The authors demonstrate that fecal CD14 increases in response to bacterial intestinal infection and that CD14 single nucleotide polymorphisms modulate susceptibility and inflammatory response to enteric pathogens in US visitors to Mexico at risk for travelers' diarrhea.
Abstract
Background. Under normal conditions, the expression of CD14, which is the principal receptor for bacterial lipopolysaccharide, is down-regulated in the intestinal mucosa but increases in response to inflammatory stimuli. The aim of the present study was to investigate whether fecal CD14 levels increased in response to infection with diarrheagenic Escherichia coli and whether single nucleotide polymorphisms (SNPs) in the CD14 gene were associated with an increased susceptibility to traveler's diarrhea (TD) in US visitors to Mexico.
Methods. Six SNPs located at the promoter, exon, and untranslated regions of CD14 were typed in a prospective cohort study of 1360 visitors to Mexico at risk for TD. Stools from visitors with TD were studied for enteric pathogens by culture, colony hybridization, and polymerase chain reaction. Fecal soluble CD14 (sCD14) was measured in a subgroup of 203 adults with diarrhea and 66 healthy controls by enzyme-linked immunosorbent assay.
Results. The minor allele frequencies for CD14 SNPs were significantly different among the various racial and ethnic groups studied. Two SNPs in the promoter region of CD14 (-159 C > T; rs2569190 and -4191 C > T; rs5744441) were found to be associated with TD in White visitors. The -159 TT genotype was associated with a higher risk for TD (Relative risk [RR], 1.21; 95% confidence interval [CI], 1.05–1.38; P = .008), whereas individuals with the -4191 TT genotype were protected from infection (RR, 0.82; 95% CI, 0.71–0.92; P = .006). Subjects with TD excreted higher levels of fecal CD14 than did healthy controls (33,480 pg/mL vs 6178 pg/mL; P < .02). Fecal sCD14 levels were higher in stool samples from visitors with TD and the -159 TT genotype than they were in visitors with the CC/CT genotypes (P = .02), and stool samples from subjects with the -4191 CC genotype had higher fecal sCD14 levels than did stool samples from visitors with the CT/TT (P = .005) genotype. In a multivariate analysis with haplotypes constructed with the 6 SNPs studied, subjects with the haplotype containing the -159 C and the -4191 T allele were less likely to acquire TD (P = .015).
Conclusions. Our study suggests that CD14 levels increase in response to bacterial diarrhea and that polymorphisms in the CD14 gene influence susceptibility to TD. Intestinal CD14 plays an important role in the innate immune response to enteric pathogens.
When lipopolysaccharide (LPS) from gram-negative bacteria interacts with the innate immune system, it induces one of the most robust inflammatory responses known. When present systemically in humans, LPS induces the release of cytokines that result in fever, tachycardia, and vasodilatation, leading in some instances to septic shock. In the gut, enteropathogens use LPS as phage receptors, to adhere to the intestinal epithelium, to form biofilm, to bind enterotoxins, and to induce chemotaxis via complement activation (reviewed in Chatterjee and Chaudhuri [1]). In addition, LPS elicits protective immune responses to enteropathogens. As examples, antibodies to LPS increase in response to infection with enterotoxigenic Escherichia coli (ETEC) [2], enteroaggregative E. coli (EAEC) [3], Shiga toxin–producing E. coli (STEC) [4], and Vibrio cholerae [5], all of which can cause diarrhea in children in developing countries and cause traveler’s diarrhea (TD) in visitors from developed countries to regions of endemicity.
At a molecular level, LPS initially binds to lipid-binding protein (LBP) and then to its primary receptor, CD14 [6]. Through additional binding with MD-2 on the cell surface, the complex interacts with Toll-like receptor 4 (TLR-4), initiating the signal transduction necessary for innate and acquired immune responses to LPS. Despite the high concentration of LPS in the microbiota colonizing the intestinal lumen, the mucosa tolerates bacterial commensals by down-regulating receptors for LPS in intestinal epithelial cells and gut-associated immune cells. At the same time, the mucosa must maintain the ability to respond to enteropathogens or invading organisms in the event that the mucosal barrier function is breached. In animal models, CD14 −/− mice are unresponsive to LPS administered systemically [7], and recombinant CD14 prevents sepsis [8]. Therapy with anti-CD14 monoclonal antibodies decreases clearance of E. coli in rabbit models of infection [9]. In the respiratory mucosa, CD14 is important in the regulation of tolerance and immunity [10] to respiratory pathogens and also serves as a bridge for adaptive immunoglobulin (Ig) E immunity [11].
CD14 is a 55-kDa glycosyl phosphatidylinositol-anchored (GPI) glycoprotein expressed primarily on the surface of monocytes, macrophages, and neutrophils. CD14 also exists as a soluble molecule devoid of the GPI anchor in serum, urine, saliva, and milk. Soluble CD14 (sCD14) has an important role in LPS-mediated activation of CD14-negative cells, such as endothelial and intestinal epithelial cells [12]. Serum sCD14 increases in autoimmune disease, septic shock, brucellosis, and tuberculosis [13–16].
In humans, CD14 is found in 5q23−q31 as a single-copy gene 88 bp in size [17, 18]. Several single nucleotide polymorphisms (SNPs) in CD14 have been associated with human diseases [14, 19–23]. A SNP in the promoter region (-159 C > T) that results in increased CD14 transcription has been associated with Crohn's disease [24] and ulcerative colitis [25].
Aside from studies in Helicobacter pylori, Shigella, and necrotizing enterocolitis, there are few studies that have examined the role of CD14 in intestinal infections. We hypothesized that, because CD14 is the primary LPS receptor and gastrointestinal infections with diarrheagenic E. coli result in LPS seroconversion, that CD14 would be important in the pathogenesis of bacterial diarrhea in travelers at risk for TD. In this study, we examined the production of fecal CD14 in response to TD and the influence that CD14 SNPs have on susceptibility to infection with diarrheagenic E. coli.
SUBJECTS, MATERIALS, AND METHODS
Human Subjects
Students (≥16 years of age) from the United States or Canada enrolled in summer language educational programs in Mexico were invited to participate in the study if they were otherwise in good health and had not traveled to an area of risk for TD within the previous 6 months. Students were enrolled within 5 days of their arrival in Mexico and maintained diaries of their bowel habits and abdominal symptoms during their stay (range, 5–42 days). TD was defined as the passage of ≥3 unformed stools in a 24-h period accompanied by at least 1 gastrointestinal symptom (nausea, vomiting, abdominal pain, cramping, urgency, and excess flatus). In case of TD, a stool sample was collected for microbiological analysis. This study was approved by the University of Texas Health Science Center (Houston) Committee for the Protection of Human Subjects.
