 2). Combination of the B2000mut and B600wt fractions still did not support transcription of the rDNA template (Figure 2B, lane 3). On the other hand, the B600mut fraction showed rDNA transcription activity when mixed with the B2000wt fraction (Figure 2B, lane 4). Finally, addition of the B2000mut fraction to an active transcription system reconstituted with the B600wt and B2000wt fractions did not inhibit specific transcription (Figure 2B, lane 5). Thus, the B2000mut fraction was not functional although it did not contain an inhibitor of rDNA transcription. On the other hand, the B600mut fraction was as active as B600wt in supporting specific transcription of rDNA.
Two complexes required for specific in vitro transcription of rDNA are present in the B2000 fraction: the Pol I–Rrn3 complex and a large TBP-containing complex, which can be separated by gel filtration (Milkereit and Tschochner, 1998). We analysed which of these two complexes purified from a wild-type strain could restore the transcriptional activity of the B2000mut fraction. As shown in Figure 2C, efficient rDNA transcription could be achieved when the B2000mut fraction was complemented with the wild-type Rrn3–Pol I complex (Figure 2C, lane 3), whereas addition of the TBP-containing complex did not complement the B2000mut fraction (Figure 2C, lane 4). This result demonstrated that the Pol I–Rrn3 complex was the unique component that was inactive in the B2000mut fraction.
Altogether, our data indicate that Pol I containing the mutant A43-6 subunit was impaired in specific transcription of rDNA in vitro. Because Pol I present in B2000mut and B2000wt displayed indistinguishable subunit compositions, the specific functional defect of the Pol I–Rrn3 complex was likely to be due to the mutations present in A43.
The A43 subunit interacts with Rrn3
The defect in the mutant A43 subunit could result in inactivation or dissociation of the Pol I–Rrn3 complex. In the wild-type B2000 fraction the Rrn3 molecules are all engaged within the Pol I–Rrn3 complex (Milkereit and Tschochner, 1998). To compare the amount of assembled Pol I–Rrn3 complex in the B2000wt and B2000mut fractions we searched for the presence of Rrn3 in these two fractions by western blot analysis using anti-Rrn3 antibodies. As shown in Figure 3A, Rrn3 was detected in the B2000wt fraction but not in the B2000mut fraction (Figure 3A, compare lanes 3 and 4). Therefore, the B2000mut fraction was devoid of Pol I–Rrn3 complex. The absence of Rrn3 in the B2000mut fraction was not due to a lesser amount of Pol I since the amounts of B2000wt and B2000mut fractions used in this experiment were normalized to contain the same amount of Pol I (Figure 3A, lanes 3 and 4). Rrn3 was not limiting in the mutant cells since equivalent amounts of free Rrn3 were detected in the B600wt and B600mut fractions (Figure 3A, lanes 5 and 6). Altogether, these data demonstrate that the inability of the B2000mut fraction to support rDNA transcription was due to absence of the Pol I–Rrn3 complex and thus strongly suggest that the A43 subunit plays a critical role in formation and/or stabilization of the Pol I–Rrn3 complex.
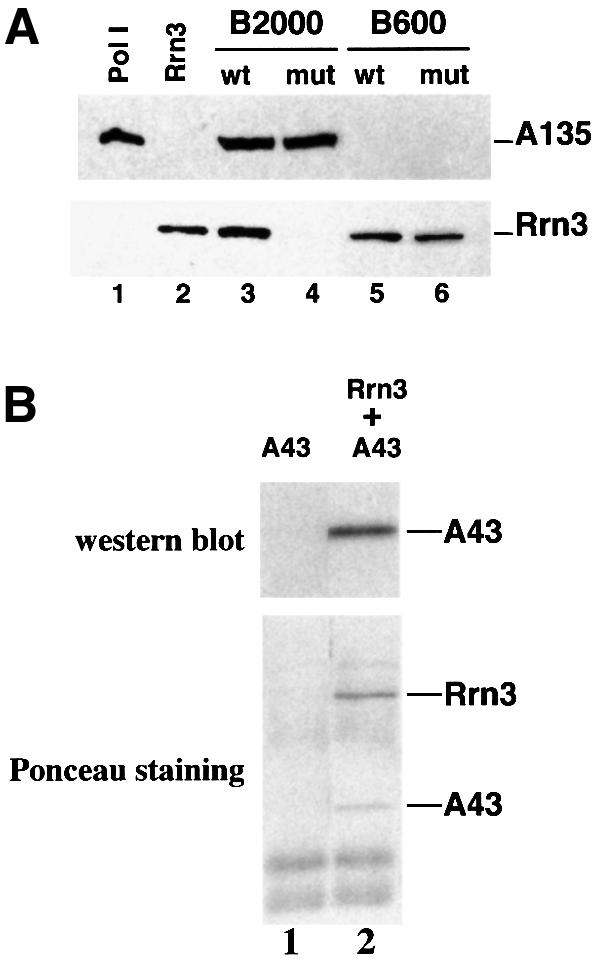
Fig. 3. Subunit A43 is an interaction target of Rrn3. (A) The Pol I–Rrn3 complex is unstable in the rpa43-6 mutant. The B2000 and B600 fractions purified from the rpa43-6 mutant (mut) and a wild-type (wt) strain were analysed for their Pol I and Rrn3 content by western blotting using antibodies directed to the A135 Pol I subunit or to Rrn3. The B2000 fractions were normalized by their activity in a non-specific in vitro transcription assay and identical amounts of proteins of the wild-type and mutant B600 fractions were used. Purified Pol I and Rrn3 are indicated. (B) Subunit A43 and Rrn3 form a stable binary complex. Escherichia coli extracts from cells expressing either subunit A43 (lane 1) or HA-(His)6-Rrn3 and subunit A43 (lane 2) were loaded onto a protein G–Sepharose column coated with a monoclonal antibody raised against the HA epitope. Proteins bound to the column were eluted by competition with the HA peptide. Eluted proteins were subjected to SDS–PAGE and the presence of A43 subunit was analysed by western blotting using polyclonal anti-A43 antibodies and by Ponceau Red staining.
