Abstract
T and B lymphocytes are developmentally and functionally related cells of the immune system, representing the two major branches of adaptive immunity. Although originating from a common precursor, they play very different roles: T cells contribute to and drive cell-mediated immunity, while B cells secrete antibodies. Because of their functional importance and well-characterized differentiation pathways, T and B lymphocytes are ideal cell-types with which to understand how functional differences are encoded at the transcriptional level. Although there has been a great deal of interest in defining regulatory factors that distinguish T and B cells, a truly genome-wide view of the transcriptional differences between these two cells types has not yet been taken. To obtain a more global perspective of the transcriptional differences underlying T and B cells, we exploited the statistical power of combinatorial profiling on different microarray platforms, and the breadth of the Immunological Genome Project (ImmGen) gene-expression database, to generate robust differential signatures. We find that differential expression in T and B cells is pervasive, the majority of transcripts showing statistically significant differences. These distinguishing characteristics are acquired gradually, though all stages of B and T differentiation. On the other hand, very few T vs. B signature genes are uniquely expressed in these lineages, but are shared throughout immune cells.
INTRODUCTION
T and B lymphocytes are closely related cell lineages of the immune system, having the unique ability to somatically rearrange gene segments encoding receptors for antigen, the key molecules of the adaptive immune system. Both lineages are thought to arise from the same bone marrow precursors, the nature of which is somewhat debated at present. They complete remarkably parallel stages of differentiation and selection before reaching morphologically similar mature states, as “naïve” lymphocytes resting in secondary lymphoid organs, from which activation by cognate antigen will provoke their terminal differentiation to effector or memory states.
Although T and B lymphocytes broadly share a role in the adaptive immune system, their functions within this responsive structure are entirely different: T cells participate primarily in cell-mediated immunity and in orchestrating cellular responses, while B cell production of antibodies is the hallmark of humoral immunity. As these functional differences are usually assumed to be underpinned by differences in the basic cell biology of these lymphocytes, there has been some interest in determining what, beyond the antigen receptors and their ancillary factors, distinguishes B and T lymphocytes. In particular, how differently B and T lymphocytes utilize the blueprint of genes encoded in the genome.
A notable early study used cDNA subtractive hybridization, where cDNA from T and B cells were isolated and subjected to exhaustive subtraction, to estimate that T and B cells differ by only 2% of their mRNA (1,2), among which TCR-encoding genes were eventually isolated. Since then, several key regulators have been found, through knockout studies, to be necessary for the differentiation of either the T or B lineages: Pax5, Ebf1 or Sfpi1 (PU.1) for B cells, Notch1 and Gata3 for T cells (3-7). While identifying such lineage-specification factors is of course essential, viewing the differences between lineages solely through the lens of a few control factors necessarily overlooks the complex transcriptional programs present in any given cell. The development of microarray technologies, and the continued improvements in microarray platforms and their annotations, have allowed a perspective on the transcriptome that is global and also more quantitatively nuanced. A few early studies used this approach to compare T and B lymphocytes (8-11), identifying sets of genes that are differentially expressed in B and T cells, as well as more generally shared sets; as might be expected, transcripts that varied during T or B lymphocyte differentiation showed more inter-lineage differential than invariant housekeeping genes (8).
While generating such data for transcripts that are strongly expressed and/or clearly differential is straightforward, there is difficulty in arriving at more general conclusions for the entire transcriptome in such comparisons. These problems lie in the confidence one can have in calls that a transcript is present or absent in a given dataset, given the difficulty in distinguishing true signals from noise due to false-negatives (non-performing features on a microarray, sub-threshold detection) or false-positives (cross-hybridizing microarray features), both of which are poorly controlled on any one microarray (12,13). In addition, the use of arbitrary thresholds to define expression differentials tends to create overly simplistic distinctions. In the present study, we have attempted to robustly define the transcriptome differences underlying T and B lymphocytes by exploiting the unique datasets generated in the pilot phases of the Immunological Genome Project (ImmGen)3. ImmGen is a collaborative group of immunology and computational biology laboratories aiming to decipher, on a broad scale, the patterns of gene expression and genetic regulatory networks of the immune system of the mouse (14). We used the cross-verifying power of expression profiling on independent microarray platforms, as well as the breadth of gene-expression datasets available in the ImmGen database, to robustly explore what distinguishes T and B lymphocytes at the transcriptional level, and to analyze when these distinctions are acquired during T and B lineage differentiation.
MATERIALS AND METHODS
Mice
6-week old C57BL/6J mice were bred in specific pathogen-free conditions, under Institutional Animal Care and Use Committee protocol (protocol 02954).
Cell Sorting and Flow Cytometry
All cells were purified using the sorting protocol and mAbs listed on www.ImmGen.org.
Microarray Analysis
For multi-platform microarrray profiling, RNA was prepared from sorted CD4+ T cell and CD19+ B cell populations from C57BL/6J mice using Trizol reagent as described (15). RNA was amplified and hybridized on the Affymetrix Mouse Gene 1.0 ST, Agilent Mouse GE 1-Color, Illumina Mouse-6 v1.1 BeadChip, and Nimblegen Mouse X12 arrays according to the procedures specific to each platform. Raw data were pre-processed using software compatible for each platform, and all normalized using the RMA algorithm. Thresholds on expression values above which a gene was considered expressed were derived for each platform by one of two distribution-based approaches. For platforms with well-defined negative control probesets (Illumina Mouse-6 and Nimblegen X12), the threshold for greater-than-chance expression was defined as expression values greater than or equal to the 95% quantile of expression values in the negative controls. The “negative controls” for Agilent and Affymetrix arrays, however, exhibited notably different behavior in relation to non-control probes (likely due to the inclusion of intronic probes with some degree of expression), and thus did not allow for the same type of control-based analysis as Illumina and Nimblegen. For these samples, a Gaussian Mixture Model (GMM) was used to arrive at thresholds consistent with a controls-based approach. GMM is an Expectation-Maximization algorithm, the aim of which is to optimize the likelihood that a set of data points is generated by a mixture of Gaussian distributions. In this case, the MATLAB software “fit” function with parameter “gauss3” was used to the model the observed chip-wide expression distribution profile of all non-control probe-sets, such that each Gaussian component of the mixture corresponded to a different source of signal (i.e. background and genuine expression). Thresholds for greater-than-chance expression were then empirically defined as the value above which there is an equal probability that the signal is part of either distribution. This setting was validated on the Illumina and Nimblegen arrays by a good fit with thresholds derived from true negative controls. Specifically, the average percentage of genes in the 4-platform common genome expressed above the GMM-derived thresholds for Affymetrix and Agilent were 50.5% and 42.7%% respectively, which is concordant with the controls-derived thresholds used for Nimblegen and Illumina (47.7%-46.4%). Conversely, the equivalent controls-derived thresholds for Affymetrix and Agilent were highly discordant, with averages of 15.5% and 84.8% respectively (data not shown).
