Abstract
Roles for Wnt9b in craniofacial development are indicated by the cleft lip mutant phenotype observed in the A/WySn mouse strain,1 caused by a retrotransposon insertion mutation at the Wnt9b locus. Analyses of the zebrafish Wnt9b ortholog, wnt9b, were pursued to provide insight into early vertebrate craniofacial patterning events mediated by Wnt9b signaling. Zebrafish wnt9b cDNA clones were isolated and found to encode an open reading frame of 358 amino acids, with 68% amino acid identity to mouse Wnt9b and 70% amino acid identity to human WNT9B. Syntenic analyses demonstrated that wnt9b and wnt3 exist as a contiguous pair in amniote vertebrate species, and that these genes are separate in the zebrafish and Takifugu genomes. During the pharyngula period, a time of extensive growth and morphogenesis, zebrafish wnt9b exhibits discrete expression in dorsal and ventral first and second branchial arch tissues, the heart, and pectoral fin buds. These analyses suggest that in zebrafish, as in humans, wnt9b plays distinct roles in directing morphogenetic movements of developing branchial arch elements, and identify the zebrafish as a useful developmental model for the study of human craniofacial cleft lip and palate.
Introduction
Cleft lip and palate are common human birth defects that occur at an incidence of 1–2 out of every 1000 births worldwide.2,3 The mouse Wnt9b gene maps to the cleft1 (clf1) locus4 in the A/WySn mouse strain, which exhibits a high rate of cleft lip caused by disruption of the mammalian primary palate. In early mouse development, wnt9b exhibits peak expression levels at 10.5 days postcoitum,4 which corresponds to the time of murine craniofacial development and morphogenesis. In particular, wnt9b is highly expressed in the surface ectoderm of the maxillary and mandibular facial primordia,5 implicating wnt9b in craniofacial patterning and development.
The wnt9b mouse knockout exhibits cleft lip and palate,1 caused by disruption of both the primary and secondary palate, as well as posterior gut defects.6 Recently, an intracisternal-A particle (IAP) transposon insertion has been identified in the mouse wnt9b gene at a position just 7 kilobases downstream of the last exon of mouse wnt9b, at the clf1 locus. This insertion was present in the A/WySn clf1 mouse strain, but not in the ancestral wild-type CBA/J strain. Complementation analyses of heterozygous wnt9b-mutant mouse males and homozygous A/WySn females (a/a) demonstrated that compound wnt9b-A/WySn mutants exhibited significant cleft lip phenotype, identifying the clf1 mutation as an alternative wnt9b mutant allele.
It has been shown that hypomethylated IAP transposon insertions can allow for antisense transcription from adjacent promoters.1 Based on this observation, it has been speculated that IAP transposon mediated wnt9b antisense promoter transcripts might interfere with the functions of conserved noncoding sequence elements regulating the wnt9b gene.1 Demonstration of this type of regulation would further implicate wnt9b as a critical regulator of craniofacial patterning and development.
To better understand the etiopathogenesis of mammalian orofacial clefting and to elucidate the role of wnt9b in craniofacial patterning and development, we began characterizations of wnt9b in an experimentally tractable vertebrate, the zebrafish. We anticipate that functional characterization of wnt9b in zebrafish craniofacial development will provide an increased understanding of the molecular signaling pathways regulating the formation of the face, and may eventually provide tools for better diagnosis, as well as ultimately better prevention strategies, for human cleft lip and palate birth defects.
