Abstract
M cells are responsible for uptake of mucosal antigens in Peyer's patches (PPs). Differentiation of M cells is thought to be induced by interactions between follicle-associated epithelium and PP cells; however, it remains elusive what types of immune cells function as M-cell inducers. Here, we attempted to identify the cells that serve as an M-cell inducer in PP. We found that a unique B-cell subset characterized by CCR6hiCD11cint resided in the subepithelial dome (SED) in mouse PP. CCR6hiCD11cint B cells showed chemotactic migration in response to CCL20. Furthermore, this unique B-cell subset substantially decreased in PP of CCR6-deficient mice, indicating that the SED localization of CCR6hiCD11cint B cells is most likely regulated by the CCL20–CCR6 system. Concomitantly, CCR6 deficiency caused remarkable decrement of M cells. Moreover, adoptive transfer of CCR6hiCD11cint B cells from wild-type mice restored the M-cell decrement in CCR6-deficient mice. Collectively, the spatial regulation of CCR6hiCD11cint B cells via the CCL20–CCR6 system may play a vital role in M-cell differentiation in mice.
Keywords: CCR6, CD11cint B cells, cell differentiation, M cells, Peyer's patch
Introduction
The gastrointestinal mucosa is continuously exposed to a vast number of food-borne pathogens as well as commensal bacteria. To evoke appropriate immune responses, the external antigens on the mucosal surface must be transported across the epithelial barrier into the gut-associated lymphoid tissue (GALT) such as Peyer's patches (PPs) and isolated lymphoid follicles. This transportation is mainly attributable to M (microfold or membranous) cells in the follicle-associated epithelium (FAE) that covers the lymphoid follicles of GALT. M cells are atypical epithelial cells specialized for uptake of luminal antigens, including phagocytosis of viruses and bacteria, to deliver them to dendritic cells (DCs) accumulating underneath FAE (1).
Although M cells are widely accepted to originate from the intestinal epithelial stem cells residing in the bottom of crypts, distinct mechanisms must direct their differentiation compared with other intestinal epithelial cell lineages (2). A recent report clearly demonstrates that stromal cells at the subepithelial dome (SED) of PP control M-cell differentiation by production of receptor activator of NF-κB ligand (RANKL) (3). On the other hand, restricted localization of M cells in FAE suggests the involvement of immune components underneath the FAE in M-cell development and/or maintenance. In support of this view, co-culture of intestinal epithelial cell lines with PP immune cells gave rise to M-like cells (4, 5). Such lymphoepithelial interaction is likely facilitated by FAE-derived chemokines regulating the spatial distribution of immune cells in PP. CCL20 is regarded as a crucial chemokine for M-cell differentiation since mice lacking CCR6, the sole receptor for CCL20, showed a significant reduction in M cells (6, 7). Given that various PP immune cells including B cells, CD11b+ DCs and CD4+CD45Rblow T cells express CCR6 (6, 8), it should be determined whether these CCR6+ subset(s) are indeed involved in M-cell induction.
To investigate M-cell differentiation, detection of M cells is vital. Until recently, binding of Ulex europaeus agglutinin I lectin and/or absence of brush border enzymes such as alkaline phosphatase have been widely applied for the detection of M cells. These criteria, however, is also applicable to goblet cells. Scanning electron microscopy is another popular method for morphological discrimination between M cells and other intestinal epithelial cells, although it is difficult to obtain highly quantitative data with this method. On the basis of these problems in M-cell detection, previous reports on M-cell differentiation have been controversial. For example, B-cell depletion by deletion of Ig genes resulted in a marked reduction in M cells (9), while Rag1-deficient mice lacking both B and T cells showed a substantial number of remaining M cells (10). The discrepancy on the contribution of B cells in M-cell differentiation may have partly resulted from the lack in M-cell-specific surface markers, which makes it difficult to accurately estimate the number of M cells.
Recently, we have shown that glycoprotein 2 (GP2) is a novel-specific surface marker for M cells (11, 12). By taking advantage of specific detection of M cells with the anti-GP2 antibody, we confirmed the significance of FAE-specific chemokine CCL20 in M-cell differentiation. FACS analysis of PP cells revealed that a novel subset of B cells characterized by CCR6hiCD11cint is substantially detected in wild-type (WT) mice but decreased in CCR6-deficient mice. Reconstitution of CCR6hiCD11cint B cells restored M-cell differentiation in CCR6-deficient mice, indicating that CCR6hiCD11cint B cells have potential to act as M-cell inducer. Thus, we propose that CCR6hiCD11cint B cells facilitate M-cell differentiation in the context of lymphoepithelial interaction.
Methods
Animals
C57BL/6J mice were purchased from CLEA Japan Inc. (Tokyo, Japan) and used as WT mice. CCR6-EGFP knock-in mice (13) were backcrossed onto C57BL/6J mice for eight generations. All mice used in this study were maintained under specific pathogen-free condition in the RIKEN animal facility. All animal experiments were approved by the Animal Research Committee of the RIKEN Yokohama Research Institute.
Immunohistochemistry
Immunohistochemical detection of CCL20 was performed as previously described (14). Briefly, paraffin sections of mouse PP were dehydrated and treated with goat polyclonal antibodies specific for CCL20 (R&D Systems, Minneapolis, MN, USA). The binding of primary antibodies was visualized with Tyramide Signal Amplification Fluorescence Systems (Perkin Elmer, Foster City, CA, USA) according to the manufacturer's instructions. To detect expression of RANKL, frozen sections of PP were treated with a mAb to mouse RANKL (IK22/5; eBioscience, San Diego, CA, USA). The binding of primary antibodies was visualized with Alexa Fluor 555-conjugated anti-rat IgG antibody (Invitrogen, Carlsbad, CA, USA). Subsequently, the slide is treated with biotinylated mAb against CD45 (30F-11; BD Biosciences, San Jose, CA, USA) in combination with FITC-conjugated streptavidin.
For whole-mount staining, PPs were dissected from small intestine of CCR6-defficient and WT mice. The whole-mount specimens of PP were fixed with BD Cytofix/Cytoperm (BD Biosciences) and were incubated with a rat anti-mouse GP2 mAb (2F11-C3) (12), followed by Alexa Fluor 488-conjugated anti-rat IgG antibody and subsequently Alexa Fluor 647-conjugated phalloidin (Invitrogen) for counterstaining.
The immunostained specimens were analyzed using a DM-IRE2 confocal laser scanning microscope and Leica confocal software (Leica Microsystems, Mannheim, Germany).
