Abstract
The molecular mechanisms directing Foxp3 gene transcription in CD4+ T cells remain ill defined. We show that deletion of the inhibitory helix-loop-helix (HLH) protein Id3 results in defective Foxp3+ Treg cell generation. We identified two transforming grothw factor-β1 (TGF-β1)-dependent mechanisms that are vital for activation of Foxp3 gene transcription, and are defective in Id3−/− CD4+ T cells. Enhanced binding of the HLH protein E2A to the Foxp3 promoter promoted Foxp3 gene transcription. Id3 was required to relieve inhibition by GATA-3 at the Foxp3 promoter. Further, Id3−/− T cells increased differentiation of Th17 cells in vitro and in a mouse asthma model. A network of factors therefore act in a TGF-β-dependent manner to control Foxp3 expression and inhibit Th17 cell development.
Regulatory T (Treg) cells expressing the transcription factor Foxp3+ are instrumental in the induction and maintenance of peripheral immune tolerance, and regulation of tumor immunity and infection1-8. Foxp3+ Treg cell differentiation requires transforming growth factor-β (TGF-β) signaling 9-13, however the molecular pathways transducing this signal remain largely unknown. The TGF-β receptor regulated adapter molecules Smad2 and 3 and the common Smad4 are involved in Foxp3 induction14-16. However, the Foxp3 promoter lacks Smad-binding sequences. In addition, the time lag between Smad2 or 3 activation (minutes) and Foxp3 mRNA expression (>12 hours) following TGF-β1 stimulation suggests that there are intermediate factors between the TGF-β1-mediated Smad signaling and Foxp3 gene transcription. Thus, understanding this is not only essential for understanding Treg cell generation, but also important for the treatment of autoimmune diseases, infection and cancer17, especially considering the reciprocal differentiation of TGF-β1-induced Foxp3+ Treg cells and Th17 cells18-23.
Here we here show that inhibitory helix-loop-helix (HLH) protein 3 (Id3), a transcription factor involved in T cell development24, 25, growth inhibition of a B cell progenitors26, and protection of mice against autoimmune-like Sjögren's syndrome27, regulates the TGF-β1-mediated reciprocal differentiation of Treg cells and Th17 cells in mice. Deletion of Id3 blocked TGF-β1-induced Foxp3+ Treg cell generation. This was attributed to a failure to enrich the binding of the basic HLH protein E2A and an inability to suppress GATA-3 binding at the Foxp3 promoter in Id3−/− T cells. These knockout T cells showed an increased differentiation of Th17 cells in vitro and in vivo. Id3-dependent reciprocal regulation of Treg cells and Th17 differentiation also occurred in an experimental model of house dust mite (HDM) -induced allergic asthma in mice.
Results
Reduction of CD4+Foxp3+ Treg cells in Id3−/− mice
Id3−/− mice have been shown to develop a T cell-dependent autoimmune-like Sjögren's syndrome27, we therefore reasoned that these knockouts might exhibit a defect in the generation and/or function of CD4+Foxp3+ Treg cells. Young Id3−/− mice (3-weeks-old) had significantly fewer CD4+Foxp3+ Treg cells in the spleen (Fig. 1a-d, Supplementary Fig. 1a) and peripheral lymph nodes (data not shown) than wild-type (WT) C57BL/6 mice, both in frequency and absolute number (Fig. 1,a-d). The numbers of CD4+Foxp3+ thymocytes were also reduced in these young Id3−/− mice (Fig. 1a-d). Helios has been used as a marker distinguishing natural from induced Treg cells (nTreg and iTreg respectively)28 and we observed that both Foxp3+Helios+ and Foxp3+Helios− Treg cells were reduced in Id3−/− mice (Fig. 1b). Therefore in terms of Helios expression, both nTreg and iTreg are affected by the absence of Id3.
Figure 1. Id3 regulates Foxp3+ Treg cell generation.
a, Flow cytometry of CD4+CD8− T cells in the thymi and the spleens in WT (Id3+/+) and Id3−/− mice (3 weeks old). Numbers in quadrants indicate percent Foxp3+CD25− cells (top left) or Foxp3+CD25+ cells (top right). Each plot is of one mouse representative of four per group. b, Numbers indicate percent Foxp3+Helios−(top left) or Foxp3+Helios+(top right) cells in CD4+CD8− T cells. c,d, Frequencies (c) and total number (d) of CD4+Foxp3+ Treg cells in the thymus (Thy) and spleens (Spl) (mean ± s.d.) of mice in a,b. (n = 7 mice). e, Id3−/− Treg cells are defective in suppressing WT T cell proliferation in cultures (mean c.p.m. ± s.d. in triplicate wells), representative of four independent experiments. White and black squares indicate the proliferation of Id3+/+ or Id3−/− Treg cells, respectively. f-h, Flow cytometry of thymocytes and splenocytes in Rag1−/− mice 4 weeks after transfer of bone marrow from Id3+/+ or Id3−/− (CD45.2+) mixed with C57BL/6 (CD45.1+) mice at a ratio of 1:2. (f) Gated CD4+CD8−CD45.1− T cells in the thymi and the spleens of one mouse representative of five in each group (Rag1−/− recipients). Numbers in quadrants indicate percent Foxp3+Helios− (top left) or Foxp3+Helios+ cells (top right). g,h, Frequencies (g,) and total number (h) of CD4+ Foxp3+ T cells in the thymocytes and spleens (mean ± s.d.) of mice in f. * P < 0.05; ** P < 0.01; *** P < 0.001.
In contrast to the reduction of Treg cells in young Id3−/− mice, the frequency of CD4+Foxp3+ Treg cells in the spleen (Supplementary Fig. 1a), lymph nodes and thymus (data not shown) of older Id3−/− mice was gradually recovered by 6-7 weeks-old and became even higher after 3-4 months. The recovery of Treg cells in older Id3−/− was primarily due to increased expansion of Treg cells, as knockout Treg cells showed significantly more Ki67+ dividing cells ex vivo than did WT Treg cells (Supplementary Fig. 1). Even in young Id3−/− mice (3-weeks-old) Treg cells already showed a substantially higher frequency of Ki67+ cells compared to WT Treg cells (Supplementary Fig. 1), despite a significantly fewer Treg cells in these young knockout mice (Fig. 1 a-d).
Despite the recovery and even higher frequency of Foxp3+ Treg cells in older Id3−/− mice, old Id3−/− mice showed signs of T cell activation and inflammation (Supplementary Fig. 1e)27. This suggests that the initial deficiency of Foxp3+ Treg cells in the neonatal Id3−/− mice resulted in dysregulated activation affecting homeostasis of T cells in Id3−/− mice. In addition, the suppressive function of Id3−/− CD4+CD25+ Treg cells was also severely compromised in in vitro co-culture assays (Fig. 1e, Supplementary Fig. 2a,b), whether suppressing WT (Fig. 1e) or Id3−/− (Supplementary Fig.2) CD4+CD25− T responder cells. However, the expression of Foxp3 and other Treg cell-associated molecules such as CD25, CTLA-4 and GITR was unaltered in the Id3−/− (data not shown). Treg cells from young (3 to 5 weeks-old, Fig. 1e and Supplementary Fig. 2) or older (4-6 months old, data not shown) Id3−/− mice showed compromised suppressive activity.
Next we generated mixed bone marrow chimeras to further confirm the function of Id3 in the generation of CD4+Foxp3+ Treg cells. We injected a mixture of C57BL/6 (CD45.1+) and Id3−/− (CD45.2+) bone marrow or a mixture of C57BL/6 (CD45.1+) and WT (CD45.2+) bone marrow into sub-lethally irradiated recombination-activating gene-1 (Rag1−/−) recipient mice. Both spleen and thymi reconstituted with Id3−/− (CD45.2+) bone marrow contained fewer CD4+Foxp3+ Treg cells than did those transplanted with WT (CD45.2+) bone marrow (Fig. 1, f-h). Similar to intact Id3−/− mice (Fig. 1b), Both Foxp3+Helios+ and Foxp3+Helios− Treg cells were decreased in mice reconstituted with Id3−/− bone marrow (Fig. 1f). The deletion of Id3 therefore results in a deficiency of Foxp3+ Treg cells.
