Abstract
The jawless vertebrates (lamprey and hagfish) are the closest extant outgroups to all jawed vertebrates (gnathostomes) and can therefore provide critical insight into the evolution and basic biology of vertebrate genomes. As such, it is notable that the genomes of lamprey and hagfish posses a capacity for rearrangement that is beyond anything known from the gnathostomes. Like the jawed vertebrates, lamprey and hagfish undergo rearrangement of adaptive immune receptors. However, the receptors and the mechanisms for rearrangement that are utilized by jawless vertebrates clearly evolved independently of the gnathostome system. Unlike the jawed vertebrates, lamprey and hagfish also undergo extensive programmed rearrangements of the genome during embryonic development. By considering these fascinating genome biologies in the context of proposed (albeit contentious) phylogenetic relationships among lamprey, hagfish, and gnathostomes, we can begin to understand the evolutionary history of the vertebrate genome. Specifically, the deep shared ancestry and rapid divergence of lampreys, hagfish and gnathostomes is considered evidence that the two versions of programmed rearrangement present in lamprey and hagfish (embryonic and immune receptor) were present in an ancestral lineage that existed more than 400 million years ago and perhaps included the ancestor of the jawed vertebrates. Validating this premise will require better characterization of the genome sequence and mechanisms of rearrangement in lamprey and hagfish.
Introduction
The jawless vertebrates represent ancient offshoots from the vertebrate ancestral lineage and can therefore provide insight onto the evolutionary potential and history of the vertebrate genome. Recent advances in sequence resources and embryological techniques for cyclostomes (lampreys and hagfish) have facilitated the characterization of developmental and genome biology of a few cyclostomes species (especially the sea lamprey: Petromyzom marinus) (Pancer et al. 2004a, 2005; Alder et al. 2005; Nagawa et al. 2007; Rogozin et al. 2007; Nikitina et al. 2009; Smith et al. 2009). Studies have revealed unique features of cyclostome genomes that are not observed in the jawed vertebrates, such as programmed genome rearrangement during early embryogenesis (Smith et al. 2009) and possession of novel immune receptors [variable lymphocyte receptors (VLR)] whose loci undergo receptor diversification via a recombination-activating genes (RAG)-independent mechanism (Alder et al. 2005; Pancer et al. 2005; Nagawa et al. 2007; Rogozin et al. 2007). Given the deep ancestry of hagfish and lamprey lineages, these new insights have the potential to shed light on the genome biology of the ancestral vertebrate lineage, which ultimately gave rise to the genomes of all extant vertebrates.
Interpreting the evolutionary significance of biological observations in the extant cyclostomes requires an understanding of the deep history of vertebrate evolution. The precise evolutionary relationships between lampreys, hagfish, and gnathostomes (jawed vertebrates) remain an open question, as different lines of evidence support different evolutionary scenarios. Despite their differences, all likely evolutionary scenarios share common threads, which permit some definitive interpretations of comparative studies. Other interpretations are contingent on resolving fine-scale evolutionary relationships between the lampreys, hagfish, and gnathostomes, but are nonetheless interesting and may present testable hypotheses.
Defining features of cyclostomes and vertebrates
The lampreys and hagfish are collectively known as the cyclostomes, although there has been substantial debate as to whether the term “cyclostomes” represents a phylogenetically valid grouping. Regardless of personal preference, it is generally agreed that hagfish and lampreys are the closest living relatives to the gnathostomes (jawed vertebrates). Together the hagfish, lampreys and gnathostomes constitute the craniate lineage. The cyclostomes possess all of the defining characteristics of the chordate lineage (notochord, dorsal hollow nerve tube and visceral arches; see Fig. 1), as well as key morphological innovations that are present in the jawed vertebrates [e.g. skull, thyroid and definitive neural crest, but see Jeffery et al. (2008) for ascidians]. However the cyclostomes lack jaws and other organ systems that were present in the ancestor of all jawed vertebrates, including paired fins, ribs, pelvic girdles, true teeth, true bone, and paired nasal sacs (Carroll 1988; Neidert et al. 2001). The unique evolutionary history and deep divergence of the lampreys and hagfishes has been critical for understanding the relative timing of key innovations on the chordate/vertebrate/gnathostome lineage.
Fig. 1.
