Abstract
Bigham, Abigail W., Melisa Kiyamu, Fabiola León-Verlarde, Esteban J. Parra, Maria Rivera-Ch, Mark D. Shriver, and Tom D. Brutsaert. Angiotensin-converting enzyme genotype and arterial oxygen saturation at high altitude in Peruvian Quechua. High Alt. Med. Biol. 9:167–178, 2008.—The I-allele of the angiotensin-converting enzyme (ACE) gene insertion/deletion (I/D) polymorphism has been associated with performance benefits at high altitude (HA). In n = 142 young males and females of largely Quechua origins in Peru, we evaluated 3 specific hypotheses with regard to the HA benefits of the I-allele: (1) the I-allele is associated with higher arterial oxygen saturation (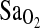 ) at HA, (2) the I-allele effect depends on the acclimatization state of the subjects, and (3) the putative I-allele effect on
) at HA, (2) the I-allele effect depends on the acclimatization state of the subjects, and (3) the putative I-allele effect on 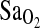 is mediated by the isocapnic hypoxic ventilatory response (HVR,
is mediated by the isocapnic hypoxic ventilatory response (HVR, 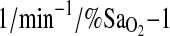 ). The subject participants comprised two different study groups including BLA subjects (born at low altitude) who were lifelong sea-level residents transiently exposed to hypobaric hypoxia (<24 h) and BHA subjects (born at HA) who were lifelong residents of HA. To control for the possibility of population stratification, Native American ancestry proportion (NAAP) was estimated as a covariate for each individual using a panel of 70 ancestry-informative molecular markers (AIMS). At HA, resting and exercise
). The subject participants comprised two different study groups including BLA subjects (born at low altitude) who were lifelong sea-level residents transiently exposed to hypobaric hypoxia (<24 h) and BHA subjects (born at HA) who were lifelong residents of HA. To control for the possibility of population stratification, Native American ancestry proportion (NAAP) was estimated as a covariate for each individual using a panel of 70 ancestry-informative molecular markers (AIMS). At HA, resting and exercise 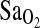 was strongly associated with the ACE genotype, p = 0.008 with ∼4% of the total variance in
was strongly associated with the ACE genotype, p = 0.008 with ∼4% of the total variance in 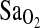 attributed to ACE genotype. Moreover, I/I individuals maintained ∼2.3 percentage point higher
attributed to ACE genotype. Moreover, I/I individuals maintained ∼2.3 percentage point higher 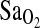 compared to I/D and D/D. This I-allele effect was evident in both BLA and BHA groups, suggesting that acclimatization state has little influence on the phenotypic expression of the ACE gene. Finally, ACE genotype was not associated with the isocapnic HVR, although HVR had a strong independent effect on
compared to I/D and D/D. This I-allele effect was evident in both BLA and BHA groups, suggesting that acclimatization state has little influence on the phenotypic expression of the ACE gene. Finally, ACE genotype was not associated with the isocapnic HVR, although HVR had a strong independent effect on 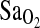 (p = 0.001). This suggests that the I-allele effect on
(p = 0.001). This suggests that the I-allele effect on 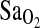 is not mediated by the peripheral control of breathing, but rather by some other central cardiopulmonary effect of the ACE gene on the renin–angiotensin–aldosterone system (RAAS).
is not mediated by the peripheral control of breathing, but rather by some other central cardiopulmonary effect of the ACE gene on the renin–angiotensin–aldosterone system (RAAS).
Key words: genetics, angiotensin-converting enzyme, renin–angiotensin–aldosterone system, polymorphism, hypoxic ventilatory response
Introduction
The human renin–angiotensin–aldosterone system (RAAS) maintains circulatory homeostasis. A central component of this system is the angiotensin-converting enzyme (ACE), which is involved in blood-pressure regulation through the conversion of angiotensin-I (AT-I) to the vasoconstrictor peptide angiotensin-II (AT-II), as well as through the degradation of the vasodilator bradykinin (Muller et al., 1997). Additionally, ACE is involved in plasma-volume regulation, as AT-II affects water and salt retention via stimulation of aldosterone release. The gene for human ACE is located on chromosome 17, and a frequently studied polymorphism is the Alu insertion/deletion (I/D) polymorphism located in intron 16 (Rigat et al., 1990). This polymorphism explains some of the variation in circulating plasma and tissue ACE levels, with the insertion (I) allele associated with lower ACE activities compared with the deletion (D) (Costerousse et al., 1993; Danser et al., 1995; Zhu et al., 2001). Hypoxia has direct effects on ACE and the RAAS, and these effects in turn may mediate many of the normal and also abnormal (i.e., pathological) cardiopulmonary responses to high altitude. For example, increases in plasma volume are considered a normal response to HA, but excessive fluid retention and increased aldosterone have also been implicated in the pathogenesis of acute mountain sickness (AMS), high altitude pulmonary hypertension (HAPH), and high altitude pulmonary edema (HAPE) (Hackett et al., 1982; Milledge et al., 1983; Bartsch et al., 1991).
A small number of recent studies have investigated the association of the ACE locus with one or more of the various high altitude pathologies, but these studies have produced either inconsistent or largely negative results (reviewed by Mortimer et al., 2004; Rupert and Koehle, 2006). In contrast, two cohort studies of the ACE locus have suggested an I-allele performance benefit for mountaineers ascending to extreme altitude (Montgomery et al., 1998; Tsianos et al., 2005). The earlier study by Montgomery et al. (1998) reported an overrepresentation of the I-allele in elite mountaineers who had ascended above 7000 m without the use of supplemental oxygen No climber in that study (out of 15) who managed to ascend beyond 8000 m without supplemental oxygen was of D/D genotype. In a follow-up prospective study with a much larger sample of recreational climbers (n = 248) attempting to ascend Mont Blanc (4807 m), the I-allele again predicted success in reaching the summit (Tsianos et al., 2005). That is, the I-allele frequency was 0.47 in those who reached the summit versus 0.21 in those who failed to reach the summit (p = 0.01). To explain this apparent performance benefit, there is evidence that individuals of I/I genotype maintain higher arterial oxygen saturation (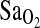 ) at rest and during exercise at HA (Woods et al., 2002). This is perhaps the result of an enhanced hypoxic ventilatory response (HVR) (Patel et al., 2003).
) at rest and during exercise at HA (Woods et al., 2002). This is perhaps the result of an enhanced hypoxic ventilatory response (HVR) (Patel et al., 2003).
