Abstract
Even closely related eukaryotic species may differ drastically in genome size. While insertion of retroelements represents a major source of genome enlargement, the mechanism mediating species- specific deletions is fairly obscure. We analyzed the formation of deletions during double-strand break (DSB) repair in Arabidopsis thaliana and tobacco, two dicotyledonous plant species differing >20-fold in genome size. DSBs were induced by the rare cutting restriction endonuclease I-SceI and deletions were identified by loss of function of a negative selectable marker gene containing an I-SceI site. Whereas the partial use of micro-homologies in junction formation was similar in both species, in tobacco 40% of the deletions were accompanied by insertions. No insertions could be detected in Arabidopsis, where larger deletions were more frequent, indicating a putative inverse correlation between genome size and the average length of deletions. Such a correlation has been postulated before by a theoretical study on the evolution of related insect genomes and our study now identifies a possible molecular cause for the phenomenon, indicating that species-specific differences in DSB repair might indeed influence genome evolution.
Keywords: C value paradox/deletions/evolution/genome size/recombination
Introduction
Double-strand breaks (DSBs) are critical lesions in genomes. Efficient repair of DSBs is therefore important for the survival of all organisms. In principle, DSBs can be repaired via illegitimate or homologous recombination. In higher eukaryotes, including plants, illegitimate recombination seems to be the main mode of DSB repair (for reviews see Puchta and Hohn, 1996; Gorbunova and Levy, 1999; Mengiste and Paszkowski, 1999; Vergunst and Hooykaas, 1999). Error prone DSB repair leads to genomic changes resulting either in deletions, insertions or various kinds of genomic rearrangements (Pipiras et al., 1998). In plants, genetic change in somatic cells is relevant for evolutionary considerations because genomic alterations in meristematic cells can be transferred to the offspring (Walbot, 1996). Thus, on the evolutionary scale somatic DSB repair might influence genome size and genome organization.
The cause for the large differences in the nuclear DNA content of eukaryotes, particularly plants, known as the ‘C value paradox’, has been a matter of debate for a long time (Cavalier-Smith, 1985; Dove and Flavell, 1988). Even closely related species with a similar phenotype may differ significantly as to their diploid genome size. One mechanism responsible for these size differences can be related to a species-specific increase/reduction of repetitive sequences. In principle, genomes may become larger via duplications and insertions or smaller via deletions. Species-specific spread of retrotransposons was postulated as a main route enlarging plant genomes (SanMiguel et al., 1996, 1998; Bennetzen and Kellog, 1997). Alternatively, deletions might reduce genome size and counterbalance enlargements (Petrov, 1997). Recently, an elegant theoretical study (Petrov et al., 2000) has demonstrated that deletions of significantly different extension within retroelements yielded species-specific genome size alterations in related insect species over evolutionary time periods. By investigating the fate of non-LTR (long terminal repeat) retrotransposons in the cricket Laupala and fruit fly Drosophila, differences in the average size of genomic deletions were elucidated. Laupala has an ∼11 times bigger genome than Drosophila and the overall rate of DNA loss is ∼40 times slower (Petrov et al., 2000). But what kind of process might be responsible for these genomic changes? Deletions may occur by different mechanisms: by replication slippage (as suggested by Capy, 2000), by unequal crossover (as suggested by Smith, 1976) or by DSB repair. Although all three processes are, due to their general nature, not expected to differ drastically between related species a priori, we decided to compare deletion formation via DSB repair in somatic cells between tobacco and Arabidopsis, two dicotyledonous plant species with a >20-fold difference in genome size (Bennett and Leitch, 1997). By using suitable restriction endonucleases for induction of breaks at specific loci in eukaryotic genomes it has been possible to characterize DSB-induced recombination pathways in detail (for reviews see Paques and Haber, 1999; Puchta, 1999a; Jasin, 2000). In our study we used a transgene harboring an 18mer recognition site for I-SceI within the negative selectable marker gene cytosine deaminase [codA (Stougaard, 1993); Figure 1A]. When a DSB is induced via Agrobacterium-mediated transient expression of I-SceI (Puchta et al., 1996), and the consecutive repair results in genomic alterations associated with the loss of marker gene function, the cells become selectable by their resistance to 5-fluorocytosine (5-FC) (Salomon and Puchta, 1998). We were able to show before that in tobacco various genomic insertions can occur during DSB repair (Salomon and Puchta, 1998), a phenomenon drastically different to yeast, where, if at all, only extrachromosomal DNA is inserted during DSB repair (Moore and Haber, 1996; Teng et al., 1996; Ricchetti et al., 1999; Yu and Gabriel, 1999). In the present study we found for the first time in eukaryotes surprisingly strong differences in DSB repair between two related species: whereas in tobacco almost every second deletion event associated with the loss of function of the marker was accompanied by the insertion of filler sequences, we were not able to detect insertions at all in the deletions isolated from Arabidopsis. Moreover, as suggested theoretically for insects, we found an inverse correlation between genome size and the average length of deletions for the two species.
