Abstract
A key step in the activation of interferon-inducible antiviral kinase PKR involves differential binding of viral double-stranded RNA (dsRNA) to its two structurally similar N-terminal dsRNA binding motifs, dsRBM1 and dsRBM2. We show here, using NMR spectroscopy, that dsRBM1 with higher RNA binding activity exhibits significant motional flexibility on a millisecond timescale as compared with dsRBM2 with lower RNA binding activity. We further show that dsRBM2, but not dsRBM1, specifically interacts with the C-terminal kinase domain. These results suggest a dynamically tuned dsRNA binding mechanism for PKR activation, where motionally more flexible dsRBM1 anchors to dsRNA, thereby inducing a cooperative RNA binding for dsRBM2 to expose the kinase domain.
Keywords: dsRNA binding domain/dynamics/NMR/PKR
Introduction
PKR is an essential component of the interferon (IFN)-mediated antiviral response, which functions by phosphorylating the α subunit of initiation factor eIF2, resulting in rapid inhibition of translation and suppression of virus spread (Stark et al., 1998; Williams, 1999). PKR is also involved in cellular signal transduction, apoptosis, growth regulation and differentiation (Meurs et al., 1990; Tan and Katze, 1999; Williams, 1999). A key step in the antiviral response and other cellular processes mediated by PKR is its activation by double-stranded RNA (dsRNA), a by-product of virus replication (Meurs et al., 1990) or cellular stress (Chu et al., 1998). dsRNA binds in a sequence-independent manner to the N-terminal dsRNA binding domain (dsRBD) of PKR, which includes two contiguous dsRNA binding motifs (dsRBM1 and dsRBM2). Despite extensive genetic and biochemical studies, the precise molecular mechanism of how dsRNA binding mediates PKR activation is not clearly understood. Two critical issues remain to be resolved. First, although both dsRBMs are involved in dsRNA binding and PKR activation, they are functionally non-equivalent, with dsRBM1 exhibiting much higher dsRNA binding activity than dsRBM2 (Green and Mathews, 1992; McCormack et al., 1994; Romano et al., 1995; Schmedt et al., 1995). Structure-based understanding of such differential RNA binding has been limited because both motifs adopt essentially the same fold with a highly conserved RNA binding surface (Nanduri et al., 1998a). It is possible that in addition to amino acid sequence, other factors such as protein flexibility may play a role in fine-tuning the RNA binding activity (Lu and Hall, 1997). Secondly, upon interaction between dsRNA and the dsRBMs, PKR has been suggested to undergo a conformational change preceding activation (Manche et al., 1992; Romano et al., 1995; Carpick et al., 1997). The molecular basis for such a conformational change has not been defined. In particular, how the two dsRBMs mediate the conformational change by differentially binding to dsRNA is not clear. In this report, we have addressed these issues using NMR spectroscopy. We have undertaken a detailed 15N relaxation analysis to determine whether structural fluctuations of dsRBMs at different timescales (picosecond– millisecond) are related to their different dsRNA binding activities. We show that regions involved in dsRNA binding on dsRBM1, as compared with dsRBM2, undergo significant millisecond–microsecond timescale motion, which reveals a strong correlation between protein mobility and the dsRNA binding activity of dsRBM. We also show that dsRBM2, but not dsRBM1, has specific intramolecular interactions with the C-terminal kinase domain, which lock the inactive PKR in a ‘closed’ conformation. These findings provide a novel view of PKR activation where dsRBM1 fluctuating with millisecond–microsecond motions serves as an anchor for dsRNA, thereby inducing a cooperative RNA binding for dsRBM2 to expose the kinase domain.