To directly evaluate if the I-allele is associated with higher 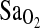 at HA as a result of increased HVR, we genotyped two groups of young male and female Peruvians (n = 142) for the ACE I/D polymorphism. One group was born-and-raised at low altitude (0 m)(BLA n = 71), and the other group was born and raised at high altitude (above 4000 m) (BHA, n = 71). For all subjects we measured pulmonary function, the submaximal and maximal exercise responses at 4338 m (including
at HA as a result of increased HVR, we genotyped two groups of young male and female Peruvians (n = 142) for the ACE I/D polymorphism. One group was born-and-raised at low altitude (0 m)(BLA n = 71), and the other group was born and raised at high altitude (above 4000 m) (BHA, n = 71). For all subjects we measured pulmonary function, the submaximal and maximal exercise responses at 4338 m (including 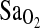 ), and the isocapnic HVR. Both subject groups were comprised of individuals of mixed European and Native American (Quechua) ancestry. The Native American ancestry proportion (NAAP) was estimated for each individual using a panel of 70 ancestry-informative markers (AIMs). While originally serving the goals of an earlier study (see Brutsaert et al., 2004), this design has some advantages that should be noted with respect to the current analysis. First, the measurement of NAAP by DNA markers allows direct covariate control for potential population stratification in the evaluation of candidate gene association. Admixture is a well-recognized problem that can produce false-positive association, but thus far no ACE–altitude study has directly addressed this issue (Pfaff et al., 2001; Hoggart et al., 2003). Another advantage of our design is that gene association can be assessed in two study groups who differed by length of exposure to HA. BHA subjects were lifelong residents of HA and fully acclimatized, whereas BLA subjects were recruited at sea level and transiently exposed to 4338 m for <24 h. By comparing these two study groups, we evaluated whether the influence of the ACE genotype on individual physiology is more important during early (i.e., acute) exposure, as has been suggested by previous investigators (Woods et al., 2002; Mortimer et al., 2004). Finally, our original study focused explicitly on the control of breathing at altitude. We assessed the isocapnic HVR in order to interrogate hypoxic chemosensitivity as mediated by the peripheral chemoreceptors. This allows direct evaluation of the hypothesis that the ACE I-allele benefit is due to increased
), and the isocapnic HVR. Both subject groups were comprised of individuals of mixed European and Native American (Quechua) ancestry. The Native American ancestry proportion (NAAP) was estimated for each individual using a panel of 70 ancestry-informative markers (AIMs). While originally serving the goals of an earlier study (see Brutsaert et al., 2004), this design has some advantages that should be noted with respect to the current analysis. First, the measurement of NAAP by DNA markers allows direct covariate control for potential population stratification in the evaluation of candidate gene association. Admixture is a well-recognized problem that can produce false-positive association, but thus far no ACE–altitude study has directly addressed this issue (Pfaff et al., 2001; Hoggart et al., 2003). Another advantage of our design is that gene association can be assessed in two study groups who differed by length of exposure to HA. BHA subjects were lifelong residents of HA and fully acclimatized, whereas BLA subjects were recruited at sea level and transiently exposed to 4338 m for <24 h. By comparing these two study groups, we evaluated whether the influence of the ACE genotype on individual physiology is more important during early (i.e., acute) exposure, as has been suggested by previous investigators (Woods et al., 2002; Mortimer et al., 2004). Finally, our original study focused explicitly on the control of breathing at altitude. We assessed the isocapnic HVR in order to interrogate hypoxic chemosensitivity as mediated by the peripheral chemoreceptors. This allows direct evaluation of the hypothesis that the ACE I-allele benefit is due to increased 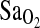 via RAAS effects on peripheral control of breathing (i.e., HVR), rather than by RAAS effects on other aspects of the central cardiopulmonary system.
via RAAS effects on peripheral control of breathing (i.e., HVR), rather than by RAAS effects on other aspects of the central cardiopulmonary system.
Materials and Methods
Subjects
One hundred forty-two Peruvian males and females (71 females and 71 males) participated in this study (Table 1).For the lowland born group (BLA), 36 females and 32 males were recruited from the Barrios Altos district of Lima, Peru. This district of Lima was chosen because approximately 10% of residents have recently downmigrated from highland Peruvian communities. All study participants from this district were born and raised at or near sea level, whereas their parents and both sets of grandparents were born >3000 m above sea level. Thus, BLA participants, although of highland origins, had no exposure to hypobaric hypoxia during growth and development. As described later, the BLA subjects were transported to 4338 meters for measurement of many of the study outcome variables. The highland-born group (BHA) was composed of 35 females and 39 males. These study subjects were recruited from Cerro de Pasco, Peru, at 4338 m above sea level. For this group, all subjects were born and raised above 3000 m, as were their parents and both sets of grandparents. All participants provided informed, written consent according to the guidelines approved by the Institutional Review Boards at the University at Albany, SUNY, and the Universidad Peruana Cayetano Heredia, Lima, Peru.
Table 1.
Genotype and Allele Frequencies for the ACE Alu Indel Polymorphism in Highland- and Lowland-Born Groups
| |
|
|
Genotype frequencya |
Allele frequencya |
|||
|---|---|---|---|---|---|---|---|
| Female | Male | I/I | I/D | D/D | I | D | |
| Lowland born (BLA) | 36 | 32 | 0.53 | 0.34 | 0.13 | 0.70 | 0.30 |
| Highland born (BHA) | 35 | 39 | 0.51 | 0.42 | 0.07 | 0.72 | 0.28 |
| Total | 71 | 71 | 0.52 | 0.38 | 0.10 | 0.71 | 0.29 |
Both groups are in Hardy Weinberg equilibrium (p < 0.05).
ACE, angiotensin-converting enzyme; I, insertion; D, deletion.
Subject screening, pulmonary function, and anthropometry
Potential subjects were identified as nonsmokers and screened via a brief clinical history and medical examination for conditions contraindicating participation in the study protocols, including chronic obstructive respiratory diseases, cardiovascular disease, and renal disease. At screening, a venous blood sample was drawn from the antecubital vein, and hemoglobin concentration [Hb] g/dL−1 was determined by a Hemocue blood hemoglobin analyzer (Angelholm, Sweden). Subjects with [Hb] less than altitude-specific cutoff values for anemia were excluded from the study. In addition, highland resident subjects with excessive polycythemia, [Hb] > 21.5, were excluded from the study. DNA was extracted from this blood sample for the genetic analyses described later.
Pulmonary function and anthropometry were assessed on each subject at the time of screening. Pulmonary function was measured using a VS400 Volumetric Spirometer (Puritan-Bennett, Mallinckrodt, Hazelwood, MO), calibrated daily with a 3-L calibration syringe. Each subject performed a maximal inspiration, followed immediately by a forced maximal expiration while in a standing position. From this procedure, the forced vital capacity (FVC) and forced expiratory volume made in 1 sec (FEV1) were determined based on the best of at least two efforts. FVC and FEV1 measures were corrected to body temperature, ambient barometric pressure, and saturation vapor pressure conditions (BTPS). Anthropomorphic measurements included weight, height, and skinfolds at subscapular, suprailiac, biceps, and triceps sites. Percent body fat was calculated from the Siri equation (Siri, 1961), and body density was calculated according to the method of Durnin and Womersley (Durnin and Womersley, 1974).
Genotyping
The ACE indel allele was amplified by polymerase chain reaction (PCR) (Evans et al., 1994). Two primers were used in each PCR, F: 5′ 3′ F: CTG GAG ACC ACT CCC ATC CTT TCT and R: 5′ 3′GAT GTG GCC ATC ACA TTC GTC AGA T. Samples were initially denatured at 94°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 65°C for 30 sec, and 72°C for 45 sec. A final extension for 5 min at 72°C completed the PCR. Genotyping was conducted by 2% agarose gel electrophoresis and visualized with ethidium bromide staining.
Estimates of Native American ancestry proportion (NAAP)
To estimate individual admixture proportions for covariate control of potential population stratification, all individuals were genotyped for 70 AIMS (rs140864, rs2065160, rs17203, MID52, rs3309, rs3340, rs2763, rs2161, rs2695, rs594689, rs1042602, rs1800498, rs1079598, rs2862, rs4646, PV92, rs2816, rs4884, MID161, rs16383, rs2814778, rs6003, rs285, rs1800404, rs1987956, rs1415878, rs1435090, rs768324, rs1980888, rs983271, rs878825, rs386569, rs1373302, rs1891760, rs963170, rs718092, rs1808089, rs1320892, rs723822, rs1881826, rs2207782, rs1112828, rs2064722, rs2351254, rs1316579, rs1935946, rs1506069, rs719776, rs1369290, rs1465648, rs2077681, rs725416, rs1594335, rs2188457, rs1861498, rs2341823, rs1327805, rs2396676, rs1461227, rs1403454, rs717962, rs1074075, rs764679, rs724729, rs717091, rs726391, rs1487214, rs725667, rs2225251, rs951784), as previously described (Shriver et al., 2003; Bonilla et al., 2004; Brutsaert et al., 2005). The AIMs were selected based on their high frequency differences between West African, Native American, and European populations, as follows: (1) 18 markers show high frequency differences between West African populations compared to Native American and European populations; (2) 13 markers show high frequency differences between European compared to West African and Native American populations, (3) 31 markers show high frequency differences between Native American compared to European and West African populations, and (4) 8 of the markers show intermediate frequency differences among all three parental populations. Based on each individual's genotypes at all 70 loci, NAAP was calculated using the program Maximum Likelihood (Hanis et al., 1986). Only a few individuals showed evidence of West African ancestry, but the inclusion or exclusion of these individuals did not affect the overall results presented in this paper. Thus, conceptually, this paper presents the proportionate European versus Native American ancestry contribution, given that historically these were the two dominant parental populations in the region.