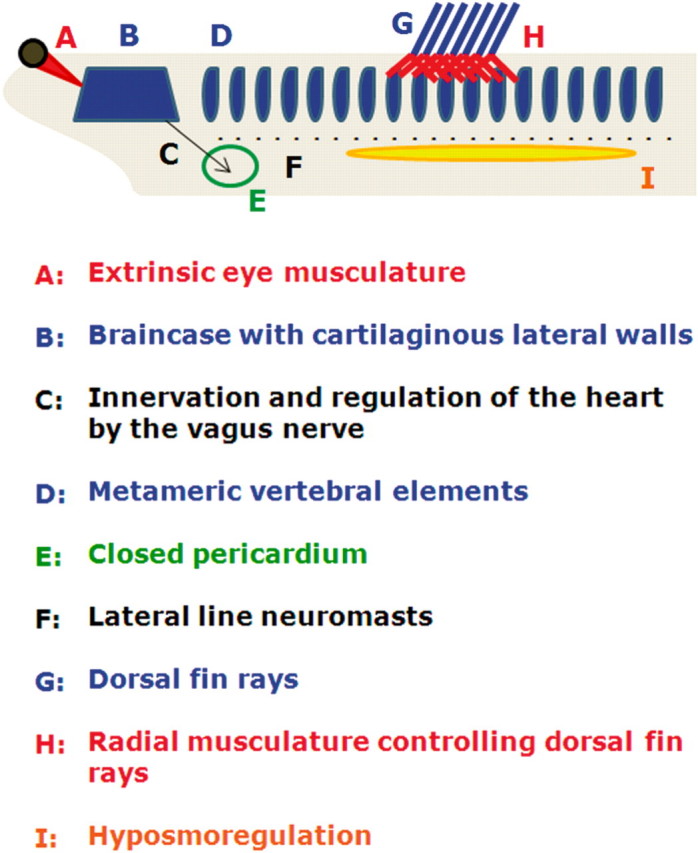
The defining morphological features of the vertebrates. All of the depicted features are considered to be defining features of the vertebrate (lamprey + gnathostomes) lineage.
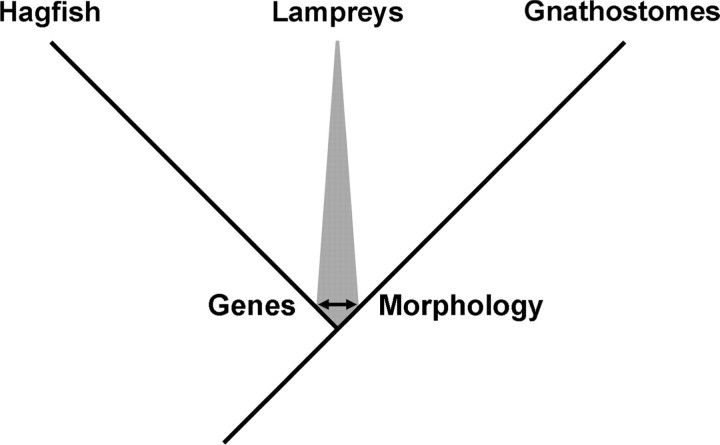
On the basis of morphology, lampreys appear to be much more closely allied to the gnathostomes than do the hagfish (Fig. 2). The lampreys and gnathostomes both possess a braincase with cartilaginous lateral walls, vertebral elements, dorsal fin rays and many other morphological and physiological features (Fig. 1), whereas hagfish lack all of these characters. These shared-derived characters have been considered by many to constitute strong evidence for a evolutionary history shared between lamprey and gnathostome lineages, exclusive of the hagfish lineage (Janvier 1995, 2008; Gess et al. 2006). However, it should be pointed out that each of these lineages has been evolving independently for ∼500 million years (MY) and one cannot ignore the possibility that hagfish have lost, or modified beyond recognition, some of the characters that unite the lampreys and gnathostomes.
Fig. 2.
Phylogeny of the basal vertebrate lineages. It is generally agreed that gnathostome (jawed vertebrates) and hagfish lineages diverged from one another following a deep split of the ancestral craniate lineage. The relative position of lamprey is not definitively resolved but the lamprey lineage is generally considered to have diverged from one of the two lineages shortly after the primary divergence. Molecular data suggest shared ancestry of lampreys and hagfish, whereas morphological data suggest shared ancestry of lampreys and gnathostomes.