Genotyping
Genomic DNA was extracted from blood (Gentra) or saliva (DNA Genotek). Six SNPs were selected for study using SNP Browser (Applied Biosystems) on the basis of the existing literature and compatibility with multiplex SNPlex platform (Applied Biosystems). Genotyping was performed with previously described methods [26].
Microbiological Examination
Fresh stool specimens were collected from subjects in case of TD and transported directly to the laboratory or transported to the laboratory in Cary-Blair transport media. Stools were studied for the presence of mucus, fecal leukocytes, occult blood, and enteropathogens as previously described [27]. ETEC were detected with oligonucleotide probes by colony hybridization [28], and EAEC was identified by HEp-2 cell adherence assays [29]. Polymerase chain reaction (PCR) with primers specific for virulence markers characteristic of EAEC, ETEC, enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), and Shiga toxin–producing E. coli (STEC) were performed as previously described [30, 31].
CD14 Enzyme-Linked Immunosorbent Assay (ELISA)
Quantification of sCD14 from fresh-frozen fecal samples was performed by ELISA (R&D Systems) using known concentrations of human recombinant CD14 as a reference along with appropriate positive and negative controls.
Statistical Analysis
Statistical analysis, including univariate and multivariate analyses, were performed using SPSS software, version 16.0 (SPSS) for Windows. The association between genotypes and phenotypes, such as microbiological and clinical data, were assessed by χ2 test. Nonparametric tests (Mann-Whitney U test and Kruskal-Wallis test) were used when fecal sCD14 levels were analyzed.
CD14 SNP Haplotyping
Haplotypes were studied using the “coalescent-based”/Bayesian algorithm [32]. (PHASE, version 2.1; http://stephenslab.uchicago.edu). Missing alleles were excluded from the analysis for haplotype frequencies. Haplotype-specific odds ratio were calculated using logistic regression, assuming an additive model that included the probability of carrying each pair of haplotypes. The model was adjusted for ethnicity, length of travel, and season of travel. All statistics were calculated with the SAS statistical package and Haploview software.
RESULTS
Study Population Characteristics and Risk Factors for TD
The epidemiologic characteristics of the study population are shown in Table 1. In this cohort study, 45% of 1360 short-term visitors to Mexico experienced TD. Most of the participants were of White origin (90%), and the proportion of subjects who experienced TD was similar in all racial and ethnic groups studied. Consistent with previous studies, the length of stay, summer-time travel, and younger age were all identified as risk factors for acquisition of TD in a univariate analysis.
Table 1.
Baseline Characteristics of Study Participants and Univariate Logistic Regression Analysis of Risk Factors for the Acquisition of Traveler’s Diarrhea in US and Canadian Visitors to Mexico
| Characteristic | All subjects | Healthy subjects | Subjects with traveler’s diarrhea | RR (95% CI) | P |
| No. (%) of subjects | 1360 (100) | 744 (54.7) | 616 (45.3) | … | |
| Male sex | 391 (28.8) | 199 (26.7) | 192 (31.2) | 1.12 (0.99–1.27) | .08 |
| Race | |||||
| White | 1222 (89.9) | 663 (89.1) | 559 (90.7) | Reference | |
| Black | 76 (5.6) | 45 (6.1) | 31 (5.0) | 1.12 (0.87–1.51) | .41a |
| Asian | 36 (2.6) | 23 (3.1) | 13 (2.1) | … | NS |
| Native American | 4 (.3) | 3 (.4) | 1 (.2) | … | NS |
| Pacific Islander | 4 (.3) | 2 (.3) | 2 (.3) | … | NS |
| Other | 18 (1.3) | 8 (1.1) | 10 (1.6) | … | NS |
| Ethnicity | |||||
| Non-Hispanic | 1197 (88.0) | 649 (87.2) | 548 (89.0) | 1.11(0.92–1.36) | .31 |
| Age, mean years ± SD | 30 ± 13 | 31 ± 14 | 28 ± 12 | … | <.001 |
| Length of stay, mean days ± SD | 25 ± 11 | 23 ± 10 | 26 ± 11 | … | <.001 |
| Season of travel | |||||
| Summer | 1080 (79.4) | 563 (52.1) | 517 (47.9) | 1.37 (1.16–1.64) | <.001 |
| All other seasons | 278 (20.4) | 181 (65.1) | 97 (34.9) | … |
NOTE. NS, not significant; RR, relative risk; SD, standard deviation.
By 2 × 2 χ2 test for the comparison with White versus Black.
Distribution of CD14 SNPs Across Racial and Ethnic Groups
We then investigated whether CD14 SNP minor allele frequency (MAF) varied according to race. We found that the MAF for the SNPs studied was significantly different in self-identified Black visitors, compared with self-identified White visitors (Supplementary Table 1).
CD14 SNPs and Risk of TD
Because length of travel, age, and season of travel influenced the occurrence of TD, and because the CD14 SNP MAF was influenced by race, we stratified the remainder of the analysis to include only White visitors (n = 959) <50 years of age who stayed in Mexico for >7 days during the summer. Two SNPs in the CD14 promoter region were associated with TD. As shown in Table 2, the CD14 -159 C > T SNP was found to confer susceptibility to TD, whereas the second SNP at position -4191 C > T was protective of TD. When we examined the association using recessive models of inheritance (Table 3), the relative risk for TD increased for subjects with the -159 TT genotype (P = .008) and decreased for travelers with the non-CC -4191 genotype (P = .006). Next, we examined the SNP allele distribution in visitors with TD who had stool samples with fecal markers of inflammation (gross blood, mucus, and white blood cells) and compared these to the allele distribution for healthy travelers (Table 3). Of interest, the CD14 -4191 SNP non-CC genotype was seen less commonly in subjects with inflammatory diarrhea than in healthy control subjects.
Table 2.
