Abstract
Laboratory models have suggested a link between metabolism and life span in vertebrates, and it is well known that the evolution of specific life histories can be driven by metabolic factors. However, little is known regarding how the adoption of specific life-history strategies can shape aging and life span in populations facing different energetic demands from either a theoretical or a mechanistic viewpoint but significant insight can be gained by using a comparative approach. Comparative biology plays several roles in our understanding of the virtually ubiquitous phenomenon of aging in animals. First, it provides a critical evaluation of broad hypotheses concerning the evolutionary forces underlying the modulation of aging rate. Second, it suggests mechanistic hypotheses about processes of aging. Third, it illuminates particularly informative species because of their exceptionally slow or rapid aging rates to be interrogated about potentially novel mechanisms of aging. Although comparative biology has played a significant role in research on aging for more than a century, the new comparative biology of aging is poised to dwarf those earlier contributions, because: (1) new cellular and molecular techniques for investigating novel species are in place and more are being continually generated, (2) molecular systematics has resolved the phylogenetic relationships among a wide range of species, which allow for the implementation of analytic tools specialized for comparative biology, and (3) in addition to facilitating the construction of accurate phylogenies, the dramatic acceleration in DNA-sequencing technology is providing us with new tools for a comparative genomic approach to understanding aging.
Introduction: energy and aging
The histories of studies on metabolic physiology and on longevity have been intertwined for more than a century since they were first linked by Rubner in 1908. He noted from a study of resting metabolic rate and longevity in five mammalian species ranging in size from guinea pigs to cows that although body size varied by 50,000-fold and longevity by 5-fold among these species, lifetime expenditure of energy per gram of body mass varied by less than 2-fold. He thus concluded that the increase in longevity accompanying increasing body size among mammalian species was likely causally associated with the concomitant decrease in expenditure of energy at the tissue level, suggesting life span itself was limited by energy expenditure (Rubner 1908). Small, short-lived animals expend their inherent allotment of energy quickly; larger, longer-lived animals expend it slowly.
Rubner’s original observation was followed up by larger studies of mammalian species’ metabolic rates and longevities and these more detailed studies reached similar conclusions (Sacher 1959; Calder 1986). Also, this observation seemed consistent with a repeated finding among invertebrates that metabolic rate correlated positively, and longevity negatively, with environmental temperature (Pearl 1928; Miquel et al. 1976). When Denham Harman, having noted these patterns, hypothesized in the mid-20th century that aging was caused by free radicals—inescapable byproducts of normal metabolism (Harman 1956)—the “rate-of-living theory” (Pearl 1928), according to which metabolic rate largely dictated longevity, seemed parsimonious, intuitively satisfying, and consistent with a mountain of supporting evidence. This metabolic determinism exhibited such a grip on the biogerontological field that Sacher, from crude back-of-the-envelope style calculations of imagined food intake and longevity, attributed the well-known life-extending effects of caloric restriction in laboratory rodents to its effect of reducing metabolic rate (Sacher 1959).
In the latter part of the 20th century, this simple, coherent picture of the determination of longevity began to come apart. One of the first pieces of contrary evidence appeared when researchers studying calorically-restricted laboratory rodents actually measured metabolic rate and found that restricted animals actually expended as much, or more, energy on a mass-specific basis than did fully-fed animals (Duffy et al. 1989; McCarter and Palmer 1992). Also, it was noted that between closely-related species or recombinant inbred strains of the same species, there was no necessary association between longevity and metabolic rate (Promislow and Haselkorn 2002; Van Voorhies et al. 2004). Then, as numerous single-gene mutations that extended life in model organisms came to light, it turned out that while some of these mutations reduced metabolic rate, others left it unchanged or even increased it (Van Voorhies 2002; Lin et al. 2004; Westbrook et al. 2009). Finally, comparative biologists noted that broad patterns of longevity among endothermic vertebrates seemed inconsistent with the rate-of-living hypothesis. Specifically, marsupials exhibit only about 75% the basal metabolic rate of similar-sized eutherians, such that the rate-of-living hypothesis would predict they should be longer-lived than eutherians, when in fact the opposite is true (Austad and Fischer 1991). Also, birds, with higher basal metabolic rates than mammals, would be predicted by the rate-of-living hypothesis to be shorter-lived than similar-sized mammals, yet the opposite is true. Birds live on average about three times as long as similar-sized mammals (Holmes and Austad 1995). Moreover, far from their being relatively constant across species, lifetime basal mass-specific metabolic expenditure varies by nearly 30-fold across just the mammals (Austad and Fischer 1991). Also, importantly, a phylogenetically-sensitive comparative analysis noted that once the impact of phylogeny was removed, there was no longer any relationship between basal metabolic rate and longvity (de Magalhaes et al. 2007). Finally, if instead of assessing basal metabolic rate, an energy state at which animals rarely if ever exist, one examines actual energy expenditure, small, short-lived species expend consistently more energy per gram of body mass per lifetime than do large, long-lived species, whether considering mammals (Speakman 2005) or birds (Furness and Speakman 2008).
Today the determinative role of the rate of energy expenditure in longevity of species has been largely discarded by biogerontological researchers as a general hypothesis. However, decades of investigation into this area, which was born from, and died by, comparative biological analyses, led to the discovery of the importance of oxygen free radicals in a variety of disease processes and possibly in aging itself (Barja 2004; Lambert et al. 2007).