Results and discussion
NMR 15N relaxation analysis of dsRBD
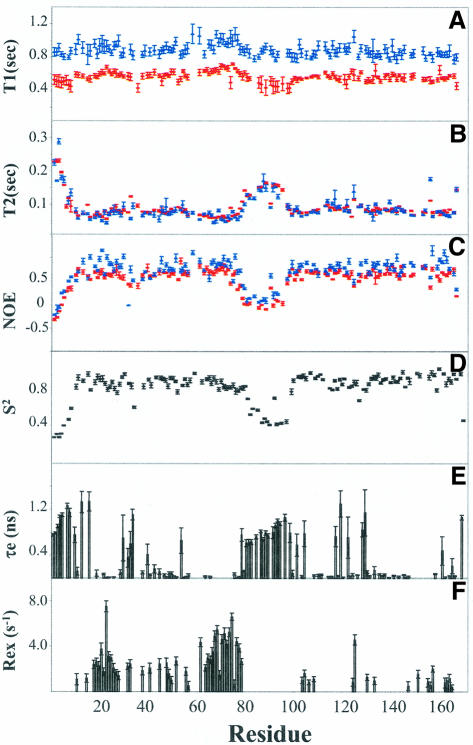
The following NMR relaxation parameters were collected in two magnetic fields to analyze the motional properties of dsRBD containing the two dsRBMs and the central linker: (i) spin–lattice relaxation time (T1) (Figure 1A); (ii) spin–spin relaxation (T2) (Figure 1B); and (iii) steady-state 1H–15N nuclear Overhauser effects (NOEs) (Figure 1C). Based on the 1H/15N chemical shift assignment of dsRBD in heteronuclear-single-quantum-correlation (HSQC) spectra (Nanduri et al., 1998b), the relaxation data for 135 out of 170 backbone 15N nuclei of dsRBD were used for analysis. Of the remaining 35 residues, six are prolines (P31, P35, P36, P53, P84 and P128); 19 residues exhibit spectral overlap (F9, E12, L14, Y17, R39, K61, L86, L96, N106, I108, R113, L114, T149, K150, A153, K154, Q155, A161 and E168); six residues exhibit severe line-broadening due to chemical exchange processes (H37, Q44, R58, A63, N91 and Y101); and four residues do not appear in the HSQC spectrum (K60, M98, A122 and Q139). Also excluded for data analysis are nine residues resulting from the cloning strategy (five at the N-terminus and four at the C-terminus) (Nanduri et al., 1998b). The average T1, T2 and NOEs for residues with regular secondary structures for dsRBM1 and dsRBM2 are compared in Table I. The values between the two motifs are similar except that the average T2 for dsRBM1 is smaller than that of dsRBM2. It is evident from Figure 1 that residues with rigid and regular secondary structures in the two motifs exhibit smaller T2 and higher NOE values than flexible loop residues. Particularly notable is the high flexibility in the 20 residue linker region (81–100) between the two dsRBMs, which exhibits large T2 values and negative NOEs (Figure 1B and C). Such high flexibility is generally considered to make the two motifs reorient essentially independently, which is similar to other cases such as calmodulin (Barbato et al., 1992) and phosphotransfer/CheY-binding domains of the histidine autokinase CheA (Zhou et al., 1996). The flexible linker may restrict reorientation of the two dsRBMs to some extent, resulting in some anisotropic motions (Zhou et al., 1996). Therefore, both isotropic (Farrow et al., 1994) and anisotropic (axially symmetric) (Tjandra et al., 1995; Lee et al., 1997) models were used for data analysis. Indeed, calculations of relaxation parameters using anisotropic models yielded a small amount of the anisotropy for dsRBD (D∥/D⊥ = 1.2), compared with individual dsRBMs, if calculated separately based on the NMR structure of dsRBD (Nanduri et al., 1998a), exhibiting negligible anisotropy (both with D∥/D⊥ ≈ 1.06). Nevertheless, the results derived from anisotropic models are consistent with those obtained from isotropic ones (see later and Materials and methods), indicating that the small amount of anisotropy imposed by the linker plays a minor role in determining the dynamics of the two dsRBMs. To be more precise, only the results obtained using anisotropic models were used for discussion throughout the text.
Fig. 1. 15N relaxation parameters of dsRBD. One hundred and thirty-five residues out of the total 170 were analyzed and plotted as a function of residue number in dsRBD (dsRBM1, 9–79; central linker, 80–100; dsRBM2, 101–170). Residues not included are described in the text. Red and blue in (A) T1, (B) T2 and (C) NOEs denote data from 500 and 600 MHz spectrometers, respectively; data for (D) S2, (E) τe and (F) Rex were derived from anisotropic models (Tjandra et al., 1995; Lee et al., 1997). Notice the dramatic difference in Rex distribution between dsRBM1 (9–79) and dsRBM2 (101–170).
Table I. Comparison of dynamics parameters for dsRBM1 and dsRBM2 of PKRa.
| dsRBM1 (9–79) | dsRBM2 (101–170) | |||
|---|---|---|---|---|
| Motional parameters for residues with regular secondary structures | ||||
| average T1 (s) | 0.5826 | 0.5647 | ||
| average T2 (s) | 0.07784 | 0.08395 | ||
| average NOEs | 0.6680 | 0.6802 | ||
| average S2 | 0.85 | 0.86 | ||
| Average S2 of individual helix, loop and sheet | ||||
| dsRBM1 | dsRBM2 | |||
| α1 (9–20) | versus | α3 (101–111) | 0.87 | 0.91 |
| loop1 (21–24) | versus | loop6 (112–114) | 0.83 | 0.88 |
| β1 (25–32) | versus | β4 (115–122) | 0.87 | 0.83 |
| loop2 (33–39) | versus | loop7 (123–129) | 0.74 | 0.79 |
| β2 (40–48) | versus | β5 (130–138) | 0.85 | 0.87 |
| loop3 (49–53) | versus | loop8 (139–143) | 0.85 | 0.87 |
| β3 (54–57) | versus | β6 (144–147) | 0.85 | 0.89 |
| loop4 (58–59) | versus | loop9 (148–149) | 0.94 | 0.86 |
| α2 (60–78) | versus | α4 (150–168) | 0.83 | 0.90 |
| Residues with Rex | 41 | 10 | ||
| Residues without Rex | 15 | 45 | ||
| Residues with exchange broadening | 5 | 3 | ||
| Residues with overlap or prolines | 10 | 12 | ||
| Total | 71 | 70 | ||
| Residues with Rex and exchange broadening involved in RNA binding regions and the surroundingsb | ||||
| dsRBM1 | dsRBM2 | |||
| region 1 N15–T16 | versus | N106-R107 | 1 (12) | 0 (4) |
| region 2 P36–R39 | versus | H126-G130 | 2 (10) | 1 (4) |
| region 3 R58–K64 | versus | S148-K154 | 6 (15) | 1 (3) |
| total | 9 (37) | 2 (11) | ||
aS2 and Rex values are derived from anisotropic models.
bNumbers in parentheses are the residues with Rex in the surroundings of dsRNA binding regions (see Figure 4).