Human CD14 Single-Nucleotide Polymorphisms (SNPs) Studied and Their Association With Traveler’s Diarrhea (TD) Among White Visitors to Mexico
| No. (%) of subjects |
|||||
| SNP, genotype | All subjects | Healthy subjects | Subjects with TD | Pa | HWE Pb |
| CD14 01 (C>T) (n = 762) | |||||
| CC | 394 (51.7) | 207 (53.9) | 187 (49.5) | ||
| CT | 295 (38.7) | 145 (37.8) | 150 (39.9) | ||
| TT | 73 (9.6) | 32 (8.3) | 41 (10.8) | .34 | .36 |
| CD14 EX02 (C>G) (n = 961) | |||||
| CC | 772 (80.3) | 397 (82.2) | 375 (78.5) | ||
| CG | 185 (19.3) | 84 (17.4) | 101 (21.1) | ||
| GG | 4 (0.42) | 2 (0.41) | 2 (0.42) | .34 | 0.27 |
| CD14 -159 (C>T) (n = 959) | |||||
| CC | 247 (25.8) | 134 (27.0) | 113 (24.4) | ||
| CT | 440 (45.9) | 240 (48.4) | 200 (43.2) | ||
| TT | 272 (28.4) | 122 (24.6) | 150 (32.4) | .02 | .48 |
| CD14 -1145 (A>G) (n = 897) | |||||
| AA | 264 (29.4) | 142 (31.5) | 122 (27.7) | ||
| AG | 424 (47.3) | 221 (48.5) | 203 (46.0) | ||
| GG | 209 (23.3) | 93 (20.4) | 116 (26.3) | .10 | .68 |
| CD14 -2839 (G>A) (n = 772) | |||||
| GG | 427 (55.3) | 220 (56.1) | 207 (54.5) | ||
| GA | 290 (37.6) | 148 (37.8) | 142 (37.4) | ||
| AA | 55 (7.1) | 24 (6.1) | 31 (8.2) | .54 | .89 |
| CD14 -4191 (C>T) (n = 897) | |||||
| CC | 538 (59.9) | 255 (55.6) | 283 (64.6) | ||
| CT | 310 (34.6) | 170 (37.0) | 140 (31.9) | ||
| TT | 49 (5.5) | 34 (7.4) | 15 (3.4) | .003 | .44 |
Healthy subjects versus subjects with TD.
HWE, Hardy-Weinberg equilibrium P value estimated for healthy control population.
Table 3.
Distribution and Association of CD14 -159 C > T and -4191 C > T Single Nucleotide Polymorphism (SNP) Genotypes and Alleles With Markers of Intestinal Inflammation in Travelers at Risk for Traveler’s Diarrhea (TD)
| Variable | Clinical outcome |
Fecal markers of intestinal inflammation No. of positive specimens/no. of stool specimens tested |
||||
| Healthy | TD | WBCs | RBCs | Mucus | Any | |
| CD14-159 C > T (n = 959) | n = 496 (52) | n = 463 (48) | n = 35/257 | n = 18/250 | n = 69/251 | n = 84 |
| Genotype | ||||||
| CC (n = 247) | 134 (27.0) | 113 (24.4) | 5 (14) | 3 (17) | 17 (25) | 19 (23) |
| CT (n = 440) | 240 (48.4) | 200 (43.2) | 17 (46) | 7 (39) | 31 (45) | 39 (46) |
| TT (n = 272) | 122 (24.6) | 150 (32.4) | 13 (37) | 8 (44) | 21 (30) | 26 (31) |
| P | .02a | .13b | .15b | .57b | .42b | |
| TT status | ||||||
| Non-TT (n = 687) | 374 (75.4) | 313 (67.6) | 22 (62.9) | 10 (55.6) | 48 (69.6) | 58 (69.0) |
| TT (n = 272) | 122 (24.6) | 150 (32.4) | 13 (37.1) | 8 (44.4) | 21 (30.4) | 26 (31.0) |
| RR (95% CI) | 1.21 (1.05–1.37) | 1.73 (0.90–3.29) | 2.36 (0.98–5.70) | 1.29 (0.79–2.06) | 1.31 (0.85–1.98) | |
| P | .008a | .11c | .09c | .3c | .2c | |
| Allele | ||||||
| C (n =934) | 508 (51.2) | 426 (46.0) | 27 (38.6) | 13 (36.1) | 65 (47.1) | 77 (45.8) |
| T (n =984) | 484 (48.8) | 500 (54.0) | 43 (61.4) | 23 (63.9) | 73 (52.9) | 91 (54.2) |
| RR (95% CI) | 0.94 (0.86–1.04) | 1.62 (1.02–2.57) | 1.82 (0.94–3.52) | 1.16 (0.85–1.58) | 1.20 (0.91–1.59) | |
| P | .23a | .04c | .09c | .41c | .21c | |
| CD14-4191 C > T (n =897) | n = 459 (51) | n = 438 (49) | n = 37/235 | n = 16/238 | n = 66/238 | n = 79 |
| Genotype | ||||||
| CC (n =538) | 255 (55.6) | 283 (64.6) | 30 (81) | 13 (81) | 48 (73) | 59 (75) |
| CT (n =310) | 170 (37.0) | 140 (31.9) | 7 (19) | 3 (19) | 16 (24) | 18 (23) |
| TT (n =49) | 34 (7.4) | 15 (3.4) | 0 (0) | 0 (0) | 2 (3) | 2 (2) |
| P | .003a | .007b | .11b | .02b | .005b | |
| CC status | ||||||
| CC (n =538) | 255 (55.6) | 283 (64.6) | 30 (81) | 13 (81) | 48 (73) | 59 (75) |
| Non-CC (n =359) | 204 (44.4) | 155 (35.4) | 7 (19) | 3 (19) | 18 (27) | 20 (25) |
| RR (95% CI) | 0.82 (0.71–0.95) | 0.31(0.14–0.70) | 0.30 (0.09–1.03) | 0.51 (0.31–0.85) | 0.47 (0.29–0.76) | |
| P | .006a | .003c | .06c | .01c | .001c | |
| Allele | ||||||
| C (n =1386) | 680 (74.1) | 706 (80.6) | 67 (90.5) | 29 (90.6) | 112 (84.8) | 136 (86.1) |
| T (n =408) | 238 (25.9) | 170 (19.4) | 7 (9.5) | 3 (9.4) | 20 (15.2) | 22 (13.9) |
| RR (95% CI) | 0.81 (0.72–0.93) | 0.32(0.15–0.68) | 0.30 (0.09–0.99) | 0.55 (0.35–0.86) | 0.51 (0.33–0.78) | |
| P | .001a | .001c | .03c | .007 c | .001c | |
NOTE. RBCs, red blood cells; WBCs, white blood cells.
2 × 2 χ2 test, comparing healthy subjects versus subjects with TD.
3 × 2 χ2 test, comparing the proportion of subjects with TD and given markers of intestinal inflammation versus healthy control subjects without TD.
2 × 2 χ2 test, comparing the proportion of subjects with TD and given markers of intestinal inflammation versus healthy control subjects without TD.