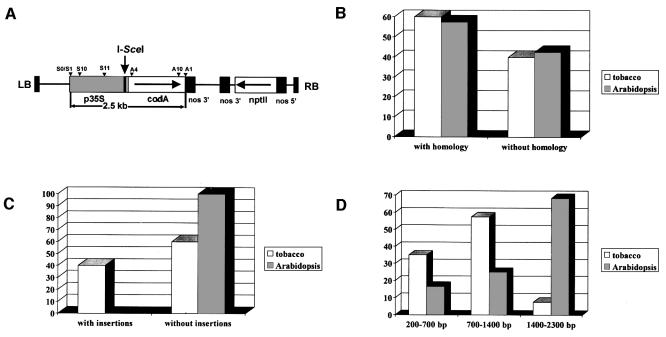
Fig. 1. (A) Schematic map of the T-DNA from the binary plasmid pBNE3I inserted into the plant genome. An I-SceI site is integrated between the codA ORF and 35S promoter. In addition, the T-DNA contained a kanamycin resistance gene (nptII). S0, S1, S10, S11, A1, A4 and A10 represent primer binding sites for the PCR amplification of the recombined junctions. The arrangement of genes on the T-DNA of the binary vector pCNE3I is identical to pBNE3I. RB, right border; LB, left border. (B–D) Comparison of the molecular properties of sequenced recombination junctions in tobacco (white boxes) and Arabidopsis (gray boxes); ordinate, events in %. (B) Involvement of homology (≥2 bp) in junction formation during the process of DSB repair. (C) Inclusion of filler sequences into the newly formed junctions. (D) Length distribution of the deletion events obtained.
Results
Tobacco and Arabidopsis plants were transformed with either of the binary vectors pBNE3I or pCNE3I. Transgenic plants with single copy insertions were identified by segregation analysis and Southern blotting. To avoid misinterpretations that might be due to potential genomic position effects, several transgenic lines were included in the experiments: for tobacco the lines B9, C15 and C19 (Salomon and Puchta, 1998), for Arabidopsis the lines B1, B2, C1, C2, C3, C4, C5 and C6. After transient transformation of transgenic plant cells with an I-SceI open reading frame (ORF) for DSB induction, double selection for kanamycin and 5-FC resistance was applied to isolate recombinant calli in which the function of the codA gene was lost. From these, DNA was prepared and the recombined junctions were amplified by PCR and sequenced. According to the primer binding sites (Figure 1A) deletions of up to 2.5 kb could be detected. As shown in Tables I and II, 40 deletions between 200 and 2300 bp were isolated for each of the two plant species (including 10 junctions of tobacco described before in Salomon and Puchta, 1998).
Table I. Compilation of deleted transgene junctions in tobacco.