The role of comparative biology in research on mechanistic aging
As shown by the examples above, comparative biology can be used as a powerful tool to investigate broad hypotheses about mechanisms of aging. In fact, one could argue that a comparative approach is the only way that broad hypotheses about mechanisms of aging can be rigorously evaluated. Studies of single species yield information about single species. It is only by comparing multiple, phylogenetically diverse species that broad hypotheses can be supported or rejected. In this sense, the most striking evidence that insulin-like signaling broadly and causally modulates longevity is the observation that mutations reducing insulin-like signaling in Caenorhabditis elegans, Drosophila melanogaster, and house mice (Mus musculus)—three laboratory species separated by a billion years of evolutionary history—all significantly extend life (Tatar et al. 2003).
If comparative biology excels at supporting or rejecting broad mechanistic hypotheses about the modulation of longevity, it also can also suggest new general hypotheses. For instance, a recent unbiased screen for orthologous genes that when suppressed extend lifespan in both budding yeast and the nematode, C. elegans, led to the general hypothesis that reduced ribosomal biogenesis slows aging (Smith et al. 2007). These two species diverged about 1.5 billion years ago and have exceedingly different basic biology and life histories, suggesting whatever the mechanistic link between suppressed ribosomal biogenesis and aging, it may be an exceptionally general phenomenon. Further investigation in these and other species support the hypotheses that reduced ribosomal biogenesis may have its effect by reducing protein synthesis, which may in turn diverts more cellular energy to maintenance activities such as DNA repair or the degradation of damaged or misfolded proteins or leads to the synthesis of fewer damaged or misfolded proteins (Kaeberlein and Kennedy 2008). This finding and its interpretation are consistent with Kirkwood’s Disposable Soma model for the evolution of aging (Kirkwood 1985). This indirect but provocative link between integrity of the proteome and longevity was further supported by two separate comparative studies in mammals. In one of these, protein oxidation and stability was compared between naked mole-rats (Heterocephalus glaber) which can live nearly 30 years (Buffenstein 2008) and same-sized house mice which live no more than 4 years (Turturro et al. 1999). Livers of naked mole-rats accumulated fewer damaged proteins with age than did mouse livers and general protein stability, in the face of induced unfolding stress, was substantially greater in naked mole-rats than in mice (Pérez et al. 2009). In the other study, mice were compared to two species of bats (Brazilian free-tailed bats, Tadarida brasiliensis, and cave bats, Myotis velifer) both of which live up to 12 years in the wild (Salmon et al. 2009). In this case, basal damage to liver proteins as measured by carbonylation was less in free-tailed bats than in mice, but this was not true of cave bats. However, when animals were subjected to radiation stress (9 Gy γ-irradiation), then both bat species exhibited lower levels of oxidation compared to mice. Furthermore, when exposed to increasing levels of unfolding stress (increasing concentrations of urea), proteins both from bat livers and naked mole-rat livers were more stable than those from mice.
The third role that comparative biology can play in research on aging is identifying species of exceptional gerontological interest, because of their exceptionally long or short lives (Austad 2010). Although the vast majority of studies on aging employs model laboratory species notable for their short lives, it is conceivable that modulation of aging in short-lived species involves different mechanisms than those involved in modulating aging in long-lived species, such as humans, or that modulators of aging between species differ from modulators within species. One suggestion that this might be so is the conflicting body-size relationship between and within species. As has been previously mentioned, larger species of birds and mammals are typically longer-lived than are smaller species (Calder 1986; Austad and Fischer 1991); however, smaller animals within species of house mice, dogs, horses and possibly others, live longer (Miller and Austad 2006). It may be worth noting that this within-species pattern of an inverse relation between size and longevity has been reported only in species subjected to strong artificial selection during the course of domestication. Thus, it could represent the disruption of evolved physiological systems and feedback mechanisms by selection for extreme traits and may not be more generally relevant.
The discovery of exceptionally long-life in a wide variety of species has accelerated recently (Austad 2010). For instance, it is only in the past few years that it has become known that naked mole-rats live nearly ten times as long as similar-sized house mice (Buffenstein and Jarvis 2002) or that some small bats can live >40 years in the wild (Podlutsky et al. 2005).
Of course, tales of exceptional longevity among animals are not new. Francis Bacon collected many examples in his History of Life & Death nearly 400 years ago. Unfortunately, these examples were usually apocryphal. For instance, elephants as reported by Bacon lived 200 years, camels 100 years. Among birds, ravens and swans were also reported to live a century (Bacon 1638). Although Bacon was mistaken about these longevities, he was the first person, to my knowledge, to correctly determine that birds are longer-lived on average than are mammals.
So what is new in the comparative biology of aging is solid documentation of species’ longevity, sometimes because of better and more extensive record-keeping in zoos and research facilities, but also because of improved husbandry or new and better techniques for determining animals’ ages. For instance, the bowhead whale (Balaena mysticetus) has now been estimated to live as long as 200 years based on the degree of racemization of aspartic acid residues in its eye lens crystallins (George et al. 1999). Although this evidence by itself may provoke skepticism because the racemization process is affected by temperature and is very indirect, the estimate is given some credence by the fact that traditional whaling implements not in use for well more than a century have been recovered from bowhead whales landed in recent years.