Ventilatory control studies
Ventilatory control studies were conducted at sea level for the BLA group and at 4338 m for the BHA group. It is important to note that the end-tidal forcing technique used to assess ventilatory control (described later) imposes the same partial pressure profiles for O2 and CO2, independent of altitude. Thus, the ventilatory control tests did not depend on test location per se. It should also be noted that HVRs in males and females were assessed via two slightly different protocols according to the goals of our original study. However, both protocols yield fully comparable measures of the HVR. That is, we measured 32 subjects using both protocols, and this produced identical mean values of HVR and a correlation between protocols of 0.76. This correlation is near the limits of repeatability for HVR protocols generally, as we measured a correlation of 0.84 for the same protocol administered on 2 separate days. More importantly, a Bland–Altman analysis (1986) reveals no bias between protocols across the range of HVRs measured. Nevertheless, to avoid potential bias, we were careful to control for both sex (i.e., test protocol) and test location in statistical analyses of HVR (see Data Analysis).
For all subjects, HVR measurement began with a preliminary procedure in which we determined the end-tidal carbon dioxide partial pressure (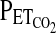 ) and end-tidal oxygen partial pressure (
) and end-tidal oxygen partial pressure (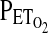 ) using a fine nasal catheter so as to disturb the subject as little as possible. An instantaneous value for the respiratory quotient was calculated to ensure that the subject was not hyperventilating due to anxiety with the protocol. After the preliminary procedure, HVR was measured by imposing specific profiles for
) using a fine nasal catheter so as to disturb the subject as little as possible. An instantaneous value for the respiratory quotient was calculated to ensure that the subject was not hyperventilating due to anxiety with the protocol. After the preliminary procedure, HVR was measured by imposing specific profiles for 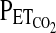 and
and 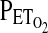 using an end-tidal forcing system (see later). In males, we used the protocol devised by Mou et al. (1995) and validated further by Zhang and Robbins (2000). This protocol, as well as the protocol used for females, is sufficiently brief so as to minimize the confounding effects of hypoxic ventilatory decline on the HVR. Throughout the protocol, the
using an end-tidal forcing system (see later). In males, we used the protocol devised by Mou et al. (1995) and validated further by Zhang and Robbins (2000). This protocol, as well as the protocol used for females, is sufficiently brief so as to minimize the confounding effects of hypoxic ventilatory decline on the HVR. Throughout the protocol, the 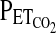 was held at ∼2 torr above the subjects' natural resting value. A settling period of 5 min was employed during which the
was held at ∼2 torr above the subjects' natural resting value. A settling period of 5 min was employed during which the 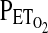 was held at 100 torr. After this settling period,
was held at 100 torr. After this settling period, 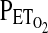 was lowered stepwise in a series of seven steps from 100 to 45 torr, with each step lasting for 50 sec. Values for
was lowered stepwise in a series of seven steps from 100 to 45 torr, with each step lasting for 50 sec. Values for 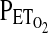 for the five intervening steps had been calculated so as to provide approximately even reductions in
for the five intervening steps had been calculated so as to provide approximately even reductions in 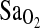 between steps and, consequently, approximately even increases in VE between steps. In females, the settling period was 10 min, again at
between steps and, consequently, approximately even increases in VE between steps. In females, the settling period was 10 min, again at 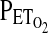 of 100 torr, followed by a single-step drop to 50 Torr for the next 5 min (hypoxia). In both protocols, HVR was calculated from the average values for ventilation (VE in BTPS units) and
of 100 torr, followed by a single-step drop to 50 Torr for the next 5 min (hypoxia). In both protocols, HVR was calculated from the average values for ventilation (VE in BTPS units) and 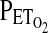 . The latter were then converted into calculated values for arterial saturation using the equation of Severinghaus (Severinghaus, 1979). HVR is given as the absolute value of the slope term (HVR,
. The latter were then converted into calculated values for arterial saturation using the equation of Severinghaus (Severinghaus, 1979). HVR is given as the absolute value of the slope term (HVR, 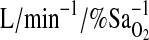 ) calculated via linear regression from the relationship between VE and desaturation.
) calculated via linear regression from the relationship between VE and desaturation.
The end-tidal forcing technique
In both protocols, the technique of end-tidal forcing was used to generate the desired profiles in 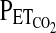 and
and 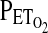 . In this technique, a computational model of the cardiorespiratory system and gas stores is first used to calculate the profiles for inspiratory
. In this technique, a computational model of the cardiorespiratory system and gas stores is first used to calculate the profiles for inspiratory 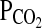 and
and 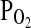 that are likely to generate the desired
that are likely to generate the desired 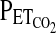 and
and 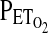 . Once these values have been calculated, the experiment begins when a computer connected to a fast gas-mixing system generates the inspiratory gas mixtures. In general, these predicted inspiratory gas mixtures will not of themselves generate the desired end-tidal values with sufficient precision, because the physiology of the individual deviates from the assumptions of the cardiorespiratory model. To overcome this problem, these predicted inspiratory values are modified during the course of the experiment by using breath-by-breath feedback from the measured
. Once these values have been calculated, the experiment begins when a computer connected to a fast gas-mixing system generates the inspiratory gas mixtures. In general, these predicted inspiratory gas mixtures will not of themselves generate the desired end-tidal values with sufficient precision, because the physiology of the individual deviates from the assumptions of the cardiorespiratory model. To overcome this problem, these predicted inspiratory values are modified during the course of the experiment by using breath-by-breath feedback from the measured 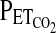 and
and 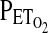 . These measured values of
. These measured values of 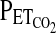 and
and 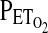 are compared with the desired values, and an integral-proportional feedback control algorithm is used to calculate the actual adjustments required for the inspiratory
are compared with the desired values, and an integral-proportional feedback control algorithm is used to calculate the actual adjustments required for the inspiratory 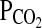 and
and 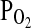 . Details of the forcing procedure and the gas-mixing system used in Peru have been described previously (Mou et al., 1995; Brutsaert et al., 2005). In all experiments, the subject sat upright and breathed to and from a gas-mixing chamber via a mouthpiece while wearing a nose clip. A pulse oximeter monitored
. Details of the forcing procedure and the gas-mixing system used in Peru have been described previously (Mou et al., 1995; Brutsaert et al., 2005). In all experiments, the subject sat upright and breathed to and from a gas-mixing chamber via a mouthpiece while wearing a nose clip. A pulse oximeter monitored 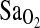 (Ohmeda 5740, Soma Technology, Chester, Connecticut). Respiratory volumes were measured using a turbine volume-measuring device (VMM 400, Interface Associates, Laguna Niguel, CA) fixed in series with the mouthpiece. Gas was sampled continuously from the mixing chamber (100 mL) close to the mouth at a rate of 20 mL/min and analyzed using a gas analyzer (Datex Normocap 200-Oxy, Meda, SA). All experimental data were recorded by computer at a sampling rate of 50 Hz using real-time, data-acquisition software written in LabView (National Instruments, Austin, TX).
(Ohmeda 5740, Soma Technology, Chester, Connecticut). Respiratory volumes were measured using a turbine volume-measuring device (VMM 400, Interface Associates, Laguna Niguel, CA) fixed in series with the mouthpiece. Gas was sampled continuously from the mixing chamber (100 mL) close to the mouth at a rate of 20 mL/min and analyzed using a gas analyzer (Datex Normocap 200-Oxy, Meda, SA). All experimental data were recorded by computer at a sampling rate of 50 Hz using real-time, data-acquisition software written in LabView (National Instruments, Austin, TX).
Exercise testing
All exercise testing was conducted in Cerro de Pasco at 4338 m. BLA subjects traveled from sea level to Cerro de Pasco where they were tested within a few hours of their arrival to our laboratory. Subjects came to the laboratory in small groups over about a 1-week period, and so general travel times were as follows: Cerro de Pasco is a 7 to 10-h bus ride from Lima on paved road. The first ∼4 to 6 hours of the trip involve a steady gain in altitude to a high mountain pass (∼4800 m). The road then descends to the Peruvian Altiplano (3600–4300 m) for the next 3 to 4 hours of the trip. Subjects rested in the laboratory for 2 to 4 h before studies were initiated and therefore were studied after 9 to 14 h of acute exposure to hypobaric hypoxia. Of 68 subjects recruited and measured in Lima, four were not measured in Cerro de Pasco. One was unable to make the trip for personal reasons, and three were diagnosed with acute mountain sickness on arrival to Cerro de Pasco and returned to the lowlands.