We next checked for a direct interaction between A43 and Rrn3. A43 and HA-His6-Rrn3 were co-expressed in Escherichia coli and crude cell extract was loaded onto a protein G–Sepharose column coated with a monoclonal antibody raised against the HA epitope. Tagged Rrn3 and associated proteins, eluted by competition with the HA peptide, were analysed by immunodetection with polyclonal anti-A43 antibodies following SDS–PAGE. As shown in Figure 3B (lane 2), A43 was detected in the eluted fraction. A43 polypeptide and tagged Rrn3 were eluted in approximately stoichiometric amounts as judged by Ponceau Red staining of the proteins on the membrane (Figure 3B, lane 2). The band corresponding to tagged Rrn3 (Figure 3B, lane 2) was identified by immunodetection with monoclonal anti-HA antibodies following SDS–PAGE (data not shown). The two protein bands corresponding to A43 and tagged Rrn3 were not found in the elution fraction from the bacterial extract containing only A43 (Figure 3B, lane 1). These results demonstrate a direct interaction between Rrn3 and the A43 subunit and indicate that the two proteins are able to form a stable binary complex.
Genetic interactions between RPA43 and RRN3
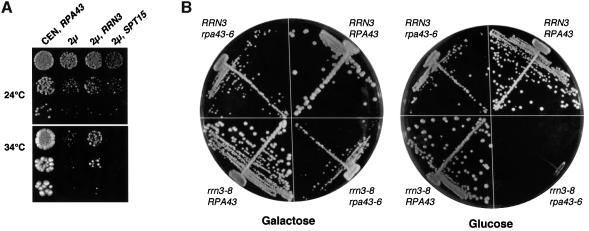
To investigate whether overexpression of Rrn3 could correct the growth defect of the rpa43 mutants, these strains were transformed with a high copy number plasmid carrying or not carrying RRN3. As shown in Figure 4A, rpa43-6 mutant cells transformed with a plasmid harbouring RRN3 grew at the restrictive temperature of 34°C, albeit somewhat more slowly than the wild-type strain. The thermosensitive phenotype was recovered upon chase of the plasmid carrying RRN3 on 5-fluoroorotic acid-containing plates (data not shown). In addition, overexpression of Rrn3 induced a very weak suppression of the rpa43-18 mutant phenotype (data not shown). None of the mutant strains were rescued by overexpression of SPT15, encoding TBP (see Figure 4A). These data suggest that the thermosensitivity of the rpa43-6 mutant resulted, at least partially, from a specific defect in in vivo interaction of the A43 subunit with Rrn3.
Fig. 4. In vivo interactions between subunit A43 and Rrn3. (A) The rpa43-6 mutant strain was transformed with a 2µ plasmid carrying either the RRN3 or SPT15 gene (encoding TBP) or with a control vector. Growth of serial dilutions of the transformants at the permissive (24°C) and non-permissive (34°C) temperatures was monitored. (B) A centromeric plasmid carrying either RPA43 or rpa43-6 was introduced into GPy44 (rrn3-8, rpa43::LEU2, pNOY102) or D128 cells (RRN3, rpa43::LEU2, pNOY102). Growth of the double mutant strain (rrn3-8 rpa43-6), of the single mutant strains (rrn3-8 RPA43 and RRN3 rpa43-6) and of the wild-type strain was examined on galactose and glucose medium at 24°C.
Synthetic lethality is a powerful genetic tool that has been used to confirm or to reveal protein–protein interactions (Doye and Hurt, 1995). We therefore investigated whether the rrn3-8 mutant allele (Cadwell et al., 1997) was genetically interacting with rpa43-6. We constructed a yeast strain harbouring the chromosomal rrn3-8 mutant allele and the rpa43-6 gene borne on a centromeric plasmid. To ensure that rRNA could be synthesized independently of Pol I, this strain was also transformed with plasmid pNOY102, which harbours an rDNA gene under the control of the Pol II galactose-inducible GAL7 promoter (Nogi et al., 1991). We analysed whether galactose-dependent synthesis of 35S rRNA by Pol II was dispensable for growth of the rpa43-6 rrn3-8 double mutant. As shown in Figure 4B, this strain was unable to grow on glucose medium at 24°C but grew on galactose medium, whereas at the same temperature each single mutant strain, RRN3 rpa43-6 and rrn3-8 RPA43, grew on glucose and galactose medium. Thus, rpa43-6 and rrn3-8 are synthetic lethal and this co-lethality is restricted to a defect in rDNA transcription. Altogether, these genetic links between A43 and Rrn3 strongly confirm the importance of the A43–Rrn3 interaction in vivo.
The A43 subunit and Rrn3 co-localize within the Pol I–Rrn3 complex
Location of A43 and Rrn3 within the Pol I–Rrn3 complex was analysed by immunoelectron microscopy. Briefly, purified Pol I and Pol I–Rrn3 complex were decorated with polyclonal antibodies raised against subunit A43 (Buhler et al., 1980) or Rrn3 (Milkereit and Tschochner, 1998), respectively. The binding sites of the antibody could be identified after comparison of the images of the decorated and the non-decorated enzyme, allowing localization of the antibodies on a previously determined three-dimensional enzyme model (Klinger et al., 1996).