The Microbiology of TD Relates to CD14 SNPs
A stool specimen was obtained from 294 (63%) of the subjects with TD. We identified a potential pathogen in 191 (65%) of the stool specimens by fecal PCR or culture. Bacterial pathogens were identified in 178 (61%) of the stool specimens (Table 4). The proportion of visitors with the CD14 -159 TT genotype was higher among visitors with TD and a bacterial pathogen identified than among visitors who remained healthy (34% vs 25%; P = .01). The CD14 -159 TT genotype was also more commonly identified among subjects with TD due to EAEC and ETEC than among healthy visitors (38% vs 25% [P = .009] for EAEC and 34% vs 25% [P = .05] for ETEC). In keeping with the MAF distribution patterns that we observed for susceptibility to TD and the markers of intestinal inflammation, we also noted that the proportion of individuals with the CD14 -4191 non-CC genotype was lower in the group of subjects in whom an enteropathogen was identified (33%) than in the group of healthy subjects (44%) (Table 4).
Table 4.
Distribution and Association of CD14 -159 C > T and -4191 C > T Single Nucleotide Polymorphism (SNP) Genotypes and Alleles With the Identification of Enteric Pathogens
| Variable | Enteropathogen identified, no. of positive specimens/no. of stool specimens tested |
||||||
| Healthy subjects | Bacterial pathogen | EAEC | ETEC | EPEC | STEC | EIEC | |
| CD14-159 C > T (n = 959) | n = 496 | n = 178/294 | n = 110/292 | n = 117/289 | n = 70/163 | n = 18/162 | n = 14/163 |
| Genotype | |||||||
| C/C n =247 | 134 (27.0) | 34 (19.1) | 20 (18.2) | 22 (18.8) | 15 (21.4) | 3 (16.7) | 6 (42.6) |
| C/T n =440 | 240 (48.4) | 83 (46.6) | 48 (43.6) | 55 (47.0) | 33 (47.1) | 8 (44.4) | 4 (28.6) |
| T/T n =272 | 122 (24.6) | 61 (34.3) | 42 (38.2) | 40 (34.2) | 22 (31.4) | 7 (38.9) | 4 (28.6) |
| P | .01a | .009a | .05a | .39a | .38a | .29a | |
| TT status | |||||||
| Non-TT (n =687) | 374 (75.4) | 117 (65.7) | 68 (61.8) | 77 (65.8) | 48 (68.6) | 12 (63.2) | 10 (71.4) |
| TT (n =272) | 122 (24.6) | 61 (34.3) | 42 (38.2) | 40 (34.2) | 22 (31.4) | 7 (36.8) | 4 (28.6) |
| RR (95% CI) | 1.40 (1.0–1.80) | 1.66 (1.18–3.32) | 1.45 (1.03–2.01) | 1.34 (0.86–2.12) | 1.74 (0.72–4.21) | 1.30 (0.41–3.60) | |
| P | .01b | .006b | .03b | .24b | .27b | .75b | |
| Allele | |||||||
| C (n =1008) | 508 (51.2) | 151 (42.4) | 88 (40.0) | 99 (42.3) | 63 (45.0) | 14 (38.9) | 16 (57.1) |
| T (n =910) | 484 (48.8) | 205 (57.6) | 132 (60.0) | 135 (57.7) | 77 (55.0) | 22 (61.1) | 12 (42.9) |
| RR (95% CI) | 1.3 (1.08–1.56) | 1.45 (1.14–1.85) | 1.34 (1.06–1.70) | 1.24 (0.91–1.70) | 1.62 (0.85–3.10) | 0.79 (0.38–1.63) | |
| P | .004b | .003b | .01b | .17b | .17b | .57b | |
| CD14-4191 C > T (n = 897) | n = 459 | n = 166/294 | n = 93/291 | n = 92/291 | n = 60/135 | n = 15/134 | n = 13/135 |
| Genotype | |||||||
| CC (n =538) | 255 (55.6) | 111 (66.9) | 68 (73.1) | 63 (68.5) | 43 (71.7) | 8 (53.3) | 7 (53.8) |
| CT (n =310) | 170 (37.0) | 50 (30.1) | 22 (23.7) | 28 (30.4) | 14 (23.3) | 6 (40.0) | 4 (30.8) |
| TT (n =49) | 34 (7.4) | 5 (3.0) | 3 (3.2) | 1 (1.1) | 3 (5.0) | 1 (6.7) | 2 (15.4) |
| P | .01a | .006a | .01a | .07a | .97a | .54a | |
| CC status | |||||||
| CC (n =538) | 255 (55.6) | 111 (66.9) | 68 (73.1) | 63 (68.5) | 43 (71.7) | 8 (53.3) | 7 (53.8) |
| Non-CC (n =359) | 204 (44.4) | 55 (33.1) | 25 (26.9) | 29 (31.5) | 17 (28.3) | 7 (46.7) | 6 (46.2) |
| RR (95% CI) | 0.70 (0.5–0.98) | 0.52 (0.34–0.79) | 0.63 (0.42–0.94) | 0.53 (0.31–0.90) | 1.09 (0.40–2.95) | 1.06 (0.36–3.13) | |
| P | .01b | .002b | .03b | .02b | 1b | 1b | |
| Allele | |||||||
| C (n =1386) | 680 (74.1) | 272 (81.9) | 158 (84.9) | 154 (83.7) | 100 (83.3) | 22 (73.3) | 18 (69.2) |
| T (n =408) | 238 (25.9) | 60 (18.1) | 28 (15.1) | 30 (16.3) | 20 (16.7) | 8 (26.7) | 8 (30.8) |
| RR (95% CI) | 0.70 (0.55–0.90) | 0.56 (0.38–0.81) | 0.61 (0.42–0.84) | 0.64 (0.38–0.96) | 1.04 (0.47–2.30) | 1.26 (0.36–2.86) | |
| P | .004b | .001b | .005b | .03b | .92b | .6b | |
NOTE. CI, confidence interval; EAEC, Enteroaggregative Escherichia coli; EIEC, Enteroinvasive E. coli; EPEC, Enteropathogenic E. coli; ETEC, Enterotoxigenic E. coli (producing ST, LT, or LT/ST); RR, relative risk; STEC, Shiga toxin–producing E. coli.
3 × 2 χ2 test, for comparing with enteropathogens versus healthy control subjects.
2 × 2 χ2 test, for comparing with enteropathogens versus healthy control subjects.