| Callus | Deletion total (bp) | Within 35S promoter (bp) | Within codA (bp) | Insertion (bp) | Homology at junction (bp) |
|---|---|---|---|---|---|
| B9-73 | 222 | 1 | 221 | – | 1 |
| C15-71 | 236 | 236 | – | – | – |
| B9-48 | 236 | 236 | – | – | – |
| B9-842 | 257 | 170 | 87 | – | 1 |
| C15-51 | 262 | 227 | 35 | 19 | n.t. |
| B9-74 | 290 | 2 | 288 | – | 1 |
| C15-67 | 305 | 260 | 45 | – | 3 |
| B9-67 | 309 | – | 309 | 58 | n.t. |
| B9-50 | 395 | 290 | 105 | 32 | n.t. |
| B9-46 | 458 | 439 | 19 | 121 | n.t. |
| C15-471 | 549 | 539 | 10 | 98 | n.t. |
| C15-18 | 549 | 533 | 16 | – | 5 |
| C15-39 | 609 | 13 | 596 | – | 5 |
| B9-300 | 661 | 659 | 2 | – | 1 |
| B9-87 | 736 | – | 736 | – | 1 |
| B9-80 | 783 | 781 | 2 | – | 3 |
| C15-59 | 822 | 518 | 304 | – | 8 |
| C15-19 | 862 | 14 | 848 | – | 4 |
| B9-69 | 901 | 877 | 24 | 25 | n.t. |
| B9-76 | 984 | 984 | – | – | – |
| C19-11 | 1055 | 1047 | 8 | – | 2 |
| C15-44 | 1055 | 1047 | 8 | – | 2 |
| B9-300 | 1060 | 1057 | 3 | – | 2 |
| B9-16 | 1097 | – | 1097 | – | 3 |
| B9-842 | 1122 | 1100 | 22 | 2 | |
| B9-561 | 1144 | 1144 | 6 | 6 | n.t. |
| C15-29 | 1152 | – | 1152 | 13 | n.t. |
| B9-21 | 1173 | 1136 | 37 | – | 4 |
| B9-8 | 1175 | 1142 | 33 | – | 1 |
| C19-5 | 1218 | 390 | 828 | – | 4 |
| B9-33 | 1223 | 1223 | – | 11 | n.t. |
| C15-12 | 1236 | 1223 | 13 | 63 | n.t. |
| B9-17 | 1255 | 1239 | 16 | 76 | n.t. |
| B9-841 | 1275 | 1231 | 44 | – | 1 |
| B9-88 | 1287 | 1281 | 6 | 5 | n.t. |
| C15-193 | 1294 | 1282 | 12 | 4 | n.t. |
| C15-22 | 1322 | 1294 | 30 | 13 | n.t. |
| C15-191 | 1912 | 921 | 991 | – | 3 |
| C15-192 | 2023 | 1319 | 704 | – | 4 |
| B9-58 | 2294 | 1197 | 1097 | 88 | n.t. |
n.t., not testable.
Table II. Compilation of deleted transgene junctions in Arabidopsis.
| Callus | Deletion total (bp) | Within 35S promoter (bp) | Within codA (bp) | Insertion (bp) | Homology at junction (bp) |
|---|---|---|---|---|---|
| C2-607 | 212 | – | 212 | – | 2 |
| C6-179 | 233 | 233 | – | – | 1 |
| C5-241 | 240 | – | 240 | – | – |
| B1-28 | 247 | 247 | – | – | 1 |
| C2-181 | 248 | – | 248 | – | 1 |
| C6-210 | 249 | – | 249 | – | – |
| C6-916 | 860 | 860 | – | – | 2 |
| B1-230 | 929 | 144 | 785 | – | – |
| C5-150 | 967 | 277 | 690 | – | 2 |
| C6-09 | 1110 | 1048 | 62 | – | 1 |
| C1-181 | 1122 | 39 | 1083 | – | 2 |
| B1-150 | 1164 | 217 | 927 | – | 2 |
| B2-150 | 1207 | 279 | 928 | – | 3 |
| C2-10 | 1235 | 273 | 926 | – | – |
| B1-99 | 1256 | 40 | 1216 | – | 1 |
| C4-179 | 1281 | – | 1281 | – | 1 |
| C3-179 | 1493 | 840 | 653 | – | – |
| B1-50 | 1497 | 1057 | 440 | – | 3 |
| C3-17 | 1498 | 840 | 658 | – | 1 |
| C6-111 | 1499 | 1055 | 444 | – | 2 |
| B2-7 | 1501 | 1055 | 448 | – | 3 |
| B1-286 | 1503 | 1056 | 447 | – | 3 |
| C2-108 | 1525 | 929 | 596 | – | 1 |
| C1-30 | 1539 | 1129 | 410 | – | 2 |
| C2-15 | 1545 | 888 | 657 | – | 3 |
| C6-215 | 1553 | 1027 | 526 | – | 2 |
| C2-2 | 1556 | 407 | 1149 | – | 2 |
| C2-29 | 1588 | 979 | 609 | – | – |
| C3-10 | 1637 | 1194 | 443 | – | 2 |
| C1-20 | 1647 | 651 | 996 | – | 1 |
| C3-12 | 1658 | 659 | 999 | – | 2 |
| C1-21 | 1740 | 654 | 1086 | – | – |
| C3-13 | 1889 | 893 | 996 | – | 2 |
| C5-241 | 1915 | 934 | 981 | – | 3 |
| C2-5 | 1916 | 922 | 988 | – | 3 |
| C2-812 | 1958 | 945 | 1013 | – | 3 |
| C2-18 | 2025 | 1088 | 937 | – | 2 |
| C1-18 | 2093 | 1064 | 1029 | – | 3 |
| C1-185 | 2095 | 1070 | 1025 | – | 6 |
| B2-1 | 2207 | 1195 | 1012 | – | – |
Hitherto, for eukaryotes two pathways of illegitimate recombination have been postulated: junctions without homologies were explained by simple ligations, whereas small patches of homologous nucleotides (two or more) within these junctions were considered to be a prerequisite for the operation of a single-strand annealing mechanism (Nicolas et al., 1995). For tobacco we found 1.5 times more junctions with small homologies than without, similar to the ratio of 1.35 determined for Arabidopsis (Figure 1B). Both species do not differ significantly (p >0.8 in a goodness of fit test) as to the occurrence of the two junction classes. However, in two other aspects