Preparation of PP lymphocytes
Mouse PP lymphocytes were prepared as described previously (15). Briefly, PPs were dissociated with 0.5 mg ml−1 collagenase (Nitta Gelatin, Osaka, Japan), 0.5 mg ml−1 DNase I (Roche, Indianapolis, IN, USA), 2% fetal bovine serum (FBS), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 20 mM HEPES (pH 7.2) in RPMI 1640 medium (Sigma-Aldrich, St Louis, MO, USA) at 37°C until single-cell suspension was obtained. The single-cell suspensions were subsequently pooled in RPMI 1640 medium supplemented with 2% FBS.
CD11cintCD19+ and CD11c-CD19+ cells were isolated from PP of WT mice by cell sorting using a FACS Aria (BD Biosciences). For cDNA preparation, total RNA was extracted with an RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and 1 μg of total RNA was reverse-transcribed using a ReverTra Ace-α kit (TOYOBO, Osaka, Japan).
Flow cytometric analysis
PP lymphocytes were incubated in 2% FBS/PBS containing an anti-CD16/32 antibody (94; eBioscience) for 15 min to block Fcγ receptors. The cells were then stained with FITC-conjugated goat anti-IgM (Southern Biotech, Birmingham, AL, USA); FITC-conjugated mAbs against CCR6 (140706; R&D Systems), MHC class II (RA3-6B2; eBioscience), CD8α (53–6.7) or CD19 (1D3); PE-conjugated mAbs against CD11b (M1/70), CD11c (HL3), IgD (11-26; eBioscience) or PDCA1 (JF05-1C2.4.1; Milteny Biotec, Bergisch Gladbach, Germany); PE-Cy7-conjugated mAbs against CD3ε (145-2C11); allophycocyanin (APC)-conjugated B220 (RA3-6B2) and a biotinylated mAb against CD11c (HL-3) or Gr-1 (RB6-8C5) in combination with PE-, PE-Cy7- or APC-conjugated streptavidin.
Surface expression of LTα1β2 was detected with LTβR-Ig (16), followed by PE-conjugated anti-human IgG polyclonal antibodies (eBioscience). Free IgG-binding sites were blocked by incubation with mouse and rat serum (1:25 dilution). Subsequently, the cells were treated with FITC-conjugated anti-CD11c, PE-Cy7-conjugated anti-CD3ε (145-2C11; eBioscience) and APC-conjugated anti-CD19 (6D5; Milteny Biotec) mAbs.
All mAbs and streptavidins were obtained from BD Biosciences unless specified otherwise. The stained cells were subjected to flow cytometric analysis using a FACS calibur with a CellQuest software (BD Biosciences), and the obtained data were further analyzed with a FlowJo software version 8.6 (Tree Star, Ashland, OR, USA).
Sequence analysis of transcripts for mouse Igμ
To detect rearranged B-cell receptor (BCR) gene products, PCR was conducted using cDNAs from CD11c+CD19+ or CD11c−CD19+, and specific primers for the variable (V) region of Igμ as previously reported (17). The PCR products subcloned into pGEM-T easy vector (Promega, Madison, WI, USA) were sequenced using an ABI 310 (ABI, Foster City, CA, USA). The obtained sequences were subjected to IMGT/V-QUEST (the International ImmunoGeneTics Information System/V-QUEry and STandardization; http://imgt.cines.fr/) (18) for alignment with sequence information of Ig genes deposited in IMGT as the reference.
Chemotaxis assay
Chemotaxis assays were performed with Transwell chambers (pore size, 3 μm; Corning Coster Corp., Corning, NY, USA) as described previously (14). Briefly, CD11cintCD19+ cells were sorted from PP of WT mice using a FACS Aria. The sorted cells were suspended at 1 × 106 cells ml−1 in RPMI 1640 medium supplemented with 2% BSA, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 20 mM HEPES (pH 7.2). The cell suspension and mouse CCL20 (10–300 ng ml−1; Peprotech, Rocky Hill, NJ, USA) was added to the upper and lower chambers, respectively. After incubation in 5% CO2 at 37°C for 4 h, the cells migrated into the lower chamber were counted using a FACS calibur.
Adoptive transfer experiments
To trace the migration of CD11cintCD19+ cells, the cells sorted from WT mice were labeled with a PKH26 red fluorescent cell linker kit (Sigma-Aldrich) and then injected (1 × 105 cells per mouse) intravenously through tail vein into WT mice. After 48 h, PPs from the mice were embedded in O.C.T compound (Sakura, Tokyo, Japan). Frozen sections (3 μm) of PP were mounted with Vectashield containing DAPI (Vector Laboratories Inc., Burlingame, CA, USA). To examine requirement of CD11cint B cells in M-cell differentiation, CD11cint B or CD11c− B cells were isolated from PP of C57BL/6J mice by a FACS Aria and were adoptively transferred to CCR6-deficient mice (2–3 × 105 cells per mouse). After the adoptive transfer of 72 h, PPs were subjected to whole-mount staining with an anti-GP2 mAb as described above.
Quantitative analysis of fluorescent beads uptake by M cells
The uptake of 200-nm diameter fluorescent polystyrene latex nanoparticles (Fluoresbrite YG; Polysciences, Warrington, PA, USA) by M cells was assessed as described previously (3). Briefly, 1 × 1010 of the nanoparticles in a volume of 200 μl PBS were orally administrated to mice fasted for overnight. The mice were euthanized 4 h after the administration. Two PPs from the ileal end were cut out and embedded in O.C.T. compound. Ten frozen sections from each PP were examined by microscopy after counterstaining with DAPI.
Real-time PCR
Isolation of total RNA from FAE and subsequent cDNA synthesis were described previously (15). Quantitative reverse transcription (RT)-PCR analysis for Gp2 expression was performed using Thermal Cycler Dice Real Time System (Takara Bio Inc., Otsu, Japan) with SYBR® Premix Ex Taq™ (Takara Bio). mRNA level of Gp2 was calculated by the comparative Ct method using glyceraldehyde 3-phosphate dehydrogenase expression as a reference. The primer sets for each gene are available upon request.
Statistical analyses
Differences between two groups were analyzed by Student's t-test. When variances were not homogeneous, the data were analyzed by Mann–Whitney U-test. Differences among more than two groups were analyzed by one-way analysis of variance followed by Tukey's test.