Fecal CD14 Increases During TD and Levels Correlate With SNPs Studied
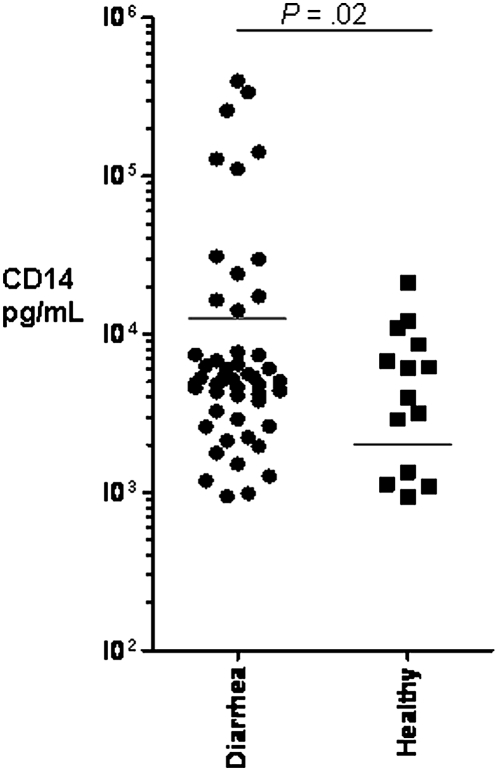
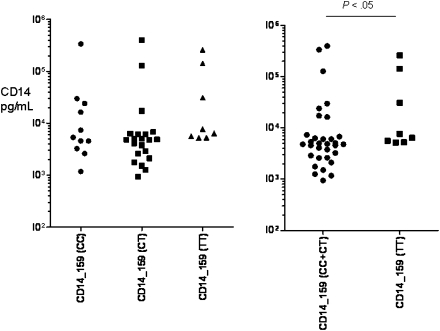
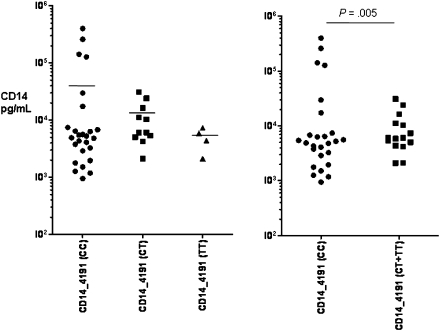
To determine genotype and phenotype correlates between the SNPs studied and the production of CD14 in response to infection, we studied the sCD14 concentrations in the stool specimens of a randomly selected subgroup of 269 Whites with and without TD. Overall, sCD14 was detectable (lower limit of detection, 125 pg/mL) in 64 (24%) of 269 stool samples; 14 (21%) of 66 of healthy subjects and in 50 (25%) of 203 of subjects with TD. However, the fecal CD14 levels were higher in those with TD cases than in healthy travelers (33,480 vs 6178 pg/mL; P = .02), as shown in Figure 1. Although the mean concentration of fecal CD14 from stool specimens of subjects with TD due to ETEC (13,307 pg/mL), EAEC (11,073 pg/mL), EPEC (12,997 pg/mL), and STEC (12,094 pg/mL) infections were higher than in the stool specimens of healthy control subjects (6178 pg/mL), this difference was not statistically significant. We then investigated the associations between the CD14 SNPs genotypes and the levels of fecal sCD14 among travelers with diarrhea due to all causes. The level of sCD14 was higher in visitors with TD and the TT genotype than in those with CT and TT genotypes of CD14 -159 SNP, but this difference did not reach statistical significance. Statistical significance was found only when those with CC and CT genotypes were grouped together and compared with those with the homozygous TT genotype (P < .05) (Figure 2). In the case of the CD14 -4191 SNP, subjects with TD and the non-CC SNP genotype were found to excrete lower levels of CD14, compared with subjects carrying the homozygous CC genotype (P = .005) (Figure 3).
Figure 1.
Fecal CD14 levels as determined by enzyme-linked immunosorbent assay in healthy travelers and in subjects with TD acquired in Mexico.
Figure 2.
Fecal CD14 levels in subjects with diarrhea and genotyped for the -159 C > T single nucleotide polymorphisms in the CD14 gene. Data are presented in pg/mL for CC (n = 11), CT (n = 21), and TT (n = 8) genotype groups. Fecal sCD14 in stool specimens assayed for visitors with the -159 CC genotype ranged from 1182 to 337,990 pg/mL (mean level ± standard deviation [SD], 39,750 ± 99,367 pg/mL); for those with the CT genotype, it ranged from 947 to 400,360 pg/mL (mean level ± SD, 29,367 ± 89,223 pg/mL); and for those with the TT genotype, it ranged from 5212 to 260,316 pg/mL (mean value ± SD, 57,969 ± 94,300 pg/mL).
Figure 3.
Fecal CD14 levels in subjects with diarrhea associated with CD14 -1491 C > T single nucleotide polymorphisms in the CD14 gene. Data are presented in pg/mL for CC (n = 26), CT (n = 11), and TT (n = 4) genotype groups. Fecal sCD14 in the stool specimens determined for patients in the -4191 CC genotype group ranged from 947 to 400,360 pg/mL (mean level ± standard deviation [SD], 40,697 ± 94,486); for visitors in the CT genotype group, it ranged from 2118 to 31,079 pg/mL (mean value ± SD, 11,162 ± 10,271); and for visitors in the TT genotype group, it ranged from 4383 to 7361 pg/mL (mean value ± SD, 5872 ± 1489).
CD14 SNP Haplotype and Risk for TD
Twenty-six haplotypes were constructed with data from the 667 Caucasian subjects who were genotyped for all of the 6 SNPs (Table 5). Four haplotypes accounted for 92% of the potential permutations. Only the haplotype containing the -159 C and the -4191 T allele (CCCAGT) was found to be associated with TD, demonstrating a protective effect in univariate (P = .009) and multivariate analyses (P = .015). Linkage disequilibrium analysis showed that the rs2569190 and rs5744441 were not in significant linkage disequilibrium (D’93 and r2=23).
Table 5.