we detected significant differences. In 40% of the cases the linkage of the DNA ends in tobacco was associated with the insertion of filler sequences, whereas in Arabidopsis we were not able to detect any such event (Figure 1C), indicating a dramatic difference between the two species (p >0.0005). In addition, the average extension of detectable deletions differed between the two species. The average deletion size in tobacco was 920 bp (without consideration of the inserted sequences of 4–188 bp) whereas in Arabidopsis it was 1341 bp. The distribution of individual sizes of the deletions differs strongly in a direct comparison (see Figure 1D). If the data were pooled into two different size classes (above and below 1400 bp) the difference was highly significant (p <0.0005 in a goodness of fit test).
Discussion
In the current study a specific class of repair events (DSB-induced deletions that result in a loss of function of a marker gene) was compared between two related dicotyledonous plant species. Owing to the selection on 5-FC and kanamycin and the PCR primer binding sites used, deletion size in our experiments could only be analyzed in the range 0.2–2.5 kb. Although other kinds of repair events, like changes that are not linked to the loss of the marker gene or bigger deletions, could not be addressed with the current experimental set-up, the presented data reveal a surprisingly strong difference in DSB repair between the two species. Whereas the mechanism of junction formation itself was not different between the two species (Figure 1B), the size classes of deletions in Arabidopsis and tobacco differed remarkably (Figure 1D). This is reminiscent of theoretical calculations for two insect species (Petrov et al., 2000) in which an inverse correlation between the genome size and the average length of deletions was suggested. We were now able to supply in our experimental study a putative molecular mechanism of the phenomenon. Although just two species were compared and a final conclusion on the matter is of course not justified yet, it is nevertheless tempting to speculate that species-specific differences in DSB repair pathways may indeed contribute significantly to the evolution of eukaryotic genome size. As Arabidopsis underwent, in comparison to the much larger soybean genome, a number of segmental duplications or possibly a complete genome duplication during evolution (Grant et al., 2000), deletion formation must have played a prominent role, resulting in the small size of the present day genome of the plant species.
Beside deletions that resulted in a loss of function of the marker gene, we were also able to isolate from tobacco insertions that led to an enlargement of the PCR product (described before in Salomon and Puchta, 1998). In our previous study we were able to demonstrate that these insertions are of nuclear origin. We were not able to detect such kinds of products in Arabidopsis at all (A.Kirik and H.Puchta, unpublished observations) and therefore concentrated our present study on the comparison of deletions associated with the loss of function of the marker gene. Thus, in tobacco almost every second DSB repair event is associated with an insertion into the break site. Interestingly, insertion of filler sequences has also been reported to accompany deletions in the large maize genome (Wessler et al., 1990). Earlier studies indicated that the frequency of insertion of filler sequences as well as their origin seem to differ strongly between lower and higher eukaryotes. In yeast, DSB repair rarely leads to the insertion of filler sequences; if present, insertions were of non-nuclear origin [cDNAs of retrotransposons or mitochondrial DNA (Moore and Haber, 1996; Teng et al., 1996; Ricchetti et al., 1999; Yu and Gabriel, 1999)]. In contrast, like in tobacco, in hamster cells insertion of nuclear sequences is more common (Liang et al., 1998). On the evolutionary scale, repeated insertion of filler sequences is expected to increase the complexity of genome organization.