For data analysis using ImmGen datasets, raw data for all populations were normalized using the RMA algorithm (16) implemented in the “Expression File Creator” module in the GenePattern suite (17). Differential signatures were visualized using the “Multiplot” module. Signature transcripts were clustered using the “Hierarchical Clustering” module, using Pearson's correlation as a metric and visualized using the “Hierarchical Clustering Viewer” heatmap module.
To display the expression of transcripts during differentiation, a modified K-means algorithm was used to cluster the B and T cell signatures in order to represent the developmental activation of their respective genes. Unlike the traditional K-means approach of clustering observations around randomly determined centroids, this analysis used predefined, theoretical centroids, each characterized by a stepwise expression profile corresponding to successive stages of activation. Consequently, n-1 centroids were used to cluster a signature comprised of n stages of development. Pearson's correlation coefficient was used as the distance metric. This results in the clustering of probesets around the single-stage activation exemplar to which it is most correlated.
The “Population Plots” position cell populations in a two-dimensional frame of reference, created using the expression values of sets of genes that most distinguish two reference populations. The X and Y axes (“B-ness” and ”T-ness”, respectively, in Fig. 4) were defined by expression values for the signature genes over-expressed in one reference population relative to the other: expression values of these genes were normalized relative to the reference populations (scaled to 0 and 1, where 0 is the expression value in the “low” population and 1 the value in the “high” population); scaled values for all signature genes were then averaged to yield the x and y coordinates of the populations tested.
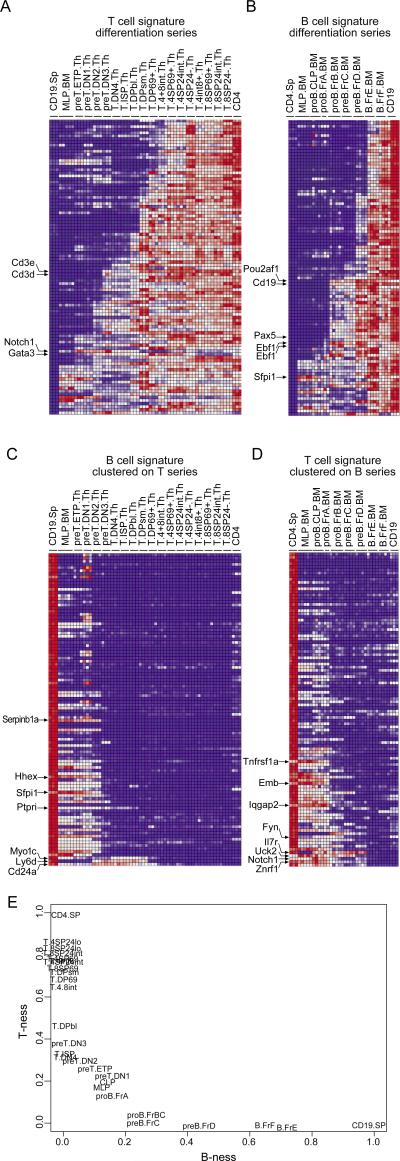
Figure 4. The transcripts that most distinguish T and B cells are acquired, or lost, in stages throughout differentiation.
Heat-map representations of the expression of the “Top 100” T cell of B cell genes during T cell differentiation in the thymus (A,C) or during B cell differentiation in the bone marrow (B,D). Cell-types have been arranged according to their sequence during differentiation and genes were clustered using an ordering algorithm according to the stage at which they are expressed. E): Population plot in which cell-types have been positioned according to their “T-ness” and “B-ness”, defined from the aggregate expression values of genes most differentially expressed in mature B and T cells (see Materials and Methods).
For cluster analysis, expression values were normalized to the mean expression for each gene, and a partition clustering algorithm (pam, S-Plus) was applied to the expression values in the T cell differentiation series. This cluster composition was then applied to expression values within nonT/nonB datasets within ImmGen (precursors, myeloid, NK cells).
All datasets have been deposited at NCBI/GEO under accession # GSE15907 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15907).
RESULTS
Defining gene expression in T and B cells from the four-platform data
As part of the evaluation process to select a microarray platform most compatible with the ImmGen project, bulk CD4+ T cells and CD19+ B cells were sorted from spleen suspensions of 6 week-old C57Bl/6J mice, for RNA preparations that were used to probe microarrays from four different commercial sources (Affymetrix Mouse Gene 1.0 ST array, Agilent Mouse GE 1-Color Array, Illumina Mouse-6 v1.1 Expression Beadchip Array and Nimblegen Mouse X12 array). Three replicate datasets were generated for each cell type and each array (except one technical failure for Agilent), and the data were used for a comparative assessment of reproducibility and noise, of importance in the context of the ImmGen program (data not shown). Relevant to the present project, we used the combined datasets to address the depth and variation of gene expression in B and T lymphocytes, under the assumption that comparable signals obtained in independent microarrays would be highly confirmatory, particularly since the various arrays use fundamentally different oligonucleotide probes (multiple 22mers for Affymetrix, single long nucleotides for others) and probe/label chemistries (cDNA or cRNA). We generated a “Common Gene Table” which included 12,299 genes represented in at least 3 out of 4 arrays (full data listed in Table SI). We then defined, for each array, threshold expression values above which a probe was scored as showing significant expression (at a probability of P <0.05, as detailed in Supplementary Material; because reliable negative controls are only present on two of the arrays, these thresholds for significant expression were based on those negative controls when present, and on a Gaussian deconvolution of expression profiles similarly applied to all four platforms). This analysis showed excellent agreement between the platforms: the expression patterns in either T or B cells proved quite reproducible overall, being between 43% and 50% of the genes represented (Table IA), with only a low proportion of false-positives (signals detected on one array but absent on all others, and thus likely to represent spurious noise) and false-negatives (signals absent on a given array but present on at least 2 others). Combining the results from all 4 arrays, and scoring those genes found to be expressed in at least 2 of the platforms, showed that a very similar proportion of the genome (49.7 %) is active in both B and T cells (Table IB).