To begin, 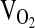 was measured at rest (5 min) with the subject seated. Following resting measurements,
was measured at rest (5 min) with the subject seated. Following resting measurements, 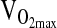 was measured while subjects operated a mechanically braked Monarch 818e research ergometer. Male and female subjects started with workloads of 1.0- to and 0.5-kg resistance, respectively, and were instructed to maintain 60 rpm. Resistance was incremented every 3 min by 0.5 and 0.25 kg for males and females, respectively, until subject volitional fatigue.
was measured while subjects operated a mechanically braked Monarch 818e research ergometer. Male and female subjects started with workloads of 1.0- to and 0.5-kg resistance, respectively, and were instructed to maintain 60 rpm. Resistance was incremented every 3 min by 0.5 and 0.25 kg for males and females, respectively, until subject volitional fatigue. 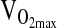 was defined as the highest level of oxygen consumption averaged over the final minute of the test concomitant with at least one of the following conditions: (1) a nonlinear increase in exercise ventilation resulting in a respiratory exchange ratio greater than 1.10, (2) a plateau in the
was defined as the highest level of oxygen consumption averaged over the final minute of the test concomitant with at least one of the following conditions: (1) a nonlinear increase in exercise ventilation resulting in a respiratory exchange ratio greater than 1.10, (2) a plateau in the 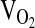 –work rate relationship, or (3) a maximal heart rate within 10% of the age-predicted maximum. In this paper we present maximal exercise response variables, as well as exercise response variables during the first two levels of submaximal exercise, when subjects were more likely to be at steady state.
–work rate relationship, or (3) a maximal heart rate within 10% of the age-predicted maximum. In this paper we present maximal exercise response variables, as well as exercise response variables during the first two levels of submaximal exercise, when subjects were more likely to be at steady state.
During 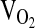 testing, subjects breathed through a low-resistance breathing valve. The expired ventilation (VE, L/min−1 BTPS), as well as the fractional concentrations of O2 and CO2 in the expired air, were processed by a Parvomedics True Max metabolic measuring system (Sandy, Utah) to produce 1-minute interval calculations of
testing, subjects breathed through a low-resistance breathing valve. The expired ventilation (VE, L/min−1 BTPS), as well as the fractional concentrations of O2 and CO2 in the expired air, were processed by a Parvomedics True Max metabolic measuring system (Sandy, Utah) to produce 1-minute interval calculations of 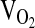 , carbon dioxide production (
, carbon dioxide production (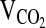 ), the respiratory exchange ratio (RER), and the ventilatory equivalents for oxygen and carbon dioxide (
), the respiratory exchange ratio (RER), and the ventilatory equivalents for oxygen and carbon dioxide (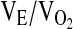 and
and 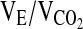 , respectively). Gas analyzers were calibrated with standard gases before each exercise test. The pneumotach used to measure ventilatory flow was also calibrated prior to each test with a 3-L calibration syringe. Heart rate (HR) was continuously monitored via telemetry (Polar Electric Oy, Kempele, Sweden) interfaced with the metabolic measuring system.
, respectively). Gas analyzers were calibrated with standard gases before each exercise test. The pneumotach used to measure ventilatory flow was also calibrated prior to each test with a 3-L calibration syringe. Heart rate (HR) was continuously monitored via telemetry (Polar Electric Oy, Kempele, Sweden) interfaced with the metabolic measuring system. 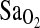 by pulse oximetry was continuously monitored by an Ohmeda 5740 pulse oximeter using a fingertip sensor (subjects were instructed not to grip with that finger). The pulse oximetry signal was acquired by an REM/400M data acquisition system (CB Sciences, Dover, New Hampshire) and recorded every 15 sec during
by pulse oximetry was continuously monitored by an Ohmeda 5740 pulse oximeter using a fingertip sensor (subjects were instructed not to grip with that finger). The pulse oximetry signal was acquired by an REM/400M data acquisition system (CB Sciences, Dover, New Hampshire) and recorded every 15 sec during 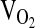 measurements.
measurements.
Data analysis
Allele and genotype frequencies for the BLA- and BHA-born groups were determined by gene counting and compared by a 2 × 2 contingency table. A χ-squared test was used to compare genotype frequencies with those expected under the Hardy–Weinberg equilibrium (HWE). ANOVA/ANCOVA was used to test for sex, group (i.e., altitude of birth), and ACE genotype effects on study outcome variables using the general linear model procedure of the SPSS statistical software package, version 10.0. Repeat measures ANCOVA was used to test for ACE genotype effects on sub-maximal exercise response variables from rest through work levels 1 and 2. In analyses of ACE allele association involving the HVR measure, it should be emphasized that sex and study group were included as control variables to account for any potential differences in HVR by protocol (i.e., males and females were measured by different protocols) and by test location (i.e., HVR was measured at sea-level and at altitude in BHA vs. BLA subjects, respectively), as previously described. Although the different protocols yield fully comparable values of HVR, statistical control in this manner ensures a nonbiased test of association between ACE and HVR. Statistical significance criteria was p < 0.05 for all tests.
Results
Allele and genotype frequencies are shown in Table 1. There were no significant differences in allele or genotype frequencies between the BLA and BHA subject groups. The ACE genotype frequencies for the BLA and BHA groups did not deviate from HWE. Table 2 gives subject characteristics organized by ACE genotype and study group. In general, the numbers of males and females were equally distributed within genotypes, except for BLA subjects of D/D genotype, where females (n = 7) exceeded males (n = 2). This distribution difference is a factor of small sample size for D/D genotypes in the population given the relatively low frequency of the D-allele in the Andes. This discrepancy does not affect the results of this study, as sex was controlled when testing for genotype effects by ANCOVA to account for well-known differences between males and females in height, weight, body fat percent, FVC, [Hb], and 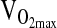 mL/min−1/kg−1.
mL/min−1/kg−1.
Table 2.
Subject Characteristics (Mean ± SD) by Altitude of Birth and ACE Alu Indel Polymorphism Genotype
| I/I | I/D | D/D | |
|---|---|---|---|
| Lowland born (BLA) | M = 20, F = 16 | M = 10, F = 13 | M = 2, F = 7 |
| Age,a yr | 25.3 ± 3.8 | 23.4 ± 3.8 | 27.7 ± 5.8 |
| Height,b cm | 158.3 ± 8.7 | 154.4 ± 7.1 | 154.0 ± 11.7 |
| Weight,b kg | 61.2 ± 9.6 | 59.1 ± 7.3 | 59.0 ± 13.8 |
| Fat,b % | 28.8 ± 9.3 | 30.4 ± 9.7 | 32.4 ± 6.9 |
| FVCb | 4.26 ± 0.89 | 3.94 ± 0.79 | 3.59 ± 0.71 |
| [Hb],b g/dL−1 | 14.3 ± 0.9 | 14.1 ± 1.4 | 14.2 ± 1.2 |
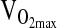 ,b mL/min−1/kg−1 ,b mL/min−1/kg−1
|
31.6 ± 8.6 | 31.0 ± 10.0 | 24.0 ± 5.1 |
| NAAP | 0.90 ± 0.08 | 0.89 ± 0.11 | 0.85 ± 0.15 |
| Highland born (BHA) | M = 19, F = 19 | M = 17, F = 14 | M = 3, F = 2 |
| Age, yr | 25.9 ± 4.4 | 24.3 ± 4.9 | 25.8 ± 5.8 |
| Height, cm | 155.3 ± 8.0 | 155.5 ± 8.4 | 155.4 ± 8.0 |
| Weight, kg | 56.2 ± 6.8 | 57.1 ± 6.8 | 51.9 ± 7.0 |
| Fat, % | 24.7 ± 7.6 | 25.4 ± 10.1 | 21.2 ± 8.6 |
| FVC | 4.56 ± 0.91 | 4.64 ± 0.98 | 4.81 ± 1.14 |
| [Hb], g/dL−1 | 17.5 ± 1.8 | 17.8 ± 1.8 | 17.6 ± 2.3 |
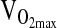 , mL/min−1/kg−1 , mL/min−1/kg−1
|
39.0 ± 10.2 | 38.8 ± 10.9 | 46.6 ± 13.7 |
| NAAP | 0.90 ± 0.10 | 0.90 ± 0.08 | 0.90 ± 0.07 |
Variable differs significantly by genotype controlling fo sex, p ≤ 0.05.