CD14 Haplotypes and Their Association With Traveler's Diarrhea (TD)
| No. (%) of subjects |
|||||||
| Haplotype | Total | Healthy | TD | OR (95% CI) | Pa | OR (95% CI) | Pb |
| CCTGGC | 304 (45.5) | 153 (43.4) | 151 (47.9) | 1.22 (.98–1.52) | .07 | 1.19 (.96–1.46) | .11 |
| CCCAGT | 135 (20.2) | 80 (22.7) | 55 (17.3) | 0.69 (.51–.91) | .009 | 0.71 (.54–.94) | .015 |
| TCCAAC | 106 (15.9) | 57 (16.2) | 49 (15.6) | 0.93 (.69–1.25) | .62 | 0.95 (.72–1.28) | .76 |
| TGCAAC | 69 (10.5) | 37 (10.5) | 32(10.5) | 0.94 (.64–1.39) | .77 | 0.96 (.66–1.39) | .82 |
| Other | 53 (7.9) | 25 (6.9) | 28 (8.7) | 1.20 (.77–1.87) | .41 | 1.16 (.76–1.78) | .49 |
NOTE. CI, confidence interval; OR, odds ratio. Underlined alleles represent CD14 -159 (C) and CD -4191 (T), respectively.
Univariate analysis.
Multivariate analysis.
DISCUSSION
CD14 plays multifunctional roles during the innate immune response to LPS. These roles are influenced by gene-environment interactions, by the anatomical site of infection, by the timing of the LPS stimulation, and by whether CD14 is membrane bound or in soluble form. It is well established that, during the initial stages of a systemic infection, LPS binds to LPS-binding protein (LBP) and membrane CD14 in monocytes to initiate an innate immune response to LPS via TLR-4 [6]. Soluble CD14 in turn can control the intensity of the response by dampening the response to LPS by transferring CD14 membrane-associated LPS to circulating lipoprotein [33] or by forming complexes with LPS and lactoferrin that can result in LPS inactivation [34].
In the intestinal mucosa, the role of CD14 is less well understood. CD14 and TLR-4 expression in the mucosa is down-regulated, presumably to prevent an exuberant immune response to the LPS-rich content of the human gut under normal conditions. In the case of the inflammatory bowel diseases ulcerative colitis and Crohn's disease, which are hypothesized to be due to aberrant inflammatory responses to the intestinal microbiota, CD14 is over-expressed in the gut, and gene polymorphisms associated with increased CD14 have been associated with inflammatory bowel disease risk and disease progression. In this study, we noted higher levels of CD14 in the stool specimens of subjects with TD than in the specimens from control subjects, which is consistent with other studies that have shown elevations of CD14 levels in response to a variety of systemic infections and which may in this case indicate a stereotypical response to a number of enteropathogens.
There is little information on the role of intestinal CD14 and its interaction with LPS during acute bacterial diarrhea. A study conducted in the 1960s first demonstrated that parenteral administration of LPS in mice resulted in proliferation of bacteria in the intestinal lumen [35]. Subsequent work demonstrated that this effect was due to the availability of fluid that exuded into the gut lumen [36], likely to provide additional nutrients. It was later shown that the systemic but not the internal administration of LPS resulted in the accumulation of luminal fluid in rat intestines [37]. These findings are in keeping with observations on the polarized distribution of TLR-4, which is absent in intestinal epithelial cells but is present in deeper layers of the intestinal mucosa.
The fact that intestinal epithelial cells retain the ability to become sensitized to LPS in the presence of sCD14 [38], the heightened inflammatory response to Shigella seen in rabbits treated with monoclonal antibodies to CD14 [39], and the observation on the development of LPS serotype-specific antibodies following infection may indicate a role for CD14 in acute intestinal infections. The fecal CD14 response seen in our travelers could have originated from neutrophils and monocytes mobilized to the intestinal mucosa during infection; however, the presence of fecal leukocyte cells did not correlate with fecal CD14 levels, suggesting other sources for the fecal CD14. It is plausible that fecal CD14 originated from intestinal epithelial cells, which have recently been found to also produce CD14 in response to LPS stimulation [40], or originated from serum that crossed into the intestinal lumen using breaches in the intestinal barrier function caused by infection, in an effect similar to that seen in the LPS rat model.
The function of CD14 in the intestinal mucosa in response to acute bacterial diarrhea is unknown. It has been proposed that CD14 has a dual role in the intestine, initially sensitizing to infection and then inducing tolerance [41]. In this study, 2 SNPs in the promoter region of the CD14 gene were associated with TD risk. One SNP (-159 C > T) has previously been shown to increase the risk for gram-negative sepsis [42], inflammatory bowel disease, and H. pylori infection–related gastric carcinoma [43]. We also identified a novel association for a protective effect against TD in a SNP located in position -4191 C > T. This SNP has not previously been associated with disease in humans. In our studies, the alleles associated with an increased risk of TD were associated with higher fecal CD14 concentrations, which is in keeping with a previous report that showed higher CD14 transcriptional activity in individuals with the -159 TT genotype [44]. On the basis of our findings, we hypothesize that, when confronting enteropathogens, the mucosa of subjects who are genetically determined to produce increased levels of CD14 mount a more vigorous innate immune response to enteropathogens. It is possible that this leads to sensitizing the intestinal epithelial cells, monocytes, and other cells in the gut to produce cytokines, chemokines, and other mediators (such as prostaglandins and adenosine) of inflammation in response to LPS, which in turn results in an acute diarrheal response. Conversely, subjects with less CD14 tolerate infection with these pathogens more readily.
Our work has several limitations. First, we were only able to collect stool specimens from some of the visitors with diarrhea, and we were unable to evaluate sCD14 levels in all cases of TD. sCD14 was only measurable in a minority of subjects. It is plausible that sCD14 degraded in the stool specimens or was in complexes with other proteins in the intestinal lumen and could therefore not be measured by the ELISA used in this study. Also, subjects who perceived their risk for infection to be higher or less likely may have opted not to participate in the study, which could have resulted in enrollment bias. Finally, the exposure to pathogenic agents of diarrhea may have not been uniform among visitors. As in all exploratory SNP association studies, additional work is needed to validate these observations in other populations of travelers and in persons living in areas that are endemic for diarrheagenic E. coli.
The magnitude of the effect of CD14 -159 and CD14 -4191 on TD was modest but is similar to the effect of SNPs in other genes (interleukin [IL] 8, IL-10, lactoferrin, and osteoprotegerin) and is similar to the effect on TD that we have observed in other studies [26, 45–47]. The occurrence of TD that is caused by a variety of microbial pathogens, of which bacterial agents are most important, is influenced by a number of definable heritable factors linked to disease pathogenesis. The modest contribution of individual gene variants suggests that the host genetic component to diarrheal disease susceptibility is complex and multigenic.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://www.oxfordjournals.org/our_journals/cid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Acknowledgments
We thank Dorothy Ruelas, Judy Guillen, Margaret DuPont, David Huang, Jackie Vaca, Stephanie A. Lee, and the administration and staff of Universidad Internacional in Cuernavaca, Morelos, Mexico, for their assistance with this project.