Id3 is required for TGF-β1-induced Foxp3 expression
Next, we determined whether the Id3 deficiency affected the generation of Foxp3+ Treg cells in vitro. We cultured peripheral naïve CD4+CD25−Foxp3− T cells with CD3- and CD28-specific antibodies in the presence of TGF-β1 to induce Foxp3+ Treg cells 9. TGF-β1 induced profoundly lower levels of Foxp3 mRNA and protein in naïve Id3−/− compared to WT CD4+CD25− T cells (Fig. 2a-c) over a range of TGF-β1 concentrations, although the defect was partially reversed with high doses of TGF-β1 (Supplementary Fig. 3). Addition of exogenous IL-2 did not correct the defect, eliminating the lack of IL-2 as the cause (Fig. 2d). Inclusion of retinoic acid (RA) in the cultures only slightly increased the percentage of Id3−/− Foxp3+ T cells, whereas RA exhibited the reported synergistic effect on TGF-β1-dependent Foxp3+ Treg differentiation in WT CD4+ T cells29 (Fig. 2e). Analysis of proximal Smad activation downstream of TGF-β1 signaling revealed no significant difference in TGF-β receptor-activated Smad2 and 3 phosphorylation (represented by P-Smad 2) between Id3−/− and WT CD4+ T cells following TGF-β1 treatment (Supplementary Fig. 4a). Smad7, an inhibitory Smad that antagonizes Smad2- and 3-mediated TGF-β signaling30, was lower in Id3−/−compared to WT CD4+ T cells following TGF-β1 treatment (Supplementary Fig. 4b), ruling-out Smad7 as a factor in causing reduced Foxp3-induction in Id3−/− cells. Id3 is therefore a critical component of TGF-β1 mediated Foxp3+ Treg generation, but it likely functions downstream of P-Smad2- and P-Smad3-mediated signal pathways.
Figure 2. Id3−/− T cells fails to generate Foxp3+ Treg cells in response to TGF-β.
a, Quantitative analysis of Foxp3, present as mRNA expression relative to HPRT (mean ± s.d. of duplicate wells) in naïve CD4+CD25− T cells overnight after TCR stimulation with or without TGF-β. Data shown is of one experiment representative of at least five. b, Flow cytometry of naïve CD4+CD25− T cells cultured with TCR (with or without TGF-β) for three days. Numbers in the quadrants indicate Foxp3+ Treg cells. Each plot is of one experiment representative of five. c, Percent Foxp3+ Treg cells (mean ± s.d. of five experiments) in cultured cells in a. ** P < 0.01. d,e, Flow cytometry of CD4+CD25− T cells cultured with indicated reagents for 3-5 days. Numbers in the quadrants indicate CD25+Foxp3+ Treg cells of Id3+/+ (top row) or Id3−/− (bottom row) mice. Each plot is of one experiment representative of at least two. RA, retinoic acid.
E2A enrichment at the Foxp3 promoter activates Foxp3
We next studied the molecular mechanisms by which Id3 is involved in TGF-β1-induced Foxp3+ Treg cell generation. Freshly isolated CD4+CD25+ Treg cells from the spleen of WT mice showed higher levels of Id3 mRNA and protein (Supplementary Fig. 5 a,b) than CD4+CD25− T cells. We then investigated whether TGF-β signaling affected Id3 expression in naïve CD4+ T cells following TCR stimulation. TGF-β1 increased the levels of Id3 mRNA during the first 1-2 h compared to TCR treatment alone (Supplementary Fig. 5c), consistent with findings in B cells26. However, by 12-24 h, TGF-β1 treated T cells showed lower levels of Id3 mRNA than T cells without TGF-β1 treatment (Supplementary Fig. 5c). A role for TGF-β1 in the regulation of Id3 expression was further verified in T cell specific TGF-β receptor I conditional knockout mice (Tgfbr1f/fCd4-cre+)12, as CD4+CD25− T cells from these knockout mice showed no difference of Id3 expression with or without TGF-β1 (data not shown). TGF-β1 regulation of Id3 required Smad3 and/or Smad4, as either Smad3 single knockout (Smad3−/−) (Supplementary Fig. 5 d), or Smad3 and Smad4 double knockout (Smad3−/−Smad4f/fLck-cre+) (data not shown) CD4+CD25− T cells lost the aforementioned biphasic effects induced by TGF-β1. Thus, Treg cells express higher levels of Id3 and TGF-β1 regulates Id3 expression in naïve CD4+ T cells in a time-dependent manner: with a transient increase during the first 2 h followed by a considerable downregulation by 12-24 h.
Id3 lacks the basic region needed for DNA binding but retains the functional dimerization domain, and therefore is regarded as an inhibitor for basic HLH protein E2A binding to its target genes by forming inactive heterodimers24. We therefore hypothesized that TGF-β1 might influence E2A binding to the Foxp3 promoter and thence regulate gene transcription. E2A proteins (E47 and E12) possess a DNA-binding domain and bind the CANN TG (E-box) DNA motif as protein dimmers to regulate gene transcription during the development of both T and B cells24. We first analyzed Foxp3 with the help of Genomatix® matinspector software and found that the sequence between −1.1kb and +1.1kb with respect to the transcription initiation site (+1), contained multiple E-box elements with a canonical nucleotide sequence (CANNTG), which are expected to bind members of the E protein family of transcription factors31-34 (Fig. 3a). Using naïve CD4+CD25− T cells isolated from spleens of WT mice, we analyzed whether TGF-β1 influenced E2A binding to the Foxp3 promoter. We treated CD4+ CD25− T cells with CD3- and CD28-specific antibodies and TGF-β1 overnight and determined E2A binding to the Foxp3 promoter with chromatin immunoprecipitation (ChIP)-coupled quantitative PCR (qPCR) assay. ChIP-qPCR was performed with an antibody against E47, a major E2A protein component24, to precipitate chromatin from CD4+CD25− T cells, followed by qPCR analysis of the precipitated DNA. To determine where E2A binds in the Foxp3 gene, we analyzed three different regions of the Foxp3 gene; 1) the 5′ promoter region (amplicon 1; −995/−798), 2) the proximal promoter plus a region comprising the untranslated 5′mRNA (amplicon 2; +327/+513), and 3) the region located next to the enhancer containing the Smad-binding domain (amplicon 3; +2335/+2532)15 (Fig. 3a). We observed increased binding of E2A to a regulatory region of Foxp3 comprising the proximal promoter and its untranslated 5′ mRNA region (+327/+513, Fig. 3b). We then quantitatively compared the relative E2A binding to this region in the Foxp3 gene in T cells treated with TCR alone and TCR plus TGF-β1 and found that TGF-β1 significantly enhanced E2A binding to the promoter compared to the control (Fig. 3c). TGF-β1-driven enhancement of E2A binding to the Foxp3 promoter occurred 12-24 h following TGF-β1 stimulation, which correlated with the detected expression of Foxp3 mRNA (>12h) in TGF-β1 treated T cells (data not shown). This enrichment of E2A binding is probably not due to the changes in protein levels, as neither treatment significantly affected whole E2A protein expression levels (Supplementary Fig. 6a). Increased E2A binding was associated with TBP (TATA binding protein) binding, a marker of active transcription, within the Foxp3 promoter, strongly suggesting that both factors were recruited to this Foxp3 gene regulatory region (Fig. 3d). We validated the essential role of TGF-β signaling in promoting E2A binding to the Foxp3 promoter by utilizing Tgfbr1f/fCd4-cre+ CD4+CD25− T cells that lack TGF-β signaling. There was no upregulation of E2A binding to the Foxp3 promoter in response to TGF-β1 treatment (Supplementary Fig. 6b). In addition to TGF-β1-treated naïve CD4+ T cells, freshly isolated CD4+CD25+ Treg cells from WT spleen also showed enrichment of E2A binding at the Foxp3 promoter compared to CD4+CD25− T cells (Fig. 3e). These data collectively support a positive role of E2A in the activation of Foxp3 transcription in response to TGF-β1 treatment, which can be explained by the direct binding of E2A to the E-box rich region within the regulatory region of the Foxp3.