Molecular phylogeny
In contrast to morphological studies, molecular studies strongly suggest that lampreys are more closely allied to the hagfish than they are to the gnathostomes (Fig. 2). Recent studies have taken advantage of emerging genomic datasets for these species and have incorporated large numbers of molecular characters from both nuclear and mitochondrial genes (Takezaki et al. 2003; Blair and Hedges 2005b; Delsuc et al. 2006; Kuraku and Kuratani 2006; Near 2009). These studies have invariably found support for shared ancestry of the lampreys and hagfish, exclusive of the gnathostomes. As with morphological studies, the deep branch lengths between lamprey, hagfish, and gnathostomes have the potential to severely influence inferences that are made from the datasets. This is because of the general tendency for the degradation of molecular-phylogenetic signals over long evolutionary distances, and the relatively short time over which these three groups diverged from their common ancestor. The potential influence of these factors on the inferred topology of the craniate tree has been discussed in depth (Near 2009). Despite the apparent disparity of molecular and morphological studies, the inferred phylogenies are identical over more than 90% of their length (Fig. 2) and this fact permits informative interpretations from studies of comparative cyclostome biology.
Resolution from an unresolved phylogeny
Regardless of one's preference of phylogeny, morphological and genetic features that are shared by lamprey and hagfish still shed light on the biology of a vertebrate ancestor that lived nearly 500 MY ago (MYA). The dates of divergence among the three major craniate clades have been estimated by analyzing the degree of molecular change and calibrating rates against the fossil record. The time between the cyclostome/gnathostome split and the lamprey/hagfish split (inferred by all molecular phylogenies) is estimated to have occurred over a window of 15–100 MY (Donoghue et al. 2000; Blair and Hedges 2005b; Kuraku and Kuratani 2006). For comparison, the most basal split of the eutherian lineage is thought to have occurred 130 MYA and the basal divergence of the human/ape clade is thought to have occurred around 15 MYA (Kumar and Hedges 1998). Relative to the length of the lamprey-hagfish-gnathostome tree, the hypothetical cyclostome ancestral lineage represents only 1–7% of the combined evolutionary history of the three lineages. This short (or perhaps non-existent) branch is consistent with observations of the fossil record that suggest that these groups likely diverged some time after the Cambrian explosion [but see Blair and Hedges (2005a)] and must have diverged well before the appearance of species in the Devonian that are strikingly similar to modern day lampreys (Gess et al. 2006) and species in the Pennsylvanian that are similar to modern-day hagfish (Bardack 1991). On the basis of fossil, molecular, and morphological data, it seems relatively safe to infer that the divergence of hagfish, lampreys, and gnathostomes occurred in rapid succession (in geological terms). Thus, an interpretation can be made that is relevant to traits that are observed in lamprey and hagfish: presence of shared traits in lamprey and hagfish indicate that the trait was present in the common ancestor of all craniates, or was present in a lineage that was very similar to the craniate ancestor (having diverged from this ancestor for only a few million years).
Evolution of early vertebrate genomes
The invertebrate chordates (amphioxus and ascidians) are the closest living outgroups to the craniates, and may therefore shed some light onto the biology of the ancestral genome that gave rise to the craniate lineage. The genomes of three invertebrate chordates have been sequenced and assembled: amphioxus [Branchiostoma floridae (Putnam et al. 2008)] and two ascidians [Ciona savignyi (Vinson et al. 2005; Kim et al. 2007) and Ciona intestinalis (Dehal et al. 2002)]. The genome of Ciona experienced significant gene loss and rearrangement after the split from the chordate stem lineage, and has not been used extensively for large-scale reconstructions of ancestral chordate genomes. However, it has shed some insight into evolution at smaller scales (Olinski et al. 2006) and evolution of overall gene content (Dehal et al. 2002).
Analyses of the amphioxus genome have proven more insightful (Hufton et al. 2008; Putnam et al. 2008). These indicate that two duplication events that are thought to have occurred in the craniate ancestor (1R and 2R, discussed in more detail below) are likely to have occurred in rapid succession after divergence of the cephalochordate lineage (Putnam et al. 2008). Moreover, it seems that the duplication and subsequent rediploidization of genomes occurred very close to, and may have overlapped with, the splits that separated the major craniate groups (Putnam et al. 2008). The amphioxus genome also provides insight into rates of rearrangement prior to and following the brief period of time that included 1R, 2R, and the radiation of the three primary craniate lineages (Hufton et al. 2008). It is proposed that rearrangement rates were highest on the lineage that immediately predated this key period, and were substantially lower after rediploidization. This is considered evidence that some aspect of the craniate ancestor’s genome biology may have predisposed it to duplication.