Unquestionably more precise than amino acid racemization is the assessment of age by growth rings in the otholith of fishes (Cailliet et al. 2001). Otoliths are calcareous structures used in balance and hearing and found in the heads of all bony fishes. Seasonal growth zones can often be visualized in otoliths like growth rings in trees, particularly when the otolith is sectioned, polished, and stained. Extensive validation of analysis of the use of otolith growth-zones to estimate age has been performed by marking the otolith via oxytetracycline injection, followed by recapture of the animal and visualizing the mark at a later date, or for longer-lived fishes validating the age by radioactive decay series. Using these radiometric methods, 47 rockfish species in the genus Sebastes were discovered to range in maximum observed longevity from as little as 12 years in the Callico rockfish (S. dalli) to 157 years in the Shortraker rockfish (S. borealis), to a remarkable 205 years in the Rougheye rockfish (S. aleutianus) (Cailliet et al. 2001).
Annual counts of growth rings validated by radiometric analysis has also led to the discovery of exceptional longevity among the bivalve mollusks (clams, mussels, oysters) (Abele et al. 2008). In fact, bivalve mollusks likely represent the longest-lived clade among metazoans, with >10 species well-documented to live at least a century and some species such as the ocean quahog (Arctica islandica) surpassing four centuries (Wanamaker et al. 2008). Some species of bivalves may live even longer (Wisshak et al. 2009).
It may well be worth establishing some of these exceptionally long-lived species in the bestiary of model aging organisms. For that to occur, several things must take place. First, multiple laboratories must be interested in studying them. Second, large numbers of known-age individuals must become available. Third, their genome, transcriptome, and proteome must be described in detail and methods developed to manipulate each. Dramatic advances in high throughput sequencing and major advances in cell biology will soon allow this third event (see below). Probably the most likely species in which this might occur is the naked mole-rat, which is already being investigated in multiple laboratories, exists in large captive colonies, and currently has a rough draft of its genome being sequenced.
Short-lived species also have a valuable role in research on aging. Most obviously, if they can be mass-cultured in the laboratory and are genetically tractable, they are valuable because the impact of genetic, dietary, or pharmacological manipulations on lifespan can be easily assessed. Thus, the vast majority of research on aging to date has used short-lived nematodes, insects, or mammals. However, short-lived species are also valuable for comparative studies, in that they can provide informative contrasts to long-lived species for evaluating mechanistic hypotheses about the evolution of long life. For instance, the contrast between the laboratory mouse and the naked mole-rat has already provided a wealth of information concerning hypotheses about the evolution of long-life. In fact, this comparison has led to a major re-evaluation of the role of oxidative stress in the modulation of aging (Andziak et al. 2005, 2006). For both these reasons, the discovery of new, exceptionally short-lived species, particularly among groups in which exceptionally long life has been documented, is also exciting.
Of particular interest from this perspective is the recent discovery and subsequent laboratory development of a short-lived killifish, Nothobranchius furzeri, which has a median longevity of ∼9 weeks (Genade et al. 2005; Terzibasi et al. 2007)—about the same as a fruit fly. Several species of killifishes have been proposed as useful laboratory models for research on aging (Herrera and Jagadeeswaran 2004), but this species seems to be the most promising because it is the shortest-lived. A necessary caveat when employing putatively short-lived species is determination that their short lives are not a consequence of inadequacies in husbandry, such that they are chronically ill, even at a young age, and short-lived for that reason. Indications of this sort of problem can be gleaned from examination of age-specific mortality parameters, which in most species increase exponentially over most adult ages (Gompertzian mortality kinetics) under adequate husbandry (Finch 1990). Indications that husbandry of N. furzeri is well-developed include Gompertzian mortality kinetics plus the appearance of various indicators of aging only in later life (Terzibasi et al. 2007).