Results
CCL20–CCR6 signaling affects M-cell differentiation
Although the CCL20–CCR6 signaling is proposed to participate in M-cell development (6, 7), precise distribution of CCL20 protein in the small intestine has not been fully addressed. Thus, we first attempted to detect CCL20 in the small intestine with an anti-CCL20 antibody. As shown in Fig. 1A, immunofluorescent signals for CCL20 were exclusively detected in FAE but not in intestinal villi or crypts. To evaluate if CCL20 is actually involved in M-cell differentiation, we counted the number of M cells in PP of CCR6-deficient mice. Whole-mount staining of PP with an anti-GP2 mAb indicated that M cells were sparsely observed in CCR6-deficient mice compared with WT mice (Fig. 1B). The frequency of M cells in the FAE was less than half in CCR6-deficinent mice compared with that in WT mice (Fig. 1C), confirming the requirement of the CCL20–CCR6 system in M-cell differentiation.
Fig. 1.
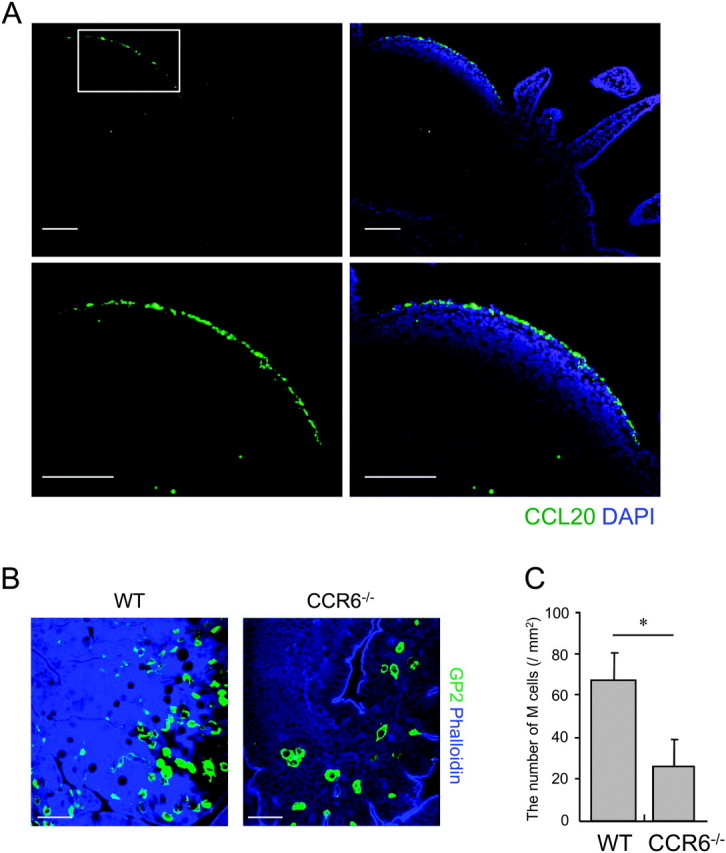
An important role of the CCL20–CCR6 system in differentiation of M cells. (A) Sections of the mouse small intestine including PP were stained with anti-CCL20 antibody (green) and DAPI (blue) for a nuclei counter stain. Lower pictures are higher magnification of the FAE dome region indicated by a square in upper pictures. Scale bars represent 100 μm. (B) Whole-mount specimens of PP from WT and CCR6-deficient mice were stained with anti-GP2 mAb and phalloidin to visualize M cells and F-actin, respectively. Scale bars represent 40 μm. (C) Number of M cells in WT and CCR6-deficient mice were quantified. The number was normalized to the area of observation field. Values are mean ± SD (n = 5). *P < 0.05 (Student's t-test). Results are representative of three separate experiments.
CCR6 deficiency causes decrement of CCR6hiCD11cintCD19+ cells in PP
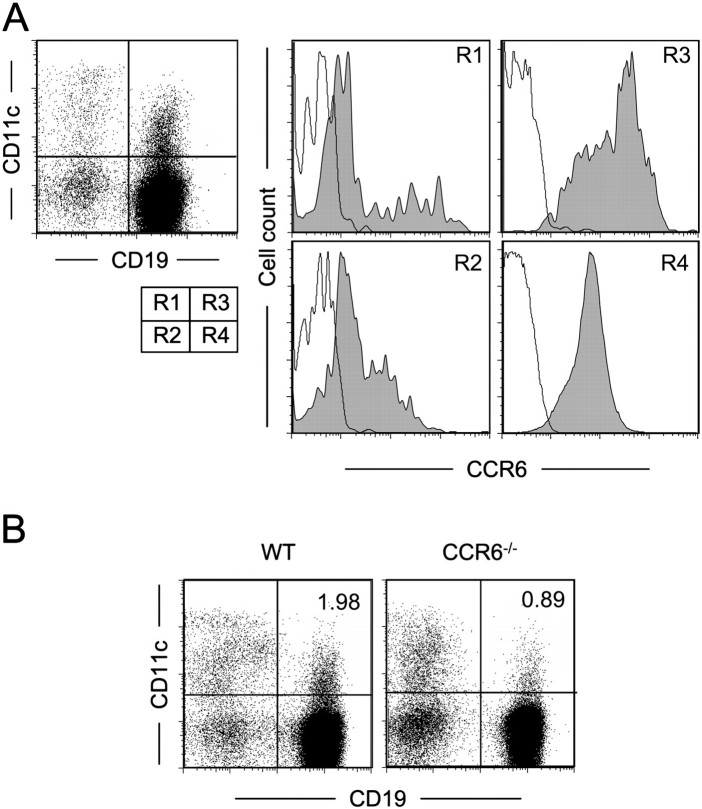
The impairment of M-cell differentiation in CCR6-deficient mice implicates chemotaxis of CCR6+ immune cells toward CCL20 to serve as an M-cell inducer in FAE. To first identify CCR6+ cell subsets in PP, we examined the surface expression of CCR6 on PP cells from WT mice. FACS analysis classified PP cells into four populations based on surface expression of CD11c and CD19 (Fig. 2A, R1 to R4). In R1 (CD11c+CD19−, conventional DC) and R2 (CD11c−CD19−) populations, only a small number of cells expressed a substantial level of CCR6 on their cell surface. In contrast, almost all R3 (CD11cintCD19+) and R4 (CD11c−CD19+, conventional B cell) cells expressed intermediate to high levels of CCR6. The expression level of CCR6 was higher in CD11cintCD19+ (R3) cells than in CD11c−CD19+ (R4) cells (Fig. 2A, histograms). We subsequently examined the influence of CCR6 deficiency on the cellular composition of PP. Among the populations, only the frequency of CD11cintCD19+ (R3) cells was prominently reduced in CCR6-deficient mice compared with that in WT mice (2.06 ± 0.10% and 0.95 ± 0.04% of total cell population, in WT and CCR6-deficient mice, respectively. Mean ± SD of three different preparations (P < 0.05) (Fig. 2B and Table 1), suggesting that CD11cintCD19+ cells migrate into PP in a CCR6-dependent manner.