Table I. Summary of multi-platform gene-expression data.
A) Splenic CD4+ T cells and CD19+ B cells were profiled on Affymetrix, Agilent, Nimbelgen and Illumina whole-genome microarrays. Resulting gene-expression data from each platform were analyzed to yield the percentage of expressed probes, percentage of false-positives (defined as a probe being expressed on one platform, but not the other three), percentage of false-negatives (defined as the absence of a probe's expression in one platform but present in the other three) and overall concordance (defined as the overall percentage of probes whose expression or absence is in agreement with the majority of platforms). B) The overall expression of the genome in T and B cells was calculated based on the number of genes registering as significantly expressed for each platform with concordance being defined as a given gene's expression or absence in 2, 3 or 4 out of 4 platforms (rows).
| Table IA | ||||
|---|---|---|---|---|
| Sample | Expressed genes (%) | False-positives (%) | False-negatives (%) | Overall Concordance (%) |
| Affymetrix CD19 | 51 | 8 | 2 | 84 |
| Affymetrix CD4 | 50 | 8 | 2 | 84 |
| Agilent CD19 | 43 | 4 | 7 | 92 |
| Agilent CD4 | 43 | 4 | 7 | 92 |
| Illumina CD19 | 47 | 9 | 8 | 84 |
| Illumina CD4 | 47 | 10 | 8 | 83 |
| Nimblegen CD19 | 46 | 5 | 4 | 89 |
| Nimblegen CD4 | 46 | 4 | 4 | 89 |
| Table IB | ||
|---|---|---|
| Concordant Chips | Expressed in CD4 (%) | Expressed in CD19 (%) |
| 2 of 4 | 49.74 | 49.67 |
| 3 of 4 | 43.26 | 43.35 |
| 4 of 4 | 32.41 | 32.06 |
Next, we generated a robust signature of differential T vs. B expression, again harnessing the combinatorial power of the multi-platform measurement to determine with a high degree of confidence the differences in transcript abundance. The data in the Common Gene Table described above were filtered for transcripts scoring positively in at least one cell-type (8411 genes) and subsequently used to generate Fold Change (FC) estimates of the T/B ratio of expression for each of the four microarrays (calculated from the mean of the triplicate expression values). There was, for the most part, very good concordance between the FC values on different platforms, consistent with results from previous microarray comparison projects (18), as illustrated for one comparison in Fig. 1A (all comparisons are shown in Fig. S1, all data listed in Table SII). We then generated consensus FCs by averaging the FCs measured on each microarray (the most diferential transcripts are listed in Table II, all data in Table SII). To avoid spurious effects due to aberrant values on any one microarray platform, an outlier elimination procedure was implemented, where the FC value from one platform was disregarded if it fell more than 3 standard deviations away from the mean of the other three platforms. T vs. B differential expression ranged up to 633-fold (for an Ig-V region), with 174/8411 transcripts showing a differential of 20-fold or greater, and 1364/8411 a differential of 2-fold or greater.
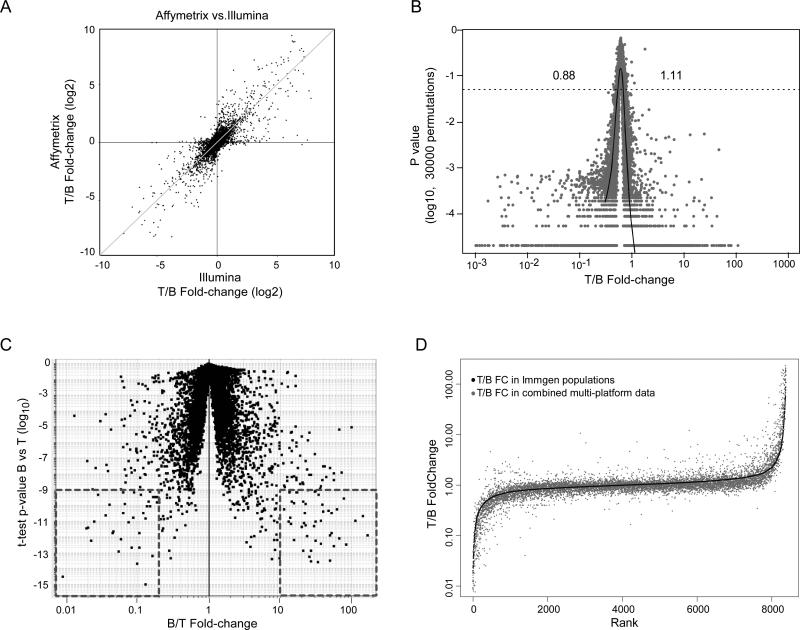
Figure 1. Defining T vs. B differential signatures.
A): RNA preparations from CD4+ cells and CD19 B cells were profiled on Affymetrix and Illumina whole-genome microarrays, and the T vs. B FoldChange was calculated for the same genes on both microarrays. B): Consensus T vs. B cell expression ratios were calculated by combining information from four different microarray platforms, and a false-discovery rate on these FoldChange values was estimated by repeated randomization of the datasets, testing how often the FoldChange observed for a given gene could be observed by chance. The threshold FoldChange values which reached statistical significance were estimated at <0.88 and > 1.11, for a genome-wide p=0.05 C): Datasets from several populations of mature T cells (whole CD3+CD4+ splenocytes, naïve CD4+ and CD8+ cells from spleen and LN, CD44hi CD4+ and CD8+ splenocytes) and B cells (whole CD19+ splenocytes, mature bone marrow “Fraction F” cells, T3 splenic subset, follicular B from spleen and peritoneal cavity, marginal zone B), all profiled on the Affymetrix MuGeneST1.0 platform, were analyzed in combination to generate consensus measures of differential expression. The aggregate T vs. B expression ratios are plotted against the Student's t-test p-value. “Top 100” signature genes for B and T are outlined. D): Comparison of T/B FoldChanges determined from the multiplatform data (black dots) or from the combined ImmGen datasets (grey dots).
Table II. Multi-platform T vs. B differential signature genes.