The relaxation data were fitted to model-free spectral density functions using five possible sets of motional parameters: (1) S2; (2) S2, τe; (3) S2, Rex; (4) S2, τe, Rex; and (5) S2f, τs, S2. The overall correlation time of dsRBD was initially estimated from the T1/T2 ratio of the residues whose NOE values are at least 0.6 so as to make sure that they are rigid and have no internal motions. The initial estimate of the overall correlation time (τm) from 96 residues with regular secondary structures is 9.53 ± 0.03 ns. The number of residues that fit to models 1–5 are 16, 26, 29, 31 and 33, respectively. Relaxation parameters calculated from the spectral density functions were compared with the experimental values to optimize the model-free parameters. The detailed internal motions on the picosecond timescale were obtained from the internal correlation time, τe in model 2. However, when residues have internal motions of significant amplitude on both the nanosecond and picosecond timescale, the extended model-free approach identified such motions (model 5). Residues having contributions from microsecond–millisecond motions require the conformational exchange term, Rex, to fit their relaxation data (model 3 or 4). The results are illustrated in Figure 1D–F, which provides information on order parameters (S2) for each N–H vector, fast internal motions (τe or τs) on the picosecond–nanosecond timescale, and slow conformational exchange (Rex) on the microsecond–millisecond timescale.
The order parameters S2 generally reflect the rigidity of regions in secondary structure that have restricted motions on the picosecond–nanosecond timescale (Figure 1D). The average order parameters of the individual helices and β-strands in the two motifs are all >0.8 (Table I). The average S2 for individual α-helices and β-strands in dsRBM1 is smaller than those in dsRBM2 except for β1 in dsRBM1 (0.87) versus β4 in dsRBM2 (0.83) (Table I), indicating that corresponding regions in the two motifs have slightly different rigidity. The order parameters of the loops/turns in dsRBMs range from 0.74 to 0.94 (Table I). Loop 2/loop 7, which are involved in dsRNA binding but poorly defined in the NMR structure (Nanduri et al., 1998a), consistently display the lowest order parameters in dsRBMs (Table I). As expected, much smaller order parameters were observed for the unstructured terminal regions and central loop (residues 80–100 with average S2 0.57) (Figure 1D), which is consistent with the small T2 and small/negative NOEs in those regions (see above; Figure 1B and C).
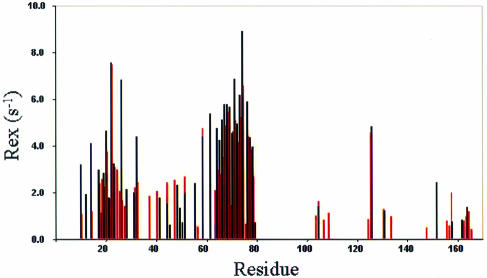
Figure 1E shows the regions in dsRBD with fast internal motions at the picosecond–nanosecond timescale. These regions include unstructured or loop residues as well as residues with regular secondary structures. The relaxation data for these regions were generally fitted to either model 2 or 5. All the residues in the unstructured terminal regions (residues 1–8 and 169–170) and the central loop were fitted to model 5 with two timescale spectral density functions, which revealed motions ranging from 128.5 ps to 1.26 ns (Figure 1E). Model 5 was also required to fit the data for residues with fast internal motions; in dsRBM1: T16 (α1), G34 (β1–β2 loop) and E54 (at the beginning of β3); and in dsRBM2: Y118 (α3), Q120, S123, E129 and G130 (β4–β5 loop). While almost all the loop residues in dsRBM2 were fitted to model 2 or 5, most of the loop residues in dsRBM1 require model 3 or 4, which include the conformational exchange term Rex (Figure 1F). Moreover, a significant number of helix and β-strand residues in dsRBM1 also required Rex to fit the data. In total, 41 residues in dsRBM1 and 10 residues in dsRBM2 have Rex values >1, which is considered to be significant conformational exchange (Figure 1F). Results derived from isotropic models revealed very similar Rex distribution, although the Rex values based on anisotropic models are in general smaller than the correponding ones obtained from isotropic models (Figure 2). The residues with Rex in dsRBD are located in dsRBM1: α1 (M11, N15, R18, Q19, K20 ), α1–β1 loop (Q21, G22, V23, V24), β1 (L25, K26, Y27, Q28, N32, S33), β1–β2 loop (D38), β2 (F41, T42, V45), β2–β3 loop (D48, G49, R50, F52), β3 (E56), β3–α2 loop (S59) and α2 (K62, K64–L75, K77–K79); and dsRBM2: α3 (L104, I105, A109), β4–β5 loop (C126), beginning of β5 (F131, K134) and α4 (E152, A158, Q164, I165).