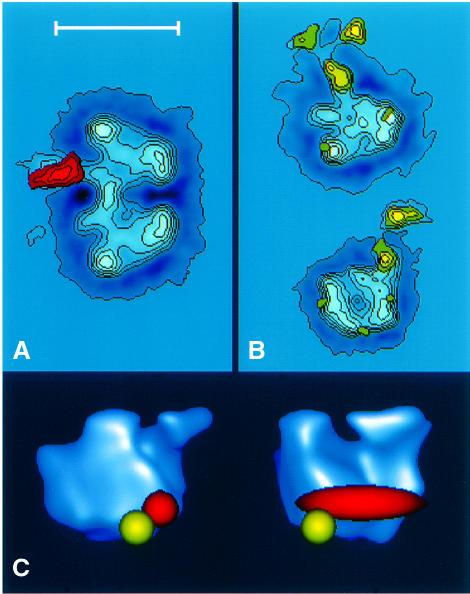
Purified Pol I adopts two oligomeric states in solution, monomers and dimers (Milkereit et al., 1997). Image analysis of the dimeric Pol I labelled with A43-specific polyclonal antibodies resulted in an averaged image in which the binding site of the probe mapped close to the dimerization interface, in a region situated at the end of a groove leading to a channel. Because the interaction site could be determined only from a single orientation of the enzyme, the localization of the A43 subunit was not resolved in the viewing direction (Figure 5A).
Fig. 5. Immunolocalization of subunit A43 and of the interaction site of Rrn3 in the Pol I three-dimensional structure. (A) Image analysis of RNA Pol I dimers labelled with anti-A43 polyclonal antibodies obtained upon analysis of 186 molecular images. The stain-excluding protein densities are outlined by contours of equal density. The difference image between the labelled and unlabelled molecules is shown in red where the contours correspond to positive differences at a threshold significance level of 5%. (B) Image analysis of RNA Pol I monomers labelled with anti-Rrn3 polyclonal antibodies. A total of 836 labelled RNA polymerase monomers was selected out of the original micrographs and 50.3% of the molecular images corresponded to two characteristic views: the –90° view, where the groove is located in the left part and the channel region in the lower left (upper); and the 0° view, where the groove is facing the observer and the channel region maps in the lower part (lower). The stain-excluding protein densities are outlined by contours of equal density. The significant differences between the labelled and unlabelled molecules are shown in yellow. (C) Surface representation of the three-dimensional RNA Pol I model showing the position of subunit A43 (red) and the interaction site of Rrn3 (yellow). The bar represents 20 nm in (A) and (B) and 12 nm in (C).
Pol I associated with Rrn3 is exclusively monomeric (Milkereit and Tschochner, 1998). Electron micrographs of antibody-labelled RNA polymerase corresponded to two well-defined molecular views. In each view the antibodies interacted with a stalk of protein density located close to the end of the groove (Figure 5B).
When projected on the three-dimensional enzyme model (Schultz et al., 1993), A43 and Rrn3 localized in close proximity at the end of a 3 nm wide groove opposite a finger-like structure that defined a channel (Figure 5C). These data therefore indicate that the A43 subunit and the Rrn3 factor are in close proximity within the Pol I–Rrn3 complex, confirming the proposal that A43 is an interaction target of Rrn3 within Pol I.
Physical interaction of Rrn3 with the core factor subunit Rrn6
The biochemical, genetic and structural studies described above revealed that one function of subunit A43 in Pol I is to participate in Pol I–Rrn3 complex formation. How the Pol I–Rrn3 complex is recruited to the promoter by the CF has still not been elucidated. One simple hypothesis is that the Pol I–Rrn3 complex interacts with one or several components of the CF or with a putative mediator linking the two complexes together. To address this question, a two-hybrid screen using Rrn3 as bait was set up. Briefly, tester strain Y190, which contains two reporter genes, lacZ and HIS3, was transformed with a plasmid harbouring a gene encoding the fusion protein Rrn3–Gal4 DNA binding domain (Rrn3–DBD) and with a genomic library containing randomly sheared DNA fragments fused to the sequence coding for the activation domain (AD) of Gal4 (Fromont-Racine et al., 1997). Twelve transformants for which activation of both reporter genes was observed were selected and the preys were identified by sequencing. They corresponded to 11 different proteins. Remarkably, one prey corresponded to a C-terminal fragment of Rrn6 (Rrn6[865–894]), one of the three subunits of the CF. We obtained a stronger activation of the two reporter genes with a larger C-terminal fragment of Rrn6 (hereafter named fragment C1), encompassing the last 123 residues of the protein (Rrn6[772–894]) (Figure 6A). These results prompted us to investigate the interaction between Rrn3 and full-length Rrn6. Indeed, when tester strain Y190 was transformed with two plasmids driving, respectively, expression of Rrn3–DBD and Rrn6–AD fusion proteins, both the lacZ and HIS3 reporter genes were activated. In all cases no activation was detected when a single fusion protein was expressed in strain Y190 (Figure 6A). Altogether, these results suggest that Rrn3 interacts with Rrn6 in vivo and that the C-terminal domain of Rrn6 is sufficient for this interaction.
Fig. 6. Rrn3 interacts with Rrn6. (A) Two-hybrid interactions between Rrn3 and Rrn6. Strain Y190 was transformed with two plasmids, one allowing expression of Rrn3 fused to the Gal4 DBD, the other expression of Rrn6, Rrn6[772–894] or Rrn6[865–897] fused to the Gal4 AD. Activation of the lacZ and HIS3 reporter genes was monitored by staining the cells in the presence of XGal or replicating the cells on 3AT-containing medium, respectively. (B) The C-terminal domain of Rrn6 binds to immobilized Rrn3. Escherichia coli extracts containing (+) or not (–) recombinant HA-tagged Rrn3 were loaded onto protein G–Sepharose columns coated with 12CA5 anti-HA antibodies, then in vitro synthesized [35S]Rrn6[C1] (Load) were applied to the columns. Proteins eluted by competition with purified HA-peptide were subjected to SDS–PAGE and analysed by autoradiography. (C) Full-length Rrn3 binds to the immobilized C-terminal domain of Rrn6. Escherichia coli extracts containing GST (–) or GST–Rrn6[772–894] fusion protein (+) were bound to glutathione–Sepharose columns. After extensive washing, partially purified recombinant HA-tagged Rrn3 (Load) was chromatographed onto the columns. Proteins eluted with glutathione were subjected to SDS–PAGE and the presence of HA-Rrn3 analysed by western blotting using monoclonal anti-HA antibodies.