Consensus T vs. B FoldChange values (calculated as the average of all four platforms, eliminating outliers) along with false discovery rate (FDR) for the ‘top 25’ most differentially expressed genes for CD4+ T and CD19+ B cells.
| Gene Symbol | Combined multiplatform T/B ratio | FDR |
|---|---|---|
| IGL-V1 | 0.002 | <0.00003 |
| H2-AB1 | 0.002 | <0.00003 |
| LY6D | 0.002 | <0.00003 |
| MS4A1 | 0.002 | <0.00003 |
| H2-AA | 0.002 | <0.00003 |
| H2-EB1 | 0.003 | <0.00003 |
| SCD1 | 0.003 | 0.000166667 |
| CD74 | 0.003 | <0.00003 |
| BLNK | 0.004 | <0.00003 |
| H2-DMB2 | 0.004 | 0.0006 |
| LY86 | 0.005 | 0.000366667 |
| CR2 | 0.005 | <0.00003 |
| H2-DMB1 | 0.005 | <0.00003 |
| LYN | 0.005 | 0.0002 |
| PLAC8 | 0.005 | <0.00003 |
| STK23 | 0.005 | 6.66667E-05 |
| FCER2A | 0.005 | <0.00003 |
| NAPSA | 0.005 | 3.33333E-05 |
| RASGRP3 | 0.006 | <0.00003 |
| FAIM3 | 0.006 | 0.0001 |
| 2010001M09RIK | 0.006 | 3.33333E-05 |
| CD79B | 0.006 | 0.000666667 |
| HHEX | 0.006 | 6.66667E-05 |
| BANK1 | 0.007 | <0.00003 |
| TNFRSF13C | 0.007 | 3.33333E-05 |
| CD3G | 177.559 | <0.00003 |
| CD247 | 131.154 | <0.00003 |
| CD3D | 125.911 | <0.00003 |
| IL7R | 117.127 | <0.00003 |
| TCRA | 98.672 | <0.00003 |
| TRAT1 | 96.180 | <0.00003 |
| IGFBP4 | 88.251 | <0.00003 |
| 2610019F03RIK | 84.180 | <0.00003 |
| E430004N04RIK | 80.586 | <0.00003 |
| A530021J07 | 76.378 | <0.00003 |
| PRKCQ | 76.298 | 0.002433333 |
| 2310032F03RIK | 70.026 | 6.66667E-05 |
| ITK | 68.390 | <0.00003 |
| PRKCH | 60.929 | <0.00003 |
| TCF7 | 56.097 | 3.33333E-05 |
| BCL11B | 55.890 | <0.00003 |
| LAT | 55.061 | 0.0002 |
| TCRB-V13 | 45.987 | <0.00003 |
| THY1 | 44.725 | <0.00003 |
| 1700025G04RIK | 44.512 | 6.66667E-05 |
| TNFRSF7 | 43.149 | <0.00003 |
| FYB | 43.011 | <0.00003 |
| BC021614 | 40.585 | 0.000133333 |
| CD6 | 40.556 | <0.00003 |
| AMPD1 | 40.043 | <0.00003 |
We estimated the significance of these aggregate FCs by a data randomization procedure: triplicate expression values for CD4+ T cells and CD19+ B cells were scrambled for each gene and each platform, and the aggregate fold change was recalculated from this randomized data as before (again applying the outlier elimination procedure). The procedure was repeated 30,000 times, counting the number of times the mock FC value for a given gene was equal or greater to that observed, yielding an estimate of the probability that the observed FC could be due to chance. As shown in Fig. 1B, most of the changes were highly significant. The range of fold change values that reached significance at p<0.05 was estimated from the FC vs. p-value scatter plot with a locally smoothed regression (loess; dark line on Fig. 1B). Significance was observed at very low FC values (> 1.11 or <0.86), involving 5671 of the 8411 commonly expressed genes analyzed. From a technical standpoint, these data confirm the notion that combinatorial microarray profiling can reliably report on minute differences in expression (19). Overall, these data indicate that the difference between T and B lymphocytes involves a relative minority of transcripts with large differences in expression, but that a large fraction (at least 65%) of transcripts are subtly but significantly different in B and T cells.
Defining a T vs. B consensus signature from the broader ImmGen data
While using multi-platform microarray profiling provided a technically robust T vs. B signature, it was limited to bulk CD4+ and CD19+ splenocytes, which do not necessarily represent the broader range of T and B lymphocytes. Thus, to complement this signature, we thought it worthwhile to create a T vs. B signature that would encompass a wider range of T and B cell subpopulations, but on a single microarray platform. The datasets of mature B and T lymphocytes available on the ImmGen database should enable the definition of differential signatures of ‘T-ness’ and ‘B-ness’ across more subpopulations. We selected datasets from a wide range of mature T and B cells, including CD4+ and CD8+ T cells from the spleen, lymph node and thymus as well as B cells of different subtypes (follicular, marginal zone, B1) from the spleen, peritoneal cavity and bone marrow. A composite T vs. B signature was calculated by averaging across the two groups of populations, and the significance of these Fold Change values was estimated with a simple Welsh's t-test (the most differential transcripts are listed in Table III). As shown in Fig. 1C, many genes were differentially expressed to a highly significant degree: 1078 genes, or 3% of the genes on the microarray attained significance at a p-value of less than 10-5 (a conservative threshold for corrected genome-wide significance), for FC values ranging from 1.2 to 180 (given the increased variance, this comparison is less effective at ascribing significance to the numerous but subtle differences described above).
Table III. Multi-platform T vs. B differential signature genes.