The prediction of a heightened propensity toward rearrangement in the craniate ancestor is intriguing, given the discoveries of dynamic genome biology in two of the three extant craniate lineages: hagfish (Kohno et al. 1986; Kubota et al. 1992, 1993, 1997, 2001) and lampreys (Smith et al. 2009). Notably, data on large-scale gene linkages are not yet available for lamprey or hagfish, and therefore the post-2R deceleration of rearrangement is only resolved for the gnathostome lineage. Ongoing improvement of the lamprey genome assembly should provide new insights into evolutionary rates for at least one cyclostome species.
In addition to the deep evolutionary distance separating cyclostomes and gnathostomes, a further complication in phylogenetic analysis is the presence of duplicated genes. While this has been somewhat difficult to assess in hagfish due to a paucity of data, there is no question that the genome of lampreys contains a wealth of duplicate genes. Many lamprey genes appear to possess at least one duplicate copy (Escriva et al. 2002; Irvine et al. 2002; Sauka-Spengler et al. 2007; Rahimi et al. 2009; Tank et al. 2009) but it is very difficult to assess whether the duplicate copies are orthologous with those of gnathostomes, or whether they are paralogous copies. By implication, those that are orthologous would have occurred prior to the lamprey-gnathostome divergence whereas those that are paralogous occurred after the lamprey-gnathostome divergence. Examples of both are known, making for difficulty in assessing the timing of the 1R and 2R WGD events. Based on analysis of selected families of gene duplicates which satisfy strict criteria, Kuraku and colleagues suggested that the data favor the scenario whereby both the 1R and 2R WGD events occurred prior to the lamprey-gnathostome split (Kuraku 2008; Kuraku et al. 2009). Improvements of the lamprey genome (notably, longer synteny relationships) will be required to better resolve this issue. Genomic data from hagfish, when they become available, will also help to resolve issues related to the 1R/2R WGD and monophyly of the cyclostomes.
Developmentally programmed restructuring of cyclostome genomes
Throughout the tree of life, there are a few species that are known to undergo large-scale reorganization of their genome during development. These reorganization events result in differentiation of somatic lineages (at the sequence and structural level) from their germline progenitors, which presumably pass through development and from generation to generation in a largely unaltered form. For more than two decades, the only known deuterostomes to undergo such large-scale genomic reorganization were hagfish. It was observed that repetitive elements were present in the germline of the hagfish Eptatretus burgeri that were absent from somatic cells of the same species and that these changes were associated with a reduction in chromosome number (Kohno et al. 1986; Nakai et al. 1991; Kubota et al. 2001). Subsequently, similar eliminated repeats and changes in karyotype were discovered in the hagfish species: Eptatretus okinoseanus (Nakai et al. 1991; Kubota et al. 1992, 1993), Paramyxine atami (Nakai et al. 1991; Kubota et al. 1997), Myxine garmani (Nakai et al. 1991), M. glutinosa (Nakai et al. 1995), Eptatretus cirrhatus (Goto et al. 1998; Nakai et al. 1995).
Recently, a similar phenomenon has been discovered in the sea lamprey, P. marinus (Smith et al. 2009). Advances in embryological and genomic resources for this species (Washington University Genome Sequencing Center 2007; Nikitina et al. 2009) have permitted characterization of the types of sequences that are lost as well as the developmental context of loss. These losses are initiated at approximately the transition from blastula to gastrula and in addition to elimination of highly repetitive sequences, as was observed in hagfish, single-copy and transcribed sequences are also lost (Smith et al. 2009). Given our current understanding of craniate phylogeny, the observation of programmed genome reorganization in both lamprey and hagfish could be considered evidence that chromatin diminution was characteristic of the common ancestor of all craniates, or of a basal taxon with a genome biology that was otherwise very similar to the craniate ancestor.
One pressing question is whether loss of DNA in the cyclostomes represents a truly ancestral condition or whether these similar phenomena evolved in parallel over nearly a billion years of independent evolution on the branches to P. marinus and the common ancestor of modern day hagfish. Resolving this will require analyses of other lamprey species to determine if elimination of DNA is widespread among lampreys and further the trace of the ancestry of DNA elimination within the lineage. Perhaps more critical to this question is resolving the mechanisms whereby loss of DNA occurs in both hagfish and lamprey. It is expected that recent and ongoing advances in genomic and developmental resources for lamprey will yield answers for these questions in the near future. The pending initiation of a hagfish genome project (http://www.genome.gov/10002154) and the recent production of embryos of the hagfish, E. burgeri, in the laboratory (Holland 2007; Janvier 2007; Ota et al. 2007) bring to light the possibility of characterizing the precise genetic changes, developmental timing of loss of DNA, and mechanisms of loss in a hagfish species.