Next, the interaction between the two polypeptides was analysed in vitro using affinity chromatography. Escherichia coli extracts containing or not recombin ant HA-His6-Rrn3 protein were bound to a protein G–Sepharose column coated with a monoclonal antibody raised against the HA epitope (Figure 6B). The 35S-labelled C-terminal fragment of Rrn6 (fragment C1) was then applied to the columns. Rrn3 and the proteins interacting with Rrn3 were eluted by competition with the HA peptide and analysed by SDS–PAGE. As shown in Figure 6B, the labelled C1 polypeptide was specifically recovered from the column of immobilized Rrn3. In a complementary experiment, E.coli cell extracts containing glutathione S-transferase (GST) or C1–GST fusion protein were loaded onto a glutathione–Sepharose column. Partially purified HA-tagged Rrn3 was then applied to the columns. Proteins were eluted by competition with glutathione and recombinant Rrn3 was detected by western blotting with anti-HA antibodies following SDS–PAGE. As shown in Figure 6C, recombinant Rrn3 bound specifically to the column of immobilized C1 fusion protein. These data support a direct interaction between Rrn3 and the C-terminus of Rrn6.
Finally, we investigated whether the C-terminus of Rrn6, which is sufficient in vitro to interact with Rrn3, was essential in vivo. To analyse the viability of haploid cells harbouring the rrn6-Δ1 mutant allele that codes for a truncated version of Rrn6 lacking the C1 fragment, a heterozygous rrn6-Δ1/RRN6 diploid strain was constructed. Tetrads resulting from sporulation of this strain were dissected. All the tetrads examined produced two viable spores, none of which contained the rrn6-Δ1 allele (data not shown). This result demonstrates that the C-terminal C1 fragment of Rrn6 is essential for cell viability and suggests that this domain is involved in vivo in a critical step of rDNA transcription.
Discussion
Data presented in this paper strongly suggest that the role of the Pol I A43 subunit in the formation of a transcriptionally competent Pol I–Rrn3 complex accounts for its essential character in vivo. We suggest that Rrn3 participates in recruitment of the Pol I–Rrn3 complex onto the pre-initiation complex by simultaneously contacting the enzyme through the A43 subunit and the CF bound to the promoter via the Rrn6 subunit.
The Pol I–Rrn3–CF connection
Previous results obtained in our laboratory demonstrated that the essential A43 subunit is not required for the core catalytic activity of Pol I (Lanzendörfer et al., 1997). In order to unravel the function of A43, we examined the properties of Pol I partially purified from yeast cells expressing the rpa43-6 allele. This enzyme was unable to transcribe rDNA specifically in vitro. Because the subunit composition and the basal catalytic activity of the mutant enzyme were indistinguishable from those of the wild type, the transcription defect directly correlated with the mutations in subunit A43. Biochemical analyses indicated the absence of detectable amounts of Pol I–Rrn3 complex in the B2000mut fraction, indicating that the mutations introduced in A43 impaired stability of the complex. A direct interaction between recombinant Rrn3 and A43 could be demonstrated, underscoring the direct role of the A43 subunit in Pol I–Rrn3 complex formation.
These results were further supported in vivo by genetic data. Overexpression of Rrn3 suppressed the phenotype conferred by the rpa43-6 allele, probably by driving the equilibrium towards formation of the Pol I–Rrn3 complex, and the rpa43-6 and rrn3-8 mutant alleles are synthetic lethal.
A direct interaction between subunit A43 and Rrn3 in the Pol I–Rrn3 complex was confirmed by immunoelectron microscopy. The A43 subunit and Rrn3 were found to co-localize on the three-dimensional envelope of Pol I. The position of A43 at the dimerization interface of Pol I may explain why Rrn3 is found exclusively associated with monomeric Pol I, whereas the free enzyme is found in two oligomeric states (monomers and dimers) (Milkereit et al., 1997). The interaction of Rrn3 with subunit A43 may dissociate the enzyme dimers or alternatively impede their formation. These observations reinforce the idea, previously suggested (Milkereit et al., 1997), that a Pol I monomer–dimer equilibrium might be one parameter regulating rDNA transcription, as in the case of TBP activity, which is regulated by its monomeric/dimeric state (Jackson-Fisher et al., 1999). Altogether, our results indicate that a major function of subunit A43 within Pol I is to contact Rrn3 directly to form the Pol I–Rrn3 competent complex.
Nomura and co-workers demonstrated that three general transcription factors, UAF, CF and TBP, are assembled onto the rDNA promoter to form a stable pre-initiation complex. In the present work we show that the Rrn6 subunit of CF interacts directly with Rrn3, strongly suggesting that the Rrn3–Rrn6 interaction belongs to an interaction network leading to recruitment of the Pol I–Rrn3 complex to the promoter. Our data underscore the importance of the C-terminal domain of Rrn6 for this interaction. This conclusion is strengthened by the lethal phenotype of deletion of the 123 C-terminal residues of Rrn6. It has previously been shown that Rrn6 simultaneously contacts the Rrn11 subunit of CF and TBP within the pre-initiation complex, likely to allow UAF-induced activation (Steffan et al., 1996). The observation that the N-terminus of Rrn6 interacts with Rrn11 (Lin et al., 1996), whereas its C-terminus contacts Rrn3, suggests the existence of discrete functional domains within Rrn6.
Altogether, our results underscore the essential role of Rrn3 in rDNA transcription initiation by directly linking Pol I to CF at least via the A43–Rrn3–Rrn6 interaction. This conclusion is consistent with previous observations showing that CF alone is able to drive in vitro basal transcription of rDNA in the presence of the Pol I–Rrn3 complex (Keys et al., 1996). One cannot exclude the possibility that other components of the pre-initiation complex may also interact with and stabilize the Pol I–Rrn3 complex within the initiation complex.