Consensus T vs. B FoldChange values (calculated as the average of all four platforms, eliminating outliers) along with false discovery rate (FDR) for the ‘top 25’ most differentially expressed genes for CD4+ T and CD19+ B cells.
| Gene Symbol | Combined multiplatform T/B ratio | FDR |
|---|---|---|
| Igl-V1 | 0.002 | <0.00003 |
| H2-Ab1 | 0.002 | <0.00003 |
| Ly6d | 0.002 | <0.00003 |
| Ms4a1 | 0.002 | <0.00003 |
| H2-Aa | 0.002 | <0.00003 |
| H2-Eb1 | 0.003 | <0.00003 |
| Scd1 | 0.003 | 0.000166667 |
| Cd74 | 0.003 | <0.00003 |
| Blnk | 0.004 | <0.00003 |
| H2-Dmb2 | 0.004 | 0.0006 |
| Ly86 | 0.005 | 0.000366667 |
| Cr2 | 0.005 | <0.00003 |
| H2-Dmb1 | 0.005 | <0.00003 |
| Lyn | 0.005 | 0.0002 |
| Plac8 | 0.005 | <0.00003 |
| Stk23 | 0.005 | 6.66667E-05 |
| Fcer2a | 0.005 | <0.00003 |
| Napsa | 0.005 | 3.33333E-05 |
| Rasgrp3 | 0.006 | <0.00003 |
| Faim3 | 0.006 | 0.0001 |
| 2010001m09rik | 0.006 | 3.33333E-05 |
| Cd79b | 0.006 | 0.000666667 |
| Hhex | 0.006 | 6.66667E-05 |
| Bank1 | 0.007 | <0.00003 |
| Tnfrsf13c | 0.007 | 3.33333E-05 |
| Cd3g | 177.559 | <0.00003 |
| Cd247 | 131.154 | <0.00003 |
| Cd3d | 125.911 | <0.00003 |
| Il7r | 117.127 | <0.00003 |
| Tcra | 98.672 | <0.00003 |
| Trat1 | 96.180 | <0.00003 |
| Igfbp4 | 88.251 | <0.00003 |
| 2610019f03rik | 84.180 | <0.00003 |
| E430004n04rik | 80.586 | <0.00003 |
| A530021j07 | 76.378 | <0.00003 |
| Prkcq | 76.298 | 0.002433333 |
| 2310032f03rik | 70.026 | 6.66667E-05 |
| Itk | 68.390 | <0.00003 |
| Prkch | 60.929 | <0.00003 |
| Tcf7 | 56.097 | 3.33333E-05 |
| Bcl11b | 55.890 | <0.00003 |
| Lat | 55.061 | 0.0002 |
| Tcrb-V13 | 45.987 | <0.00003 |
| Thy1 | 44.725 | <0.00003 |
| 1700025g04rik | 44.512 | 6.66667E-05 |
| Tnfrsf7 | 43.149 | <0.00003 |
| Fyb | 43.011 | <0.00003 |
| Bc021614 | 40.585 | 0.000133333 |
| Cd6 | 40.556 | <0.00003 |
| Ampd1 | 40.043 | <0.00003 |
We then asked whether this second signature derived from multiple B and T cell populations within the ImmGen datasets would compare to that derived above by multi-platform analysis of CD4+ and CD19+ splenocytes. The majority of each signature's ‘Top 100’ most distinguishing transcripts are shared, with 64% of T cell transcripts and 52% of B cell transcripts being present in both the multi-platform and ImmGen determinations. A ranked plot of the T vs. B FCs in the two signatures reveals good overall matching across the differential ranking (Fig. 1D and Table SII). Some differences between the two signatures were observed, however, which are to be expected as the ImmGen determination used a broad array of T and B populations while the multi-platform determination used solely CD4+ and CD19+ splenocytes (for instance, CD4 itself ranks differently in the two signatures).
Are the transcripts that distinguish T and B cells specific to these lymphoid lineages?
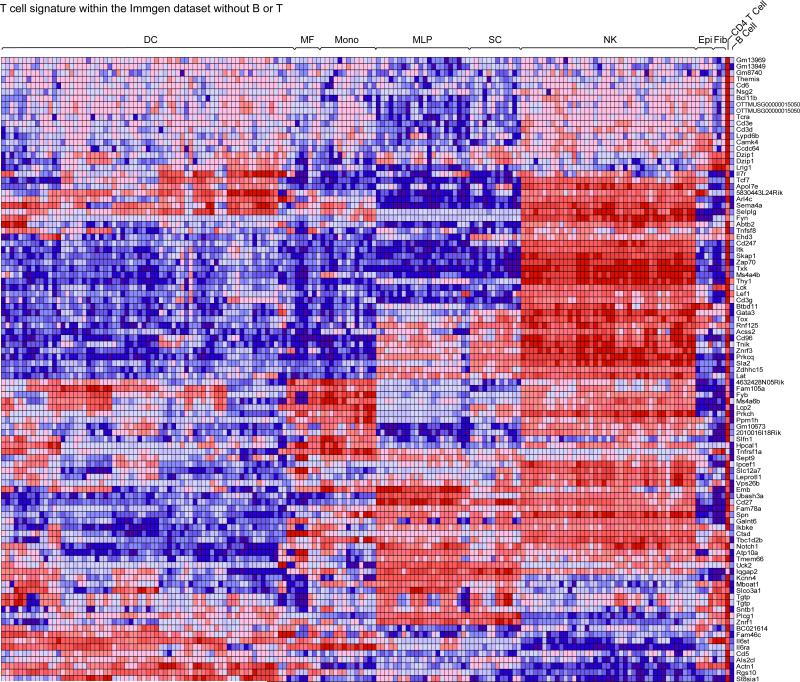
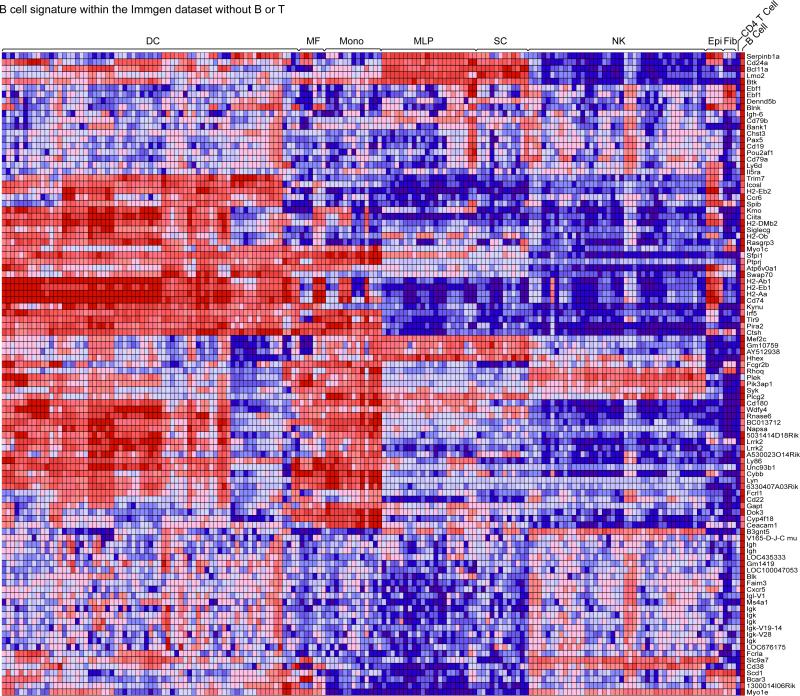
Having generated these robust T vs. B differential signatures, we next asked whether the transcripts that most distinguish T and B cells are unique to these cells, or whether their expression is also shared with cells of other nonT/nonB lineages. Since, in most schemas of hematopoietic cell differentiation, B and T lymphocytes represent terminal splits of the same lymphocyte branch, one might expect that the transcripts that sharply distinguish them may be uniquely expressed, solely present there and not in any other lineage (as are TCR and Ig transcripts for instance). More generally, it is of interest to ask how many transcripts uniquely define a particular cell-type, and how many truly T- or B-specific genes actually exist, other than the antigen-specific receptors that defined these cells. To address this question, we mapped the expression of the 100 genes that most strongly differentiate T or B cells across the other immune-cell populations of the ImmGen database (DCs and macrophages, NK cells, stem cells; γδT cells were not considered because too similar to αβT cells). As shown in the heat-map representations of Figs. 2 and 3, T and B signature transcripts were shared extensively with other lineages. As might be expected, T cell transcripts were more frequently shared with NK cells, B cell transcripts with dendritic or other myeloid cells, but this was not an absolute rule, and there were significant clusters of T signature transcripts present in myeloid cells, and B signature transcripts in NK cells. Even stromal cells and monocytes expressed some B or T cell genes. These data indicate that the transcripts that most distinguish T and B lymphocytes are broadly expressed in other immune cells, and hardly any transcripts fall into the category of being absolutely specific to B or T lymphocytes.