Fig. 2.
Identification of CCR6hiCD11cint B cells. (A) The dot plot shows expression of CD19 and CD11c of PP cells. The histograms show CCR6 expression of the cells in R1–R4 gates. Filled and blank areas show staining with anti-CCR6 mAb and isotype control, respectively. A representative result of three independent experiments is shown. (B) PP cells from WT and CCR6-deficient mice were monitored for surface expression of CD19 and CD11c. Percentage of CD11cintCD19+ cells in total cell population is shown. The result is a representative of four independent experiments.
Table 1.
The cellular composition of PP in WT and CCR6 KO mice
| Cellular composition (%)a |
Relative ratio (CCR6 KO/WT, %) | ||
| WT | CCR6 KO | ||
| CD11c+CD19− (R1) | 11.9 ± 2.04 | 13.0 ± 3.04 | 109 |
| CD11c−CD19− (R2) | 23.6 ± 2.70 | 32.2 ± 1.73 | 136 |
| CD11cintCD19+ (R3) | 2.06 ± 0.10 | 0.95 ± 0.04 | 46 |
| CD11c−CD19+ (R4) | 62.4 ± 2.63 | 53.8 ± 1.28 | 86 |
Lymphocytes from PP of WT and CCR6 KO mice were stained with anti-CD11c and anti-CD19 mAbs and were analyzed by a FACS calibur. PP cells were divided into four populations based on expression of CD11c and CD19. Values are mean ± SD from three independent experiments.
CCR6hiCD11cintCD19+ cells belong to B-cell lineage
To further characterize CD11cintCD19+ cells, we analyzed more in detail the expression of surface markers in this cell population. The CD11cintCD19+ cells were positive for MHC class II, B220, IgM and IgD, whereas they were negative for CD4, CD8α, Gr-1 and PDCA1 (Fig. 3A). The expression profiles of these markers were compatible between CD11cintCD19+ cells and CD11c−CD19+ conventional B cells (data not shown), suggesting that the former cell population belongs to B-cell lineage. To address this more directly, we examined whether CD11cintCD19+ cells carry a rearranged BCR gene. RT-PCR analysis for the V region of Igμ was performed on total RNA samples from CD11cintCD19+ cells, in addition to conventional B cells (CD11c−CD19+) and DC (CD11c+CD19−) as positive and negative references, respectively. PCR products with the anticipated size were detected in CD11cintCD19+ and conventional B cells but not in DC (Fig. 3B). Sequence analysis of the PCR products from CD11cintCD19+ cells with IMGT (18) data confirmed the presence of VDJ rearranged BCR gene in these cells. The sequences of the V region of Igμ from CD11cintCD19+ cells were heterogeneous (data not shown). Collectively, CD11cintCD19+ cells in PP were classified as a novel sub-population of B-lineage cells. Hereafter, we refer to CD11cintCD19+ cells as CD11cint B cells in this paper.
Fig. 3.
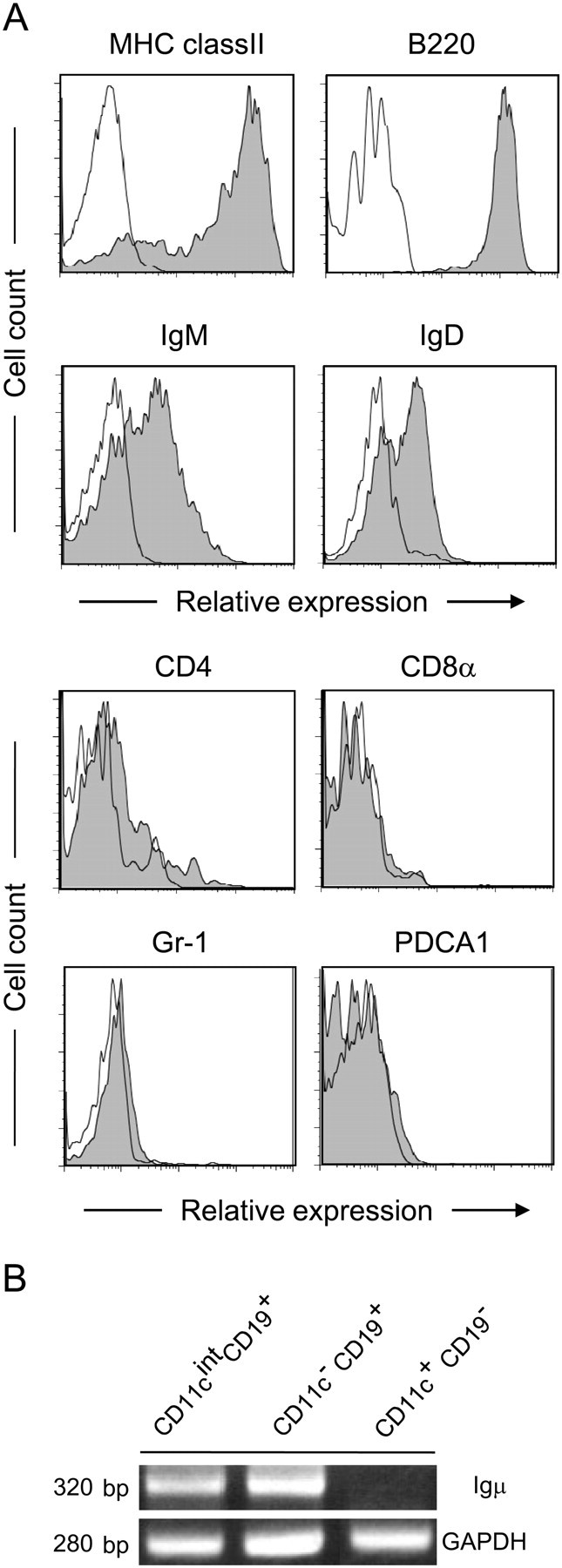
Expression profiles of surface marker on CD11cintCD19+ cells. (A) PP cells from WT mice were stained with indicated specific antibodies (gray area) or isotype controls (blank area). The cells in the CD11cintCD19+ gate were shown. Three independent experiments gave similar results and the representative data are shown. (B) PCR was performed with cDNAs derived from CD11cintCD19+, CD11c−CD19+ or CD11c+CD19− cell fraction and primer sets specific for the rearranged V region of Igμ (30 cycles) or glyceraldehyde 3-phosphate dehydrogenase (25 cycles) as an internal control.