A model of Pol I recruitment and regulation
Data presented in this paper and by others (Steffan et al., 1996; Keener et al., 1998; Milkereit and Tschochner, 1998) suggest that activation of Pol I by Rrn3 might be a three step process: (i) Rrn3 interacts with Pol I subunit A43, converting inactive Pol I monomers or dimers into competent Pol I–Rrn3 complexes (other subunits of the enzyme may participate in this step); (ii) Rrn3 triggers recruitment of the Pol I–Rrn3 complex by contacting the CF within the pre-initiation complex (via at least the Rrn6 subunit); and (iii) as in the case of the Pol III–TFIIIB interaction (Brun et al., 1997; Kassavetis et al., 1998), there is likely to be a post-recruitment step leading to open complex formation and initiation by Pol I. These three steps leading to initiation of rDNA transcription are possible regulatory targets to tune the cell growth rate but the simplest model is that regulation of Pol I transcription is achieved at the level of formation of the Pol I–Rrn3 complex. Transcription of the rDNA unit is a highly regulated event. In S.cerevisiae, Acanthamoeba castellanii and murine cells transcription of the rDNA precursor gradually decreases when cells enter stationary phase (Tower and Sollner-Webb, 1987; Schnapp et al., 1993; Clarke et al., 1996). Biochemical studies demonstrated that this down-regulation is caused by disappearance of the active, initiation-competent form of Pol I (named Pol I-i in yeast, Pol A in A.castellanii and Factor C in mouse) (Paule et al., 1984; Bateman and Paule, 1986; Tower and Sollner-Webb, 1987; Riggs et al., 1995; Milkereit and Tschochner, 1998). In yeast, transcriptionally inactive extracts from stationary phase cells contain substantial amounts of Rrn3 and Pol I, but lack the Pol I–Rrn3 complex (Milkereit and Tschochner, 1998). Therefore, formation and disruption of the Pol I–Rrn3 complex appear to constitute a molecular switch for regulating rRNA synthesis. A post-translational modification of Rrn3 and/or A43 orthologues may be involved in this regulation, which implies the participation of regulatory factors that remain to be identified. The stable interaction of subunit A43 with Rrn3 using E.coli recombinant proteins suggests that phosphorylation of A43, shown in vivo in yeast (Bréant et al., 1983), is not required for this interaction.
Despite a similar configuration of the rDNA promoters in yeast and higher eukaryotes, the factors involved in pre-initiation complex formation are globally unrelated. UBF, found in different mammalian systems, has no structural homologue in yeast and no structural homologues of yeast UAF or CF subunits have been found so far in the genomes of higher eukaryotes. In addition, mammalian systems display functional differences: TIF-IB/SL1, a multimeric complex containing TBP as a bona fide subunit, is strictly required in higher eukaryotes for rDNA transcription (Zomerdijk and Tjian, 1998), whereas in yeast TBP interacts with both UAF and CF but is not a subunit of these two complexes and is not required for in vitro basal transcription of rDNA (Steffan et al., 1996). Given these differences, it is remarkable that the A43 subunit is conserved from yeast to human (see Figure 1B). Importantly, a recent paper demonstrates that human Rrn3 can functionally replace yeast Rrn3 in S.cerevisiae cells (Moorefield et al., 2000), showing that this factor has been structurally and functionally conserved through evolution. Recent data demonstrate that the murine factor TIF-IA corresponds to Rrn3 (Bodem et al., 2000). The conservation of both A43 and Rrn3 through evolution leads us to speculate that the mechanisms of Pol I recruitment on the rDNA promoter are conserved in higher eukaryotes.
Materials and methods
Plasmids, strains and media
Plasmids and yeast strains used in this study are listed in Tables I and II, respectively. Standard yeast genetic techniques and media were used (Sherman et al., 1979).
Table I. Plasmids used in this study.
| Plasmid | Description | Vector | Reference/donor |
|---|---|---|---|
| Yeast plasmids | |||
| pGP5 | CEN, TRP1, RPA43 | pRS314 | this study |
| pGP5-4 | CEN, TRP1, rpa43–4 | pRS314 | this study |
| pGP5-6 | CEN, TRP1, rpa43–6 | pRS314 | this study |
| pGP5-14 | CEN, TRP1, rpa43–14 | pRS314 | this study |
| pGP5-18 | CEN, TRP1, rpa43–18 | pRS314 | this study |
| pGP5-24 | CEN, TRP1, rpa43–24 | pRS314 | this study |
| pGP32 | 2µ, LEU2, GAL4[768–881]-RRN6[865–894] | pACT2 | this study |
| pGP40 | 2µ, LEU2, GAL4[768–881]RRN6[772–894] | pACT2 | this study |
| pGP44 | 2µ, LEU2, GAL4[768–881]RRN6 | pACT2 | this study |
| pAS-RRN3 | 2µ, TPR1, GAL4[1-147]-RRN3 | pASΔ | J.Steffan |
| yCPA43 | CEN, URA3, RPA43 | yCP | Thuriaux et al. (1995) |
| pSYCYES2 | 2µ, URA3, RRN3 | pSYC | Cadwell et al. (1997) |
| pL1 | 2µ, URA3, SPT15 | pFL44 | Cavallini et al. (1989) |
| pNOY102 | 2µ, URA3, RDN under control of the GAL7 promoter | pC1/1 | Nogi et al. (1991) |
| pSIRT | mini 26S | Ye30-Δ6 | Musters et al. (1989) |