Figure 2. The transcripts that most distinguish T and B cells are expressed throughout immune cells.
Heat-map representations of the expression of the “Top 100” T cell signature genes across the immune cell populations contained in the ImmGen database. Genes are arranged by hierarchical clustering.
Figure 3. The transcripts that most distinguish T and B cells...continued.
Heat-map representations of the expression of the “Top 100” B cell signature genes across the immune cell populations contained in the ImmGen database. Genes are arranged by hierarchical clustering.
We cannot completely rule out the possibility that this conclusion is influenced by spurious lymphocyte contamination in some datasets, but this seems unlikely since, if a given dataset were contaminated with T or B lymphocytes, one would expect that all of the T or B specific signature would appear expressed. It is clear from Figs. 2 and 3, however, that only distinct modules of the T or B signatures are expressed within a given population.
How are transcriptional characteristics of mature T and B cells acquired during differentiation?
The differentiation of T and B lymphocytes is a well-characterized process marked by distinct stages that can be tracked by the expression of various cell-surface molecules (20,21). As such, T and B cells are attractive lineages in which to ask how the ‘identity’ of mature cells is acquired. While a good deal is known about the timing of expression of various transcription factors during the differentiation of these two cell-types, (3,22,23), differentiation along the T and B lineages involves many other transcripts (24). We thus asked how the identity of mature T and B cells, as reflected in their above-defined distinguishing transcripts, is acquired during differentiation. In other words, when does a B cell become a B cell, or a T cell become a T cell? To address this question, we used an ordering algorithm to arrange T and B signature transcripts according to the stage at which they are induced during differentiation. As shown in the heat-map representations of Figs. 4A-B, we found that signature transcripts are acquired in a sequential manner, evenly through several steps of differentiation rather than being coordinately turned on at one particular stage. These steps do not particularly coincide with the rearrangement of antigen-receptor genes, but occur through the Double-Negative and Double-Positive stages for thymic T cell precursors, and through the transitions of pro- and pre-B cells in the bone marrow. In this respect, the full identity of T and B cells is realized gradually, and not fully attained until maturity. This finding goes against the notion that expressing a TCR is what makes a T cell, or a BCR a B cell.
Conversely, we asked when signature transcripts of the ‘other’ lineage were switched off, plotting the expression of T cell signature genes during B cell differentiation and vice-versa. As illustrated in Figs. 4C-D, signature genes of the other lineage are turned off quite early during differentiation, faster than the defining signature transcripts are acquired. In T cells, most B cell signature transcripts are turned off by the Double Negative 2 stage, while in B cells most T cell signature transcripts are turned off by the Fraction B, pro-B cell stage.
This progression of “identity acquisition” through the early lineages is reflected in the population plots of Fig. 4E, where populations are positioned according to their expression of T- and B-defining transcripts, and where the sequence of differentiation is clearly delineated.
Do the same regulatory modules control signature genes in T or B lineages and in nonT/nonB cells?