Figure 3. Enrichment of E2A binding to the Foxp3 promoter.
a, Schematic analysis of E protein binding sites (E-Boxes) at the Foxp3 promoter. b, Relative E2A binding ability (E47/control IgG, ChIP-qPCR assay) to the indicated E boxes in TCR and TGF-β treated CD4+CD25− T cells. E2A shows strongest binding to the E-boxes located at +327/+513. c, Analysis of relative E2A binding (E47/control IgG, ChIP-qPCR assay at +327/+513) to Foxp3 promoter in WT CD4+CD25− T cells 12-24 h after TCR stimulation with or without TGF-β. Data represent mean ± s.d. of E2A binding of four independent experiments. ** P < 0.01. d, A positive correlation between E2A binding and TBP binding to the Foxp3 promoter in WT CD4+CD25− T cells at 12-24 h after TCR and TGF-β treatment. White bar indicates the cells treated with CD3- and CD28-antibodies alone; black bar indicates plus TGF-β. Data are displayed as normalized ratios of E47/control IgG or TBP/Control IgG (mean ± s.d. of duplicate wells in one representative experiment of two, ChIP-qPCR assay at +327/513). e, Relative E2A binding ability (E47/control IgG, at +327/+513) in purified CD4+CD25+ (CD25+) Treg cells and CD4+CD25− (CD25−) T cells. (mean ± s.d. of triplicate wells in one representative experiment of two).
To study whether E2A binding to the Foxp3 promoter has a causative function in Foxp3 transcription, we challenged the ability of TGF-β1 to activate Foxp3 transcription after reducing E2A expression. We first performed gene knock-down experiments in mouse EL4 T lymphoma cells (subclone EL4 LAF), as TGF-β1 and TCR stimulation induce Foxp3 expression in these cells15, 35. Using shRNA, we knocked-down the expression of E2A (Fig. 4a, insert). Reduction of E2A protein in EL4 LAF cells blocked TGF-β1 induction of Foxp3 expression (Fig. 4a). To validate an intrinsic effect of E2A protein binding on Foxp3 transcription, we used a reporter Foxp3 construct containing the Foxp3 proximal promoter and the Smad enhancer (which responds to TGF-β1 directly15) to engineer E-box mutants (Fig. 4b, left panel) and challenged the ability of TGF-β1 to activate Foxp3 promoter activity. Mutation in the E-boxes diminished the activation of the Foxp3 promoter in response to TGF-β1 and TCR stimulation (Fig. 4b, right panel). Furthermore, Foxp3 induction in naïve CD4+CD25− T cells from Tamoxifen-induced conditional E2A and HEB (Transcription factor 12) double-knockout mice (E2af/fHebf/f Er-cre+) with CD3- and CD28-specific antibodies plus TGF-β36 (Supplementary Fig. 6 d,e). We used these double knockout mice because they largely eliminated the possible compensatory effect of HEB in single E2A conditional knockout mice, and T cell-specific E2A-HEB double knockout mice could not be utilized as they showed an almost complete deficiency of CD4+ T cells, including Treg cells37. These Tamoxifen-induced E2A-HEB double-deficient T cells generated significantly fewer Foxp3+ Treg cell than did control T cells (Fig. 4 c,d). The Foxp3 induction defect was most profound at low concentration of TGF-β1 (0.2 ng/ml) (Fig. 4 c,d). Furthermore, we knocked-down E2A with siRNA in WT primary naïve CD4+CD25− cells, and showed that reduction of E2A resulted in defective Foxp3 induction in response to TGF-β1 (Fig. 4e). Thus, our findings reveal that E2A protein binding to the Foxp3 promoter represents a key step (i.e. enhancer) in directly turning on Foxp3 gene transcription in naïve CD4+ T cells in response to TGF-β1.
Figure 4. E2A binding to the Foxp3 promoter is required for Foxp3 gene activation by TGF-β.
a, Foxp3 mRNA expression relative to Hprt (mean ± s.d. of three experiments) in EL-4 LAF cells with (shRNA) or without (Scram, Scrambled-control RNA) deficiency of E2a at 12h after activation with CD3- and CD28- antibodies with or without TGF-β. The insert shows the reduction for E47 protein. b, Mutation of E-boxes in the Foxp3 construct with enhancer (left panel) reduced TGF-β-induced Foxp3 gene activity (right panel) (mean ± s.d. of the triplicate measurements in one experiment representative of three). c,d, E2af/f/Hebf/f ER-cre+ CD4+ T cells showed defect of Foxp3+ Treg cell induction to anti-CD3+CD28 and TGF-β (0.2 ng/ml). E2af/f Hebf/f Er-cre+ mice were treated with sunflower seed oil (control) or Tamoxifen and splenic CD4+CD25− T cells were cultured for 3 days. c, flow cytometry of CD4+ T cells in one representative mouse. d, frequencies of CD4+Foxp3+ Treg cells in individual mice (two independent experiments). e, Foxp3 mRNA relative to HPRT (mean ± s.d. of four samples in two experiments) in naïve CD4+CD25− T cells with (siRNA) or without (Scram) E2A deficiency after TGF-β and TCR stimulation. The insert indicates E2a mRNA reduction by siRNA. f, Deletion of Id3 blocked TGF-β-mediated enrichment of E2A binding to the Foxp3 promoter in CD4+CD25− T cells (mean ± s.d. of E2A binding in three independent experiments). n,s, not statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
We then examined E2A binding to the Foxp3 promoter in Id3−/− CD4+CD25− T cells which exhibited a severely reduced frequencies of Foxp3+ Treg cells in response to TGF-β1 treatment (Fig. 2). The Id3−/− CD4+CD25− T cells had slightly but reproducibly higher basal levels of E2A binding to the Foxp3 promoter in response to TCR stimulation alone than did WT control T cells (data not shown and Fig. 5f), which could indicate an inhibitory function of Id3 for E2A protein binding to E boxes on target genes24. However, TGF-β1 treatment failed to enhance E2A binding to the Foxp3 promoter in Id3−/− T cells over the cells treated with TCR alone (Fig. 4 f). Freshly isolated CD4+CD25+ Treg cells from the spleens of Id3−/− mice showed no significant enrichment of E2A binding at the Foxp3 promoter compared to CD4+CD25− T cells in the same mice (Supplementary Fig. 6 c). The data were consistent with the positive function of E protein binding to E boxes at the Foxp3 promoter, but it raised an intriguing question as to why TGF-β1 treatment failed to upregulate E2A protein binding to the Foxp3 promoter and consequent Foxp3 expression in the Id3−/− T cells.
Figure 5. Id3 deficiency increases GATA-3.
a, GATA-3 mRNA expression relative to HPRT in naïve CD4+CD25− T cells at 12 h after activation with TCR and indicated reagents (mean ± s.d. of duplicate measurements in one experiment). b, Flow cytometry of CD4+CD25− T cells at 24 h after activation. Numbers in quadrants indicate percent Foxp3+GATA-3−(top left), Foxp3+GATA-3+(top right) or Foxp3−GATA3+ (bottom right) cells. c, IL-4 mRNA relative to HPRT in CD4+CD25− T cells at 2 h after activation (mean ± s.d. of duplicate measurements in one representative experiment). d, Flow cytometry of Tgfbr1f/f Cd4-cre+ and Tgfbr1f/+Cd4-cre+ CD4+CD25− T cells cultured for 24 h. Numbers in quadrants indicate same cells as in (b). e, Knockdown of E2a with SiRNA decreased TCR-driven IL-4 gene expression in WT naïve CD4+CD25− T cells. IL-4 mRNA expression relative to HPRT is shown (mean ± s.d. of triplicate measurements in one representative experiment). *P < 0.05. f, Relative GATA-3 binding (GATA-3/control IgG) at the Foxp3 promoter in CD4+CD25− T cells (cultured for 24 h). Data shown are mean ± s.d. of duplicate wells in one representative experiment . g, Elimination of GATA-3 with neutralization of IL-4 (αIL-4) restored enrichment of E2A binding to the Foxp3 promoter induced by TGF-β in Id3−/− CD4+ CD25− T cells (cultured for 24 h). Data shown are mean ± s.d. of duplicate wells in one representative experiment. Data shown in a,b,c,d are one experiment representative of at least three, and in e,f,g representative of two.