VLR recombination versus RAG-mediated VDJ recombination of gnathostome immune receptors
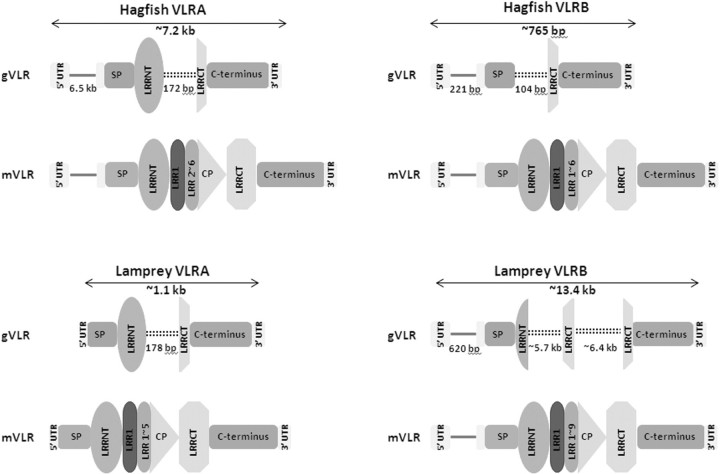
All vertebrates undergo a limited number of programmed rearrangements and thus impart their immune receptors with diverse structures [e.g. immunoglobulins (Igs), T-cell receptor (TCR), and major histocompatibility complex (MHC)] that have the ability to produce enormous numbers of receptor repertoires that recognize diverse antigens. Although Igs, TCRs, MHC, and other fundamental molecules of vertebrate adaptive immunity have not been identified in jawless vertebrates, recent findings have revealed that the lamprey and hagfish possess an alternate adaptive immune receptor system: the VLRs (Pancer et al. 2004a, 2005). The VLRs evolved independently of the Igs and utilize a RAG-independent strategy to generate receptor variants that recognize pathogens (Alder et al. 2005; Pancer et al. 2005; Nagawa et al. 2007; Rogozin et al. 2007; reviewed by Saha 2010). From the perspective of genome structure, the germline VLR loci (gVLRA and gVLRB) are incomplete and unable to achieve functionality unless they undergo rearrangement (Pancer et al. 2004a). Despite the presence of presumptive promoter, transcription initiation-termination sites and the functional start-stop codons, the “core” region between the start and stop codons is incomplete and does not encode a VLR open reading frame. Various numbers of variable leucine-rich repeats (LRRs) are recruited from genomic regions that flank the locus and their importation results in the elimination of long intervening regions that disrupt the germline locus (Fig. 3). Sequencing surveys either from reverse transcriptase assays, from clones isolated from cDNA libraries or from direct genomic PCR amplifications have revealed substantial variation in the length of rearranged VLR genes, which results from the incorporation of variable numbers of diverse LRRV cassettes (Pancer et al. 2004a, 2005; Alder et al. 2005; Nagawa et al. 2007; Rogozin et al. 2007). This incorporation occurs via an as-yet unknown recombinational system, which produces sequence signatures that are consistent with a replication-based recombinational mechanism (Pancer et al. 2005; Alder et al. 2005; Nagawa et al. 2007; Rogozin et al. 2007). Thus, the mechanisms of VLR antigen recognition and receptor diversification are substantially different from those that derive from RAG-mediated VDJ recombination (Table 1).
Fig. 3.
Schematic representations of hagfish (upper panel) and lamprey (lower panel) gVLR genes and their rearranged forms (mVLRs). Coding regions are color-coded (gray—5′ UTR; orange—signal peptide; blue—LRRNT; yellow—LRRCT, pink—C terminus). Solid red lines indicate introns that disrupt the 5′ UTR, while black double dotted lines indicate the intervening sequence. LRR1, LRR, and CP are imported into the mature VLR from flanking regions (not shown) and supplant the intervening sequence during the rearrangement process.
Table 1.