Results
Identification and characterization of the zebrafish wnt9b gene
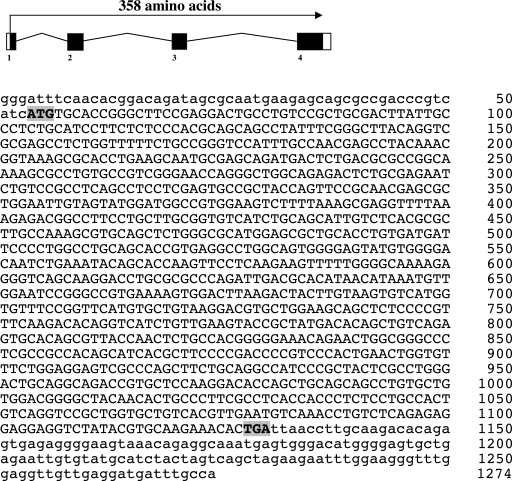
Preliminary BLAST analysis of the Sanger ENSEMBL site using a Fugu ortholog of the mouse Wnt9b protein identified a partial zebrafish wnt9b cDNA sequence (ENSDART 00000003613) within contig ctg10251. Conserved synteny between the mouse and zebrafish was revealed by the identification of the zebrafish arf2 gene at a location adjacent to the zebrafish wnt9b gene, similar to the organization of the adjacent mouse arf2 and wnt9b gene loci. CLUSTAL W phylogenetic analysis (data not shown) of the partial zebrafish wnt9b cDNA indicated significant identity to mammalian Wnt9b genes justifying further investigation of these loci. Reverse-transcription polymerase chain reaction (RT-PCR) and 5′ rapid amplification of cDNA ends (RACE) analyses were used to clone a zebrafish wnt9b cDNA of 1274 bp, containing an open reading frame of 1074 bp encoding 358 amino acids, a 5′ untranslated region (UTR) of 53 bp, and a 3′UTR of 144 bp (Fig. 1). Comparison of the zebrafish wnt9b cDNA sequence to that of human, rat, and stickleback wnt9b cDNAs revealed 71%, 66%, and 66% nucleotide sequence identities, respectively (Fig. 2).
FIG. 1.
Genomic organization and nucleotide sequence of zebrafish wnt9b. The genomic organization and cDNA sequence for the zebrafish wnt9b gene are shown. Four exons were identified by comparison of zebrafish wnt9b genomic and cDNA nucleotide sequences. RACE analysis was used to identify the transcriptional start site 54 nucleotides upstream of the start codon. To avoid gaps present in the ENSEMBL genomic sequence, primers were carefully chosen to include an open reading frame of 358 amino acids and a portion of the 3′UTR. The start and stop codons are indicated by bold highlighted typeface.
FIG. 2.
Nucleotide sequence alignment of the human, rat, stickleback, and zebrafish wnt9b mRNAs. Zebrafish wnt9b transcript was aligned with human, rat, and stickleback wnt9b RNAs using the Clustal algorithm within the MEGA 3.0 program. This analysis demonstrated that the start and stop (bolded and highlighted) codons of all of the wnt9b genes align, providing confidence that the entire zebrafish coding sequence was isolated. Species and accession numbers are Hs: Homo sapiens, NM_003396; Rn: Rattus norvegicus, ENSRNOT00000005074; Ga: Gasterosteus aculatus, ENSGACT00000018773; Dr: Danio rerio.
The predicted amino acid sequence of zebrafish Wnt9b exhibited 68%, 68%, and 87% amino acid identity to human, rat, and stickleback Wnt9b, respectively (Fig. 3), including the presence of 23 conserved cysteine residues, a conserved Asn-linked glycoslylation site, and two small coding deletions between cysteine residues 13 and 15 as compared to the Wnt1 protein sequence family, all of which are characteristics of the Wnt9 protein family.4,7 Analyses using the SignalP 3.0 signal peptide prediction algorithm (www.cbs.dtu.dk/services/SignalP/) identified a signal peptide cleavage site between amino acids 26 (alanine, A) and 27 (tyrosine, Y), which would generate a mature Wnt9b protein of 332 amino acids.
FIG. 3.
Amino acid sequence comparison of Wnt9b proteins. The most likely cleavage site for the signal peptide is found after amino acid 26, indicated by the “↓,” as determined by the SignalP 3.0 Web site algorithm. There are 27 cysteine residues highlighted in red. Those shown in bold represent the 23 that are usually found in other members of the Wnt9 family. The bold blue “N” indicates the potential N-linked glycosylation site. The positions for the two coding deletions are indicated by a “▾.” These deletions are typical for the Wnt9 protein family relative to the Wnt1 family protein sequence. Dots represent identity; dashes are missing amino acids. Species and accession numbers are Hs: Homo sapiens, NP_003387; Mm: Mus musculus, NP_035849; Ga: Gasterosteus aculatus, ENSGACP00000018735; Dr: Danio rerio.