| Plasmids for expression in E.coli | |||
| pGP4 | HA-HIS-RPA43 under control of the T7 promoter | pRSET5d | this study |
| pGP34 | HA-HIS-RRN3 under control of the T7 promoter | pRSET5d | this study |
| pGP37 | RRN6[772–894] under control of the T7 promoter | pRSET5d | this study |
| pGP45 | A43 under control of the T7 promoter | pACYC184-11b | this study |
| pGP47 | GST-RRN6[772–894] under control of the T7 promoter | pGEX3X | this study |
Table II. Strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| GPy9 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pGP5 | this study |
| GPy11-4 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pGP5-4 | this study |
| GPy11-6 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pGP5-6 | this study |
| GPy11-14 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pGP5-14 | this study |
| GPy11-18 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pGP5-18 | his study |
| GPy11-24 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pGP5-24 | this study |
| GPy21 | MATα rpa43::LEU2 rrn3-8 ura3-52 trp1 leu2 his3Δ200 his7Δ2 ade/yCPA43 | this study |
| GPy43 | MATα rpa43::LEU2 rrn3-8 ura3-52 trp1 leu2 his3Δ200 his7Δ2 ade/pNOY200 | this study |
| GPy44 | MATα rpa43::LEU2 rrn3-8 ura3-52 trp1 leu2 his3Δ200 his7Δ2 ade/pNOY102 | this study |
| GPy52 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1 rrn6-Δ1 | this study |
| MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1 RRN6 | ||
| D101-I2 | MATa rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/yCPA43 | Thuriaux et al. (1995) |
| D128 | MATα rpa43::LEU2 ade2-101 ura3-52 lys2-801 trp1-Δ63 his3Δ200 leu2-Δ1/pNOY102 | Thuriaux et al. (1995) |
| yCC95 | MATα rrn3-8 ade5 ura3-52 trp1-289 his7-2 leu2-112 | Cadwell et al. (1997) |
| Y190 | MATa ade2-101 trp1-901 his3 leu2-3,112 gal4 gal80 ura3-52::GAL1-lacZ::URA3 lys2::GAL1-HIS3 cyhR | Harper et al. (1993) |
Random mutagenesis
rpa43 mutant alleles were obtained by mutagenic PCR (200 µM MnCl2) and gap repair as described (Muhlrad et al., 1992).
Purification of the B600 and B2000 fractions
Purification of the B600 and B2000 fractions and the Pol I–Rrn3 and TBP-containing complexes was performed as previously described (Milkereit et al., 1997).
In vitro transcription assays
Non-specific activity of RNA Pol I was assayed as described (Buhler et al., 1974). Specific transcription assays were performed as described (Milkereit et al., 1997) with 40 ng of plasmid pSIRT (Musters et al., 1989). Transcripts were separated by electrophoresis on a 6% polyacrylamide gel containing 7 M urea in TBE and submitted to autoradiography.
Preparation of recombinant A43, Rrn3 and Rrn6 proteins
Recombinant Rrn3 and Rrn6 proteins expressed in E.coli. Transformed BL21(DE3) cells were grown at 24°C in LB medium supplemented with ampicillin (500 µg/ml), chloramphenicol (34 µg/ml) and sorbitol (1 M). Expression of the recombinant protein was induced at 0.4 OD600 with 250 µM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were shaken for 4 h at 24°C, collected and resuspended in breaking buffer [100 mM Tris–HCl pH 8, 20% glycerol, 400 mM KOAc, 1 mM EDTA, 5 mM Mg(OAc)2, 5 mM β-mercaptoethanol and 0.1% Tween 20] supplemented with protease inhibitors (Complete; Boehringer). Cells were lysed by two cycles of freezing and thawing in liquid nitrogen. Lysozyme (20 µg/ml) and benzonase (100 U/ml) were added and the suspension was incubated for 1 h at 4°C. Crude extract was centrifuged in a Beckman 45-Ti rotor for 1 h at 40 000 r.p.m.
Recombinant A43 and Rrn3 co-expressed in E.coli. Cell culture and induction were performed as described above. Cells were resuspended in breaking buffer [50 mM Tris–HCl pH 8, 20% glycerol, 100 mM KOAc, 5 mM Mg(OAc)2 and 0.1% Tween 20] supplemented with protease inhibitors and were lysed with an Eaton press and the crude extract was centrifuged as above.
Partial purification of rRrn3. The S100 fraction from a 12 l culture was loaded on a Q-Sepharose High-Load 20 ml column equilibrated with Q buffer [20 mM Tris–HCl pH 8, 20% glycerol, 5 mM Mg(OAc)2 and 0.1% Tween 20] containing 400 mM KOAc. After washing, proteins were eluted with a linear gradient from 400 mM to 2 M KOAc in Q buffer. Rrn3-containing fractions, as determined by western blot analysis using monoclonal anti-HA antibodies (16B12; Babco), were pooled and supplemented with 5 mM imidazole. The pool was loaded onto a 1 ml Hi-Trap nickel column equilibrated with Ni buffer [20 mM Tris–HCl pH 8, 20% glycerol, 500 mM KOAc, 5 mM Mg(OAc)2 and 0.1% Tween 20] containing 5 mM imidazole. The column was developed with a linear gradient from 5 to 200 mM imidazole in Ni buffer; Rrn3-containing fractions were pooled and dialysed against 20 mM Tris–HCl pH 8, 20% glycerol, 50 mM KOAc, 5 mM Mg(OAc)2, 5 mM β-mercaptoethanol and 0.1% Tween 20.
[35S]Rrn6[C1] synthesis. [35S]Rrn6[C1] was synthesized in a wheatgerm extract (Promega) supplemented with T7 RNA polymerase (Promega), [35S]methionine (0.8 mCi/ml) and 1 µg of purified plasmid pGP37.