Fig. 2. Comparison of Rex distributions in dsRBD. Rex derived from isotropic models is marked in red and from anisotropic models in black, showing the consistency of the two models (the anisotropy is small, as discussed in the text).
Correlation between millisecond motions and dsRNA binding
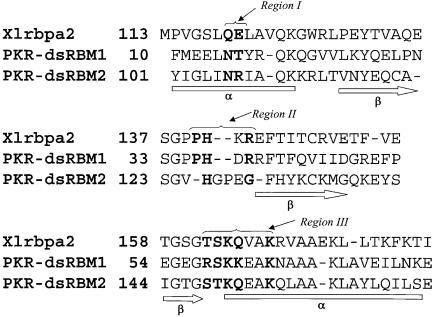
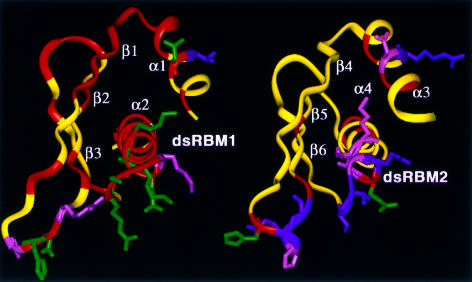
The dramatic difference in Rex distribution between the two dsRBMs of PKR allows us to establish a direct correlation between the conformational flexibility at a millisecond timescale and dsRNA binding activity. dsRBM1 and dsRBM2 exhibit 27% sequence identity and strong sequence homology with other RNA binding domains (Figure 3). Previous structural studies of dsRBD of PKR in combination with mutational results have identified a conserved RNA binding site on each dsRBM (Nanduri et al., 1998a) (Figure 3). Specific residues involved in dsRNA binding were more precisely identified in the recent crystal structure of homologous Xlrbpa-2 dsRBM complexed with a symmetric RNA duplex (Ryter and Schultz, 1998) (Figure 3). For dsRBMs of PKR, these binding residues correspond to three regions (Figure 3): N15–T16 of α1 (region 1), P36–R39 of the β1–β2 loop (region 2) and R58–K64 of the β3–α2 loop and the beginning of α2 (region 3) in dsRBM1; and N106–R107 of α3 (region 1), H126–G130 of the β4–β5 loop (region 2) and S148–K154 of the β6–α4 loop and the beginning of α4 (region 3) in dsRBM2. An NMR structure of the homologous Staufen dsRBM complexed with a stem–loop of 12 bp dsRNA was also recently reported (Ramos et al., 2000). The same RNA binding regions were identified in the NMR structure except that the α1 helix was also found to interact with the stem–loop (see discussion below). Except for T16 (fast internal motion), P36 (no amide), R39 and K61 (spectral overlap), all the residues in regions 1–3 of PKR dsRBM1 undergo slow conformational exchange either with Rex (N15, D38, S59, E62, K64) or line-broadening (H37, R58, K60, A63) (Figure 4). The rest of the residues with Rex in dsRBM1 are either directly or indirectly involved with the RNA binding regions through a network of interactions (Figure 4; Table I). These residues are mostly located in α1, β1 and α2, which pack against each other (Figure 4). Nearly three-quarters of the residues in dsRBM1 have Rex (Table I), indicating a concerted motion of the motif at the millisecond–microsecond timescale. Such concerted motion is important since multiple conformational substates of dsRBM may be required for binding to various dsRNAs (see below). In contrast, the majority of the residues in dsRBM2 undergo restricted picosecond–nanosecond motions with only a few residues having Rex (Figure 4; Table I). Interestingly, the residues with Rex in dsRBM2, although fewer, are all clustered in the dsRNA binding site and its surroundings (Figure 4; Table I).
Fig. 3. Primary and secondary structures and sequence comparison of PKR dsRBMs and Xrlbp dsRBM. Three regions involved in dsRNA binding are indicated (Ryter and Schultz, 1998). Residues directly involved in interacting with dsRNA are highlighted in bold.
Fig. 4. Three-dimensional distribution of residues with Rex (red) in dsRBM1 and dsRBM2. The side-chains of the dsRNA binding residues from regions 1–3 are displayed. The residues with Rex or exchange broadening are displayed in green; the residues without Rex are displayed in magenta; and the residues unanalyzed due to spectral overlap are displayed in pink. Notice that regions surrounding the RNA binding site of dsRBM1 are covered by residues with Rex. A network of interactions extends the exchange effects from the dsRNA binding site to the surrounding regions such as the central hydrophobic core involving residues from α1, β1, β2 and α2.