Variable is significantly different between lowland- and highland-born groups across all genotypes, p ≤ 0.05.
ACE, angiotensin-converting enzyme; FVC, forced vital capacity; NAAP, Native American ancestry proportion; I, insertion; D, deletion.
Controlling for sex, we detected no significant genotype differences within study groups for any of the subject characteristics except for age, which was slightly higher in the D/D BLA group (p = 0.029). When BLA and BHA groups were combined for analysis, again only age was significantly different by genotype (slightly higher in D/D, p = 0.026). Notably, substantial group differences related to the altitude of birth (i.e., BHA versus BLA), were observed. For example, when controlling for sex, BLA subjects were significantly taller (p = 0.048), heavier (p < 0.01), and fatter (p < 0.01) than BHA. BLA subjects also had smaller FVCs, lower [Hb], and lower 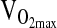 , mL/min−1/kg−1 measured at 4338 m (p < 0.01 for all comparisons). However, these differences were expected and are consistent with many previous studies comparing highland- and lowland-born groups in the Andes. These group differences are controlled for in the statistical analyses when testing for ACE genotype associations (see later). A final general consideration is that mean NAAP was relatively high in the sample (0.90) and was not significantly different by genotype, sex, or group. The high NAAP was expected based on subject-selection criteria. That is, all subjects were selected on the basis of their self-identification as indigenous Andean and the highland Andean origins of their parents and grandparents.
, mL/min−1/kg−1 measured at 4338 m (p < 0.01 for all comparisons). However, these differences were expected and are consistent with many previous studies comparing highland- and lowland-born groups in the Andes. These group differences are controlled for in the statistical analyses when testing for ACE genotype associations (see later). A final general consideration is that mean NAAP was relatively high in the sample (0.90) and was not significantly different by genotype, sex, or group. The high NAAP was expected based on subject-selection criteria. That is, all subjects were selected on the basis of their self-identification as indigenous Andean and the highland Andean origins of their parents and grandparents.
Genotype differences in the HVR and exercise response at 4338 m were tested by ANCOVA or repeated measures ANCOVA using the entire study sample available, which ranged from n = 138 to 142 depending on the outcome measure. Our approach was to test covariate effects on a particular study outcome first, including age, sex, birthplace, and sex-by-birthplace interactions. If a covariate was significant as an independent effect, it was retained in the ANCOVA model testing for a genotypic effect. A final step was to rerun the analysis, including NAAP as an additional covariate in order to control for possible population stratification. As shown in Table 3, there were no significant differences by genotype for HVR, 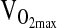 ,-L/min−1,
,-L/min−1, 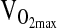 , mL/min−1/kg−1, or
, mL/min−1/kg−1, or 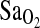 at maximal work output, although for the latter (
at maximal work output, although for the latter (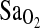 ) the p-value approached significance at 0.071. NAAP was retained as a covariate testing for ACE effects on
) the p-value approached significance at 0.071. NAAP was retained as a covariate testing for ACE effects on 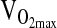 given a marginally significant p-value (i.e., higher
given a marginally significant p-value (i.e., higher 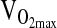 with higher NAAP, p = 0.059), and given results published previously showing smaller
with higher NAAP, p = 0.059), and given results published previously showing smaller 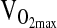 decrement with increasing NAAP in subjects tested at sea level and again at high altitude (Brutsaert et al., 2003). However, it is important to note that for all variables tested ACE genotype analyses were similar with or without NAAP included as a covariate, which indicates that there is no problem with population stratification in this study sample.
decrement with increasing NAAP in subjects tested at sea level and again at high altitude (Brutsaert et al., 2003). However, it is important to note that for all variables tested ACE genotype analyses were similar with or without NAAP included as a covariate, which indicates that there is no problem with population stratification in this study sample.
Table 3.
Adjusted Mean Values ± SE by ACE Genotype from ANCOVA for Various Physiologic Measures
| |
Adjusted mean values ± SE |
|
|
|
|
||
|---|---|---|---|---|---|---|---|
| I/I (n = 72) | I/D (n = 52) | D/D (n = 14) | S | CovariatesaBP | NAAP | p-value, ACE effect | |
| HVR, L/min−1/%−1 | 0.71 ± 0.05 | 0.67 ± 0.06 | 0.55 ± 0.13 | 0.456 | |||
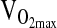 , L/min−1 , L/min−1
|
2.08 ± 0.04 | 2.04 ± 0.04 | 2.06 ± 0.08 | x | x | x | 0.673 |
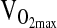 , L/min−1/kg−1 , L/min−1/kg−1
|
35.3 ± 0.6 | 34.9 ± 0.8 | 36.0 ± 1.5 | x | x | x | 0.663 |
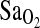 @ max, % @ max, % |
82.7 ± 0.5 | 82.0 ± 0.6 | 80.0 ± 1.1 | x | x | 0.071 | |
Covariates are S = sex, BP = birthplace, and NAAP = Native American ancestry proportion. A covariate was retained in ANCOVA when it had an independent effect on an outcome variable (P ≤ 0.05), or as indicated in the text.
ACE, angiotensin-converting enzyme; I, insertion; D, deletion; HVR = hypoxic-ventilatory response.
In repeated measures analysis of the submaximal exercise response at 4338 m (Table 4), there were no significant differences by genotype for 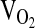 , VE,
, VE, 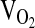 , HR, or RER. However,
, HR, or RER. However, 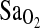 levels at rest and across different levels of sub-maximal exercise were significantly higher in subjects of I/I genotype (Table 4 and Fig. 1, p = 0.008). Again, inclusion/exclusion of NAAP made no difference to these analyses. However, NAAP was retained as a covariate in the final analysis of VE or VE-derived variables, as we have previously shown that subjects of high NAAP have lower VE at altitude (Brutsaert et al., 2005). It should be noted that the ACE I/I genotype effect on
levels at rest and across different levels of sub-maximal exercise were significantly higher in subjects of I/I genotype (Table 4 and Fig. 1, p = 0.008). Again, inclusion/exclusion of NAAP made no difference to these analyses. However, NAAP was retained as a covariate in the final analysis of VE or VE-derived variables, as we have previously shown that subjects of high NAAP have lower VE at altitude (Brutsaert et al., 2005). It should be noted that the ACE I/I genotype effect on 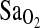 was evident in both highland- and lowland-born subjects, although no significant interaction between ACE genotype and birthplace was detected. Combining I/D and D/D genotypes into one group for comparison with I/I yielded nearly identical results. That is, individuals with I/I genotype did not differ for any measured phenotype against the combined I/D + D/D subjects except for
was evident in both highland- and lowland-born subjects, although no significant interaction between ACE genotype and birthplace was detected. Combining I/D and D/D genotypes into one group for comparison with I/I yielded nearly identical results. That is, individuals with I/I genotype did not differ for any measured phenotype against the combined I/D + D/D subjects except for 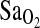 , where the difference by genotype was highly significant, large, and consistent from rest to submaximal exercise.
, where the difference by genotype was highly significant, large, and consistent from rest to submaximal exercise.
Table 4.