Syntenic analysis of the zebrafish wnt9a/b loci
The wnt9 and wnt3 paralogs are paired in both the human and mouse genomes, with Wnt9b located adjacent to Wnt3, and Wnt9a located next to Wnt3a. The contiguous pairing of Wnt9-type (Wnt9x) and Wnt3-type (Wnt3x) paralogs is also observed in the cow genome and in the more distantly related chicken genome, as found at the UCSC Genome Browser (www.genome.cse.ucsc.edu/). This conserved paired organization suggests that the Wnt9x and Wnt3x genes may be coordinately regulated, as has been demonstrated for other digene clusters.8,9 To further investigate the conserved organization of Wnt9 and Wnt3 paralogs, we examined Wnt gene organization in the zebrafish (Danio rerio) and Takifugu genomes.
Zebrafish wnt9a (Chr2:1.97–1.99 Mb within Zv6_sc171) is located more than 1 Mb upstream of wnt3 (Chr2:32 Mb within Zv6_sc177), both on chromosome 2. The Takifugu wnt9b and wnt3 genes map to separate scaffolds in the pufferfish genome, as viewed at the ENSEMBL genome database. The only Wnt9x/Wnt3x contiguous pair that we were able to identify was the Takifugu wnt9a and wnt3a digene cluster, located as adjacent genes at approximately 1.15 to 1.2 Mb on scaffold 68. The positive identification of each zebrafish and Fugu wnt gene paralog was confirmed by BLAST analyses and by the presence of conserved adjacent gene paralogs. For example, in both zebrafish and Takifugu genomes, the wnt9b gene was located between the gosr2 and arf2 genes. Even when the wnt9x and wnt3x genes mapped to two separate chromosomal locations, it was still possible to identify conserved, adjacent paralogous genes that apparently moved with them to their new position.
Early expression of wnt9b in early heart development
We next examined the expression of wnt9b throughout early zebrafish development. Whole mount hybridization (WISH) analyses revealed that from 24 to 26 hours postfertilization (hpf), wnt9b was highly expressed in the forming heart tube. At 24 hpf, wnt9b is expressed in the heart tube (Fig. 4A, arrow, he), located to the left of the zebrafish midline consistent with the documented leftward “jogging” of the early heart tube.10 By 26 hpf, the head has extended along the rostral caudal axis, and has increased in height along the dorsal–ventral axis. The heart tube has also extended, maintaining wnt9b expression only in the most anterior aspect (Fig. 4B, arrow, he).
FIG. 4.
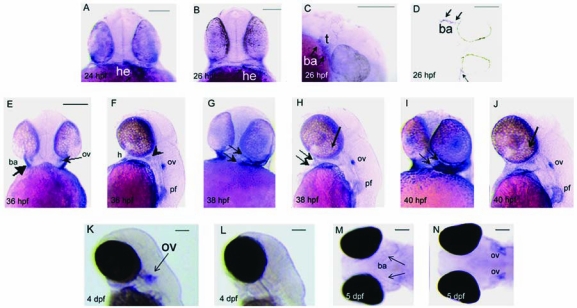
WISH analysis of wnt9b in developing embryos. (A) Frontal view, 24 hpf embryo. Expression of wnt9b in the early heart tube (arrow, he). (B) Frontal view, 26 hpf embryo. wnt9b expression is restricted to the anterior portion of the heart tube (arrow, he). (C) Lateral view at 26 hpf, anterior to the right. wnt9b is expressed in first and second arch tissues (arrows, ba) surrounding the trigeminal (t) ganglion. (D) Oblique longitudinal sectioned 26 hpf embryo reveals wnt9b expression in branchial arch ectoderm (arrows, ba). (E, F) 36 hpf embryo. Frontal view reveals wnt9b expression in anterior branchial arch tissues (E, arrow, ba), and in the otic vesicles (ov). (F) Lateral view reveals wnt9b expression in the heart (h), otic vesicles (ov), pectoral fin bud (pf), and anterior arch tissue (arrowhead). (G) Frontal view of 38 hpf embryo reveals distinct wnt9b expression in anterior ba tissues (arrows), and lateral view (H) reveals wnt9b expression in ba (arrows), ov, and pectoral fin (pf). (I, J) Branchial arch, ov, and pf wnt9b expression persists at 40 hpf. At 4 dpf (K, arrow) and 5 dpf (M, N), wnt9b is detected in otic vesicles, but is reduced or absent in branchial arch tissues. (L) All sense controls were negative. Scale bars, 100 μm.