Affinity chromatography
Chromatography of [35S]Rrn6[772–894] was on a column of Rrn3 immobilized to protein G–Sepharose beads (Pharmacia) coated with 45 µg of purified 12CA5 antibodies, equilibrated in breaking buffer with 1% (w/v) low fat milk. S100 containing or not HA-tagged Rrn3 (1 ml) was then passed five times through the beads. The beads were extensively washed with breaking buffer supplemented with 1% (w/v) low fat milk then with binding buffer [20 mM Tris–HCl pH 8, 20% glycerol, 50 mM KOAc, 5 mM Mg(OAc)2, 5 mM β-mercaptoethanol and 0.1% Tween 20]. An aliquot of 50 µl of the [35S]Rrn6[772–894] mixture, diluted in 300 µl of binding buffer, was then loaded five times onto the same column. After extensive washing with binding buffer, proteins were eluted at 37°C with 1 ml of binding buffer containing 1 µg/µl purified HA-peptide. Proteins were subjected to SDS–PAGE and analysed by autoradiography on BiomaxMS films (Amersham).
Chromatography of rRrn3 was on a column of immobilized GST–Rrn6[772–894]. Glutathione–Sepharose beads were prepared as above and 150 µl of partially purified HA-His6-Rrn3 were loaded on the column. Proteins were eluted with 1 ml of binding buffer supplemented with 30 mM glutathione and analysed by western blotting using monoclonal anti-HA antibodies (16B12; Babco).
Two-hybrid interaction between Rrn3 and Rrn6
The two-hybrid screening was performed essentially as described (Flores et al., 1999). Strain Y190 transformed with pAS-Rrn3 was transformed with a DNA genomic library (Fromont-Racine et al., 1997). Transformants were selected on 50 and 75 mM 3AT-containing medium and tested for activation of the lacZ reporter gene.
Specimen preparation for electron microscopy and image processing
Formation of immune complexes and image analysis of the data were performed as described by Klinger et al. (1996).
Acknowledgments
Acknowledgements
We greatly acknowledge Emmanuel Favry for invaluable technical help. We are very grateful to Valérie Goguel and Carl Mann for critical reading of the manuscript. We thank Anne Peyroche for helpful advice in genetics and for careful reading of the manuscript. We thank Irwin Davidson (Strasbourg, France) and Joan Steffan (University of California, Irvine, CA) for providing plasmids. This work was supported by a Human Frontier Science Program Organization grant. G.P. was supported by a short-term grant from the Formation pour la Recherche Médicale and was a recipient of a FEBS short-term fellowship.
References
- Bateman E. and Paule,M.R. (1986) Regulation of eucaryotic ribosomal RNA transcription by RNA polymerase modification. Cell, 47, 445–450. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Learned,R.M., Jantzen,H.M. and Tjian,R. (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science, 241, 1192–1197. [DOI] [PubMed] [Google Scholar]
- Bodem J., Dobreva,G., Hoffmann-Rohrer,U., Iben,S., Zentgraf,H., Delius,H., Vingronss,M. and Grummt,I. (2000) TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep., 1, 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréant B., Buhler,J.-M., Sentenac,A. and Fromageot,P. (1983) On the phosphorylation of yeast RNA polymerases A and B. Eur. J. Biochem., 130, 247–251. [DOI] [PubMed] [Google Scholar]
- Brun I., Sentenac,A. and Werner,M. (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J., 16, 5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R.P., Ryan,K. and Sollner-Webb,B. (1994) Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol. Cell. Biol., 14, 5010–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler J.-M., Sentenac,A. and Fromageot,P. (1974) Isolation, structure and general properties of yeast ribonucleic acid polymerase A (or I). J. Biol. Chem., 249, 5963–5970. [PubMed] [Google Scholar]
- Buhler J.-M., Huet,J., Davies,K.E., Sentenac,A. and Fromageot,P. (1980) Immunological studies of yeast nuclear RNA polymerases at the subunit level. J. Biol. Chem., 255, 9949–9954. [PubMed] [Google Scholar]
- Cadwell C., Yoon,H.J., Zebarjadian,Y. and Carbon,J. (1997) The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol., 17, 6175–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C. and Riva,M. (1998) Yeast RNA polymerase I subunits and genes. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, Berlin, Germany, pp. 9–38. [Google Scholar]
- Cavallini B., Faus,I., Matthes,H., Chipoulet,J.M., Winsor,B., Egly,J.M. and Chambon,P. (1989) Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc. Natl Acad. Sci. USA, 86, 9803–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke E.M., Peterson,C.L., Brainard,A.V. and Riggs,D.L. (1996) Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem., 271, 22189–22195. [DOI] [PubMed] [Google Scholar]
- Clos J., Normann,A., Ohrlein,A. and Grummt,I. (1986) The core promoter of mouse rDNA consists of two functionally distinct domains. Nucleic Acids Res., 14, 7581–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V. and Hurt,E.C. (1995) Genetic approaches to nuclear pore structure and function. Trends Genet., 11, 235–241. [DOI] [PubMed] [Google Scholar]
- Eberhard D., Tora,L., Egly,J.M. and Grummt,I. (1993) A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res., 21, 4180–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A. et al. (1999) A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA, 96, 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain,J.C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Gadal O., Mariotte-Labarre,S., Chedin,S., Quemeneur,E., Carles,C., Sentenac,A. and Thuriaux,P. (1997) A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol., 17, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. (1998) Initiation of murine rDNA transcription. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, Berlin, Germany, pp. 135–154. [Google Scholar]
- Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hempel W.M., Cavanaugh,A.H., Hannan,R.D., Taylor,L. and Rothblum,L.I. (1996) The species-specific RNA polymerase I transcription factor SL-1 binds to upstream binding factor. Mol. Cell. Biol., 16, 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Fisher A.J., Chitikila,C., Mitra,M. and Pugh,B.F. (1999) A role for TBP dimerization in preventing unregulated gene expression. Mol. Cell, 3, 717–727. [DOI] [PubMed] [Google Scholar]