Adjusted Mean Values of Exercise Response at 4338 m by ACE Genotype
| |
|
Adjusted mean values ± SE |
|
|
|
|
||
|---|---|---|---|---|---|---|---|---|
| I/I (n = 72) | I/D (n = 52) | D/D (n = 14) | S | CovariatesaBP | NAAP | p-value, ACE effect | ||
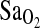 , % , % |
R | 86.49 ± 0.46 | 85.41 ± 0.53 | 82.61 ± 1.03 | x | 0.008 | ||
| E1 | 83.45 ± 0.50 | 81.04 ± 0.59 | 81.27 ± 1.14 | |||||
| E2 | 82.60 ± 0.51 | 81.30 ± 0.60 | 80.53 ± 1.16 | |||||
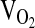 , L/min−1 , L/min−1
|
R | 0.33 ± 0.01 | 0.32 ± 0.01 | 0.32 ± 0.01 | x | 0.396 | ||
| E1 | 1.05 ± 0.01 | 1.06 ± 0.01 | 1.04 ± 0.02 | |||||
| E2 | 1.38 ± 0.01 | 1.38 ± 0.01 | 1.33 ± 0.03 | |||||
| VE L/min−1 | R | 6.5 ± 0.1 | 6.1 ± 0.2 | 5.6 ± 0.3 | x | x | x | 0.307 |
| STPD | E1 | 16.9 ± 0.3 | 16.9 ± 0.4 | 16.0 ± 0.7 | ||||
| E2 | 23.8 ± 0.4 | 23.8 ± 0.5 | 22.7 ± 0.9 | |||||
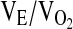 |
R | 19.9 ± 0.4 | 19.9 ± 0.5 | 17.9 ± 1.0 | x | x | 0.583 | |
| E1 | 16.5 ± 0.2 | 16.2 ± 0.3 | 16.1 ± 0.6 | |||||
| E2 | 17.8 ± 0.3 | 17.5 ± 0.3 | 18.3 ± 0.7 | |||||
| HR, bpm | R | 87.4 ± 1.3 | 88.1 ± 1.5 | 87.4 ± 3.0 | x | x | 0.402 | |
| E1 | 123.9 ± 1.4 | 126.5 ± 1.6 | 124.4 ± 3.2 | |||||
| E2 | 144.2 ± 1.7 | 148.0 ± 1.9 | 141.1 ± 3.8 | |||||
| RER | R | 0.84 ± 0.01 | 0.84 ± 0.01 | 0.78 ± 0.02 | x | x | 0.563 | |
| E1 | 0.85 ± 0.01 | 0.84 ± 0.01 | 0.86 ± 0.01 | |||||
| E2 | 0.93 ± 0.01 | 0.93 ± 0.01 | 0.94 ± 0.02 | |||||
Covariates are S = sex, BP = birthplace, and NAAP = Native American ancestry proportion. A covariate was retained in repeated measures ANCOVA when it had an independent effect on an exercise variable (p ≤ 0.05) or as indicated in the text.
Values are from R through two-levels of submaximal exercise (E1 and E2).
STPD, standard temperature, pressure, dry; VE, expired ventilation; HR, heart rate; RER, respiratory exchange rate.
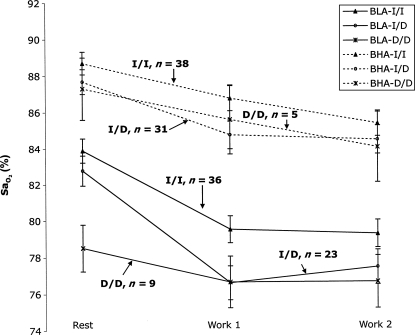
FIG. 1.
Mean 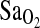 shown across the three stages of submaximal exercise: rest,k work1, and work2, for ACE genotype and groups BLA and BHA. Subjects with the I/I genotype have higher mean
shown across the three stages of submaximal exercise: rest,k work1, and work2, for ACE genotype and groups BLA and BHA. Subjects with the I/I genotype have higher mean 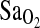 than either I/D or D/D individuals for both BLA and BHA groupings.
than either I/D or D/D individuals for both BLA and BHA groupings.
To more fully explore the hypothesis that the I-allele effect on 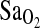 works through an increased HVR, we ran multivariate models with resting and exercise
works through an increased HVR, we ran multivariate models with resting and exercise 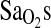 as dependent variables and ACE genotype, study group, and HVR as independent variables. The results of these statistical models were similar for all
as dependent variables and ACE genotype, study group, and HVR as independent variables. The results of these statistical models were similar for all 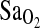 outcome measures, including resting and exercise measures. To simplify presentation, only results for
outcome measures, including resting and exercise measures. To simplify presentation, only results for 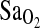 measured at work level 1 are given in Table 5. In addition, ACE was entered as a dichotomous variable comparing the I/I genotype effect against the combined I/D + D/D genotypes. This is justified based on the results presented in Table 4 and Fig. 1. Model 1 shows significant independent effects of birthplace (i.e., BHA vs. BLA), ACE genotype, and HVR on the
measured at work level 1 are given in Table 5. In addition, ACE was entered as a dichotomous variable comparing the I/I genotype effect against the combined I/D + D/D genotypes. This is justified based on the results presented in Table 4 and Fig. 1. Model 1 shows significant independent effects of birthplace (i.e., BHA vs. BLA), ACE genotype, and HVR on the 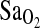 , explaining about 52% of the total variance. All possible two- and three-way interactions among independent variables were tested, but none of these were significant. As expected, the group effect on the
, explaining about 52% of the total variance. All possible two- and three-way interactions among independent variables were tested, but none of these were significant. As expected, the group effect on the 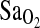 was quite large, accounting for most of the variance. Also as expected, higher HVR predicted higher
was quite large, accounting for most of the variance. Also as expected, higher HVR predicted higher 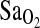 . However, independent of these effects, ACE I/I genotype was associated with a 2.3 percentage point higher
. However, independent of these effects, ACE I/I genotype was associated with a 2.3 percentage point higher 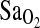 compared to the combined I/D and D/D genotypes. Models 2 and 3 were run without the ACE and HVR variables, respectively. The ACE genotype and the HVR each explained approximately the same amount of the variance in
compared to the combined I/D and D/D genotypes. Models 2 and 3 were run without the ACE and HVR variables, respectively. The ACE genotype and the HVR each explained approximately the same amount of the variance in 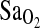 , that is, ∼4%. However, the most important consideration is that the elimination of either ACE (model 2) or HVR (model 3) from a statistical model did not greatly modify the effect of the remaining independent variables when compared to model 1. Thus, ACE genotype and HVR have independent effects on
, that is, ∼4%. However, the most important consideration is that the elimination of either ACE (model 2) or HVR (model 3) from a statistical model did not greatly modify the effect of the remaining independent variables when compared to model 1. Thus, ACE genotype and HVR have independent effects on 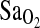 , and there is no direct evidence that the ACE genotype works through the HVR to affect the
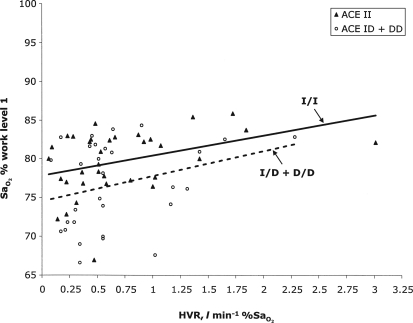
, and there is no direct evidence that the ACE genotype works through the HVR to affect the 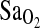 . This can also be seen in Fig. 2, which shows higher overall
. This can also be seen in Fig. 2, which shows higher overall 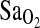 in I/I individuals, but a similar positive relationship between HVR and
in I/I individuals, but a similar positive relationship between HVR and 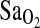 within genotypes.
within genotypes.
Table 5.
Multivariate Models Testing for Effects on the 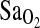 Measured During Submaximal Exercise at 4338 m
Measured During Submaximal Exercise at 4338 m
| Model 1 | Model 2 | Model 3 | ||||
|---|---|---|---|---|---|---|
| Variable | β | p-value | β | p-value | β | p-value |
| BHA vs. BLA | −7.62a | <0.001 | −7.61a | <0.001 | −7.65a | <0.001 |
| HVR | 2.61 | <0.001 | 2.80 | <0.001 | — | <0.001 |
| ACE | 2.33b | <0.001 | — | <0.001 | 2.42b | <0.001 |
| R2 = 0.52 | R2 = 0.48 | R2 = 0.48 | ||||
β-value represents the difference in mean 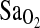 (i.e., lower
(i.e., lower 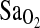 ) for BLA subjects tested at 4338 m compared with BHA subjects.
) for BLA subjects tested at 4338 m compared with BHA subjects.
β-value represents the difference in mean 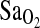 for I/I subjects compared with combined I/D and D/D subjects.
for I/I subjects compared with combined I/D and D/D subjects.
BHA, high altitude birth; BLA, low altitude birth; HVR, hypoxic ventilatory response; ACE, angiotensin-converting enzyme; I, insertion; D, deletion.
FIG. 2.
The relationship between 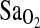 and HVR shown by genotype. I/I individuals are compared to I/D and D/D individuals combined.
and HVR shown by genotype. I/I individuals are compared to I/D and D/D individuals combined.