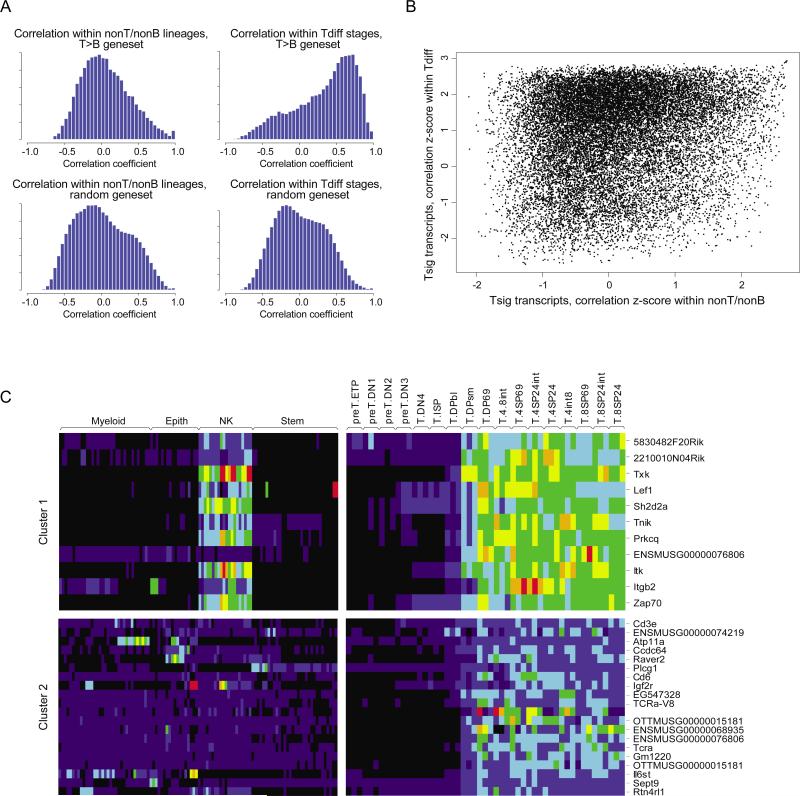
The expression signatures that distinguish T cells from B cells are acquired through distinct steps of T or B cell differentiation, and their expression is also shared with other nonT/nonB lineages along distinctive patterns (Figs. 2-4). It was thus of interest to ask whether the same regulatory influences operate in both contexts, or whether transcripts obey different regulators (or combinations thereof) during T cell differentiation and when they are active outside the T lineage. Transcriptional regulation operates on modules of co-regulated transcripts, which are similarly controlled by shared regulators; strongly correlated expression throughout a panel of cell populations is an indicator of such co-regulation. By extension, common regulatory influences (transcription factors, miRNAs) operating within stages of T differentiation and through nonT/nonB lineages should be reflected as pair-wise correlations that exist in both contexts. To address this question, we measured the pair-wise correlation coefficients between transcripts of the “Top200” T signature, across both the T-differentiation and nonT/nonB datagroups. A Pearson correlation coefficient was used as a metric. As a reference, pair-wise correlation coefficients across the same two datagroups were also computed for a randomly selected set of transcripts. As illustrated in Fig. 5A, correlations between T signature transcripts within the T-differentiation datagroup showed a skewed distribution, with a much greater proportion of high correlation coefficients than within the reference gene-set. In contrast, this bias was far more modest within the nonT/nonB datagroup. The different distribution of pair-wise correlations for T signature genes within the T and nonT/nonB datagroups was compared directly in the scatter plot of Fig. 5B (after transformation to a z-score, to normalize against the distributions of correlation coefficients within the reference gene-set). As expected, most pairs of transcripts correlated strongly within the T lineage, but showed little or no correlation within nonT/nonB lineages. On the other hand, some transcript pairs did show strong correlation across both datagroups (mapping to the top right quadrant of Fig. 5B). This distribution suggests that the majority of co-regulatory relationships that operate within stages of T cell differentiation are not maintained in other lineages, although a few are.
Figure 5. Partial sharing of co-regulated gene clusters within T cell differentiation and outside the T cell lineage.
To determine which transcripts exhibit coordinated expression, as a reflection of possible shared regulatory mechanisms, pair-wise correlation coefficients were calculated for all transcripts of the “Top 200” T cell signature genes, within all ImmGen datasets except for T and B cells (“nonT/nonB”) or within the T cell differentiation datasets. As a reference, the same coefficients were calculated on a set of 2000 transcripts picked at random. A): distribution of the correlation coefficients; note that there is a very significant skewing of the distribution of correlation coefficients between T signature genes in the T-differentiation datagroup (top left), far less marked within the nonT/nonB datagroup (top right). B): Scatter plot comparison of all pair-wise correlations between T signature genes within the nonT/nonB (X-axis) or T-differentiation (Y-axis) datagroups; to avoid artifacts due to the different sizes and composition of the nonT/nonB and T-differentiation datasets, the primary correlation coefficients were transformed to a z-score by reference to the mean and standard deviation of the correlation coefficients for the randomly picked reference gene-set. Note that the majority of transcript pairs that show strong correlation within the T-differentiation datagroup (z-score > 2) show no correlation within the nonT/nonB populations (z-scores distributed around 0), although there is a distinct “shoulder” of gene pairs that do show some correlation across both conditions (top right of the plot). C): A k-means clustering algorithm was used to partition T-signature genes into distinct clusters based on their correlation within the T-differentiation datagroup. Transcript levels for representative clusters are shown as a heat-map for the nonT/nonB (left) and T-differentiation (right) datagroups. A few clusters showed consistent expression across both datagroups (e.g. Cluster 1, top, primarily reflecting shared expression with NK cells), while many were only co-regulated within the T-differentiation datagroup.
To investigate this point further, we used a simple sequential clustering algorithm to parse the T-signature transcripts into distinct co-regulated clusters, according to their expression patterns through T cell differentiation, and identifying the sub-clusters that did or did not show correlation within the nonT/nonB datagroup. As shown in Fig. 5C, some sub-clusters did show good homogeneity of expression in both datagroups (e.g. cluster #1, which corresponded to a set of genes predominantly activated in the late stages of thymic T cell differentiation and quite uniquely co-expressed in NK cells) while others showed no preserved pattern of expression in nonT/nonB cells (e.g. cluster #2, also activated late in T differentiation but which showed no consistent expression pattern outside the T lineage). Thus, only a minority of the transcripts that characterize T lymphocytes belong to co-regulated gene clusters that are reutilized in different cell types.
DISCUSSION
A central goal of this work was to define, from a genome-wide perspective, the transcriptional differences that underlie T and B lymphocytes. We used the power of combinatorial microarray profiling as well as the breadth of cell populations available from the ImmGen project to explore the transcripts that provide their identities to T and B lymphocytes, in a more robust and in-depth perspective than could be provided in the comparisons preformed previously (8-11). The results show that transcriptional differences between B and T cells are very broad, not solely limited to a few specific markers commonly used to distinguish them by flow cytometry. On the other hand, there are very few transcripts uniquely specific to B and T cells, most being shared with other cell-types in the immune system.
Combinatorial microarray profiling to describe the transcriptome of a cell has several distinct advantages over gene-expression profiling with a single array. First, this approach eliminates any probe biases inherent to a particular chip's design. It is likely that this “cross-checking” resulted in our finding no difference in the overall number of genes expressed in T cells compared with B cells, which had been suggested by Hoffman et al (8). In addition, combining platforms avoids the false-positives and false-negatives that commonly affect 5-10% of the probe-sets on any one microarray support. Finally, combinatorial profiling allows for discovery of differential gene expression at greater depth and confidence. Thus, in contrast to previous studies, we estimate that at least 65% of the transcripts expressed in T and B cells are differential, most of which at very subtle FC values. In fact, had we compared even more datasets, it is plausible that every single gene expressed in T and B cells would be found to be significantly different.