Financial support. This work was supported by the National Institutes of Health (R01 AI54948-01 to P. C. O.), the University of Texas Center for Clinical and Translational Sciences (UL1RR024148) of the University of Texas Medical School at Houston, and DK56338, which funds the Texas Gulf Coast Digestive Diseases Center.
Potential conflicts of interest. P. C. O. has received payments for lectures and speakers’ bureau from Salix Pharmaceuticals, Pfizer, Cubist, and Theravance and has received payment through his institution for consultation work with Intercell unrelated to this article. All other authors: no conflicts.
References
- 1.Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae: III. Biological functions. Biochim Biophys Acta. 2006;1762:1–16. doi: 10.1016/j.bbadis.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Stoll BJ, Svennerholm AM, Gothefors L, Barua D, Huda S, Holmgren J. Local and systemic antibody responses to naturally acquired enterotoxigenic Escherichia coli diarrhea in an endemic area. J Infect Dis. 1986;153:527–534. doi: 10.1093/infdis/153.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Gomez HF, Mathewson JJ, Johnson PC, DuPont HL. Intestinal immune response of volunteers ingesting a strain of enteroadherent (HEp-2 cell-adherent) Escherichia coli. Clin Diagn Lab Immunol. 1995;2:10–13. doi: 10.1128/cdli.2.1.10-13.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins C, Chart H, Smith HR, et al. Antibody response of patients infected with verocytotoxin-producing Escherichia coli to protein antigens encoded on the LEE locus. J Med Microbiol. 2000;49:97–101. doi: 10.1099/0022-1317-49-1-97. [DOI] [PubMed] [Google Scholar]
- 5.Qadri F, Ryan ET, Faruque AS, et al. Antigen-specific immunoglobulin A antibodies secreted from circulating B cells are an effective marker for recent local immune responses in patients with cholera: comparison to antibody-secreting cell responses and other immunological markers. Infect Immun. 2003;71:4808–4814. doi: 10.1128/IAI.71.8.4808-4814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 7.Haziot A, Ferrero E, Lin XY, Stewart CL, Goyert SM. CD14-deficient mice are exquisitely insensitive to the effects of LPS. Prog Clin Biol Res. 1995;392:349–351. [PubMed] [Google Scholar]
- 8.Haziot A, Rong GW, Lin XY, Silver J, Goyert SM. Recombinant soluble CD14 prevents mortality in mice treated with endotoxin (lipopolysaccharide) J Immunol. 1995;154:6529–6532. [PubMed] [Google Scholar]
- 9.Opal SM, Palardy JE, Parejo N, Jasman RL. Effect of anti-CD14 monoclonal antibody on clearance of Escherichia coli bacteremia and endotoxemia. Crit Care Med. 2003;31:929–932. doi: 10.1097/01.CCM.0000054870.25767.EE. [DOI] [PubMed] [Google Scholar]
- 10.Kabesch M. A glitch in the switch? Of endotoxin, CD14, and allergy. Am J Respir Crit Care Med. 2006;174:365–366. doi: 10.1164/rccm.2604006. [DOI] [PubMed] [Google Scholar]
- 11.Vercelli D, Baldini M, Stern D, Lohman IC, Halonen M, Martinez F. CD14: a bridge between innate immunity and adaptive IgE responses. J Endotoxin Res. 2001;7:45–48. [PubMed] [Google Scholar]
- 12.Pugin J, Heumann ID, Tomasz A, et al. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 13.Ayaslioglu E, Tekeli E, Birengel S. Significant elevation of serum soluble CD14 levels in patients with brucellosis. Jpn J Infect Dis. 2005;58:11–14. [PubMed] [Google Scholar]
- 14.Hoheisel G, Zheng L, Teschler H, Striz I, Costabel U. Increased soluble CD14 levels in BAL fluid in pulmonary tuberculosis. Chest. 1995;108:1614–1616. doi: 10.1378/chest.108.6.1614. [DOI] [PubMed] [Google Scholar]
- 15.Oesterreicher C, Pfeffel F, Petermann D, Muller C. Increased in vitro production and serum levels of the soluble lipopolysaccharide receptor sCD14 in liver disease. J Hepatol. 1995;23:396–402. doi: 10.1016/0168-8278(95)80197-9. [DOI] [PubMed] [Google Scholar]
- 16.Wuthrich B, Kagi MK, Joller-Jemelka H. Soluble CD14 but not interleukin-6 is a new marker for clinical activity in atopic dermatitis. Arch Dermatol Res. 1992;284:339–342. doi: 10.1007/BF00372036. [DOI] [PubMed] [Google Scholar]
- 17.Ferrero E, Hsieh CL, Francke U, Goyert SM. CD14 is a member of the family of leucine-rich proteins and is encoded by a gene syntenic with multiple receptor genes. J Immunol. 1990;145:331–336. [PubMed] [Google Scholar]
- 18.Goyert SM, Ferrero E, Rettig WJ, Yenamandra AK, Obata F, Le Beau MM. The CD14 monocyte differentiation antigen maps to a region encoding growth factors and receptors. Science. 1988;239:497–500. doi: 10.1126/science.2448876. [DOI] [PubMed] [Google Scholar]
- 19.Boniotto M, Braida L, Ventura A, Percopo S, Amoroso A, Crovella S. Promoter polymorphisms of the CD14 gene in Italian patients with coeliac disease. J Med Genet. 2003;40:e108. doi: 10.1136/jmg.40.9.e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koenig W, Khuseyinova N, Hoffmann MM, et al. CD14 C(-260)–>T polymorphism, plasma levels of the soluble endotoxin receptor CD14, their association with chronic infections and risk of stable coronary artery disease. J Am Coll Cardiol. 2002;40:34–42. doi: 10.1016/s0735-1097(02)01937-x. [DOI] [PubMed] [Google Scholar]
- 21.Litonjua AA, Belanger K, Celedon JC, et al. Polymorphisms in the 5' region of the CD14 gene are associated with eczema in young children. J Allergy Clin Immunol. 2005;115:1056–1062. doi: 10.1016/j.jaci.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Raunio T, Knuuttila M, Karttunen R, Vainio O, Tervonen T. Serum sCD14, polymorphism of CD14(-260) and periodontal infection. Oral Dis. 2009;15:484–489. doi: 10.1111/j.1601-0825.2009.01573.x. [DOI] [PubMed] [Google Scholar]
- 23.Virta M, Pessi T, Helminen M, et al. Interaction between CD14-159C>T polymorphism and Helicobacter pylori is associated with serum total immunoglobulin E. Clin Exp Allergy. 2008;38:1929–1934. doi: 10.1111/j.1365-2222.2008.03103.x. [DOI] [PubMed] [Google Scholar]