How does the conformational flexibility of dsRBM1 contribute to its higher dsRNA binding affinity than dsRBM2? The crystal structure of the Xlrbpa-2 dsRBM–dsRNA complex revealed that the interactions of dsRNA with dsRBM are essentially RNA sequence independent, primarily involving 2′-OH groups in the minor groove and phosphodiester backbone oxygens in the major groove (Ryter and Schultz, 1998). However, the structure of a symmetric dsRNA in the complex deviates from the ideal A form duplex with some backbone distortion and a widened major groove (Ryter and Schultz, 1998), whereas the NMR structure of the Staufen dsRBM–dsRNA complex revealed that a 12 bp dsRNA connected to a helix-stabilizing tetraloop preserves the A form double-helical structure (Ramos et al., 2000). These results indicate that variable dsRNAs may bind to dsRBM with slightly different conformations. Indeed, PKR is known to recognize a variety of viral dsRNAs, including structured RNAs that contain only short segments of uninterrupted Watson–Crick base pairing, such as HIV-Tar, adenovirus VAI RNA, EBV RNAs, EBER-1 and EBER-2, etc. (Clemens et al., 1994). In fact, a more recent systematic study has documented that PKR can bind dsRNAs with secondary structure defects such as a GA:AG mismatch and non-contiguous helix (Bevilacqua et al., 1998). These dsRNAs with structures that deviate from the ideal A form apparently require fine-tuning of the dsRBM conformations at the RNA binding site for optimal binding. Indeed, the binding modes of Staufen dsRBM and Xlrbp-2 dsRBM to different dsRNAs differ slightly, in particular the α1 helix of the former was found to interact with the stem–loop structure (Ryter and Schultz, 1998). Since dsRBM1 of PKR is capable of binding to the structurally variable dsRNAs much more favorably than dsRBM2 (Green and Mathews, 1992; McCormack et al., 1994; Romano et al., 1995; Schmedt et al., 1995), multiple substates of dsRBM1 fluctuating at the millisecond– microsecond timescale may confer the flexibility for these slightly distorted dsRNAs. Consistently, dsRBM1 binds dsRNA with a Kd in the micromolar–subnanomolar range (Schmedt et al., 1995), indicating the off rate of binding in the millisecond–microsecond timescale. On the other hand, dsRBM2 has more restricted motions on its dsRNA binding site, which may prevent its optimal binding to dsRNAs with deviated A form structures. We propose that dsRBM1 has the flexibility to adapt to a variety of viral dsRNAs with imperfect A form structures, thereby facilitating the cooperative dsRNA binding of dsRBM2 to achieve a total high affinity.
Millisecond–microsecond motion in the protein binding interface in relation to multiple protein–target interactions has also recently been shown in a few other examples (Lu and Hall, 1997; Wyss et al., 1997; Feher and Cavanagh, 1999). In the case of dsRBM–RNA interactions, it should be pointed out that in addition to motional flexibility, variations of amino acids in the dsRNA binding site may play an important role in determining the RNA binding specificity. However, we noticed that the variations in the RNA binding site in PKR are hydrophilic in nature and hence may not be the only factor for specificity (Figure 3). More importantly, as shown in the crystal structure of the dsRBM–dsRNA complex, six out of a total of 10 RNA binding amino acids use their backbones for interactions and >90% of side-chains interact with dsRNA through water and 2′-OH groups (Ryter and Schultz, 1998). Therefore, protein backbone flexibility is likely to play a critical role in fine-tuning the dsRNA binding activity of dsRBD. That >50% of the binding residues use their backbone for RNA binding in fact emphasizes the importance of millisecond backbone motion as derived from our study. Knowledge of a more quantitative dynamics–RNA affinity correlation requires detailed analyses of the dsRBD–RNA complex and dsRBD mutants with altered RNA binding activities. While such studies are being pursued by our laboratory, the results in this report have demonstrated a clear dynamic difference between the two dsRBMs, which correlates well with their differential RNA binding activities.