Branchial arch and pectoral fin wnt9b expression domains
At 26 hpf, discrete bilateral wnt9b expression was observed in first and second branchial arch primordia (Fig. 4C, arrows). A slightly oblique frontal section at 26 hpf revealed distinct wnt9b expression in the distinctly invaginated branchial arch ectoderm (Fig. 4D, arrow). Later, 36 hpf, low pectoral fin stage embryos exhibit wnt9b expression in branchial arch surface ectoderm and in the otic vesicle (Fig. 4E, F). Distinct wnt9b expression is detected in the heart, in anterior arch, otic, and pectoral fin bud tissue (Fig. 4F). At 38 hpf, frontal view revealed anterior arch wnt9b expression as a broad band that bifurcated into dorsal and ventral domains surrounding the primordial mouth (Fig. 4G, arrows), while lateral view revealed branchial arch, otic, and pectoral fin bud expression (Fig. 4H). At 40 hpf, robust wnt9b expression is maintained in anterior arch tissue (Fig. 4I), with lateral views revealing branchial arch, otic, and pectoral fin bud expression (Fig. 4J). By 4 dpf, wnt9b expression is significantly reduced in branchial arch tissue, but is highly expressed in the developing otic vesicles (Fig. 4K, ov, arrow). Reduced branchial arch and robust otic vesicle expression is maintained in 5 dpf embryos (Fig. 4M, N). RT-PCR was used to further define wnt9b expression in developing zebrafish, confirming its expression at all times examined, from 1 hpf to adulthood (Fig. 5).
FIG. 5.
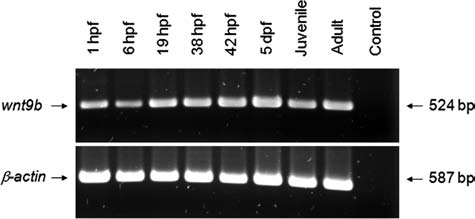
RT-PCR analysis of wnt9b expression in developing zebrafish. RT-PCR was used to detect wnt9b mRNA in developmentally staged zebrafish, as indicated. wnt9b transcripts were present at all developmental times examined, complementing and expanding the less sensitive WISH analyses shown in Figure 4. β-Actin primers were used as positive controls.
Discussion
Conservation of wnt3x/9x digene pairs
The fact that wnt9b/wnt3 and wnt9a/wnt3a gene sets have been evolutionarily conserved as digene pairs in several amniote vertebrate species suggests that they might be coordinately regulated. In contrast, wnt9x/wnt3x gene pairing is not evident in either the zebrafish or the pufferfish. In these two fish genomes, most of the wnt9x and wnt3x genes were mapped to distinct chromosomal locations, with the exception of the wnt9a and wnt3a genes, which map as a digene pair in the Takifugu genome, similar to the wnt gene pairing found in higher vertebrates. This indicates that the Wnt9x/Wnt3x digene cluster is at least as old as the time of divergence from a common ancestor of ray finned and lobe finned fish, approximately 450 million years ago.
Zebrafish wnt9b exhibits discrete branchial arch expression
In the zebrafish, the branchial arches form during the pharyngula period, 24–48 hpf.11 Premigratory neural crest cells (NCCs), which will eventually migrate into and populate the branchial arches, are established at approximately 12 hpf, as somitogenesis begins. Anterior NCC streams begin to advance at approximately 15 to 16 hpf, followed by that of progressively more posterior NCC streams, such that by 24 hpf, all five migrating pharyngeal arch streams are evident. Segmentation of branchial arch primordia is more apparent by 30 hpf, particularly as revealed by distinct mesenchymal and endodermal marker gene expression patterns.12,13
The expression of zebrafish wnt9b suggests very early roles in patterning first and second pharyngeal arch tissues. In particular, wnt9b exhibits dynamic and discrete expression during a critical time in early branchial arch development when significant morphogenetic movements are occurring. Zebrafish branchial arch wnt9b expression was first observed in surface ectoderm intimately associated with NCC migration,14,15 indicating the potential instructive roles for wnt9b in guiding the migration and/or differentiation of cranial NCCs, and of NCC-derived craniofacial structures. Importantly, the results shown here in the zebrafish model are consistent with previously recognized roles for wnt9b in first and second branchial arch development in higher vertebrates. Very similar branchial arch wnt9b expression patterns were recently characterized in Xenopus.16 Interestingly, wnt9b has recently been reported to be expressed in zebrafish median fin radials using RT-PCR.17 Future detailed functional studies of wnt9b in the regulation of craniofacial morphogenesis, easily performed in the tractable zebrafish model system, are likely to improve our current understanding of critical molecular events regulating early craniofacial development, and may eventually help to clarify the pathogenesis of human orofacial clefting diseases.