But what are the molecular causes for the differences in DSB repair between Arabidopsis and tobacco? The average length of deletions in tobacco might be smaller either due to lower exo- or endonuclease activities attacking the break ends or, alternatively, to a better protection of the broken ends against degradation. Furthermore, it has been postulated that insertion of filler sequences occurs via a DNA synthesis-dependent strand annealing-like mechanism (Gorbunova and Levy, 1997; Salomon and Puchta, 1998), similar to a mechanism that plays a prominent role in homologous DSB repair in somatic plant cells (Rubin and Levy, 1997; Puchta, 1998, 1999b; Reiss et al., 2000). Such a pathway would also require the protection of invading strands during the annealing and copying process. Thus, both the different sizes of deletions as well as different frequencies of insertion of filler sequences could eventually be due to a single genetic difference between the two plant species.
Materials and methods
Plant transformation
The binary vectors pBNE3I (Figure 1A) and pCNE3I carrying the recombination substrate, and pCI-SceI carrying the I-SceI expression cassette as well as the production of the transgenic tobacco lines B7, C15 and C19, were described previously (Salomon and Puchta, 1998). Transgenic tobacco plantlets were transiently transformed with pCI-SceI (Puchta et al., 1996), and recombinant calli were selected and propagated as described (Salomon and Puchta, 1998). Arabidopsis plants of the cultivar Columbia were transformed with the Agrobacterium strains harboring either the binary vector pBNE3I or pCNE3I via vacuum infiltration as described by Bechtold and Pelletier (1998). From transgenic Arabidopsis lines, roots were produced and transformed as described (Valvekens et al., 1992). After vacuum infiltration with agrobacteria, roots were placed for 2 days on callus induction medium (CIM) containing phytohormones and then transferred onto shoot induction medium (SIM) containing 50 mg of kanamycin sulfate, 200 mg of vancomycin and 500 mg of cefotaxin per liter as well as varying concentrations of FC: 200 mg/l for the first 5 days, 150 mg/l for another 7 days, 100 mg/l for the next 14 days and 50 mg/l for further propagation.
DNA analysis
DNA extraction from leaf tissues and calli, and Southern blotting for the determination of the number of copies of the integrated transgenes were performed as described (Salomon and Puchta, 1998). Genomic DNA was analyzed via PCR using the primers S0 (5′-pCCAATCCCACAAAAA TCTGAGC-3′), S1, S10, S11, A1, A4 and A10 as described (Salomon and Puchta, 1998). The amplification products were cloned into the pCR 2.1-Topo vector using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, USA) and propagated in TOP10 One Shot Cells (Invitrogen) according to the manufacturer’s instructions. Sequence analysis was performed with the automatic DNA-sequencer AFL-Express (Pharmacia, Uppsala, Sweden). Standard M13-20Forward, M13Reverse, T3 and T7 primers were used for the sequencing reaction. Goodness of fit tests were performed as described (Simpson et al., 1960).
Acknowledgments
Acknowledgements
We would like to thank Ingo Schubert, Waltraud Schmidt-Puchta and Ulrich Wobus for useful criticism of the manuscript and Susanne König for sequence analysis. The studies were funded by grants from the Deutsche Forschungsgemeinschaft and the Biotechnology program of the Land Sachsen-Anhalt.