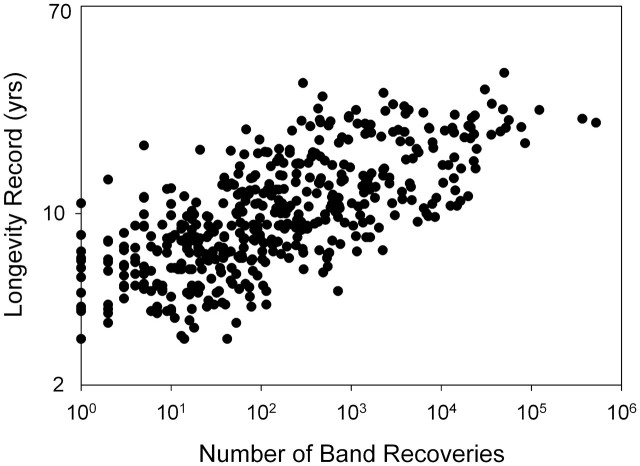
In comparative research on aging, informative contrasts may also be made among species exhibiting divergent longevities in the wild. Although in the past it was commonly believed that animals in the wild did not suffer appreciable senescence, this is now known to have been a misconception due to lack of information. Senescence of various types has now been documented in a wide variety of birds and mammals (Promislow 1991; Brunet-Rossinni and Austad 2006; Monaghan et al. 2008). In comparative studies, phylogenetic affinity is an important concern, because the more closely related species are, the fewer are the likely functional and genetic differences among them that are unrelated to differences in longevity. Thus, it would likely be more informative to contrast long-lived and short-lived bats rather than long-lived bats with short-lived rodents. However, it is not trivial to establish that species in the wild are short-lived, because lack of sufficient data on completed life spans may make species appear shorter-lived than they really are. For instance, a number of comparative analyses have been performed on broad patterns of birds’ longevity, in which species’ longevity was determined by recoveries of previously banded birds (Calder, 1986; Furness and Speakman, 2008). However, it is worth noting that published longevity records, at least of North American birds, from such banding records are strongly affected by the number of bands recovered. Specifically, nearly half (49%) of the variation in maximum reported field longevity can be explained by the sample size of recovered bands alone (Fig. 1). This is a substantially greater fraction of the variation than is explained by body size (32%). As a specific illustrative example, the longevity record for the Eastern bluebird (Sialis sialis) is 10.5 years compared to 6.1 years for the closely-related Western bluebird (S. mexicana). This may represent a real difference in survival, but on the other hand it may not. The former record is based on more than 1600 recoveries of bands, whereas the latter is based on only 114. As sample size of completed life spans increases, this potential problem abates. Maximum longevity of Herring gulls, for instance, was reported to be 27.25 years based on 35,000 recovered bands (Clapp et al. 1982), and 7 years later had increased only slightly (to 28.0 years) based on 1500 additional recoveries (Klimkiewicz and Futcher, 1989). The ultimate message is that in either laboratory of field studies, considerable care should be taken to ensure that putative short-lived species are truly, rather than only apparently, short-lived.
Fig. 1.
Relationship between the published longevity records of North American bird species based on the recovery of bands and the number of bands recovered (r2 = 0.49, N = 475, P < 0.0001). Note that the number of bands recovered explains more of the variation in longevity than does the species’ body size (r2 = 0.32, N = 486, P < 0.0001, data not shown). Data from Clapp et al. (1982, 1983), Klimkiewicz et al. (1983), Klimkiewicz and Futcher (1987, 1989).
True field “short-gevity” can be established, however, For instance, a potentially useful contrast may be made among long-lived versus short-lived vespertilionid bats, such as the little brown bat (Myotis lucifugus) with a longevity record of 34 years (Davis and Hitchcock 1995) and the evening bat (Nycticeus humeralis) with a reported longevity of 6 years (Wilkinson and South 2002). Evidence that the evening bat really is this short-lived includes that Wilkinson banded more than 1300 females of this highly philopatric species in the 1980s and 1990s and followed their annual survival, accruing more than 1100 recaptures of banded bats. No banded bat was determined to be >6-years old. Moreover, Watkins banded numerous bats in the same colonies 10–20 years earlier and Wilkinson found no banded bats from Watkins’ study (G.S. Wilkinson, personal communication). Finally, as might be expected in a bat species that is exceptionally short-lived, evening bats have a considerably higher reproductive rate than do most bats. Whereas most bat species bear one pup annually, evening bats bear two, or even occasionally three, young (Watkins 1972; Wilkinson 1992). Thus, a diversity of evidence indicates that evening bats are truly short-lived (for a bat). Finally, it is worth noting that AnAGE, a well-curated, broad database of species longevities, is now available on-line (http://genomics.senescence.info/species).
Comparative biology of aging in the 21st century
As informative as comparative biology has been in assessing patterns and processes of aging in the past, the future promises to be even more revealing. I say this because of three important, relatively recent trends in biological science.
First, new techniques in cell biology and biochemistry are leading to more refined, species-independent, tools for investigating cellular processes. To focus on one example from a large number of possible examples, measurement of in vivo oxidative damage has been a mainstay of research on aging for several decades as the oxidative stress hypothesis of aging held sway. Yet quantitation of oxidative damage has been fraught with technical problems, particularly when employing noninvasive approaches (Halliwell and Gutteridge 2007). For instance, measurement of lipid peroxidation by quantification of malondialdehyde (MDA) in tissues and body fluids, such as plasma and urine, has been a common measure of generalized oxidative stress. However, this assay suffers from lack of specificity, poor sensitivity due to its rapid metabolism, its chemical instability and reactivity, and its generation of artifacts (Roberts and Milne 2008). More recently, these problems have been largely overcome by the discovery and development of straightforward gas chromatography/mass spectrometry assays for F2-isoprostanes. These prostaglandin-like compounds are produced by free radical-catalyzed peroxidation of arachidonic acid. Unlike MDA, they are metabolically stable and nonreactive and their auto-oxidation can be easily prevented by rapid storage at −70°C or below (Arneson and Roberts 2007). Thus, quantitation of F2-isoprostanes is not only one of the most accurate methods for detecting oxidative injury, it can be employed with equal ease to assess samples obtained in either laboratory or field. Arachidonic acid is distributed in membranes rather uniformly throughout the body, so measurement of F2-isoprostanes is a good indicator of generalized lipid peroxidation. A second class of isoprostanes, F4-isoprostanes or neuroprostanes, with similar chemical properties to F2-isoprostanes are derived from peroxidation of docosahexaenoic acid (DHA), which is highly enriched in the vertebrate brain. DHA accounts for 25–35% of total fatty acids of neuronal membranes (Skinner et al. 1993). Thus F4-isoprostanes are excellent markers of oxidative damage to the brain (Arneson and Roberts 2007).