Comparison of VDJ and VLR rearrangement
| Feature | VDJ recombination (mammals) | VLR recombination (cyclostomes) |
|---|---|---|
| Gene organization | Tandemly-arrayed variable (V), diversity (D) and joining (J) segments. | The VLR genes are organized with sequences encoding invariable LRR modules at the 5′- and 3′- ends of the encoded ectodomain followed by a stretch that codes for the long stalk region and GPI cleavage site. Extensive numbers of LRR cassettes and other VLR modules flank the locus at its 5′ and 3′ ends. |
| Mechanism of rearrangement | RAG proteins associate with the recombination signaling sequences (RSS) to initiate DNA cleavage and recombination. | An unknown replication-based mechanism that uses short stretches of homology among flanking LRR sequences (5–30 bp) to build a sequence that encodes a functional ectodomain and to remodel the germline locus. |
| Deployed proteins | Membrane-bound and secreted dimers. | Membrane-bound monomers and secreted pentamers. |
| Cellular context of rearrangement | B lymphocytes rearrange and deploy BCR; T lymphocytes rearrange and deploy TCR. | VLRA and VLRB genes are rearranged and deployed by distinct cell populations that exhibit characteristics similar to T and B lymphocytes, respectively. |
| Function of rearranged genes | Recognition of diverse foreign antigens | Recognition of diverse foreign antigens |
| Magnitude of receptor repertoire diversity | Thought to be more than 1 × 1014 | Thought to be more than 1 × 1014 |
Although the cyclostomes and gnathostome systems result in superficially similar outcomes of diversified immune receptors that were both shaped through evolution within very similar lymphocyte cell populations (reviewed by Saha et al. 2010), the two systems apparently represent convergent solutions to the same problem of antigen recognition. On the other hand, it seems extremely unlikely that the VLR-based immune systems of lamprey and hagfish evolved independently, as both rearrange and deploy orthologous/homologous genes. Thus, it seems abundantly clear that a rearranging VLR molecule and some as-yet unknown rearrangement mechanism were present in the common ancestor of all craniates, or in an organism with a genome biology and immunology that were very similar to the craniate ancestor.
The VLRs in hagfish and lamprey apparently represent orthologous receptor systems, however, other evolutionary aspects of the adaptive immune system are much less clear. From the standpoint of the gnathostome system, bona fide functional homologs of RAG, Ig, TCR, and MHC have not been identified in the lamprey genome despite substantial efforts to find them over the past several decades (Uinuk-Ool et al. 2002; Cooper and Alder 2006; Amemiya et al. 2007). Molecules exhibiting some structural similarities to TCR and B-cell receptor (Ig) have been identified in the cyclostomes (Pancer et al. 2004b; Cannon et al. 2005; Suzuki et al. 2005; Haruta et al. 2006); however, none of these possess gene organizations or transcriptomic profiles indicating that they are capable of generating a diversified immune repertoire. From the perspective of the cyclostome/ancestral system, a fuller understanding of immune receptor diversification will require identification of the molecules that mediate VLR rearrangement and the rearrangement status of VLR homologs in gnathostome genomes. Putative gnathostome homologs of VLR have been proposed (Rogozin et al. 2007), although these are highly divergent from VLR and other similar molecules are now being discovered that may show signatures of past rearrangements (Saha et al. 2010). It seems ever clearer that resolving germline genome sequences for lamprey and hagfish will be necessary to elucidate the evolutionary history of VLR and VJD systems.
Conclusions
A complete understanding the evolutionary relationships between hagfish, lampreys, and gnathostomes will be critical for resolving the precise evolutionary history of extant vertebrate genomes and dissecting cellular mechanisms of genome biology. However, by taking into account the general consensus of data suggesting ancient and rapid evolutionary divergence of these three lineages, we can gain powerful insight into the biology of very old genomes, even at the current level of understanding. The key evolutionary position, and fascinating developmental and genome biologies of the jawless vertebrates argue strongly for the continued development of genome resources for these groups, especially targeted at their germline genomes. Ultimately, the development of germline genome assemblies for lamprey and hagfish should yield comparative synteny information that will provide highly-informative evolutionary characters for phylogenetic reconstruction and aid in the resolution of relationships among 1R/2R paralogy groups. Moreover, it will facilitate the identification of the genes that mediate programmed genome rearrangements and rearrangement of the VLR locus, thereby permitting more rigorous analyses of the presumed shared ancestry of lamprey’s and hagfish’s rearrangement mechanisms, and resolving the evolutionary fates of ancestral rearrangement-gene homologs within the gnathostome lineage.