Although this breadth is impressive, what does it mean that such a large percentage of genes is differentially expressed in such subtle manner, when thinking of the physiology of T and B lymphocytes? One perspective is that these broadly distributed but subtle levels of differential expression actually have little or no functional impact on the cell. One can imagine that a transcriptional regulator activates or represses the expression of a particular gene or module that specifies an important function in either T or B cells but that, in doing so, it also creates transcriptional or post-transcriptional perturbations that ripple at low levels throughout the genetic regulatory network of the cell. These small expression variations across the genome would essentially be an unavoidable reverberation accompanying a larger and more meaningful variation, but have no functional consequences in themselves, if the key networks that regulate metabolic homeostasis or cell proliferation and survival are sufficiently robust in the context of such variation. There would thus be no need to guard against such changes. A similar argument has been made for the impact of microRNAs, each of which can have mild but widespread effects, but with perhaps only a few truly meaningful and evolutionarily selected targets. On the other hand, these variations between B and T cells are so pervasive that it is difficult to believe that they are not meaningful in some way. In addition, microarrays tend to compress and under-represent differences in transcript abundance, relative to quantitative PCR. Differences of 1.2-1.3 fold by microarray are often closer to 2-fold when measured by real-time PCR. Such differences may thus be in a range that influences many genetic or molecular systems (e.g. copy-number-dependence in heterozygous mutations, metabolic regulation, etc). Of course, testing the significance of many minor variations is not experimentally tractable today.
We also found that the vast majority of these T/B differential transcripts are not specific to either of these lineages, but are widely represented throughout immune system cell types. Some of this shared expression might have been expected based on known physiology (e.g. antigen presentation pathways active in both B cells and DCs, cytotoxic effector molecules in NK and T cells), but other elements were less predictable. Again, some of these shared expression patterns may be unintended side-effects of transcriptional control pathways, but these data suggest that there is much re-utilization of functional proteins across cell types. There is precedent for cross-lineage sharing of gene products, even if their activity varies with context. For instance, the transcription factor, Tbx21 (aka Tbet) controls different specialized functions in different cells, favoring Th1 effector functions in T cells, promoting class switching to IgG2a in B cells, and necessary for induction of Type-1 interferons in dendritic cells by TLR9 ligands (25). Similarly, Blimp-1 was originally discovered as a transcriptional repressor of INF-β in human HeLa cells, then found to be required for the differentiation and maintenance of immunoglobulin-secreting B cells and plasma cells, and later identified as impacting T cell differentiation at several stages (in the thymus, during Th1/2 specification, and in Treg cells) (26).
Overall, the picture painted by these studies of the relationship between T and B lymphocytes departs somewhat from prior notions, with very few transcripts that are exquisitely specific of either cell, but with differences in transcriptome distributions that are very broad but also quite nuanced.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Vladimir Jojic and Mark Davis for comments, and eBiosciences, Affymetrix and Expression Analysis for their support of the ImmGen Project.
Footnotes
This work has been supported by the National Institute of Allergy and Infectious Disease of the National Institute of Health (R24 AI072073 to CB and DM).
List of abbreviations used in this paper: FC, Fold Change.
REFERENCES
- 1.Davis MM, Cohen DI, DeFranco AL, Paul WE. The isolation of B and T cell-specific genes. B and T Cell Tumors, UCLA Symposia on Molecular and Cellular Biology. 1982;24:215–220. [Google Scholar]
- 2.Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg EV. Cell lineage regulators in B and T cell development. Nat. Immunol. 2007;8:441–444. doi: 10.1038/ni1461. [DOI] [PubMed] [Google Scholar]
- 4.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat. Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 5.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 6.Busslinger M. Transcriptional control of early B cell development. Annu Rev. Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 7.Hagman J, Lukin K. Transcription factors drive B cell development. Curr. Opin. Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann R, Bruno L, Seidl T, Rolink A, Melchers F. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J Immunol. 2003;170:1339–1353. doi: 10.4049/jimmunol.170.3.1339. [DOI] [PubMed] [Google Scholar]
- 9.Kluger Y, Tuck DP, Chang JT, Nakayama Y, Poddar R, Kohya N, Lian Z, Ben Nasr A, Halaban HR, Krause DS, Zhang X, Newburger PE, Weissman SM. Lineage specificity of gene expression patterns. Proc Natl. Acad Sci U S. A. 2004;101:6508–6513. doi: 10.1073/pnas.0401136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutton JJ, Jegga AG, Kong S, Gupta A, Ebert C, Williams S, Katz JD, Aronow BJ. Microarray and comparative genomics-based identification of genes and gene regulatory regions of the mouse immune system. BMC. Genomics. 2004;5:82. doi: 10.1186/1471-2164-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren CM, Godowski P, Williams PM, Chan AC, Clark HF. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes. Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 12.Kothapalli R, Yoder SJ, Mane S, Loughran TP., Jr. Microarray results: how accurate are they? BMC. Bioinformatics. 2002;3:22. doi: 10.1186/1471-2105-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C, Carta R, Zhang L. Sequence dependence of cross-hybridization on short oligo microarrays. Nucleic Acids Res. 2005;33:e84. doi: 10.1093/nar/gni082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 15.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat. Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de LF, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venanzi ES, Melamed R, Mathis D, Benoist C. The variable immunological self: genetic variation and nongenetic noise in Aire-regulated transcription. Proc Natl Acad Sci U S A. 2008;105:15860–15865. doi: 10.1073/pnas.0808070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowlkes BJ, Pardoll DM. Molecular and cellular events of T cell development. Adv. Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- 21.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 22.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northrup DL, Allman D. Transcriptional regulation of early B cell development. Immunol. Res. 2008;42:106–117. doi: 10.1007/s12026-008-8043-z. [DOI] [PubMed] [Google Scholar]
- 24.Mick VE, Starr TK, McCaughtry TM, McNeil LK, Hogquist KA. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J Immunol. 2004;173:5434–5444. doi: 10.4049/jimmunol.173.9.5434. [DOI] [PubMed] [Google Scholar]
- 25.Peng SL. The T-box transcription factor T-bet in immunity and autoimmunity. Cell Mol. Immunol. 2006;3:87–95. [PubMed] [Google Scholar]
- 26.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.