Another example in which techniques now allow important cellular processes to be investigated without specialized species-specific tools such as antibodies is protein misfolding. Intracellular proteins must be folded precisely to function correctly and many factors from oxidation, to excess heat, to fatty acids, to other misfolded proteins, can interfere with the maintenance of proper conformation. Improperly folded proteins may become aggregated and/or toxic and consequently a complex cellular machinery for degrading misfolded proteins has evolved (Goldberg 2003). It is now relatively straightforward to monitor the conformational status of a complex mixture of proteins such as found in specific tissues by measuring changes in the surface hydrophobicity of the proteins. The compound BisANS (4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid) binds reversibly to hydrophobic pockets on the surface of proteins and this binding can be visualized fluorometrically. This binding becomes covalent after exposure to longwave UV light, thus allowing the screening of the proteome for changes in surface hydrophobicity associated with misfolding (Pierce et al. 2006). This technique was employed in the previously-mentioned observations that global protein stability appears tightly linked to species’ longevity in a selection of mammals (Pérez et al. 2009; Salmon et al. 2009).
Possibly the most important technical advance in the past decade for both individualized regenerative medical therapy as well as for comparative studies of aging is the development of induced pluripotent stem cells (iPSCs). These iPSCs can be generated from a variety of differentiated cell types using a cocktail of transcription factors, and are similar in their properties to embryonic stem cells, in that when appropriately stimulated they can be transformed into virtually any other type of cell (Takahashi and Yamanaka 2006). Proof-of-principle for this claim was the creation of adult mice from reprogramming of differentiated mouse cells (Boland et al. 2009). IPSCs can be created even using transcription factors from other mammalian species—mouse cells can use human transcription factors, for instance—and have now been created from at least six mammalian species (humans, rhesus macaques, marmosets, dogs, mice, and pigs) (Liu et al. 2008; Nakagawa et al. 2008; Li et al. 2009; Trounson 2009; Wu et al. 2010).
The advantage of iPSCs is that to the extent that such cells exist for a diversity of species, it allows in vitro investigation of nearly any type of cell. For instance, for nearly three decades researchers have been evaluating hypotheses about mechanisms of aging by comparing cultured cells, typically fibroblasts, from species of varying longevities (Röhme 1981; Kapahi et al. 1999; Lorenzini et al. 2005). However, fibroblasts are disposable cells, easily destroyed and replaced if they become damaged. It might be that permanent cells such as neurons or cardiomyocytes would be more informative than fibroblasts about critical species’ protective mechanisms that combat fundamental processes of aging. The existence of iPSCs, which can be differentiated into neurons or cardiomyocytes in culture, would allow comparative investigation of cell types that are not otherwise available. An example of how iPSCs could be employed might be in the investigation of causes of Alzheimer’s Disease. Alzheimer’s Disease appears to occur only in humans but not even in humans’ close relatives, such as chimpanzees (Finch and Stanford 2004). One hypothesis for the development of Alzheimer’s Disease is that oligomers of beta amyloid are toxic to human neurons (Geula et al. 1998). A reasonable experimental approach to evaluating this idea would be to determine whether beta amyloid was less toxic to neurons from a range of nonhuman primates, particularly humans’ close relatives, like chimpanzees, that do not get the disease. However, for ethical reasons, isolated chimpanzee neurons are difficult to obtain. However, it would be possible to differentiate chimpanzee iPSCs into neurons and study their properties in that manner.
In addition to the development of the these cellular and biochemical techniques that mitigate some of the difficulties in implementing research on nontraditional species, a second major advance facilitating comparative research is the emergence of a vastly improved understanding of the phylogenetic relationships among species at both large and small scales. The analysis of comparative data can be greatly enhanced by taking account of the phylogenetic relationships among the species studied rather than by treating each species as an independent data point (Felsenstein 1985; Harvey and Pagel 1991). However, these phylogenetically-sensitive methods require accurate phylogenies. The great wealth of DNA-sequence data that have accumulated in recent years, combined with better techniques for detecting evolutionary divergence patterns using sequence differences, have revolutionized our understanding of the evolutionary relationships among species. For instance, the two major superphyla of protostome invertebrates—the Ecdysozoa and the Lophotrochozoa were only recognized in the late 1990s (Aguinaldo et al. 1997) and confirmed in detail even more recently (Dunn et al. 2008). Such alterations in our view of evolutionary history can significantly alter inferences from experimental results. To choose one example, in a traditional coelomic view of evolutionary history the common ancestor of nematodes and insects is also the common ancestor of mammals. Thus, if one observed the same phenomenon—such as reduced insulin/IGF signaling leading to longer life—in both nematodes and insects then a reasonable inference would be that the trait is ancestral in mammals as well. According to the newer Ecdysozoan phylogeny, nematodes and insects diverged from one another after diverging from the clade leading to mammals; therefore a trait shared by the conventional laboratory research models of aging, C. elegans and D. melanogaster, is not necessarily ancestral to mammals (Austad and Podlutsky 2006). Thus, on that basis, a mechanism modulating aging in C. elegans and D. melanogaster cannot be assumed to be generalizable.