CD11cint B cells show migratory response to CCL20
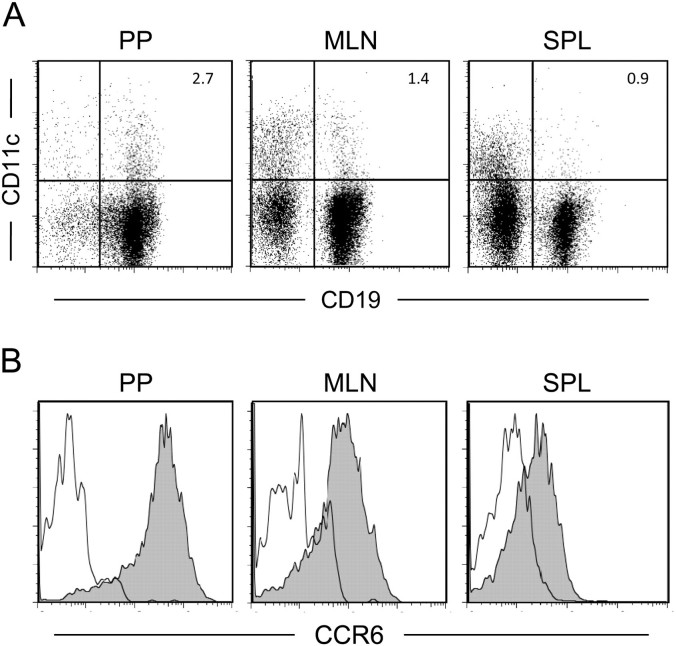
Since CCR6 deficiency brought about decrement of CD11cint B cells in PP, these cells seem to migrate into PP in response to CCL20 secreted from FAE. To assess correlation between CCR6 expression and distribution of CD11cint B cells, we conducted FACS analysis of CD11c and CCR6 on B cells in different lymphoid organs. In addition to PP, CD11cint B cells were observed in the mesenteric lymph node (MLN) and spleen, albeit at a lesser extent (Fig. 4A). Remarkably, the expression level of CCR6 in CD11cint B cells was different among them. In PP, most of CD11cint B cells abundantly expressed CCR6, while CD11cint B cells exhibited an intermediate level of CCR6 in MLN. Splenic CD11cint B cells scarcely expressed CCR6 (Fig. 4B). These results are consistent with the notion that CCR6hiCD11cint B cells might be recruited to PP in response to FAE-derived CCL20.
Fig. 4.
Detection of CD11cint B cells in PP, MLN and the spleen (SPL). (A) Cells from PP, MLN and the SPL of WT mice were analyzed for surface expression of CD11c and CD19. (B) CD11cint B cells were gated and analyzed for surface expression of CCR6. Filled and blank histograms show staining with anti-CCR6 mAb and isotype control, respectively. Three independent experiments gave similar results and the representative data are shown.
To gain further evidence for the roles of CCL20 on the migration of CD11cint B cells to PP, we next performed in vitro chemotaxis assay. CCL20 induced chemotactic response by CD11cint B cells in a concentration-dependent manner (Fig. 5A). We also assessed migration of CD11cint B cells in vivo using adoptive transfer experiment. CD11cint B cells from WT mice were labeled with PKH26 and were adoptively transferred to WT mice. After 48 h, a substantial number of the transferred CD11cint B cells migrated into the SED region of PP (Fig. 5B), although a part of them were also observed in the intestinal lamina propria (data not shown). In agreement with this observation, whole-mount staining with anti-CD11c and anti-CD19 mAbs confirmed the localization of endogenous CD11cint B cells in SED region of PP from WT mice (Fig. 5C). Taken together, these data suggest that CCL20 derived from FAE attracts migration of CD11cint B cells to PP and further to the SED region of PPs.
Fig. 5.
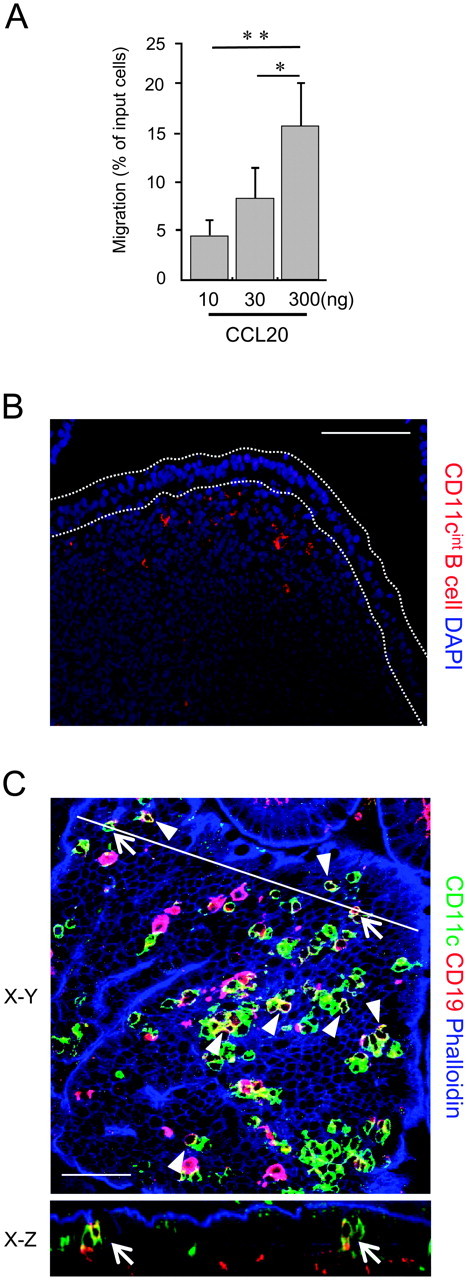
Distribution of CD11cint B cells in PP. (A) Chemotactic activity of CD11cint B cells was examined with various concentrations of CCL20. Values are mean ± SD (n = 3). *P < 0.05, **P < 0.01 (one-way analysis of variance followed by Tukey's post hoc test). A representative result of three independent experiments is shown. (B) CD11cint B cells prepared from PP of WT mice were labeled with PKH26 red fluorescent dye and intravenously injected into recipient WT mice. PP tissue sections were prepared 48 h after the injection. The sections were counterstained with DAPI. Scale bar represents 100 μm. (C) Whole-mount specimens of PP were stained with an anti-CD11c (green) and CD19 (red) mAbs and phalloidin (blue). Arrows and Arrowheads represent CD11cint B cells. The X–Z image of FAE at the position indicated by a solid line in the X–Y image is shown. Scale bars represent 40 μm.