dsRBM2–kinase domain interaction and the model for PKR activation
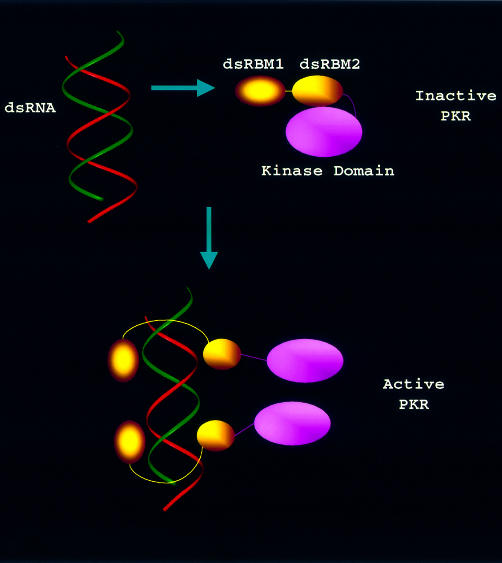
How then would the dynamically tuned dsRNA binding to the two N-terminal dsRBMs induce the activation of the C-terminal kinase domain? Biochemical and biophysical studies suggested that dsRNA binds to the dsRBMs in a cooperative fashion, inducing a conformational change of PKR for its activation, and such conformational change may be accompanied by dimerization and autophosphorylation (Carpick et al., 1997; Williams, 1999). We hypothesized that dsRBD may be a negative regulator by masking the dimerization site and the kinase domain. To investigate this, we prepared the purified PKR kinase domain fused to the maltose binding protein (MBP) and titrated the kinase domain into the 15N-labeled dsRBD. Interestingly, monitoring of the 1H–15N correlation spectrum revealed that dsRBM2, but not dsRBM1 (Figure 5), is engaged in interacting with the C-terminal kinase domain. Surface plasmon resonance experiments showed that the binding affinity between dsRBD and the kinase domain is ∼500 nM (data not shown). On the other hand, titration of full-length PKR or putative dimerization domain (Tan et al., 1998) (244–296) into dsRBD did not result in any spectral change, indicating that dsRBM2 keeps the full-length PKR in a ‘closed’/‘inactive’ conformation. This is strongly supported by biochemical data showing that while intact PKR is latent, deletion of the entire dsRBD constitutively activates PKR (Wu and Kaufman, 1997), and swapping dsRBM1/dsRBM2 substantially increases the kinase activity (Romano et al., 1995). The data are also consistent with the earlier report that ATP becomes associated with PKR only after exposure to dsRNA (Galabru and Hovanessian, 1987). Thus, the cooperative binding of dsRNA to dsRBM1 and dsRBM2 releases the restraint of dsRBM2 on the kinase domain, leading to an ‘open’/‘active’ conformation of PKR. The structures of dsRBD (Nanduri et al., 1998a) and dsRBM–dsRNA complexes (Ryter and Schultz, 1998; Ramos et al., 2000), and biochemical studies (Green and Mathews, 1992; Bevilacqua and Cech, 1996) have indicated that one dsRBM can cover 6–8 bp of dsRNA with the whole monomer dsRBD containing two dsRBMs covering 12–16 bp of dsRNA. On the other hand, it has been shown that PKR binds minimally to 30 bp dsRNA to reach full capacity of activity (Manche et al., 1992). These results indicate that one PKR molecule may bind to part of a longer dsRNA while leaving the adjacent space for another PKR molecule (Figure 6). Such an arrangement would greatly facilitate the PKR dimerization that is required for its activation. Based on a combination of our dynamics and structural data as well as previously reported biochemical data, we propose a model for dsRNA-mediated PKR activation (Figure 6). In this model, dsRBM2 locks the kinase domain in a ‘closed’ conformation in the latent PKR whereas dsRBM1 is unconstrained by the rest of the protein, fluctuating with millisecond motions. Viral dsRNA is first anchored to the free dsRBM1, inducing the cooperative binding to dsRBM2, which then exposes the kinase domain. This would also relax the major dimerization region (244–296) that is otherwise restrained due to the interaction between dsRBM2 and the kinase domain. Such concerted conformational change then results in PKR dimerization, autophosphorylation and activation.
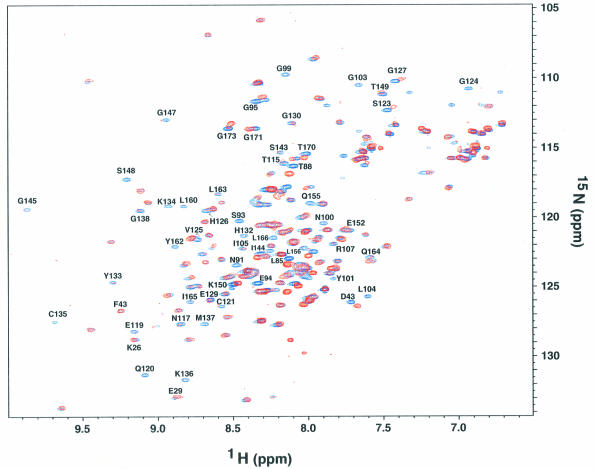
Fig. 5. 1H–15N correlation spectra of the dsRBD in the absence (blue) and presence (red) of MBP–kinase domain (dsRBD:MBP–kinase domain = 1:1.1). Note the broadening or disappearance of residues in the entire dsRBM2 due to increased tumbling time (MBP–kinase domain, ∼70 kDa). On the other hand, the independently tumbling dsRBM1 (Nanduri et al., 1998a and also see text) that has no interaction with the kinase domain is essentially unaffected.
Fig. 6. A model for dsRNA-mediated PKR activation. In the inactive PKR, dsRBM2 contacts with the kinase domain for a ‘closed’ conformation whereas dsRBM1 is unconstrained, fluctuating with millisecond motions. Viral dsRNA is first anchored to the free dsRBM1, inducing the cooperative binding to dsRBM2, which then exposes the kinase domain. This would also relax the major dimerization region (244–296), which is otherwise restrained due to the interaction between dsRBM2 and the kinase domain. Such concerted conformational change then results in PKR dimerization, autophosphorylation and activation.