A role for Id3 in regulation of GATA-3 expression
We reasoned that, besides enhancing positive regulators of Foxp3 transcription (E2A binding to the promoter), TGF-β1 must also act to remove a negative regulator(s) that represses Foxp3 transcription, and we hypothesized this would require Id3. We focused on GATA-3, a key transcription factor for Th2 cell differentiation38, reported to bind directly to the Foxp3 promoter and inhibit its transcription39. Since TGF-β inhibits Gata3 expression in CD4+ T cells39, 40, we first compared Gata3 expression in Id3−/− and WT CD4+CD25− T cells. Although naïve CD4+ T cells isolated from WT and Id3−/− mice showed undetectable Gata3 (data not shown), Id3−/− T cells showed much higher levels of Gata3 mRNA (Fig. 5a) and protein (Fig. 5b) after TCR stimulation than WT T cells. Importantly, while it almost completely eliminated Gata3 expression in the control T cells, TGF-β1 only partially inhibited Gata3 expression in Id3−/− T cells (Fig. 5a,b). The increased Gata3 expression in Id3−/− T cells was likely attributable to high IL-4 production in these T cells following TCR stimulation (Fig. 5c and data not shown). TGF-β1 failed to significantly inhibit IL-4 expression in Id3−/− T cells (Fig. 5 c). Although TGF-β1 could suppress Gata3 expression in Id3−/− T cells, the overall higher levels of GATA-3 in Id3−/− cells meant TGF-β was unable to completely suppress GATA-3 to the levels seen in WT cells (Fig. 5a,b). To validate an essential role for TGF-β1 signaling in the suppression of Gata3, we examined its expression in Tgfbr1f/f Cd4-cre+ T cells. These had higher levels of GATA-3 than did control T cells (Tgfbr1f/+Cd4-Cre+) following TCR stimulation (Fig. 5d). In these knockout T cells, addition of TGF-β1 had no inhibitory effects on GATA-3 expression and no Foxp3 induction was observed (Fig. 5d) 12. Thus, Id3 is required for regulation of Gata3 in CD4+ T cells, which is primarily through control of IL-4 production.
To understand how Id3 regulates IL-4 production in WT CD4+ T cells, we hypothesized that Id3 forms a complex with E2A and prevents it from binding to the E-boxes at the promoter of Il4. Indeed, Il4 contains at least 4 E-boxes in a highly conserved region of its promoter (400bp upstream from transcriptional starting site). We showed that knockdown of E2A with siRNA in WT CD4+CD25− T cells significantly decreased Il4 induction in response to TCR stimulation (Fig. 5e). These data suggest that the “extra” E2A in the absence of Id3 could induce large amounts of IL-4 that in turn drives GATA-3 expression.
Next, we studied whether Id3 deficiency affected GATA-3 binding to the Foxp3 promoter. Consistent with the inhibition of Gata3 expression (Fig. 5a,b), TGF-β1 reduced GATA-3 binding to the Foxp3 promoter in WT CD4+ T cells as determined by ChIP-qPCR assay (Fig. 5f). In line with the higher levels of Gata3 expression in Id3−/− T cells (Fig. 5a,b), there was a marked increase in GATA-3 binding to the Foxp3 promoter in these cells following TCR stimulation compared to WT cells (Fig. 5f). TGF-β1 however did not reduce GATA-3 binding in Id3−/− T cells to levels seen in WT cells (Fig. 5f). These data suggest that failure to remove GATA-3 from the Foxp3 promoter acted as a “brake” (silencer) on Foxp3 expression. In the absence of Id3, this “brake” rendered TGF-β1 unable to mediate the enrichment of E2A binding to Foxp3 promoter, resulting in lack of Foxp3 gene transcription in Id3−/− T cells.
To confirm this, we eliminated GATA-3 in TGF-β1-treated Id3−/− CD4+ T cells by administrating an IL-4 neutralization antibody; we had already shown that naïve Id3−/− CD4+ CD25− T cells produced large amounts of IL-4 immediately upon TCR stimulation (Fig. 5c), and IL-4 is a potent GATA-3 inducer41. Indeed, neutralizing IL-4 in TGF-β1-treated Id3−/− CD4+ T cells abrogated their GATA-3 expression (Fig. 5a,b), and permitted Foxp3+ Treg cell generation (Fig. 5b, and Supplementary Fig. 7). Consistent with the positive role of E2A binding in Foxp3 transcription, neutralizing IL-4 in the presence of TGF-β1 restored the enrichment of E2A binding to Foxp3 promoter in Id3−/− CD4+ T cells (Fig. 5g). However, inhibiting only GATA-3 in the absence of TGF-β did not result in any generation of Foxp3+ Treg cells in either Id3−/− or WT CD4+ T cells (Fig. 5b), suggesting downregulation of GATA-3 is only one element of the complex TGF-β1-mediated program of Foxp3 induction. This notion was further supported by the fact that elimination of GATA-3 with IL-4 neutralization in the presence of TGF-β1 did not result in Foxp3 induction in Tgfbr1f/f Cd4-cre+ T cells (Fig. 5d). In contrast to IL-4, neutralization of IL-6, IL-23 or IFN-γ in the TGF-β1-treated Id3−/− CD4+CD25− T cells all failed to restore Foxp3+ Treg cell generation (Supplementary Fig. 7 and data not shown), eliminating signals mediated by these cytokines as regulators in this process. Thus, our data reveal that TGF-β1 induces Foxp3 expression through at least two indispensable and interdependent molecular events: promoting E2A protein binding to the Foxp3 promoter (enhancer) and inhibiting and/or removing negative factors bound to the promoter such as GATA-3 (silencer). Both of these events are defective in Id3−/− T cells.
Deletion of Id3 increases Th17 cells
Since the Id3−/− T cells exhibited a defect in generation of CD4+Foxp3+ Treg cells, and recent evidence indicated a TGF-β1 reciprocal control of Treg and Th17 cell differentiation18, 22 and Foxp3 directly binds and inhibits the expression of the orphan nuclear receptor RORγt (encoded by Rorc) 22, we investigated possible dysregulation of Th17 cell differentiation in Id3−/− T cells. We first examined Th17 frequency in the gut of Id3−/− mice, in which Th17 cells can be detected in normal un-manipulated mice19. Id3−/− mice showed much higher frequencies and absolute numbers of Th17 cells in the lamina propia, Peyer's patches and intraepithelial lymphocytes (IEL) than did WT mice (Fig 6 a,b and data not shown). In addition, the amount of IL-17 per cell also increased in Id3−/− Th17 cells compared to that in WT Th17 cells (Fig. 6a, right panel). Th17 cells in the spleen and peripheral LNs were hardly detectable in young and non-symptomatic Id3−/− mice as in WT mice, but was significantly higher in older (> 6 months) mice compared to the age-matched WT mice (data not shown). These data suggest that loss of Id3 in mice leads to an increase in Th17 cells.