Better phylogenetic information has been particularly helpful in comparative studies of mammals. Prior to 2001, there was no generally accepted topology even of the major mammalian orders. However, since then, consistent and nicely resolved mammalian phylogenies have emerged (Murphy et al. 2004; Bininda-Emonds et al. 2007), such that now even such formerly troublesome groups as the rodents (Steppan et al. 2004) and bats (Teeling et al. 2005) have nicely resolved topologies and reasonable estimates of times of divergence. Consequently, it becomes easier to determine the number of independent evolutionary events that one encompasses in a comparative study, making statistical inference cleaner and more reliable. Such broad comparative analyses of the pattern of aging among mammals have recently been made (de Magalhaes et al. 2007; Ricklefs 2010).
The final development revolutionizing comparative studies in the 21st century, already foreshadowed in the above discussion of phylogeny, is the impact of the massive—and accelerating—increase in DNA-sequencing and RNA-sequencing power over the past decade. As of now (May 2010), there are whole genome sequences of 2× coverage or better for at least 28 species of mammals with at least 50 species approved for sequencing, many of these already in process (http://www.genome.gov/10002154). This number is expected to increase rapidly. Moreover, transcriptome sequencing will provide information, in addition to the existence and variation among genomes, but more importantly, on which genes are expressed in which tissues. Perhaps more importantly, such sequence information will allow the development of reagents for manipulating gene expression. For instance, mRNA sequence data will allow the design of small interfering RNA’s with which, using appropriate vectors, one will be able suppress the expression of specific genes in cells or in specific tissues of any species of exceptional interest.
In summary, comparative biology has been an important tool for the investigation of mechanisms of aging for nearly as long as aging has been the subject of formal study. Early work focused on the relationship between metabolism and aging, although as more and more data were assembled the idea that metabolic rate somehow determined the rate of aging fell into disfavor. In recent times, comparative research on aging largely consisted of confirming findings discovered in one of the four traditional model organisms (Saccharomyces cerevesiae, C. elegans, D. melanogaster, and M. musculus) in the others. Such confirmation indicated how broadly conserved were mechanisms such as the impact of insulin/IGF signaling on longevity. Today, and in the future, additional advances will come from broadening the bestiary of species to include those that are exceptionally long-lived in addition to current models which are all of short-lived species. A satchel full of new tools have recently become available that will allow the most sophisticated of cellular and molecular techniques to be applied to virtually any species of exceptional interest.
Funding
National Institute on Aging (K07 AG025063, R01 AG022873, and R01 AG035327); Paul Glenn Foundation for Medical Research, the Ellison Medical Foundation; the San Antonio Area Foundation for supporting my comparative research on aging.
References
- Abele D, Strahl J, Brey T, Philipp EE. Imperceptible senescence: ageing in the ocean quahog Arctica islandica. Free Radic Res. 2008;42:474–80. doi: 10.1080/10715760802108849. [DOI] [PubMed] [Google Scholar]
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–93. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Andziak B, O’Connor TP, Buffenstein R. Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech Ageing Dev. 2005;126:1206–12. doi: 10.1016/j.mad.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Andziak B, O’Connor TP, Qi W, DeWaal EM, Pierce A, Chaudhuri AR, Van RH, Buffenstein R. High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell. 2006;5:463–71. doi: 10.1111/j.1474-9726.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- Arneson KO, Roberts LJ. Measurement of products of docosahexaenoic acid peroxidation, neuroprostanes, and neurofurans. Methods Enzymol. 2007;433:127–43. doi: 10.1016/S0076-6879(07)33007-3. [DOI] [PubMed] [Google Scholar]
- Austad SN. Methusaleh's Zoo: how nature provides us with clues for extending human health span. J Comp Pathol. 2010;142(Suppl. 1):S10–21. doi: 10.1016/j.jcpa.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 1991;46:B47–53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Austad SN, Podlutsky A. A critical evaluation of nonmammalisan models for aging research. In: Masoro EJ, Austad SN, editors. Handbook of the Biology of Aging. San Diego: Academic Press; 2006. pp. 449–69. [Google Scholar]
- Bacon F. The historie of life and death. Whitefish (MT): Kessinger; 1638. [Google Scholar]
- Barja G. Free radicals and aging. Trends Neurosci. 2004;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–12. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–4. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Brunet-Rossinni AK, Austad SN. Senescence in wild populations of mammals and birds. In: Masoro EJ, Austad SN, editors. Handbook of the Biology of Aging. San Diego: Academic Press; 2006. pp. 243–66. [Google Scholar]
- Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178:439–45. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Jarvis JU. The naked mole rat–a new record for the oldest living rodent. Sci Aging Knowledge Environ. 2002 doi: 10.1126/sageke.2002.21.pe7. 2002e7. [DOI] [PubMed] [Google Scholar]
- Cailliet GM, Andrews AH, Burton EJ, Watters DL, Kline DE, Ferry-Graham LA. Age determination and validation studies of marine fishes: do deep-dwellers live longer? Exp Gerontol. 2001;36:739–64. doi: 10.1016/s0531-5565(00)00239-4. [DOI] [PubMed] [Google Scholar]
- Calder WAI. Size, function, and life history. Mineola, New York: Dover; 1986. [Google Scholar]
- Clapp RB, Klimkiewicz MK, Futcher AG. Longevity records of North American birds: Columbidae through Paridae. J Field Ornithol. 1983;54:123–37. [Google Scholar]
- Clapp RB, Klimkiewicz MK, Kennard JH. Longevity records of North American birds: Gaviidae through Alcide. J Field Ornithol. 1982;53:81–124. [Google Scholar]
- Davis WH, Hitchcock HB. A new longevity record for the bat Myotis lucifugus. Bat Res News. 1995;36:6. [Google Scholar]
- de Magalhaes JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62:149–60. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy PH, Feuers RJ, Leakey JA, Nakamura K, Turturro A, Hart RW. Effect of chronic caloric restriction on physiological variables related to energy metabolism in the male Fischer 344 rat. Mech Ageing Dev. 1989;48:117–33. doi: 10.1016/0047-6374(89)90044-4. [DOI] [PubMed] [Google Scholar]
- Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–49. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Finch CE. Longevity, Senescence, and the Genome. Chicago: University of Chicago Press; 1990. [Google Scholar]
- Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q Rev Biol. 2004;79:3–50. doi: 10.1086/381662. [DOI] [PubMed] [Google Scholar]
- Furness LJ, Speakman JR. Energetics and longevity in birds. Age. 2008;30:75–87. doi: 10.1007/s11357-008-9054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genade T, Benedetti M, Terzibasi E, Roncaglia P, Valenzano DR, Cattaneo A, Cellerino A. Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell. 2005;4:223–33. doi: 10.1111/j.1474-9726.2005.00165.x. [DOI] [PubMed] [Google Scholar]
- George JC, Bada J, Zeh J, Scott L, Brown SE, O’Hara T, Suydam R. Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can J Zoology. 1999;77:571–80. [Google Scholar]
- Geula C, Wu CK, Saroff D, Lorenzo A, Yuan M, Yankner BA. Aging renders the brain vulnerable to amyloid beta-protein neurotoxicity. Nat Med. 1998;4:827–31. doi: 10.1038/nm0798-827. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–9. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge J. Free radicals in biology and medicine. Oxford (UK): Oxford University Press; 2007. [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. Oxford (UK): Oxford University Press; 1991. [Google Scholar]
- Herrera M, Jagadeeswaran P. Annual fish as a genetic model for aging. J Gerontol A Biol Sci Med Sci. 2004;59:101–7. doi: 10.1093/gerona/59.2.b101. [DOI] [PubMed] [Google Scholar]
- Holmes DJ, Austad SN. Birds as animal models for the comparative biology of aging: a prospectus. J Gerontol A Biol Sci Med Sci. 1995;50:B59–66. doi: 10.1093/gerona/50a.2.b59. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kennedy BK. Protein translation. Aging Cell. 2008;7:777–82. doi: 10.1111/j.1474-9726.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Boulton ME, Kirkwood TB. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med. 1999;26:495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- Kirkwood TBL. Comparative and evolutionary aspectics of longevity. In: Finch CE, Schneider EL, editors. Handbook of the biology of aging. New York: Van Nostrand Reinhold; 1985. pp. 27–44. [Google Scholar]
- Klimkiewicz MK, Clapp RB, Futcher AG. Longevity records of North American birds: Remizidae through Parulinae. J Field Ornithol. 1983;54:287–94. [Google Scholar]
- Klimkiewicz MK, Futcher AG. Longevity records of North American birds: Coerebinae through Estrildidae. J Field Ornithol. 1987;58:318–33. [Google Scholar]
- Klimkiewicz MK, Futcher AG. Longevity records of North American birds: supplement 1. J Field Ornithol. 1989;60:469–94. [Google Scholar]
- Lambert AJ, Boysen HM, Buckingham JA, Yang T, Podlutsky A, Austad SN, Kunz TH, Buffenstein R, Brand MD. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–18. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–9. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–90. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Lorenzini A, Tresini M, Austad SN, Cristofalo VJ. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech Ageing Dev. 2005;126:1130–3. doi: 10.1016/j.mad.2005.05.004. [DOI] [PubMed] [Google Scholar]
- McCarter RJ, Palmer J. Energy metabolism and aging: a lifelong study of Fischer 344 rats. Am J Physiol. 1992;263:E448–52. doi: 10.1152/ajpendo.1992.263.3.E448. [DOI] [PubMed] [Google Scholar]
- Miller RA, Austad SN. Growth and aging: why do big dogs die young? In: Masoro EJ, Austad SN, editors. Handbook of the biology of aging. San Diego: Academic Press; 2006. pp. 512–33. [Google Scholar]
- Miquel J, Lundgren PR, Bensch KG, Atlan H. Effects of temperature on the life span, vitality and fine structure of Drosophila melanogaster. Mech Ageing Dev. 1976;5:347–70. doi: 10.1016/0047-6374(76)90034-8. [DOI] [PubMed] [Google Scholar]