References
- Bechtold N. and Pelletier,G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol., 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Bennett M.D. and Leitch,I.J. (1997) Nuclear DNA amounts in angiosperms—583 new estimates. Ann. Bot., 80, 169–196. [Google Scholar]
- Bennetzen J.L. and Kellog,E.A. (1997) Do plants have a one-way ticket to the genomic obesity? Plant Cell, 9, 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P. (2000) Perspectives: evolution. Is bigger better in cricket? Science, 287, 985–986. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (1985) The Evolution of Genome Size. John Wiley and Sons, Chichester, UK. [Google Scholar]
- Dove G.A. and Flavell,R.B. (1988) Genome Evolution. Academic Press, London, UK. [Google Scholar]
- Gorbunova V. and Levy,A.A. (1997) Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res., 25, 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V. and Levy,A.A. (1999) How plants make ends meet: DNA double-strand break repair. Trends Plant Sci., 4, 263–269. [DOI] [PubMed] [Google Scholar]
- Grant D., Cregan,P. and Shoemaker,R.C. (2000) Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl Acad. Sci. USA, 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. (2000) Chromosome breaks and genomic instability. Cancer Invest., 18, 78–86. [DOI] [PubMed] [Google Scholar]
- Liang F., Han,M., Romanienko,P. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T. and Paszkowski,J. (1999) Prospects for the precise engineering of plant genomes by homologous recombination. Biol. Chem., 380, 749–758. [DOI] [PubMed] [Google Scholar]
- Moore J.K. and Haber,J.E. (1996) Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature, 383, 644–646. [DOI] [PubMed] [Google Scholar]
- Nicolas A.L., Munz,P.L. and Young,C.S. (1995) A modified single-strand annealing model best explains the joining of DNA double-strand breaks in mammalian cells and cell extracts. Nucleic Acids Res., 23, 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. (1997) Slow but steady: reduction of genome size through biased mutation. Plant Cell, 9, 1900–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D.A., Sangster,T.A., Johnston,J.S., Hartl,D.L. and Shaw,K.L. (2000) Evidence for DNA loss as a determinant of genome size. Science, 287, 1060–1062. [DOI] [PubMed] [Google Scholar]
- Pipiras E., Coquelle,A., Bieth,A. and Debatisse,M. (1998) Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J., 17, 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. (1998) Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J., 13, 331–339. [Google Scholar]
- Puchta H. (1999a) Use of I-SceI to induce double-strand breaks in Nicotiana. Methods Mol. Biol., 113, 447–451. [DOI] [PubMed] [Google Scholar]
- Puchta H. (1999b) DSB-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics, 152, 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. and Hohn,B. (1996) From centiMorgans to basepairs: homologous recombination in plants. Trends Plant Sci., 1, 340–348. [Google Scholar]
- Puchta H., Dujon,B. and Hohn,B. (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl Acad. Sci. USA, 93, 5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B., Schubert,I., Köpchen,K., Wendeler,E., Schell,J. and Puchta,H. (2000) RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc. Natl Acad. Sci. USA, 97, 3358–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M., Fairhead,C. and Dujon,B. (1999) Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature, 402, 96–100. [DOI] [PubMed] [Google Scholar]
- Rubin E. and Levy,A.A. (1997) Abortive gap repair: the underlying mechanism for Ds elements formation. Mol. Cell. Biol., 17, 6294–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S. and Puchta,H. (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J., 17, 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P. et al. (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science, 274, 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel P., Gaut,B.S., Tikhonov,A., Nakajima,Y. and Bennetzen,J.L. (1998) The paleontology of intergene retrotransposons of maize. Nature Genet., 20, 43–45. [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Roe A. and Lewontin,R.C. (1960) Quantitative Zoology. Harcourt, Brace and World, New York, NY. [Google Scholar]
- Smith G.P. (1976) Evolution of repeated DNA sequences by unequal crossover. Science, 191, 528–535. [DOI] [PubMed] [Google Scholar]
- Stougaard J. (1993) Substrate-dependent negative selection in plants using a bacterial cytosine deaminase gene. Plant J., 3, 755–761. [Google Scholar]
- Teng S.C., Kim,B. and Gabriel,A. (1996) Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature, 383, 641–644. [DOI] [PubMed] [Google Scholar]
- Valvekens D., van Lijsebettens,M. and van Montagu,M. (1992) Arabidopsis regeneration and transformation (root explant system). In Lindsey,K. (ed.), Plant Tissue Culture Manual: Fundamentals and Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 1–17. [Google Scholar]
- Vergunst A.C. and Hooykaas,P.J.J. (1999) Recombination in the plant genome and its application in biotechnology. Crit. Rev. Plant Sci., 18, 1–31. [Google Scholar]
- Walbot V. (1996) Sources and consequences of phenotypic and genotypic plasticity in flowering plants. Trends Plant Sci., 1, 27–33. [Google Scholar]
- Wessler S., Tarpley,A., Purugganan,M., Spell,M. and Okagaki,R. (1990) Filler DNA is associated with spontaneous deletions in maize. Proc. Natl Acad. Sci. USA, 87, 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. and Gabriel,A. (1999) Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell, 4, 873–881. [DOI] [PubMed] [Google Scholar]