Figure 6. Id3−/− CD4+ T cells differentiate into Th17 cells in response to TGF-β in vitro.
a,b, Flow cytometry of CD4+ T cells in the lamina propia (LP) and Peyer's patches (PP) in mice (7-8-weeks old). (a) Quadrants indicate frequency (%) of intracellular IL-17+ (left) or mean fluorescence intensity (MFI) of IL-17+ (right) cells in gated CD4+ T cells in a representative mouse in each group. (b) Absolute number of IL-17+CD4+ cells (mean ± s.d, n = 3 mice) in LP and PP. Data representative of three independent experiments. c, Flow cytometry of naïve CD4+ CD25− T cells ( 3-5 weeks-old) cultured with TCR stimulation with the indicated reagents for 3-5 days. Quadrants indicate percent CD4+ versus intracellular IL-17+ cells in one experiment representative of more than five. d, ELISA analysis of cytokines in 72-h culture supernatants of CD4+CD25− T cells. Data represent mean ± s.d. of IL-17 in four independent experiments, except for IL-6 treatment (two experiments).. e, Neutralization of endogenous IL-4 abrogates TGF-β induced IL-17 production in Id3−/− naïve CD4+ T cells. Data shown indicate mean ± s.d. of quadrant measurements in two independent experiments. f, Quantitative RT-PCR analysis of RORγt, presented as mRNA expression relative to HPRT in CD4+CD25− T cells at 48 h after activation with TCR and indicated cytokines. Data shown represent one of three independent experiments. * P < 0.05; ** P < 0.01; n.s., not statistically significant
To test whether Id3 was involved in Th17 differentiation, we cultured naïve CD4+CD25− T cells with TGF-β1 alone or TGF-β1 plus IL-618, 21, 22. TGF-β1 plus TCR stimulation significantly increased IL-17 producing cells in Id3−/−, but not WT naïve CD4+CD25− T cells (Fig. 6 c,d,e). Adding IL-6 to TGF-β1 cultures failed to further enhance Th17 cells in Id3−/− T cells, but substantially upregulated IL-17+ cells in WT cultures (Fig. 6c,d) 18, 20-22. Consistent with IL-17 production, TGF-β1 plus TCR stimulation induced optimal expression of Rorc in Id3−/− T cells, and adding IL-6 provided no significant enhancement (Fig. 6f). In line with these results, neutralization of endogenous IL-6, IL-21 or IL-23 alone or in combination had no effect on TGF-β1-induced Th17 cell differentiation in Id3−/−CD4+CD25− T cells (data not shown); however, neutralizing IL-4 with an antibody abrogated the polarization of Th17 cells induced by TGF-β1 in Id3−/− T cells (Fig. 6e), possibly because Foxp3 expression had been restored (Fig. 5b, Supplementary Fig. 7b).
We next investigated whether E2A binding to the Rorc promoter could directly enhance transcription and IL-17 production in WT naïve CD4+ T cells. There are four E-boxes located in the promoter of the Rorc gene 42, 43, and binding of E-proteins to these is required for Rorc transcription in immature thymocytes during maturation43. We first determined that TGF-β1 dramatically enriched E2A binding at the Rorc promoter compared to TCR stimulation alone in WT CD4+CD25− T cells (Supplementary Fig. 8a). We knocked-down E2A expression in WT CD4+CD25− T cells with siRNA and showed that reduction of E2A substantially decreased Rorc gene transcription and IL-17 production in the CD4+ T cells in response to TGF-β1 plus IL-6 (Supplementary Fig. 8b,c). These data suggest that E2A, in combination with yet undetermined transcription factor(s) downstream of IL-6, plays a role in Rorc gene activation and consequent Th17 differentiation in normal T cells.
In addition to the in vitro studies, we also determined that naïve Id3−/− CD4+ T cells preferentially differentiated into IL-17+ following transfer into syngeneic Rag1−/− mice (Supplementary Fig. 8d). Although the percentage of Foxp3+ T cells was also higher in the transplanted Id3−/− T cells than in WT T cells, the degree of increase in Foxp3+ Treg cells was much less than that in Th17 cells. Thus, the ratio of Th17 to Foxp3+ Treg cells was substantially higher in Id3−/− T cells (Supplementary Fig. 8d). Taken together, our data demonstrate that, in the absence of Id3, CD4+ T cells preferentially differentiated into Th17 cells.
Id3 regulates Treg and Th17 cells in HDM-induced asthma
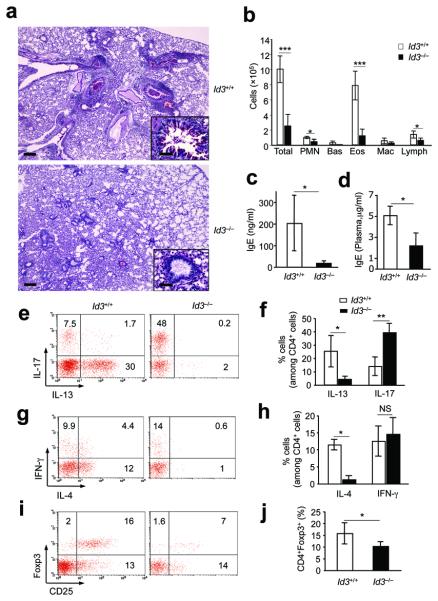
We next examined whether preferential differentiation to Th17 cells in Id3−/− cells might also occur during the course of disease in Id3−/− mice. We chose a model of HDM-induced allergic asthmatic inflammation in the lung, characterized by dominant Th2 cytokines 9. Although IL-17 has been found in experimental and human allergy and asthma, the exact function of Th17 cells in the pathogenesis and development of asthmatic lung inflammation is under debate44, 45. While control mice developed massive inflammatory cell infiltrates in the lung with mucus obstruction of the airways9, Id3−/− mice showed considerably less infiltration of inflammatory cells and reduction of mucus production in the airways (Fig. 7a). Analysis of cells from bronchioalveolar lavage (BAL) revealed significantly fewer inflammatory leukocytes, particularly eosinophils (Fig. 7b). In accordance with the decreased inflammation in the lungs, Id3−/− mice exhibited substantially lower levels of IgE in the BAL washes (Fig. 7c) and in the circulation (Fig. 7d) compared to WT mice. CD4+ T cells isolated from BAL in Id3−/− mice showed a significantly higher frequency of Th17 cells than in WT mice (Fig 7e, f), whereas the CD4+IFN-γ+ T cells were similar (Fig. 7g,h). Id3−/− CD4+ T cells in BAL had dramatically reduced IL-13+ (Fig 7e, f) and IL-4+ (Fig. 7g.h) cells compared to WT mice. Analysis of IL-4 proteins in BAL washes and plasma gave similar results (Supplementary Fig 9). CD4+Foxp3+ Treg cells were reduced in the BAL of Id3−/− mice compared to WT mice (Fig. 7 i,j). A similar increase in Th17 cells and decrease in IL-13+ and IL-4+ T cells were also revealed in the medial draining lymph nodes and spleens of Id3−/− mice (Supplementary Fig. 10), whereas CD4+ Foxp3+ Treg cells were unchanged or increased in the same tissues of these knockout mice (Supplementary Fig 10). Nevertheless, the overall ratio of Th17 cells to Foxp3+ Treg cells was considerably higher in Id3−/− than in WT mice. As a control for HDM-induced lung inflammation, we also examined the T cells in the untreated Id3−/− lungs. Un-manipulated Id3−/− mice showed higher levels of IL-17 cells in the BAL than naïve WT mice (Supplementary Fig.11). The BAL of naïve WT mice CD4+ T cells showed a high frequency of IL-17+ cells (~18%, Supplementary Fig. 11), which was higher than the asthmatic WT mice induced by HDM (Fig. 7 e). There was only slightly higher IFN-γ+ or IL-4+, but hardly detectable IL-13+ CD4+ T cells in Id3−/− compared to WT BAL (Supplementary Fig. 11). Importantly, untreated Id3−/− BAL contained substantially fewer CD4+ Foxp3+ Treg cells than did WT mice (Supplementary Fig. 11). Thus, our data demonstrated that Id3 also regulates the reciprocal differentiation of Treg and Th17 cells in vivo in naive lungs and also in response to the allergic allergen in experimental asthma.