It should be mentioned that while activated by low concentrations of dsRNA, PKR is inhibited by higher dsRNA concentrations (Williams et al., 1979; Galabru et al., 1989). However, the inhibition only occurs when high concentrations of dsRNA are added in the beginning. In other words, once activated by low concentrations of dsRNA, PKR can not be inhibited by subsequent addition of high concentrations of dsRNA (Galabru et al., 1989). Based on our structural/dynamic results, we provide the following explanations: at lower dsRNA concentrations, the flexible dsRBM1 with higher RNA binding affinity first anchors to dsRNA, and facilitates the cooperative dsRNA binding to lower affinity dsRBM2 to expose the kinase domain; however, once both dsRBMs have bound to dsRNA, dsRNA added at higher concentrations would no longer bind to PKR (Carpick et al., 1997), probably due to the saturation of the two RNA binding sites in the wrap-around conformation (Nanduri et al., 1998a). On the other hand, if high dsRNA concentrations are added at the beginning, the binding may not be cooperative; for example, the two dsRBMs in PKR may simultaneously bind to two different dsRNA molecules, which leads to a distorted overall protein conformation unfavorable for the kinase function. To this end, we have found that addition of a larger excess amount of dsRNA to dsRBD leads to protein aggregation, as judged from gel filtration and NMR experiments (not shown), which provides evidence of aggregation-based PKR inhibition with high concentrations of dsRNA. High concentrations of dsRNA may cause non-specific binding to PKR, thus resulting in its aggregation. It should be noted that inhibition of PKR can also be achieved by several kinase inhibitors that are specific to PKR, such as K3L, E3L, etc. (Williams, 1999), and these inhibitors may serve to switch off the PKR after its activation in vivo.
In summary, we have shown that the two dsRBMs in PKR exhibit significantly different backbone dynamics that correlate with their differential dsRNA binding activities. We further showed that the less well-conserved dsRBM2 with low RNA binding affinity interacts with the C-terminal kinase domain. These results suggest a dynamically tuned dsRNA binding mechanism for PKR activation where flexible dsRBM1 may first anchor to dsRNA and then induce the cooperative RNA binding of dsRBM2 to release the otherwise ‘locked’ kinase domain for catalysis.
Materials and methods
Sample preparation
Uniformly 15N-labeled dsRBD sample was prepared as previously described (Nanduri et al., 1998a). dsRBD elutes from the gel-filtration column as a monomer, which is consistent with its overall correlation time (Nanduri et al., 1998a). The PKR kinase domain (273–526) was subcloned into pMAL-C2 vector containing cDNA of MBP as a fusion (New England Biolabs). The purification of MBP–PKR kinase domain was performed according to the manufacturer’s instructions (New England Biolabs). The dimerization domain 244–296 was synthesized by the biotechnology core facility (Lerner Research Institute, Cleveland Clinic Foundation, Cleveland, OH).
NMR experiments and data analysis
All the NMR experiments were conducted at 25°C using Varian Inova 500 and 600 MHz spectrometers, each equipped with a triple-resonance probe head and a shielded z-gradient unit. 15N relaxation parameters, T1 and T2, and 1H–15N heteronuclear NOEs were obtained using the experiments described (Farrow et al., 1994). T1 experiments were collected with delay times of 11.4, 90.9, 204.5, 329.5, 454.5, 625.0, 829.6 and 1090.9 ms. The T2 measurements were performed with delay times of 16.6, 33.3, 49.9, 66.5, 83.2, 99.8, 116.4 and 133.1 ms. Two NOE experiments were collected: one with a 3.0 s recycle delay followed by a 3.0 s pre-saturation period, and the other with a 6 s recycle delay without any pre-saturation. 1H–15N HSQC were collected before and after the collection of all dynamics data. Data were processed using NMRPipe software (Delaglio et al., 1995) with T1, T2 and steady state NOEs determined as described (Tjandra et al., 1995). The standard deviations are <10% for T1 and NOE values, respectively, and <5% for T2. Model-free parameters were fitted according to the previously described procotols (Farrow et al., 1994). Analysis of the anisotropic diffusional tensors (axially symmetric) was performed using the protocols described (Tjandra et al., 1995; Lee et al., 1997) and the programs provided by Dr A.G.Palmer. Calculations show that the axially symmetric models were better than the fully anisotropic models for dsRBD (F-test for the former is 18.5 versus 4.6 for the latter) (Lee et al., 1997).
Acknowledgments
Acknowledgements
We thank Frank Delaglio for NMRPipe software, Art Palmer for the software for the dynamics studies, Lewis Kay for pulse sequences and Tom Haas for helping with the surface plasmon resonance experiment. The work was supported by NIH grants to J.Q. (HL58758) and B.R.G.W. (AI34039).