Funding
This work was supported by the National Institutes of Health [grant number GM079492] and the National Science Foundation [grant number MCB-0719558] to C.T.A. and the National Institutes of Health [grant number T32-HG00035, F32-GM087919] to J.J.S. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
References
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–3. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, Saha NR, Zapata A. Evolution and development of immunological structures in the lamprey. Curr Opin Immunol. 2007;19:535–41. doi: 10.1016/j.coi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardack D. First fossil Hagfish (Myxinoidea): a record from the Pennsylvanian of Illinois. Science. 1991;254:701–3. doi: 10.1126/science.254.5032.701. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular clocks do not support the Cambrian explosion. Mol Biol Evol. 2005a;22:387–90. doi: 10.1093/molbev/msi039. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005b;22:2275–84. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Pancer Z, Mueller MG, Skapura D, Cooper MD, Litman GW. Variable domains and a VpreB-like molecule are present in a jawless vertebrate. Immunogenetics. 2005;56:924–9. doi: 10.1007/s00251-004-0766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RH. Vertebrate paleontology and evolution. New York: W.H. Freeman & Co; 1988. [Google Scholar]
- Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–22. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–67. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–8. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Donoghue PC, Forey PL, Aldridge RJ. Conodont affinity and chordate phylogeny. Biol Rev Camb Philos Soc. 2000;75:191–251. doi: 10.1017/s0006323199005472. [DOI] [PubMed] [Google Scholar]
- Escriva H, Manzon L, Youson J, Laudet V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol. 2002;19:1440–50. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- Gess RW, Coates MI, Rubidge BS. A lamprey from the Devonian period of South Africa. Nature. 2006;443:981–4. doi: 10.1038/nature05150. [DOI] [PubMed] [Google Scholar]
- Goto Y, Kubota S, Kohno S. Highly repetitive DNA sequences that are restricted to the germ line in the hagfish Eptatretus cirrhatus: a mosaic of eliminated elements. Chromosoma. 1998;107:17–32. doi: 10.1007/s004120050278. [DOI] [PubMed] [Google Scholar]
- Haruta C, Suzuki T, Kasahara M. Variable domains in hagfish: NICIR is a polymorphic multigene family expressed preferentially in leukocytes and is related to lamprey TCR-like. Immunogenetics. 2006;58:216–25. doi: 10.1007/s00251-006-0098-1. [DOI] [PubMed] [Google Scholar]
- Holland ND. Hagfish embryos again: the end of a long drought. Bioessays. 2007;29:833–6. doi: 10.1002/bies.20620. [DOI] [PubMed] [Google Scholar]
- Hufton AL, Groth D, Vingron M, Lehrach H, Poustka AJ, Panopoulou G. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008;18:1582–91. doi: 10.1101/gr.080119.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine SQ, Carr JL, Bailey WJ, Kawasaki K, Shimizu N, Amemiya CT, Ruddle FH. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. J Exp Zool. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- Janvier P. The dawn of the vertebrates: characters versus common ascent in the rise of current vertebrate phylogenies. Paleontology. 1995;39:259–87. [Google Scholar]
- Janvier P. Evolutionary biology: born-again hagfishes. Nature. 2007;446:622–3. doi: 10.1038/nature05712. [DOI] [PubMed] [Google Scholar]
- Janvier P. Early jawless vertebrates and cyclostome origins. Zoolog Sci. 2008;25:1045–56. doi: 10.2108/zsj.25.1045. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Chiba T, Krajka FR, Deyts C, Satoh N, Joly JS. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: insights into the ancestry and evolution of the neural crest. Dev Biol. 2008;324:152–60. doi: 10.1016/j.ydbio.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Kim JH, Waterman MS, Li LM. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 2007;17:1101–10. doi: 10.1101/gr.5894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno S, Nakai Y, Satoh S, Yoshida M, Kobayashi H. Chromosome elimination in the Japanese hagfish, Eptatretus burgeri (Agnatha, Cyclostomata) Cytogenet Cell Genet. 1986;41:209–14. doi: 10.1159/000132231. [DOI] [PubMed] [Google Scholar]
- Kubota S, Ishibashi T, Kohno S. A germline restricted, highly repetitive DNA sequence in Paramyxine atami: an interspecifically conserved, but somatically eliminated, element. Mol Gen Genet. 1997;256:252–6. doi: 10.1007/s004380050567. [DOI] [PubMed] [Google Scholar]
- Kubota S, Kuro-o M, Mizuno S, Kohno S. Germ line-restricted, highly repeated DNA sequences and their chromosomal localization in a Japanese hagfish (Eptatretus okinoseanus) Chromosoma. 1993;102:163–73. doi: 10.1007/BF00387731. [DOI] [PubMed] [Google Scholar]