Figure 7. Id3 regulates Th17 cells in HDM-induced asthma.
a, Mucus production in airways and inflammation in lungs in one mouse representative of nine to ten mice (PAS staining, Scale bar, 1000μm) in each group. The insert is an enlarged illustration of indicated areas (red star, Scale bar, 250 μm). b, Numbers of inflammatory cells in BAL 4 days after last i.t. challenge with HDM (mean ± s.d. of cell numbers in four mice each group). c,d, ELISA analysis of IgE in BAL (c, mean± s.d. of IgE in 4 mice per group) or plasma (d, mean ± s.d. of IgE in 6-9 mice per group). e-h, Flow cytometry of intracellular expression of IL-17, IL-13, IFN-γ and IL-4 in CD4+ T cells in BAL. e,g, Each plot is of one mouse representative of ten (Id3+/+) or nine (Id3−/−) mice. f,h, mean ± s.d. of the data in e,g, respectively. i, Flow cytometry of gated CD4+ T cells, presented as CD25 versus intracellular Foxp3 in one mouse representative of nine to ten mice in each group. j, Mean ±s.d. of the data in (i). * P < 0.05; ** P < 0.01; *** P <0.001; n.s, not statistically significant. Data represent three independent experiments.
Discussion
We have shown that Id3 is critical for promoting Foxp3+ Treg cell generation and controlling Th17+ cell differentiation. A null mutation in Id3 leads to a severe deficit in CD4+Foxp3+ Treg cells in mice. This Treg cell deficiency occurs in both the spleen and the thymus in young Id3−/− mice, although the reduction in the former is more profound and consistent than in the latter. We show that the defective Foxp3+ Treg cell generation in Id3−/− mice is T cell intrinsic, as CD4+ T cells in chimeras reconstituted with Id3−/− bone marrow contain fewer Foxp3+ Treg cells compared to WT controls. We have excluded increased cell death and decreased cell expansion as causes for the Treg cell deficiency in young Id3−/− mice, because Id3−/− Treg cells show no increase in apoptosis (data not shown) and exhibit higher rates of proliferation in vivo. Increased expansion of Treg cells in the absence of Id3 is responsible for the higher frequency of Foxp3+ T cells in older knockout mice. Although the detailed mechanisms remain to be elucidated, the inflammation and autoimmune-like disease in older Id3−/− mice can be attributed to at least two possibilities. First, the initial deficiency of Foxp3+ Treg cells in the neonatal Id3−/− mice resulted in lack of regulation of T cell activation. Alternatively but non-exclusively, the defective suppressive function of Id3−/− Treg cells could certainly explain how an inflammatory syndrome is accompanied by higher frequencies of Treg cells in older Id3−/− mice.
Importantly, we have determined that Id3 plays a crucial role in the correct execution of TGF-β1-mediated signals in T cells. Downstream of the TGF-β1 signal, we show that increased binding of bHLH protein E2A, plays a key role in the activation of Foxp3 transcription, which was defective in Id3−/− T cells. This enrichment of E2A binding is mediated by the E-boxes that we have detected in the regulatory region of the Foxp3. This is in agreement with a recent finding46, showing that this same region (+1Kb) corresponds to an open-chromatin status (H3-K4me3) in a Foxp3 gene that is being transcribed. Our results show that after TGF-β1 treatment, TBP binds the Foxp3 promoter at the same time as E2A, further suggesting a positive role for E2A in transcriptional regulation of Foxp3. This activity is regulated through TGF-β1 signaling; regardless of the method used to disrupt E2A binding to the Foxp3 promoter, disruption impaired the ability of TGF-β1 to activate Foxp3.
Our data suggest that TGF-β1 mediates an enrichment of E2A binding at the Foxp3 promoter and that this is important in the activation of Foxp3 gene transcription. However, the molecular mechanisms by which TGF-β1 enriches E2A binding remain unknown; there is no experimental evidence that Smad2 or Smad3 directly interact with E2A proteins. TGF-β treatment downregulates Id3 expression in WT T cells at 12-24 h, which suggest potential release of E2A protein from Id3-E2A complexes, with E2A being able to bind to E-boxes at the Foxp3 promoter. Additionally, whether TGF-β1-mediated suppression of GATA-3 plays a direct role in the enrichment of E2A binding at the Foxp3 promoter also remains elusive, although inhibition of GATA-3 could restore the E2A enrichment in response to TGF-β1 in Id3−/− T cells. Notably, E2A mutation experiments suggest that TGF-β1 enhances the amount of E2A bound at the Foxp3 promoter, rather than changing the functional capacity of E2A. Lastly, TGF-β1 could influence the Foxp3 gene epigenetically by loosening the chromatin allowing more E2A access to the Foxp3 promoter. The E-proteins have been shown to complex with p300/CBP and recruit histone acetyltransferases and RNA polymerase II28, which may consequently promote Foxp3 gene transcription47. It was recently reported that intron-1 rs3761548 is related to the defective transcription of Foxp3 in psoriasis by abrogating E47 and c-Myb binding48. Thus, the detailed mechanisms by which E2A binds to E-boxes at Foxp3 promoter and how this promotes the Foxp3 gene transcription awaits further investigation.
We have further demonstrated that defective TGF-β1-mediated enrichment of E2A binding to the Foxp3 promoter in Id3−/− T cells are largely due to high levels of GATA-3 in these cells. In Id3−/− cells, uncontrolled IL-4 production promotes GATA-3 expression, which occupies the Foxp3 promoter and cannot be sufficiently suppressed by TGF-β1; thus E2A cannot be enriched at the Foxp3 promoter. Supporting this, neutralization of IL-4 in TGF-β-treated Id3−/− T cells, completely blocked Gata3 expression and restored E2A binding at the Foxp3 promoter. This consequently allowed Foxp3+ Treg cell generation.
To further address this we have attempted to knock-down Gata3 with siRNA in naïve T cells. However, Gata3 knockdown proved difficult in Id3−/−T cells (only 20-30% reduction), but was easier in WT T cells (reduction of 80-90%). Consequently, GATA-3 siRNA-treated Id3−/− T cells showed only a slight increase in TGF-β-mediated Foxp3 induction (data not shown). These data further support that over-expression of GATA-3 was a key factor in defective Foxp3 induction in Id3−/− T cells. One possible reason for insufficient Gata3 knock-down by siRNA is uncontrolled IL-4 production in Id3−/− CD4+ T cells, this would antagonize the siRNA-GATA-3 by continuously inducing more GATA-3. How Id3 deficiency leads to uncontrolled IL-4 production in T cells remains an exciting question. Here we show that E2A plays an important role in switching on the Il4 gene in TCR-stimulated WT CD4+ T cells, indicating a role for Id3 in inhibiting E2A binding to the Il4 promoter.
Based upon data presented here, we propose that TGF-β1 induces Foxp3 expression through at least two indispensable and complementary molecular events: by promoting E protein binding to the Foxp3 promoter (enhancer) and by inhibiting and/or removing the negative factors bound to the Foxp3 promoter such as GATA-3 (silencer). The delicate balance of enhancer and silencer converging at the Foxp3 promoter results in correct Foxp3 gene transcription. Id3 plays a critical role in the regulation of this process (Supplementary Fig. 12). Although it is too simplistic to state that E2A and GATA-3 are the only enhancer and silencer in Foxp3 gene transcription, our findings provided an example of a dynamic balance between positive and negative factors as the key to correct gene transcription.