References
- Barbato G., Ikura,M., Kay,L.E., Pastor,R.W. and Bax,A. (1992) Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry, 31, 5269–5278. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C. and Cech,T.R. (1996) Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry, 35, 9983–9994. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., George,C.X., Samuel,C.E. and Cech,T.R. (1998) Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry, 37, 6303–6316. [DOI] [PubMed] [Google Scholar]
- Carpick B.W., Graziano,V., Schneider,D., Maitra,R.K., Lee,K. and Williams,B.R.G. (1997) Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-1 TAR RNA. J. Biol. Chem., 272, 9510–9516. [DOI] [PubMed] [Google Scholar]
- Chu W.M., Ballard,R., Carpick,B.W., Williams,B.R. and Schmid,C.W. (1998) Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol., 18, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J., Laing,K.G., Jeffrey,I.W., Schofield,A., Sharp,T.V., Elia,A., Matys,V., James,M.C. and Tilleray,V.J. (1994) Regulation of the interferon-inducible eIF-2α protein kinase by small RNAs. Biochimie, 76, 770–778. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Farrow N.A. et al. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry, 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- Feher V.A. and Cavanagh,J. (1999) Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature, 400, 289–293. [DOI] [PubMed] [Google Scholar]
- Galabru J. and Hovanessian,A. (1987) Autophosphorylation of the protein kinase dependent on double-stranded RNA. J. Biol. Chem., 262, 15538–15544. [PubMed] [Google Scholar]
- Galabru J., Katze,M.G., Robert,N. and Hovanessian,A.G. (1989) The binding of double-stranded RNA and adenovirus VAI RNA to the interferon-induced protein kinase. Eur. J. Biochem., 178, 581–589. [DOI] [PubMed] [Google Scholar]
- Green S.R. and Mathews,M.B. (1992) Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev., 6, 2478–2490. [DOI] [PubMed] [Google Scholar]
- Lee L.K., Rance,M., Chazin,W.J. and Palmer,A.G. (1997) Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C α nuclear spin relaxation. J. Biomol. NMR, 9, 287–298. [DOI] [PubMed] [Google Scholar]
- Lu J. and Hall,K.B. (1997) Tertiary structure of RBD2 and backbone dynamics of RBD1 and RBD2 of the human U1A protein determined by NMR spectroscopy. Biochemistry, 36, 10393–10405. [DOI] [PubMed] [Google Scholar]
- Manche L., Green,S.R., Schmedt,C. and Mathews,M.B. (1992) Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol., 12, 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack S.J., Ortega,L.G., Doohan,J.P. and Samuel,C.E. (1994) Mechanism of interferon action motif I of the interferon-induced, RNA-dependent protein kinase (PKR) is sufficient to mediate RNA-binding activity. Virology, 198, 92–99. [DOI] [PubMed] [Google Scholar]
- Meurs E., Chong,K., Galabru,J., Thomas,N.S., Kerr,I.M., Williams,B.R.G. and Hovanessian,A.G. (1990) Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell, 62, 379–390. [DOI] [PubMed] [Google Scholar]
- Nanduri S., Carpick,B.W., Yang,Y., Williams,B.R.G. and Qin,J. (1998a) Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J., 17, 5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri S., Carpick,B., Yang,Y., Williams,B.R.G. and Qin,J. (1998b) 1H, 13C, 15N resonance assignment of the 20 kDa dsRNA binding domain of protein kinase PKR. J. Biomol. NMR, 12, 349–351. [DOI] [PubMed] [Google Scholar]
- Ramos A., Grunert,S., Adams,J., Micklem,D.R., Proctor,M.R., Freund,S., Bycroft,M., St Johnston,D. and Varani,G. (2000) RNA recognition by a Staufen double-stranded RNA binding domain. EMBO J., 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P.R., Green,S.R., Barber,G.N., Mathews,M.B. and Hinnebusch,A.G. (1995) Structural requirements for double-stranded RNA binding, dimerization and activation of the human eIF-2α kinase DAI in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter J.M. and Schultz,S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmedt C., Green,S.R., Manche,L., Taylor,D.R., Ma,Y. and Mathews,M.B. (1995) Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR). J. Mol. Biol., 249, 29–44. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R.G., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Tan S.L. and Katze,M.G. (1999) The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J. Interferon Cytokine Res., 19, 543–554. [DOI] [PubMed] [Google Scholar]
- Tan S.L., Gale,M.J.,Jr and Katze,M.G. (1998) Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol. Cell. Biol., 18, 2431–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N., Feller,S.E., Pastor,R.W. and Bax,A. (1995) Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J. Am. Chem. Soc., 117, 12562–12566. [Google Scholar]
- Williams B.R.G. (1999) PKR; a sentinel kinase for cellular stress. Oncogene, 18, 6112–6120. [DOI] [PubMed] [Google Scholar]
- Williams B.R.G., Gilbert,C.S. and Kerr,I.M. (1979) The respective roles of the protein kinase and pppA2′ p5′ A2′ p5 A-activated endonuclease in the inhibition of protein synthesis by double stranded RNA in rabbit reticulocyte lysates. Nucleic Acids Res., 6, 1335–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. and Kaufman,R.J. (1997) A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem., 272, 1291–1296. [DOI] [PubMed] [Google Scholar]
- Wyss D.F., Dayie,K.T. and Wagner,G. (1997) The counterreceptor binding site of human CD2 exhibits an extended surface patch with multiple conformations fluctuating with millisecond to microsecond motions. Protein Sci., 6, 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., McEvoy,M.M., Lowry,D.F., Swanson,R.V., Simon,M.I. and Dahlquist,F.W. (1996) Phosphotransfer and CheY-binding domains of the histidine autokinase CheA are joined by a flexible linker. Biochemistry, 35, 433–443. [DOI] [PubMed] [Google Scholar]