CD11cint B cells contribute to M-cell differentiation
The reduction of CD11cint B cells in CCR6-deficient mice led us to assume that these cells may contribute to M-cell differentiation. To evaluate this, CD11cint and CD11c− B cells from PP of WT mice were adoptively transferred to CCR6-deficient mice. Three days later, we collected PP from the recipient mice and counted the number of M cells stained with the anti-GP2 mAb. We found that density of M cells were 1.9-fold increased in PP of the mice receiving CD11cint B cells compared with those receiving CD11c− B cells (Fig. 6A and B). The increment of functional M cells after adoptive transfer of CD11cint B cells was further confirmed by the fluorescent nanoparticle-uptake assay (3). The number of the nanoparticles internalized into PP was significantly higher in CCR6-deficient mice receiving CD11cint B cells compared with those receiving CD11c− B cells (Fig. 6C). Collectively, these results indicate that CD11cint B cells serve as an M-cell inducer.
Fig. 6.
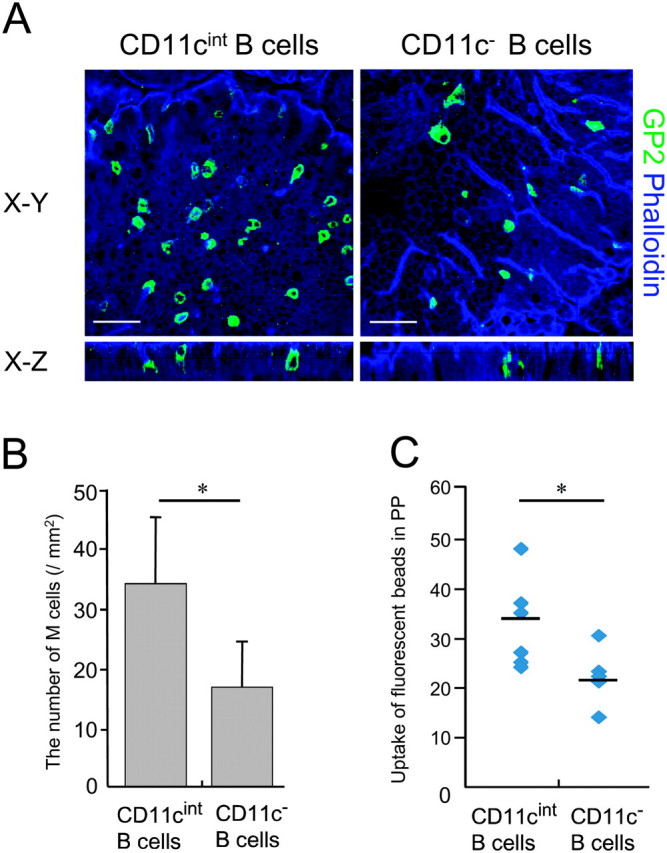
Involvement of CD11cint B cells in differentiation of M cells. (A) CD11cint and CD11c− B cells from WT mice were adoptively transferred to CCR6-deficient mice. Three days later, whole-mount specimens of PP were stained with an anti-GP2 (green) mAb and phalloidin (blue) to visualize M cells and F-actin, respectively. Scale bars represent 40 μm. (B) Quantification of M cells in FAE of CCR6-deficient mice after the adoptive transfer of CD11cint and CD11c− B cells. The number of M cells was counted in FAE region, and the value was normalized to the area of observation field. Values are mean ± SD of 15 observation fields in three mice per group. *P < 0.05 (Mann–Whitney U-test). (C) The number of nanoparticles internalized into PP was quantified in CCR6-deficient mice receiving CD11cint B cells or CD11c− B cells. Values are mean ± SD of three mice per group. At least 10 observation fields were examined per mice. *P < 0.05 (Student's t-test).
CD11cint B cells does not affect the expression of RANKL in PP
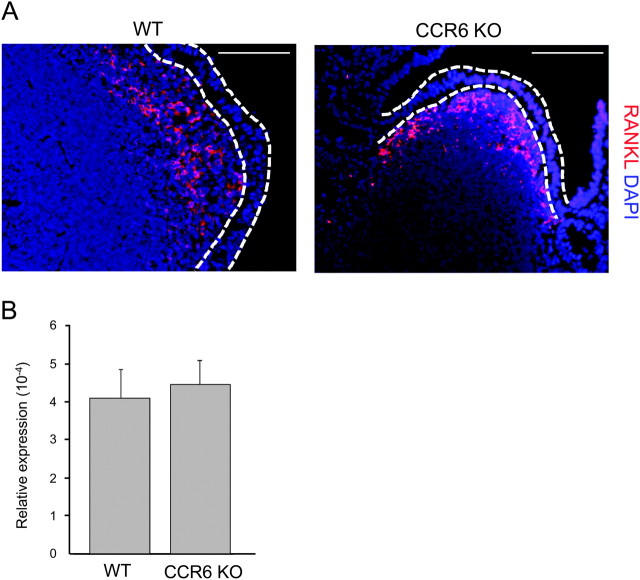
Recently, Knoop et al. showed that RANKL expressed by stromal cells in PP SED is a central molecule involved in M-cell differentiation (3). This raises the possibility that impaired CD11cint B-cell migration due to CCR6 deficiency may affect the expression of RANKL by the stromal cells in PP SED, which ultimately results in the impairment in M-cell differentiation. Thus, we examined the expression of RANKL in CCR6-deficient mice, which lack the normal migration of CD11cint B cells to PP SED region. Consistent with the previous report (3), RANKL was highly expressed in the SED region of PP from WT mice. Of note, RANKL-positive cells did not express CD45, confirming that RANKL is expressed by stromal cells (data not shown). In support of this notion, hematopoietic cell populations including CD11cint B cells expressed little, if any, Tnfsf11 mRNA encoding RANKL protein (Supplementary Figure 1 is available at International Immunology Online). We observed RANKL expression in PP SED region in CCR6-deficient mice (Fig. 7A). Furthermore, the expression level of RANKL-encoding Tnfsf11 mRNA was comparable between PP from CCR6-deficient mice and WT mice by real-time PCR analysis (Fig. 7B). Thus, ablation of CCR6 does not seem to affect RANKL expression in PP. These data suggest that RANKL expression by stromal cells in PP SED does not require the presence of CD11cint B cells in this region.
Fig. 7.