- Jantzen H.M., Chow,A.M., King,D.S. and Tjian,R. (1992) Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev., 6, 1950–1963. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Kumar,A., Letts,G.A. and Geiduschek,E.P. (1998) A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc. Natl Acad. Sci. USA, 95, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Dodd,J., Lalo,D. and Nomura,M. (1997) Histones H3 and H4 are components of upstream activation factor required for high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl Acad. Sci. USA, 94, 13458–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Josaitis,C.A., Dodd,J.A. and Nomura,M. (1998) Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem., 273, 33795–33802. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Vu,L., Steffan,J.S., Dodd,J.A., Yamamoto,R.T., Nogi,Y. and Nomura,M. (1994) RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev., 8, 2349–2362. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Lee,B.S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- Klinger C., Huet,J., Song,D., Petersen,G., Riva,M., Sentenac,A., Bautz,E.K.F., Oudet,P. and Schultz,P. (1996) Localization of yeast RNA polymerase I core subunits by immunoelectron microscopy. EMBO J., 15, 4643–4653. [PMC free article] [PubMed] [Google Scholar]
- Kuhn A. and Grummt,I. (1992) Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc. Natl Acad. Sci. USA, 89, 7340–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzendörfer M., Smid,A., Klinger,C., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (1997) A shared subunit belongs to the eukaryotic core RNA polymerase. Genes Dev., 11, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Liljelund P., Mariotte,S., Buhler,J.-M. and Sentenac,A. (1992) Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 89, 9302–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Moorefield,B., Payne,J., Aprikian,P., Mitomo,K. and Reeder,R.H. (1996) A novel 66-kilodalton protein complexes with Rrn6, Rrn7 and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 6436–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner,H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Schultz,P. and Tschochner,H. (1997) Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol. Chem., 378, 1433–1443. [DOI] [PubMed] [Google Scholar]
- Moorefield B., Greene,E.A. and Reeder,R.H. (2000) RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl Acad. Sci. USA, 97, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Hanada,K., Yamamoto,K. and Song,C.Z. (1998) PAF53: an RNA polymerase I associated factor that promotes specific ribosomal RNA gene transcription by interacting with the Upstream Binding Factor (UBF). In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, Berlin, Germany, pp. 95–105. [Google Scholar]
- Musters W., Knol,J., Maas,P., Dekker,A.F. and Van Heerikhuizen,H. (1989) Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res., 17, 9661–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Vu,L. and Nomura,M. (1991) An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 88, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M. (1998) Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, Berlin, Germany, pp. 155–172. [Google Scholar]
- Nomura M., Nogi,Y., Yano,R., Oakes,M., Keys,D., Vu,L. and Dodd,J. (1993) RNA polymerase I, the nucleolus and synthesis of 35S rRNA in the yeast Saccharomyces cerevisiae. In Nierhaus,K.H., Franceschi,F., Subramanian,A.R., Erdmann,V.A. and Wittmann-Liebold,B. (eds), The Translational Apparatus. Plenum Press, New York, NY, pp. 89–99. [Google Scholar]
- Paule M.R., Iida,C., Perna,P.J., Harris,G., Brown,S. and Kownin,P. (1984) Faithful initiation of ribosomal RNA transcription from cloned DNA by purified RNA polymerase I. Biochemistry, 23, 4167–4178. [DOI] [PubMed] [Google Scholar]
- Riggs D.L., Peterson,C.L., Wickham,J.Q., Miller,L.M., Clarke,E.M., Crowell,J.A. and Sergere,J.-C. (1995) Characterization of the components of reconstituted Saccharomyces cerevisiae RNA polymerase I transcription complexes. J. Biol. Chem., 270, 6205–6210. [DOI] [PubMed] [Google Scholar]
- Riva M., Schaffner,A.R., Sentenac,A., Hartmann,G.R., Mustaev,A.A., Zaychikov,E.F. and Grachev,M.A. (1987) Active site labeling of the RNA polymerase A, B and C from yeast. J. Biol. Chem., 262, 14377–14380. [PubMed] [Google Scholar]
- Schnapp A., Clos,J., Hadelt,W., Schreck,R., Cvekl,A. and Grummt,I. (1990a) Isolation and functional characterization of TIF-IB, a factor that confers promoter specificity to mouse RNA polymerase I. Nucleic Acids Res., 18, 1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer,C., Rosenbauer,H. and Grummt,I. (1990b) A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. Nucleic Acids Res., 18, 1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Schnapp,G., Erny,B. and Grummt,I. (1993) Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol., 13, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp G., Schnapp,A., Rosenbauer,H. and Grummt,I. (1994) TIF-IC, a factor involved in both transcription initiation and elongation of RNA polymerase I. EMBO J., 13, 4028–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P., Celia,H., Riva,M., Sentenac,A. and Oudet,P. (1993) Three-dimensional model of yeast RNA polymerase I determined by electron microscopy of two-dimensional crystals. EMBO J., 12, 2601–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Lawrence,C.W. (1979) Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Smid A., Riva,M., Bouet,F., Sentenac,A. and Carles,C. (1995) The association of three subunits with yeast RNA polymerase is stabilized by A14. J. Biol. Chem., 270, 13534–13540. [DOI] [PubMed] [Google Scholar]
- Steffan J., Keys,D., Dodd,J. and Nomura,M. (1996) The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor dependent recruitment of core factor. Genes Dev., 10, 2551–2563. [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Keys,D.A., Vu,L. and Nomura,M. (1998) Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol. Cell. Biol., 18, 3752–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P., Mariotte,S., Buhler,J.-M., Sentenac,A., Vu,L., Lee,B.S. and Nomura,M. (1995) Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J. Biol. Chem., 270, 24252–24257. [DOI] [PubMed] [Google Scholar]
- Tower J. and Sollner-Webb,B. (1987) Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell, 50, 873–883. [DOI] [PubMed] [Google Scholar]
- Warner J.R. (1989) Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev., 53, 256–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi,Y., Dodd,J.A. and Nomura,M. (1996) RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk J. and Tjian,R. (1998) Structure and assembly of human selectivity factor SL1. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, Berlin, Germany, pp. 67–74. [Google Scholar]