The inability of CD4+ T cells lacking Id3 to induce Foxp3 results in an intrinsic preference toward Th17 cell differentiation, as restoration of Foxp3 in Id3−/− T cells abrogates their IL-17 production, consistent with the finding of Foxp3 interacting and suppressing RORγt expression22. In addition, we have shown that E2A binding to the Rorγt promoter may directly enhance its transcription, and consequent IL-17 production in CD4+ T cells. Evidence supporting this conclusion include; TGF-β-enrichment of E2A binding at the Rorc promoter in TCR stimulated WT T cells, and that siRNA knockdown of E2A decreases Rorc gene transcription and IL-17 production in response to TGF-β and IL-6. One key issue is how IL-6 signals crosstalk with TGF-β-mediated E2A enrichment to promote Rorc gene transcription, and what is the missing link in this process? In contrast to the Foxp3 promoter, the Rorc promoter contains no GATA-3 binding sites. Nonetheless, these findings provide a novel cue to help decipher the mystery of Th17 differentiation.
The demonstration that deletion of Id3 enhanced Th17+ cells in an asthma model highlights a function for Id3 in the balance between Treg and Th17 cell differentiation in vivo. Notably, this preferential upregulation of Th17+ cells is not limited to allergic inflammation in Id3−/− mice. It was also observed in a model of experimental autoimmune encephalomyelitis (data not shown), suggesting a generality in the role of Id3 in the reciprocal differentiation of Th17 and Treg cells. However, increased IL-17+ cells and decreased Th2 cytokines, and lower levels of allergic inflammation in Id3−/− mice were surprising. Especially given that Id3−/− T cells produce high levels of IL-4 in vitro and in naïve Id3−/− lungs.
The mechanisms underlying decreased Th2 cytokines remain undefined, but our data could suggest a critical role of HDM treatment in the Id3−/− mice. BAL from naïve Id3−/− mice contains higher frequencies of Th17 cells compared to naïve WT mice. HDM-challenge decreased Th17 cells and increased Th2 cells in the BAL of WT mice, but further increased Th17 cells, and downregulated Th2 cells in the BAL of Id3−/− mice. Thus, it remains possible that Th17 cells may inhibit Th2 differentiation in asthmatic inflammation. Nevertheless, our data at least indicate lack of a positive correlation between Th17 cells and lung inflammation in asthma, consistent with recent published observations44. However, this does not necessarily eliminate the possibility that Th17 cells play a role in the initiation of the disease45. Taken together, our data reveal that Id3 is not only a key regulator of TGF-β-dependent immune responses, capable of promoting Foxp3+ induction and inhibiting the differentiation of Th17 cells, but also a crucial factor in diseases involving immune dysregulation.
ONLINE METHODS
Mice
C57BL/6 and Rag1−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Id3−/− mice27 (on a C57BL/6 background), Smad3−/− and littermates (on the C57BL/6x129 background), Tgfbr1f/fCd4-cre+, Tgfbr1f/+Cd4-cre+ 49, or E2af/f HEBf/fER-cre+36 were bred in our facilities under specific pathogen-free conditions. All animal studies were performed according to National Institutes of Health guidelines for use and care of live animals and approved by the Animal Care and Use Committees of NIDCR and Duke University.
Antibodies and Reagents
The following anti-murine antibodies were from BD Biosciences (San Diego, CA): PE-conjugated anti-CD45RB, anti-CD69, anti-CD62L or anti-IFN-γ, APC-conjugated anti-CD44 or anti-IL-4, purified anti-CD3 (NA/LETM), anti-CD28 (NA/LETM) and anti-FcRII/III, FITC-, PE- or APC-conjugated anti-murine CD25 (7D4 and PC61), PerCP-, or FITC-conjugated anti-CD4 or anti-CD8α, their respective isotype controls, and purified (NA/LETM) anti-IL-4, anti-IL-6, or anti-IFN-γ. The following were from eBioscience (San Diego, CA): PE-conjugated anti-GATA3, anti-RORγt, or anti-IL-13, FITC-, PE- or APC-conjugated anti-mouse/rat Foxp3 Staining Set. APC-conjugated anti-mouse IL-17 was from BioLegend (San Diego, CA). Anti-Id3 (B72-1), anti-IL-21, anti-IL-23p19, recombinant murine IL-6, IL-4, and IL-2, and human TGF-β1 were from R&D Systems (Minneapolis, MN).
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed using a Red ChIP kit (Diagenode Inc. NJ) per the manufacturer's instructions. CD4+CD25−(10×106) or CD4+CD25+ (7-10×106) T cells) were routinely used. CD4+CD25− cells were stimulated with plate-coated anti-CD3(5 μg/ml) and soluble anti-CD28- (2 μg/ml) with or without TGF-β (2 ng/ml) for 12-24 h. Equal amounts of processed chromatin were used as input controls or incubated with 4μg of anti-human/murine E47 (BD Biosciences), anti-TBP (Diagenode), anti-GATA-3 (Santa Cruz) or their respective control antibodies. Immunoprecipitated and total input DNAs were analyzed using a SYBR-Green Supermix kit and a Quantitative real-time PCR icycler iQTM detection system (BIO-RAD). The PCR primers for detecting promoters were: Foxp3-amplicon 1, fw-GGCCGCTATGTGTATGGTTT, and rev-CTGCTGCGAGTCTCTGAGTG; Foxp3-amplicon 2, fw-GCAACTCAAGATGCTGTCCA, rev-GGCTGGAAGAGACAGACAGG and Foxp3-amplicon 3, fw-GCGCTTATGTGGCTTCTTTC, and rev- GCAGATGGATGGGTCTTTGT. For Rorγt, FW: GTGCAGATCTAAGGGCTGAGGC, Rev; CATTCACTTACTTCTCATGACTG.
T cell purification and culture
CD4+ T subsets were isolated from spleens and lymph nodes with using magnetic beads (Miltenyi Biotec, Auburn, CA) 12, 50. Treg cell suppression co-culture assay was performed as described 50.
Treg and Th17 cells differentiation in vitro
Treg and Th17 cell differentiations were performed as reported 9, 18. Naïve CD4+CD25− T cells were cultured with plate-bound anti-CD3(5 μg/ml) and soluble anti-CD28 (2 μg/ml). TGF-β1 (0.002-20 ng/ml), IL-6 (50 ng/ml), IL-21 (50 ng/ml), IL-2 (5 ng/ml) or RA (100 nM) were used as indicated. In some cultures, ant-IL-4 (10μg/ml), anti-IFN-γ (10μg/ml), anti-IL-6 (10μg/ml) anti-IL-21(10μg/ml) or anti-IL-23 (10μg/ml) were included. Culture supernatants were collected at 24 (IL-4) or 72 (IL-17) h for cytokine ELISA (eBioscience and BioLegend).
Flow cytometry
The flow cytometry analysis was performed as described previously12. .
Immunoblotting
Immunoblot analysis was performed as described before12. Antibodies against following molecules were used: Smad2/3, E47, Id3, and GATA-3 (BD bioscience), P-Smad2 (Cell Signaling Technology), α-tubulin (Sigma), and β-actin (Santa Cruz).
Real-time RT-PCR
Real-time RT-PCR was performed as described previously 12, 49. Foxp3, Rorγt, Id3, Il4, Smad7, Gata3 and Hprt (hypoxanthine phosphoribosyl transferase) were analyzed using TaqMan gene expression assay kit. The primers (NM_054039.1 for foxp3, NM_021283.1 for il4, NM_013556.2 for hprt, NM_001042660.1 for smad7, NM_008091.3 for gata3) were from Applied Biosystems. The primers and probe for rorc were described previously19.
RNA interference
EL4 LAF cells15, 35 were incubated with E2A shRNA lentiviral particles (106 TU/ml, Sigma) in complete DMEM for 3 h. Puromycin (1 and 1.5 μg/ml) was added into the cell cultures at day 2 and 5, respectively. The efficiency of E2A knockdown was determined with anti-E47 antibody at day 9. Five shRNAs (all from Sigma) were tested. Two of them (XM_125750.3-438s1c1 and XM_125750.3-2060s1c1) were most effective in the knock-down of E2A, which, together with control particles (SHC003H, Sigma), were utilized for the Foxp3 induction with anti-TCR and TGF-β. The results obtained with XM_125750.3-438s1c1 are shown.