Materials and Methods
Zebrafish husbandry
Zebrafish were bred and maintained at 28°C on a 14 h light, 10 h dark cycle, as previously described.16 Embryos were generated by natural spawnings, collected, grown in a recirculating system, and staged as described.18
RT-PCR and 5′/3′ RACE analyses
Total RNA was extracted from whole zebrafish embryos using TRIzol Reagent (Invitrogen, Carlsbad, CA). Both the RT-PCR and RACE reactions were performed using the BD SMART RACE cDNA Amplification Kit (BD Biosciences, Palo Alto, CA). First-strand cDNA synthesis was performed using the modified lock-docking oligo(dT) primer and the BD SMART II oligo (BD Biosciences). The wnt9b transcriptional start site was identified and confirmed by 5′RACE using the wnt9b gene specific primer (TGAAC CGGAAACACCATGACACTTACAA) from exon 3, and the universal primer in the 5′RACE reaction. Advantage 2 Polymerase (BD Biosciences) was used to amplify 5′RACE products by PCR. PCR products containing the full coding region of zebrafish wnt9b were generated using the forward and reverse primers, GCCGACCCGTCATCATGTGC and GGCTGG CAAATCATCCTCAACA, respectively, and cloned into the pCR2.1-TOPO cloning vector (Invitrogen). Amplicons were confirmed by double-stranded nucleotide sequencing at The Forsyth Institute DNA sequencing core facility.
WISH analysis
Sense and antisense cRNA probes were transcribed in the presence of digoxigenin-11-UTP from linearized full-length zebrafish wnt9b cDNA templates, using the DIG RNA Labeling Kit (SP6/T7) (Roche, Indianapolis, IN). WISH analyses were performed at 70°C, as previously described.19 Embryos were fixed in methanol; cleared in 75% glycerol in phosphate buffered saline, triton X-100 (PBST); examined using Zeiss Stemi SV11, Leica DMRE, and Leica MZ12 microscopes; and photographed using an AxioCam HRc digital camera with Axiovision 3.1 software.
Detection of developmental stages of mRNA expression profile by reverse transcription polymerase chain reaction
Developmentally staged zebrafish embryos (1 hpf, 6 hpf, 19 hpf, 38 hpf, and 42 hpf), larva of 5 dpf, juvenile zebrafish, and adult zebrafish were collected, frozen, and stored at −80°C. Total RNA was isolated by using TRIzol Reagent (Invitrogen), and 4.5μg of each total RNA sample was used in reverse transcription reactions using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). One seventh of each reverse transcription reaction was used as template in 50-μl PCRs, which were performed with Platinum PCR SuperMix High Fidelity (Invitrogen) and the following reaction conditions: denaturing at 95°C for 5 minutes; followed by 35 cycles at 95°C for 40 seconds, 60°C for 1 minute, and 72°C for 2 minutes; and final extension at 72°C for 7 minutes. Twelve microliter aliquots of each PCR reaction were resolved by agarose gel electrophoresis, respectively, stained with ethidium bromide, and digitally photographed. PCR primers were zebrafish β-actin upstream20, 5′-GGAGAAG ATCTGGCATCACACCTTCTAC-3′; zebrafish β-actin downstream, 5′-TGGTCTCGTGGATAC CGCAAGATTCCAT-3′; Wnt9b upstream, 5′-TGGAGCGCTGCACCTGTGATGATTCC-3′; Wnt9b downstream, 5′-ACAGCACAGGCT GCTGCAGCTGG-3′.