- Kubota S, Nakai Y, Kuro-o M, Kohno S. Germ line-restricted supernumerary (B) chromosomes in Eptatretus okinoseanus. Cytogenet Cell Genet. 1992;60:224–8. doi: 10.1159/000133345. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takano J, Tsuneishi R, Kobayakawa S, Fujikawa N, Nabeyama M, Kohno S. Highly repetitive DNA families restricted to germ cells in a Japanese hagfish (Eptatretus burgeri): a hierarchical and mosaic structure in eliminated chromosomes. Genetica. 2001;111:319–28. doi: 10.1023/a:1013751600787. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–20. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kuraku S. Insights into cyclostome phylogenomics: pre-2R or post-2R. Zool Sci. 2008;25:960–8. doi: 10.2108/zsj.25.960. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zool Sci. 2006;23:1053–64. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8:206–13. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Kubota S, Goto Y, Ishibashi T, Davison W, Kohno S. Chromosome elimination in three Baltic, south Pacific and north-east Pacific hagfish species. Chromosome Res. 1995;3:321–30. doi: 10.1007/BF00713071. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Kubota S, Kohno S. Chromatin diminution and chromosome elimination in four Japanese hagfish species. Cytogenet Cell Genet. 1991;56:196–8. doi: 10.1159/000133087. [DOI] [PubMed] [Google Scholar]
- Near TJ. Conflict and resolution between phylogenies inferred from molecular and phenotypic data sets for hagfish, lampreys, and gnathostomes. J Exp Zool B Mol Dev Evol. 2009;312:749–61. doi: 10.1002/jez.b.21293. [DOI] [PubMed] [Google Scholar]
- Neidert AH, Virupannavar V, Hooker GW, Langeland JA. Lamprey Dlx genes and early vertebrate evolution. Proc Natl Acad Sci USA. 2001;98:1665–70. doi: 10.1073/pnas.98.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina N, Bronner-Fraser M, Sauka-Spengler T CSHL editors. Emerging model organisms: A laboratory manual. Cold Spring Harbor, NY, USA: CSHL Press; 2009. The Sea Lamprey Petromyzon marinus: a model for evolutionary and developmental biology; pp. 405–29. [DOI] [PubMed] [Google Scholar]
- Olinski RP, Lundin LG, Hallbook F. Conserved synteny between the Ciona genome and human paralogons identifies large duplication events in the molecular evolution of the insulin-relaxin gene family. Mol Biol Evol. 2006;23:10–22. doi: 10.1093/molbev/msj002. [DOI] [PubMed] [Google Scholar]
- Ota KG, Kuraku S, Kuratani S. Hagfish embryology with reference to the evolution of the neural crest. Nature. 2007;446:672–5. doi: 10.1038/nature05633. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004a;430:174–80. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Mayer WE, Klein J, Cooper MD. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc Natl Acad Sci USA. 2004b;101:13273–8. doi: 10.1073/pnas.0405529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102:9224–9. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–71. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rahimi RA, Allmond JJ, Wagner H, McCauley DW, Langeland JA. Lamprey snail highlights conserved and novel patterning roles in vertebrate embryos. Dev Genes Evol. 2009;219:31–6. doi: 10.1007/s00427-008-0258-4. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–56. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- Saha NR, Smith J, Amemiya CT. Evolution of adaptive immune recognition in jawless vertebrates. Semin Immunol. 2010;1:25–33. doi: 10.1016/j.smim.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–20. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA. 2009;106:11212–7. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shin I, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol. 2005;174:2885–91. doi: 10.4049/jimmunol.174.5.2885. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Figueroa F, Zaleska-Rutczynska Z, Klein J. Molecular phylogeny of early vertebrates: monophyly of the agnathans as revealed by sequences of 35 genes. Mol Biol Evol. 2003;20:287–92. doi: 10.1093/molbev/msg040. [DOI] [PubMed] [Google Scholar]
- Tank EM, Dekker RG, Beauchamp K, Wilson KA, Boehmke AE, Langeland JA. Patterns and consequences of vertebrate Emx gene duplications. Evol Dev. 2009;11:343–53. doi: 10.1111/j.1525-142X.2009.00341.x. [DOI] [PubMed] [Google Scholar]
- Uinuk-Ool T, Mayer WE, Sato A, Dongak R, Cooper MD, Klein J. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci USA. 2002;99:14356–61. doi: 10.1073/pnas.212527699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson JP, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–35. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington University Genome Sequencing Center. 2007. [(Accessed January 1, 2010)]. http://genome.wustl.edu/pub/organism/Other_Vertebrates/Petromyzon_marinus/assembly/Petromyzon_marinus-3.0/. Washington University School of Medicine. [Google Scholar]