- 24.Klein W, Tromm A, Griga T, et al. A polymorphism in the CD14 gene is associated with Crohn disease. Scand J Gastroenterol. 2002;37:189–191. doi: 10.1080/003655202753416867. [DOI] [PubMed] [Google Scholar]
- 25.Obana N, Takahashi S, Kinouchi Y, et al. Ulcerative colitis is associated with a promoter polymorphism of lipopolysaccharide receptor gene, CD14. Scand J Gastroenterol. 2002;37:699–704. doi: 10.1080/00365520212504. [DOI] [PubMed] [Google Scholar]
- 26.Mohamed JA, DuPont HL, Jiang ZD, et al. A single-nucleotide polymorphism in the gene encoding osteoprotegerin, an anti-inflammatory protein produced in response to infection with diarrheagenic Escherichia coli, is associated with an increased risk of nonsecretory bacterial diarrhea in North American travelers to Mexico. J Infect Dis. 2009;199:477–485. doi: 10.1086/596319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DuPont HL, Jiang ZD, Okhuysen PC, et al. Antibacterial chemoprophylaxis in the prevention of traveler's diarrhea: evaluation of poorly absorbed oral rifaximin. Clin Infect Dis. 2005;41(Suppl 8):S571–S576. doi: 10.1086/432954. [DOI] [PubMed] [Google Scholar]
- 28.Murray BE, Mathewson JJ, DuPont HL, Hill WE. Utility of oligodeoxyribonucleotide probes for detecting enterotoxigenic Escherichia coli. J Infect Dis. 1987;155:809–811. doi: 10.1093/infdis/155.4.809. [DOI] [PubMed] [Google Scholar]
- 29.Nataro JP, Seriwatana J, Fasano A, et al. Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect Immun. 1995;63:4721–4728. doi: 10.1128/iai.63.12.4721-4728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang DB, Mohamed JA, Nataro JP, DuPont HL, Jiang ZD, Okhuysen PC. Virulence characteristics and the molecular epidemiology of enteroaggregative Escherichia coli isolates from travellers to developing countries. J Med Microbiol. 2007;56:1386–1392. doi: 10.1099/jmm.0.47161-0. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed JA, Huang DB, Jiang ZD, et al. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol. 2007;45:121–126. doi: 10.1128/JCM.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. PHASE version 2.1, http://stephenslab.uchicago.edu. [Google Scholar]
- 33.Labeta MO, Vidal K, Nores JE, et al. Innate recognition of bacteria in human milk is mediated by a milk-derived highly expressed pattern recognition receptor, soluble CD14. J Exp Med. 2000;191:1807–1812. doi: 10.1084/jem.191.10.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baveye S, Elass E, Fernig DG, Blanquart C, Mazurier J, Legrand D. Human lactoferrin interacts with soluble CD14 and inhibits expression of endothelial adhesion molecules, E-selectin and ICAM-1, induced by the CD14-lipopolysaccharide complex. Infect Immun. 2000;68:6519–6525. doi: 10.1128/iai.68.12.6519-6525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ushiba D, Sugiyama K, Nakane M. Changes in intestinal flora of mice following administration of bacterial endotoxins. Jpn J Microbiol. 1962;6:69–78. [Google Scholar]
- 36.Rowley D, Marneerushapisal V. Local cell mediated antibacterial immunity in the intestine. Basel: Karger; 1983. [Google Scholar]
- 37.Mathan VI, Penny GR, Mathan MM, Rowley D. Bacterial lipopolysaccharide-induced intestinal microvascular lesions leading to acute diarrhea. J Clin Invest. 1988;82:1714–1721. doi: 10.1172/JCI113785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uehara A, Sugawara S, Watanabe K, et al. Constitutive expression of a bacterial pattern recognition receptor, CD14, in human salivary glands and secretion as a soluble form in saliva. Clin Diagn Lab Immunol. 2003;10:286–292. doi: 10.1128/CDLI.10.2.286-292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Wenneras C, Ave P, Huerre M, et al. Blockade of CD14 aggravates experimental shigellosis. J Endotoxin Res. 2001;7:442–446. [PubMed] [Google Scholar]
- 40.Yu LC, Turner JR, Buret AG. LPS/CD14 activation triggers SGLT-1-mediated glucose uptake and cell rescue in intestinal epithelial cells via early apoptotic signals upstream of caspase-3. Exp Cell Res. 2006;312:3276–3286. doi: 10.1016/j.yexcr.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Lin SM, Frevert CW, Kajikawa O, et al. Differential regulation of membrane CD14 expression and endotoxin-tolerance in alveolar macrophages. Am J Respir Cell Mol Biol. 2004;31:162–170. doi: 10.1165/rcmb.2003-0307OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutherland AM, Walley KR, Russell JA. Polymorphisms in CD14, mannose-binding lectin, and Toll-like receptor-2 are associated with increased prevalence of infection in critically ill adults. Crit Care Med. 2005;33:638–644. doi: 10.1097/01.ccm.0000156242.44356.c5. [DOI] [PubMed] [Google Scholar]
- 43.Zhao D, Sun T, Zhang X, et al. Role of CD14 promoter polymorphisms in Helicobacter pylori infection–related gastric carcinoma. Clin Cancer Res. 2007;13:2362–2368. doi: 10.1158/1078-0432.CCR-06-2612. [DOI] [PubMed] [Google Scholar]
- 44.LeVan TD, Bloom JW, Bailey TJ, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 45.Flores J, DuPont HL, Lee SA, et al. Influence of host interleukin-10 polymorphisms on development of traveler's diarrhea due to heat-labile enterotoxin-producing Escherichia coli in travelers from the United States who are visiting Mexico. Clin Vaccine Immunol. 2008;15:1194–1198. doi: 10.1128/CVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang ZD, Okhuysen PC, Guo DC, et al. Genetic susceptibility to enteroaggregative Escherichia coli diarrhea: polymorphism in the interleukin-8 promotor region. J Infect Dis. 2003;188:506–511. doi: 10.1086/377102. [DOI] [PubMed] [Google Scholar]
- 47.Mohamed JA, DuPont HL, Jiang ZD, et al. A novel single-nucleotide polymorphism in the lactoferrin gene is associated with susceptibility to diarrhea in North American travelers to Mexico. Clin Infect Dis. 2007;44:945–952. doi: 10.1086/512199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.