Discussion
The results of this study show an association of ACE I/I genotype with higher resting and submaximal exercise 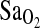 in two groups of Peruvians who differed by place of birth and who were tested at 4338 m altitude (p = 0.008). In both study groups, individuals of I/I genotype maintained a ∼2.3 percentage point higher
in two groups of Peruvians who differed by place of birth and who were tested at 4338 m altitude (p = 0.008). In both study groups, individuals of I/I genotype maintained a ∼2.3 percentage point higher 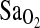 , explaining about 4% of the total variance in resting and exercise values. This finding is consistent with a previous cohort study that measured
, explaining about 4% of the total variance in resting and exercise values. This finding is consistent with a previous cohort study that measured 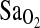 over many days in trekkers of European ancestry to >5000 meters in the Himalaya (Woods et al., 2002). In that study, individuals of I/I genotype who ascended to above 5000 m in 12 days were able to maintain higher
over many days in trekkers of European ancestry to >5000 meters in the Himalaya (Woods et al., 2002). In that study, individuals of I/I genotype who ascended to above 5000 m in 12 days were able to maintain higher 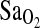 , especially as altitude increased. A similar trend was evident, but not significant, in a slower ascent group who trekked over 18.5 days. In contrast, a study by Patel el al. (2003) showed no significant association of the I-allele or I/I genotype with higher
, especially as altitude increased. A similar trend was evident, but not significant, in a slower ascent group who trekked over 18.5 days. In contrast, a study by Patel el al. (2003) showed no significant association of the I-allele or I/I genotype with higher 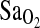 when subjects were briefly exposed to a fractional inspired oxygen concentration (
when subjects were briefly exposed to a fractional inspired oxygen concentration (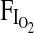 ) of 0.125 in the laboratory. Again, a nonsignificant trend for higher
) of 0.125 in the laboratory. Again, a nonsignificant trend for higher 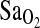 was evident in I/I study subjects.
was evident in I/I study subjects.
The results of the current study extend these previous findings in a number of important ways. First, the association of I/I genotype with higher 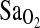 persisted even after statistical control for ancestry (NAAP). This control step accounts for the possibility of spurious association due to population stratification, a problem that is well recognized in gene association studies (e.g., (Kittles et al., 2002). Control for stratification allows strong inference that the ACE I/D locus is either functionally related to
persisted even after statistical control for ancestry (NAAP). This control step accounts for the possibility of spurious association due to population stratification, a problem that is well recognized in gene association studies (e.g., (Kittles et al., 2002). Control for stratification allows strong inference that the ACE I/D locus is either functionally related to 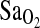 or in close linkage disequilibrium (LD) with a true causal locus affecting
or in close linkage disequilibrium (LD) with a true causal locus affecting 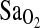 at HA. When alleles at two or more loci are inherited together they are in LD (for a review, see Ardlie et al., 2002). This is in contrast to linkage equilibrium, where there is no association of alleles between loci. Therefore, if the ACE I/D locus is in LD with the causal locus, the I-allele may always be inherited with the allelic variant at the causal locus that increases
at HA. When alleles at two or more loci are inherited together they are in LD (for a review, see Ardlie et al., 2002). This is in contrast to linkage equilibrium, where there is no association of alleles between loci. Therefore, if the ACE I/D locus is in LD with the causal locus, the I-allele may always be inherited with the allelic variant at the causal locus that increases 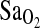 , and the D-allele may always be inherited with the allelic variant at the causal locus that does not increase
, and the D-allele may always be inherited with the allelic variant at the causal locus that does not increase 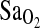 . In addition, it is also possible that the ACE I/D locus acts synergistically with another as yet unidentified unlinked locus or loci. It has been shown that polymorphic loci in the RAAS act synergistically with one another in the pathogenesis of disease (Qi et al., 2007). Moreover, the ACE I/D polymorphism has been shown to act in such a manner with genes outside the RAAS (Ryu et al., 2002; Castellano et al., 2003; Wang et al., 2006).
. In addition, it is also possible that the ACE I/D locus acts synergistically with another as yet unidentified unlinked locus or loci. It has been shown that polymorphic loci in the RAAS act synergistically with one another in the pathogenesis of disease (Qi et al., 2007). Moreover, the ACE I/D polymorphism has been shown to act in such a manner with genes outside the RAAS (Ryu et al., 2002; Castellano et al., 2003; Wang et al., 2006).
The second important result is that the association of I/I genotype with higher 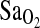 was similar in both BLA and BHA subjects, and these two study groups differed greatly in their length of exposure to hypoxia. BLA subjects were nonacclimatized, whereas BHA subjects were fully acclimatized and had lived their entire lives at HA. Thus, the I-allele effect appears to be independent of acclimatization status and penetrant, whether or not subjects have experienced a lifelong or developmental exposure to hypoxia. The finding that the I-allele effect is independent of acclimatization state is relevant to the hypothesis of Woods et al. (2002), who suggested that the I-allele benefit is short lived and more important during early-stage acclimatization to HA. According to Woods et al., this could be due to the normalization of the plasma aldosterone concentration to plasma renin activity ratio (PAC/PRA) that occurs after 12 to 20 days at HA. Our results clearly do not support this hypothesis. Rather, it is possible that the Woods et al. study was underpowered to detect a 1% to 2% difference in
was similar in both BLA and BHA subjects, and these two study groups differed greatly in their length of exposure to hypoxia. BLA subjects were nonacclimatized, whereas BHA subjects were fully acclimatized and had lived their entire lives at HA. Thus, the I-allele effect appears to be independent of acclimatization status and penetrant, whether or not subjects have experienced a lifelong or developmental exposure to hypoxia. The finding that the I-allele effect is independent of acclimatization state is relevant to the hypothesis of Woods et al. (2002), who suggested that the I-allele benefit is short lived and more important during early-stage acclimatization to HA. According to Woods et al., this could be due to the normalization of the plasma aldosterone concentration to plasma renin activity ratio (PAC/PRA) that occurs after 12 to 20 days at HA. Our results clearly do not support this hypothesis. Rather, it is possible that the Woods et al. study was underpowered to detect a 1% to 2% difference in 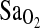 in their “slow ascenders,” given a sample size of only n = 40 (14 D/D, 22 I/D, and 4 I/I), compared with the n = 142 in this study (14 D/D, 54 I/D, and 74 I/I). For example, based on
in their “slow ascenders,” given a sample size of only n = 40 (14 D/D, 22 I/D, and 4 I/I), compared with the n = 142 in this study (14 D/D, 54 I/D, and 74 I/I). For example, based on 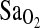 mean and variance values from the current study, we calculate a power of no more than 24% to detect a 2% difference in
mean and variance values from the current study, we calculate a power of no more than 24% to detect a 2% difference in 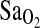 between I/I and either of the other two genotypes in the Woods et al. study. Obviously, this is an important limitation to consider for future studies with European populations, given the low frequency of the I-allele compared to the present Native American group. Additionally, there is high variance in
between I/I and either of the other two genotypes in the Woods et al. study. Obviously, this is an important limitation to consider for future studies with European populations, given the low frequency of the I-allele compared to the present Native American group. Additionally, there is high variance in 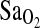 measured by pulse oximetry, and we attribute only a relatively small effect of the ACE locus on
measured by pulse oximetry, and we attribute only a relatively small effect of the ACE locus on 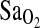 . Alternatively, the difference between our study and the negative findings in the Woods et al. and Patel et al. studies could be due to differences in the patterns of LD between populations that affect the strength of the I-allele–
. Alternatively, the difference between our study and the negative findings in the Woods et al. and Patel et al. studies could be due to differences in the patterns of LD between populations that affect the strength of the I-allele–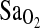 association. However, this explanation seem less likely, given the clear trend to higher
association. However, this explanation seem less likely, given the clear trend to higher 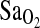 that was shown in both aforementioned studies with European groups.
that was shown in both aforementioned studies with European groups.