- Monaghan PA, Charmantier A, Nussey DH, Ricklefs RE. The evolutionary ecology of senescence. Funct Ecol. 2008;22:371–8. [Google Scholar]
- Murphy WJ, Pevzner PA, O’Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–9. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Pearl R. The rate of living. New York: A.A. Knopf; 1928. [Google Scholar]
- Pérez VI, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106:3059–64. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, deWaal E, Van RH, Richardson A, Chaudhuri A. A novel approach for screening the proteome for changes in protein conformation. Biochemistry. 2006;45:3077–85. doi: 10.1021/bi052031i. [DOI] [PubMed] [Google Scholar]
- Podlutsky AJ, Khritankov AM, Ovodov ND, Austad SN. A new field record for bat longevity. J Gerontol A Biol Sci Med Sci. 2005;60:1366–8. doi: 10.1093/gerona/60.11.1366. [DOI] [PubMed] [Google Scholar]
- Promislow DE, Haselkorn TS. Age-specific metabolic rates and mortality rates in the genus Drosophila. Aging Cell. 2002;1:66–74. doi: 10.1046/j.1474-9728.2002.00009.x. [DOI] [PubMed] [Google Scholar]
- Promislow DEL. Senescence in natural populations of mammals: a comparative study. Evolution. 1991;45:1869–87. doi: 10.1111/j.1558-5646.1991.tb02693.x. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE. Insights from comparative analyses of aging in birds and mammals. Aging Cell. 2010;9:273–84. doi: 10.1111/j.1474-9726.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LJ, Milne GL. Isoprostanes. J Lipid Res. 2008;50(Suppl):S219–23. doi: 10.1194/jlr.R800037-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhme D. Evidence for a relationship between longevity of mammalian species and life spans of normal fibroblasts in vitro and erythrocytes in vivo. Proc Natl Acad Sci USA. 1981;78:5009–13. doi: 10.1073/pnas.78.8.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubner M. Munich. Oldenbourg: 1908. Das Problem der Lebensdauer und seine Beziehungen zum Wachstum und Ernahrung. [Google Scholar]
- Sacher GA. Relation of life span to brain weight and body weight in mammals. In: Wolstenholme GEW, O’Connor M, editors. CIBA Foundation Colloquia on Ageing. London: Churchill; 1959. pp. 115–3. [Google Scholar]
- Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 2009;23:2317–26. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ER, Watt C, Besson JA, Best PV. Differences in the fatty acid composition of the grey and white matter of different regions of the brains of patients with Alzheimer's disease and control subjects. Brain. 1993;116 Pt (3):717–25. doi: 10.1093/brain/116.3.717. [DOI] [PubMed] [Google Scholar]
- Smith ED, Kennedy BK, Kaeberlein M. Genome-wide identification of conserved longevity genes in yeast and worms. Mech Ageing Dev. 2007;128:106–11. doi: 10.1016/j.mad.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Body size, energy metabolism and lifespan. J Exp Biol. 2005;208:1717–30. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–53. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–51. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–4. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Terzibasi E, Valenzano DR, Cellerino A. The short-lived fish Nothobranchius furzeri as a new model system for aging studies. Exp Gerontol. 2007;42:81–9. doi: 10.1016/j.exger.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Trounson A. Rats, cats, and elephants, but still no unicorn: induced pluripotent stem cells from new species. Cell Stem Cell. 2009;4:3,–4. doi: 10.1016/j.stem.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Van Voorhies WA. The influence of metabolic rate on longevity in the nematode Caenorhabditis elegans. Aging Cell. 2002;1:91–101. doi: 10.1046/j.1474-9728.2002.00022.x. [DOI] [PubMed] [Google Scholar]
- Van Voorhies WA, Khazaeli AA, Curtsinger JW. Testing the “rate of living” model: further evidence that longevity and metabolic rate are not inversely correlated in Drosophila melanogaster. J Appl Physiol. 2004;97:1915–22. doi: 10.1152/japplphysiol.00505.2004. [DOI] [PubMed] [Google Scholar]
- Wanamaker AD, Jr, Heinemeier J, Scourse JD, Richardson CA, Butler PG, Eiriksson J, Knudsen KL. Very long-lived molluscs confirm 17th century AD tephra-based radiocarbon reservoir ages for north Icelandic shelf waters. Radiocarbon. 2008;50:1–14. [Google Scholar]
- Watkins LC. Nycticeius humeralis. Mammalian Species. 1972;23:1–4. [Google Scholar]
- Westbrook R, Bonkowski MS, Strader AD, Bartke A. Alterations in oxygen consumption, respiratory quotient, and heat production in long-lived GHRKO and Ames dwarf mice, and short-lived bGH transgenic mice. J Gerontol A Biol Sci Med Sci. 2009;64:443–51. doi: 10.1093/gerona/gln075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GS. Communal nursing in the evening bat, Nycticeius humeralis. Behav Ecol Sociobiol. 1992;31:225–35. [Google Scholar]
- Wilkinson GS, South JM. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–31. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- Wisshak M, López Correa M, Gofas S, Salas C, Taviani M, Jakobsen J, Freiwald A. Shell architecture, element composition, and stable isotope signature of the giant deep-sea oyster Neopycnodonte zibrowii sp. n. from the NE Atlantic. Deep-Sea Res I. 2009;56:374–407. [Google Scholar]
- Wu Y, Zhang Y, Mishra A, Tardif SD, Hornsby PJ. Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res. 2010;4:180–8. doi: 10.1016/j.scr.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]