RANKL is normally expressed in PP in the absence of CD11cint B cells. (A) Frozen sections of PP from WT and CCR6-deficient mice were stained with anti-RANKL antibody (red) and DAPI (blue) for a nuclei counter stain. Scale bars represent 100 μm. (B) Total RNA samples were isolated from PP of the same mice as described in A and were subjected to real-time PCR analysis. The mRNA expression level of Tnfsf11 encoding RANKL was normalized to the value of GAPDH. Data are mean ± SD (n = 3).
Discussion
In this study, we have demonstrated that CD11cint B cells, a unique subset of B cells characterized by the B220+CCR6hiCD11cintCD19+IgM+IgD+ surface phenotype, localize in PP. Adoptive transfer of these CD11cint B cells from WT mice to CCR6-deficient mice restored decrement of M cells caused by CCR6 deficiency, indicating that CD11cint B cells serve as an M-cell inducer. CD11cint B cells were unique in their expression of CD11c. Although a subset of plasmacytoid DCs (pDCs) expressing indoleamine 2,3-dioxygenase (IDO) is also positive for CD11c, CD19 and CCR6 (19), CD11cint B cells and IDO+ pDCs likely represent distinct cell populations: CD11cint B cells do not express typical pDC markers such as PDCA-1 and Gr-1 (19), whereas they carry productively rearranged BCR genes, a hallmark of B-cell lineage. Recent report manifested that human memory B-cell sub-populations also express CD11c (20). Despite the similarity in expression of CD11c, CD11cint B cells showed little if any expression of FCRL4, CD20, CD95, RUNX2, SOX5 and RANKL, all of which are abundant in the human memory B-cell sub-populations (data not shown). Further studies are needed to clarify whether mouse CD11cint B cells belong to memory B-cell lineage or not.
RANKL expression in SED of PP is a critical factor controlling the differentiation of M cells (3). However, to date the molecular mechanism underlying the induction of RANKL in SED region has not been addressed. A previous report suggested that RANKL is mainly expressed by stromal cells in the SED region beneath the FAE of PP (21). We also confirmed that RANKL is mainly produced by CD45-negative non-hematopoietic cells, but not CD45-positive hematopoietic cells including CD11cint B cells, in PP of WT mice (Supplementary Figure 1 is available at International Immunology Online). We observed that deficiency of CCR6 does not affect the expression of RANKL in PP (Fig. 7A), and vice versa, neutralization of RANKL with anti-RANKL antibody did not influence the expression of CCL20 in FAE (data not shown). These data support the notion that the CCL20–CCR6 and RANKL–RANK systems are regulated independently of each other. Furthermore, substantial reduction of M cells in CCR6-deficient mice implies that RANKL is essential but not sufficient for induction of M cells. Thus, we propose that both RANKL and CD11cint B cells are required in M-cell induction. Compared with a profound reduction in the number of M cells in the absence of RANKL (3), we observed that the impairment in M-cell differentiation is milder in CCR6-deficient mice. This leaves the possibility that other cell subset(s) in PP partially compensates the absence of CD11cint B cells for M-cell differentiation. Further studies will be required to clarify the question.
RANKL signals through its receptor RANK and plays an important developmental role in multiple tissues (22, 23). RANK is expressed on multiple cell types including osteoclasts and their precursor, DCs, endothelial cells and mammary epithelial cells (24). This raises the possibility that RANKL signals CD11cint B cells, leading to the M-cell differentiation. Thus, we examined whether CD11cint B cells express RANK. Expression of Tnfrsf11a mRNA encoding RANK was high in intestinal epithelium, namely FAE and villous epithelium (VE), whereas it was minimal in PP lymphocytes including CD11cint B cells (Supplementary Figure 2 is available at International Immunology Online). This is consistent with the previous report showing that RANK is selectively expressed by intestinal epithelium including FAE and VE in PP (3). Together, these observations suggest that RANKL likely signals epithelial cells directly, rather than indirectly through CD11cint B cells, to promote M-cell differentiation. However, it is still possible that RANKL–RANK signaling acts indirectly on CD11cint B cells via unknown factor(s) (other than CCL20 as mentioned above) for M-cell differentiation. Additional experiments will be necessary to verify this speculation.
Signaling via LTβR is documented to be another important pathway implicated in M-cell differentiation since inhibition of this pathway with LTβR-Ig fusion protein hampered differentiation of M cells in Rag1-deficient mice (10). Our FACS analysis suggested that CD11cint B cells moderately expressed LTα1β2 (data not shown). However, a previous study has suggested that B-cell-specific deletion of LT does not affect M-cell differentiation (25), demonstrating that CD11cint B cells do not employ LT-LTβR signaling for M-cell induction. Considering that LTβR signaling is responsible for CCL20 expression in FAE (26), LTβR might indirectly promote accumulation of CD11cint B cells underneath FAE via CCL20–CCR6 system, thereby contributing to M-cell development.
Based on our data in conjunction with previous findings, we postulate a model for M-cell differentiation in PP. Initially, communication between hematopoietic cells and epithelial cells via the LTβR signaling may trigger FAE-specific CCL20 expression, which in turn attracts CD11cint B cells toward FAE. This migrating CD11cint B cells interact, either directly or indirectly, immature epithelial cells to promote their differentiation into M cells. That is to say, M-cell differentiation must be highly dependent on the proximity of CD11cint B cells and FAE, which is attributable to the lymphoepithelial interaction.
In conclusion, our data presented here uncover a new participant involved in M-cell differentiation, although we do not exclude the possibility that there might be other cell subset(s) of M-cell inducers in PP. Further analysis for elucidation of effector molecule(s) for M-cell induction produced by CD11cint B cells should shed light on the molecular mechanisms for M-cell differentiation in the context of lymphoepithelial interaction.
Supplementary data
Supplementary Figures 1 and 2 are available at International Immunology Online.
Funding
Grants-in-Aid for Young Scientists (A) (22689017 to K.H.) and (B) (18790343, 20790383 to K.H.); Scientific Research (B) (21390155 to H.O.); Scientific Research in Priority Areas (21022049 to K.H.) and Scientific Research on Innovative Areas (20113003 to H.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Sasakawa Scientific Research Grant from The Japan Science Society (to K.H.); Takeda Science Foundation (to H.O.); The Mitsubishi Foundation to H.O.
Supplementary Material
Acknowledgments
We would like to thank Dr Jeffrey L. Browning for LTβR-Ig. We also thank Takashi Kanaya, Shinji Fukuda, Shunsuke Kimura, Kazuya Kawano, Sayaka Kawano, Yumiko Fujimura and Kei-ichiro Suzuki for helpful discussion and technical assistance. The authors declare no competing financial interests.