E2A or Gata3 knockdown with siRNA in naïve CD4+ cells was performed per manufacture's instructions (Lonza, Amaxa mouse T cell nucelofector kit). WT naïve CD4+CD25− (1 × 106) cells were stimulated with anti-CD3 and anti-CD28 overnight, and transduced with E2A and/or Gata3 siRNA particles (5nM) by electropolation. After overnight, cells were re-stimulated with anti-CD3 and TGF-β for 24 h. mRNAs for foxp3, Il4, RoRγt or Il17 were determined by real-time PCR. Four siRNAs for Gata3 (SI01009771, SI01009778, SI01009792 and SI02708615) and four for E2A (SI01444359, SI01444366, SI01444373 and SI01444380) (Qiagen) were tested. The results shown were obtained with SI01009778 (GATA-3) and SI01444373 (E2A), respectively. The control siRNA was 1027280.
Mutation of E-boxes and luciferase assay
Site directed mutagenesis for all of the consensus sequence for E-box element (CANNTG to AGNNCT) on the Foxp3 promoter region was performed by Genscript Corporation (NJ) and confirmed by DNA sequencing. EL4 LAF cells (2×106) were transfected with control 15 or E-box mutated Foxp3 promoter with enhancer reporter plasmid (3μg) in 100 μl of Nucleofector Solution L (LONZA), and co-transfected with pRL-Null (Promega) as an internal control. Transfected cells were stimulated with anti-CD3 and anti-CD28 with TGF-β (2ng/ml) for 12h. The Foxp3 promoter activity was determined by a dual luciferese assay system (Promega)15, 35. The relative Foxp3 activity is determined as (Firefly luminescence - background)/(Renilla luminescence-background)
Adoptive T cell transfer
CD4+CD25−CD62L+ cells from Id3−/− or WT spleens were transferred (i.p.) into Rag1−/− mice. Four weeks later, donor CD4+ T cells were analyzed by flow cytometry.
HDM-induced asthma
HDM-induced asthma was induced as described previously9. 6–10-wk-old Id3−/− or WT control mice were used.
Mixed bone marrow chimeras
Bone marrow (BM) was isolated from 3-weeks-old CD45.2+ Id3+/+ or CD45.2+ Id3−/− mice or 5 to 6-week-old CD45.1+ C57BL/6 WT mice. Donor BM from Id3+/+ or Id3−/− mice (1.5 ×106 cells) was mixed with BM from CD45.1+ WT mice (3 ×106 cells) and injected intravenously into sublethally-irradiated (450 rads) Rag1−/−mice. Cells from thymi and spleens were analyzed by flow cytometry after 4 weeks.
Statistical analysis
Student's t-tests (two-tailed) were used to analyze the significance of data comparison, except where otherwise indicated.
Supplementary Material
Acknowledgements
We thank Drs. Y.H. Chen, Q. Ruan and M. Tone, University of Pennsylvania for providing the Foxp3 constructs and EL4/LAF cells. This research was supported by the Intramural Research Program of the NIH, NIDCR.
Footnotes
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.,com/natureimmunology/.
Note: Supplementary information is available on the Nature Immunology website.
Author Contributions T.M. and J.L. designed and did experiments, analyzed data and contributed to the writing of the manuscript; J.P.V. designed and did Chip experiments, analyzed data and contributed to the writing of the manuscript; J.K. did experiments, analyzed data and contributed to the writing of the manuscript; Y.W. designed and W.W. and D.Y. performed the luciferase, E2A knockdown and Id3 immunoblot experiments and analyzed the data; B.Z. and Y.Z. generated and identified E2A/HEB knockout mice and provided critical input; P.Z. and B.Z. performed experiments; J. S.G. supervised and designed the ChIP study and contributed to the writing of the manuscript; and W.J.C. conceptualized the research, directed the study, designed experiments, analyzed data and wrote the manuscript.
Author Information The authors declare no competing financial interests.
References
- 1.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nature reviews. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 2.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nature immunology. 2005;6:331–337. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 3.Sakaguchi S. Regulatory T cells: key controllers of immunologic self-tolerance. Cell. 2000;101:455–458. doi: 10.1016/s0092-8674(00)80856-9. [DOI] [PubMed] [Google Scholar]
- 4.Shevach EM. CD4+ CD25+ suppressor T cells: more questions than answers. Nature reviews. 2002;2:389–400. doi: 10.1038/nri821. [DOI] [PubMed] [Google Scholar]
- 5.von Boehmer H. Mechanisms of suppression by suppressor T cells. Nature immunology. 2005;6:338–344. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- 6.Waldmann H, Adams E, Fairchild P, Cobbold S. Infectious tolerance and the long-term acceptance of transplanted tissue. Immunol Rev. 2006;212:301–313. doi: 10.1111/j.0105-2896.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 7.Hill JA, Benoist C, Mathis D. Treg cells: guardians for life. Nature immunology. 2007;8:124–125. doi: 10.1038/ni0207-124. [DOI] [PubMed] [Google Scholar]
- 8.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annual review of immunology. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Kretschmer K, et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nature immunology. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nature immunology. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 13.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolting J, et al. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. The Journal of experimental medicine. 2009;206:2131–2139. doi: 10.1084/jem.20090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tone Y, et al. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nature immunology. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 16.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 18.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 20.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 21.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science (New York, N.Y. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murre C. Helix-loop-helix proteins and lymphocyte development. Nature immunology. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 25.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 26.Kee BL, Rivera RR, Murre C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nature immunology. 2001;2:242–247. doi: 10.1038/85303. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Dai M, Zhuang Y. A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity. 2004;21:551–560. doi: 10.1016/j.immuni.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 28.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science (New York, N.Y. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 30.Nakao A, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling [see comments] Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 31.Cohen-Kaminsky S, et al. Chromatin immunoselection defines a TAL-1 target gene. The EMBO journal. 1998;17:5151–5160. doi: 10.1093/emboj/17.17.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh I, Bishop P, Chmielewski J. DNA binding properties of basic helix-loop-helix fusion proteins of Tal and E47. J Pept Res. 2001;57:354–360. doi: 10.1034/j.1399-3011.2001.00846.x. [DOI] [PubMed] [Google Scholar]
- 33.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. The Journal of experimental medicine. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun XH, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 35.Ruan Q, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones ME, Kondo M, Zhuang Y. A tamoxifen inducible knock-in allele for investigation of E2A function. BMC developmental biology. 2009;9:51. doi: 10.1186/1471-213X-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 39.Mantel PY, et al. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS biology. 2007;5:e329. doi: 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol. 2000;165:4773–4777. doi: 10.4049/jimmunol.165.9.4773. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science (New York, N.Y. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 43.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 44.Schnyder-Candrian S, et al. Interleukin-17 is a negative regulator of established allergic asthma. The Journal of experimental medicine. 2006;203:2715–2725. doi: 10.1084/jem.20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakae S, et al. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 2002;17:375–387. doi: 10.1016/s1074-7613(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 46.Rudra D, et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nature immunology. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. The Journal of experimental medicine. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, Chen L, Hao F, Wang G, Liu Y. Intron-1 rs3761548 is related to the defective transcription of Foxp3 in psoriasis through abrogating E47/c-Myb binding. Journal of cellular and molecular medicine. 2010;14:226–241. doi: 10.1111/j.1582-4934.2008.00370.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Perruche S, et al. Lethal effect of CD3-specific antibody in mice deficient in TGF-beta1 by uncontrolled flu-like syndrome. J Immunol. 2009;183:953–961. doi: 10.4049/jimmunol.0804076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perruche S, et al. CD3-specific antibody-induced immune tolerance involves transforming growth factor-beta from phagocytes digesting apoptotic T cells. Nature medicine. 2008;14:528–535. doi: 10.1038/nm1749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.