Bioinformatics
The SignalP 3.0 server (www.cbs.dtu.dk) was used to predict the likely cleavage site of the Wnt9b signal sequence peptide.21 CLUSTAL W 1.82 software and TreeView 1.6.6 software (www.ebi.ac.uk/clustalw/index.html) were used to assess the phylogenetic relationships of Wnt9b protein sequences.
Acknowledgments
We wish to thank all the members of the Yelick Lab for helpful discussions, advice, and expertise, and Loic Fabricant and Seija Cope for expert zebrafish husbandry. We thank The Forsyth Institute DNA Sequencing Core staff for their expertise. This research was supported in part by NIH/NIDCR grants DE12076 (P.C.Y.), DE14528 (P.A.J.), and DE14683 (T.L.F.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Juriloff DM. Harris MJ. McMahon AP. Carroll TJ. Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76(8):574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- 2.Marazita ML. Field LL. Cooper ME. Tobias R. Maher BS. Peanchitlertkajorn S. Liu YE. Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet. 2002;71(2):349–364. doi: 10.1086/341944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- 4.Qian J. Jiang Z. Li M. Heaphy P. Liu YH. Shackleford GM. Mouse Wnt9b transforming activity, tissue-specific expression, and evolution. Genomics. 2003;81(1):34–46. doi: 10.1016/s0888-7543(02)00012-5. [DOI] [PubMed] [Google Scholar]
- 5.Lan Y. Ryan RC. Zhang Z. Bullard SA. Bush JO. Maltby KM, et al. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235(5):1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll TJ. Park JS. Hayashi S. Majumdar A. McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9(2):283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Katoh M. Comparative genomics on Wnt3-Wnt9b gene cluster. Int J Mol Med. 2005;15(4):743–747. [PubMed] [Google Scholar]
- 8.Zerucha T. Stuhmer T. Hatch G. Park BK. Long Q. Yu G, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20(2):709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghanem N. Jarinova O. Amores A. Long Q. Hatch G. Park BK, et al. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13(4):533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin AJ. Tsang M. Weinberg ES. Heart and gut chiralities are controlled independently from initial heart position in the developing zebrafish. Dev Biol. 2000;227(2):403–421. doi: 10.1006/dbio.2000.9924. [DOI] [PubMed] [Google Scholar]
- 11.Kimmel CB. Ballard WW. Kimmel SR. Ullmann B. Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 12.Schilling TF. Kimmel CB. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120(3):483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- 13.Crump JG. Maves L. Lawson ND. Weinstein BM. Kimmel CB. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development. 2004;131(22):5703–5716. doi: 10.1242/dev.01444. [DOI] [PubMed] [Google Scholar]
- 14.Akimenko MA. Ekker M. Wegner J. Lin W. Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14(6):3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller CT. Schilling TF. Lee K. Parker J. Kimmel CB. Sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127(17):3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- 16.Garriock RJ. Warkman AS. Meadows SM. D'Agostino S. Krieg PA. Census of vertebrate Wnt genes: isolation and developmental expression of Xenopus Wnt2, Wnt3, Wnt9a, Wnt9b, Wnt10a, and Wnt16. Dev Dyn. 2007;236(5):1249–1258. doi: 10.1002/dvdy.21156. [DOI] [PubMed] [Google Scholar]
- 17.Crotwell PL. Mabee PM. Gene expression patterns underlying proximal-distal skeletal segmentation in late-stage zebrafish, Danio rerio. Dev Dyn. 2007;236(11):3111–3128. doi: 10.1002/dvdy.21352. [DOI] [PubMed] [Google Scholar]
- 18.Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene: 1995. [Google Scholar]
- 19.Thisse C. Thisse B. Schilling TF. Postlethwait JH. Structure of the zebrafish snail gene and its expression in wild type spadetail and no tail mutant embryos. Development. 1993;119(4):1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 20.Fang PK. Solomon KR. Zhuang L. Qi M. McKee M. Freeman MR. Yelick PC. Caveolin-1α and −1β perform nonredundant roles in early vertebrate development. Am J Pathol. 2006;169(6):2209–2222. doi: 10.2353/ajpath.2006.060562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendtsen JD. Nielsen H. von Heijne G. Brunak S. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]