References
- 1.Kraehenbuhl JP, Neutra MR. Epithelial M cells: differentiation and function. Annu. Rev. Cell Dev. Biol. 2000;16:301. doi: 10.1146/annurev.cellbio.16.1.301. [DOI] [PubMed] [Google Scholar]
- 2.Gebert A, Fassbender S, Werner K, Weissferdt A. The development of M cells in Peyer's patches is restricted to specialized dome-associated crypts. Am. J. Pathol. 1999;154:1573. doi: 10.1016/S0002-9440(10)65410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knoop KA, Kumar N, Butler BR, et al. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol. 2009;183:5738. doi: 10.4049/jimmunol.0901563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerneis S, Pringault E. Plasticity of the gastrointestinal epithelium: the M cell paradigm and opportunism of pathogenic microorganisms. Semin. Immunol. 1999;11:205. doi: 10.1006/smim.1999.0176. [DOI] [PubMed] [Google Scholar]
- 5.Kanaya T, Miyazawa K, Takakura I, et al. Differentiation of a murine intestinal epithelial cell line (MIE) toward the M cell lineage. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;295:G273. doi: 10.1152/ajpgi.00378.2007. [DOI] [PubMed] [Google Scholar]
- 6.Lugering A, Floer M, Westphal S, et al. Absence of CCR6 inhibits CD4+ regulatory T-cell development and M-cell formation inside Peyer's patches. Am. J. Pathol. 2005;166:1647. doi: 10.1016/S0002-9440(10)62475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westphal S, Lugering A, von Wedel J, et al. Resistance of chemokine receptor 6-deficient mice to Yersinia enterocolitica infection: evidence of defective M-cell formation in vivo. Am. J. Pathol. 2008;172:671. doi: 10.2353/ajpath.2008.070393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams IR. Chemokine receptors and leukocyte trafficking in the mucosal immune system. Immunol. Res. 2004;29:283. doi: 10.1385/IR:29:1-3:283. [DOI] [PubMed] [Google Scholar]
- 9.Golovkina TV, Shlomchik M, Hannum L, Chervonsky A. Organogenic role of B lymphocytes in mucosal immunity. Science. 1999;286:1965. doi: 10.1126/science.286.5446.1965. [DOI] [PubMed] [Google Scholar]
- 10.Debard N, Sierro F, Browning J, Kraehenbuhl JP. Effect of mature lymphocytes and lymphotoxin on the development of the follicle-associated epithelium and M cells in mouse Peyer's patches. Gastroenterology. 2001;120:1173. doi: 10.1053/gast.2001.22476. [DOI] [PubMed] [Google Scholar]
- 11.Terahara K, Yoshida M, Igarashi O, et al. Comprehensive gene expression profiling of Peyer's patch M cells, villous M-like cells, and intestinal epithelial cells. J. Immunol. 2008;180:7840. doi: 10.4049/jimmunol.180.12.7840. [DOI] [PubMed] [Google Scholar]
- 12.Hase K, Kawano K, Nochi T, et al. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature. 2009;462:226. doi: 10.1038/nature08529. [DOI] [PubMed] [Google Scholar]
- 13.Kucharzik T, Hudson JT, III, Waikel RL, et al. CCR6 expression distinguishes mouse myeloid and lymphoid dendritic cell subsets: demonstration using a CCR6 EGFP knock-in mouse. Eur. J. Immunol. 2002;32:104. doi: 10.1002/1521-4141(200201)32:1<104::AID-IMMU104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Hase K, Murakami T, Takatsu H, et al. The membrane-bound chemokine CXCL16 expressed on follicle-associated epithelium and M cells mediates lympho-epithelial interaction in GALT. J. Immunol. 2006;176:43. doi: 10.4049/jimmunol.176.1.43. [DOI] [PubMed] [Google Scholar]
- 15.Hase K, Ohshima S, Kawano K, et al. Distinct gene expression profiles characterize cellular phenotypes of follicle-associated epithelium and M cells. DNA Res. 2005;12:127. doi: 10.1093/dnares/12.2.127. [DOI] [PubMed] [Google Scholar]
- 16.Browning JL, Sizing ID, Lawton P, et al. Characterization of lymphotoxin-alpha beta complexes on the surface of mouse lymphocytes. J. Immunol. 1997;159:3288. [PubMed] [Google Scholar]
- 17.Wang X, Stollar BD. Human immunoglobulin variable region gene analysis by single cell RT-PCR. J. Immunol. Methods. 2000;244:217. doi: 10.1016/s0022-1759(00)00260-x. [DOI] [PubMed] [Google Scholar]
- 18.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munn DH, Sharma MD, Mellor AL. Ligation of B7-1/B7-2 by human CD4+ T cells triggers indoleamine 2,3-dioxygenase activity in dendritic cells. J. Immunol. 2004;172:4100. doi: 10.4049/jimmunol.172.7.4100. [DOI] [PubMed] [Google Scholar]
- 20.Ehrhardt GR, Hijikata A, Kitamura H, et al. Discriminating gene expression profiles of memory B cell subpopulations. J. Exp. Med. 2008;205:1807. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor RT, Patel SR, Lin E, et al. Lymphotoxin-independent expression of TNF-related activation-induced cytokine by stromal cells in cryptopatches, isolated lymphoid follicles, and Peyer's patches. J. Immunol. 2007;178:5659. doi: 10.4049/jimmunol.178.9.5659. [DOI] [PubMed] [Google Scholar]
- 22.Kong YY, Yoshida H, Sarosi I, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 23.Kim N, Odgren PR, Kim DK, et al. Diverse roles of the tumor necrosis factor family member TRANCE in skeletal physiology revealed by TRANCE deficiency and partial rescue by a lymphocyte-expressed TRANCE transgene. Proc. Natl Acad. Sci. USA. 2000;97:10905. doi: 10.1073/pnas.200294797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leibbrandt A, Penninger JM. RANK/RANKL: regulators of immune responses and bone physiology. Ann. NY Acad. Sci. 2008;1143:123. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- 25.Tumanov AV, Kuprash DV, Mach JA, et al. Lymphotoxin and TNF produced by B cells are dispensable for maintenance of the follicle-associated epithelium but are required for development of lymphoid follicles in the Peyer's patches. J. Immunol. 2004;173:86. doi: 10.4049/jimmunol.173.1.86. [DOI] [PubMed] [Google Scholar]
- 26.Rumbo M, Sierro F, Debard N, et al. Lymphotoxin beta receptor signaling induces the chemokine CCL20 in intestinal epithelium. Gastroenterology. 2004;127:213. doi: 10.1053/j.gastro.2004.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.