The similarity between BHA and BLA subject groups is also important to consider with respect to the issue of gene–environment (GE) interaction and developmental effects. In this particular case, apparently no GE interaction is taking place over developmental time to determine 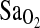 at HA, as both of our study groups show the same I-allele effect. We surmise from this that the ACE I/D polymorphism is not involved in differential regulation of gene expression during growth and development with lifelong consequences on the adult phenotype, at least not for the
at HA, as both of our study groups show the same I-allele effect. We surmise from this that the ACE I/D polymorphism is not involved in differential regulation of gene expression during growth and development with lifelong consequences on the adult phenotype, at least not for the 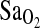 phenotype. This does not mean that this is generally the case, and indeed there are examples in the broader literature of GE interaction taking place over developmental time frames (e.g. Kajantie et al., 2004). However, in the HA literature, our study represents the first direct test for interaction between a gene with a known phenotypic effect and lifelong environmental exposure to altitude or sea-level conditions.
phenotype. This does not mean that this is generally the case, and indeed there are examples in the broader literature of GE interaction taking place over developmental time frames (e.g. Kajantie et al., 2004). However, in the HA literature, our study represents the first direct test for interaction between a gene with a known phenotypic effect and lifelong environmental exposure to altitude or sea-level conditions.
Despite a strong association of the ACE genotype with 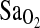 , we detected no association of the ACE genotype with the isocapnic HVR. More importantly, there was no evidence, via multivariate modeling, that the ACE I-allele works through increased HVR to increase
, we detected no association of the ACE genotype with the isocapnic HVR. More importantly, there was no evidence, via multivariate modeling, that the ACE I-allele works through increased HVR to increase 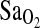 , as previously hypothesized (Patel et al., 2003; Swenson, 2004). The ACE–HVR–
, as previously hypothesized (Patel et al., 2003; Swenson, 2004). The ACE–HVR–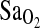 hypothesis derives in part from animal studies that have shown that the peripheral chemoreceptors contain angiotensin receptors with potential effects on the regulation of VE (Allen, 1998; Paton and Kasparov, 1999). But, support for this mechanism is based only on the results from Patel et al. (2003), who showed an increased VE response in I/I individuals when subjects were exercised in hypoxia and normoxia. However, some consideration is warranted, as the exertional VE is not the same thing as the HVR. HVR is the increase in VE per unit decrease in
hypothesis derives in part from animal studies that have shown that the peripheral chemoreceptors contain angiotensin receptors with potential effects on the regulation of VE (Allen, 1998; Paton and Kasparov, 1999). But, support for this mechanism is based only on the results from Patel et al. (2003), who showed an increased VE response in I/I individuals when subjects were exercised in hypoxia and normoxia. However, some consideration is warranted, as the exertional VE is not the same thing as the HVR. HVR is the increase in VE per unit decrease in 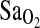 , and when measured under isocapnic conditions the measure evaluates peripheral chemosensitivity and the peripheral control of breathing. In contrast, exercise hyperpnea is measured under poikilocapnic conditions and has a much more complex regulation that is not well understood (Dempsey et al., 1995). Thus, the present study provides no support for an HVR-mediated mechanism to explain higher
, and when measured under isocapnic conditions the measure evaluates peripheral chemosensitivity and the peripheral control of breathing. In contrast, exercise hyperpnea is measured under poikilocapnic conditions and has a much more complex regulation that is not well understood (Dempsey et al., 1995). Thus, the present study provides no support for an HVR-mediated mechanism to explain higher 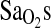 in individuals of I/I genotype. If it is true that control of breathing does not explain higher
in individuals of I/I genotype. If it is true that control of breathing does not explain higher 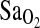 , then perhaps other central cardiopulmonary effects of ACE underlie the I-allele–
, then perhaps other central cardiopulmonary effects of ACE underlie the I-allele–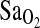 association. For example, angiotensin II modulates hypoxia-induced pulmonary vasoconstriction (Kiely et al., 1995), and differences in circulating ACE could affect ventilation–perfusion (V/Q) relationships within the lung (Woods et al., 2002; Mortimer et al., 2004). However, given the lack of information in the literature on this topic, such mechanisms remain speculative.
association. For example, angiotensin II modulates hypoxia-induced pulmonary vasoconstriction (Kiely et al., 1995), and differences in circulating ACE could affect ventilation–perfusion (V/Q) relationships within the lung (Woods et al., 2002; Mortimer et al., 2004). However, given the lack of information in the literature on this topic, such mechanisms remain speculative.
We also failed to detect an association of the ACE genotype with 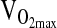 at 4338 m, despite a number of positive reports from studies conducted at sea level. For example, a study by Hagberg et al. (2002) showed 23% higher
at 4338 m, despite a number of positive reports from studies conducted at sea level. For example, a study by Hagberg et al. (2002) showed 23% higher 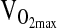 in postmenopausal women of I/I versus D/D genotype. A study by Rankinen et al. (2000) showed no association of ACE with baseline
in postmenopausal women of I/I versus D/D genotype. A study by Rankinen et al. (2000) showed no association of ACE with baseline 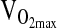 , but showed larger increases in
, but showed larger increases in 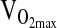 after training in subjects of D/D genotype. In the Rankinen et al. study, the effect was only evident in subjects of European ancestry, but not in subjects with significant African ancestry. This sort of ancestry dependence again raises the issue of population stratification. It is noteworthy that we did not detect an association between ACE and
after training in subjects of D/D genotype. In the Rankinen et al. study, the effect was only evident in subjects of European ancestry, but not in subjects with significant African ancestry. This sort of ancestry dependence again raises the issue of population stratification. It is noteworthy that we did not detect an association between ACE and 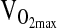 even after controlling for NAAP. By doing so, we accounted for the possibility that stratification is hiding a true association. Nevertheless, our study was not designed to specifically address the hypothesis of ACE genotype effects on
even after controlling for NAAP. By doing so, we accounted for the possibility that stratification is hiding a true association. Nevertheless, our study was not designed to specifically address the hypothesis of ACE genotype effects on 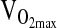 , and so we urge caution in the interpretation of this negative finding. In particular,
, and so we urge caution in the interpretation of this negative finding. In particular, 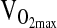 is affected by a large number of social and environmental factors, especially physical activity level. We were unable to control for physical activity level (i.e., training) nor were we able to impose a training program as was the case in the study by Rankinen et al. (2000).
is affected by a large number of social and environmental factors, especially physical activity level. We were unable to control for physical activity level (i.e., training) nor were we able to impose a training program as was the case in the study by Rankinen et al. (2000).
Given the Quechua origins and long-term HA ancestry of our study population, the results of this study should also be considered briefly from an evolutionary or adaptive perspective. If there is an I-allele benefit at HA, then the question becomes whether natural selection has increased the I-allele frequency in native groups like the Quechua. Rupert et al. (1999) were the first to document the relatively high I-allele frequency in Quechua (0.72), and our study now documents practically the same frequency (0.71, Table 1).However, the Quechua I-allele frequency does not appear to be increased relative to all other Native American populations. Among published Native American population frequencies for the ACE Indel polymorphism, the I-allele ranges from a low in Alaskan Natives (Eskimos, Native Amerindians, and Aleuts) of 0.45 to a high of 1.0 in the Ache of eastern Paraguay. Moreover, many low altitude Native Americans exhibit I-allele frequencies greater than that observed in the Quechua. However, it should be noted that the allele frequency differences observed among the Native American populations may be the result of different founder events, due to differences in genetic drift based on varying population size or a combination of the two. Perhaps the relatively high ACE-I frequency in Amerindians was due to a founder event and then later facilitated the rapid expansion into the Andes, as originally speculated by Rupert et al. (1999). If true, this would imply that subsequent selective pressure was not sufficient to increase the frequency further, but again this is speculation only.
In comparison to Asian HA populations in the Himalaya, the I-allele frequency has been documented at 0.67 in highland native Ladakhis from Northern India (Qadar Pasha et al., 2001) and ranging from 0.51 to 0.64 in native Tibetans from Lhasa, depending on whether subjects were hypertensive (Gesang et al., 2002). These frequencies are slightly higher than the mean European, African, or Asian I-allele frequencies in the literature, but there is substantial variation within regionally defined groups worldwide (Brutsaert and Parra, 2006). Indeed, in many populations from sea level, the I-allele frequency is as high or higher than the frequencies reported above for HA natives. For example, in the study by Qadar Pasha et al. (2001), the highest I-allele frequency (0.73, n = 20) was actually observed in a study population of lowland origins who were first generation migrants to the Ladakhi region. Thus, from the existing data, natural selection cannot be conclusively evoked as the explanation for the relatively high I-allele frequency in Quechua. This is similar to the conclusion reached previously by Rupert et al. (1999). The possibility of a more complex involvement of the ACE gene in human evolutionary history at HA remains. For example, it is possible that the ACE I/D polymorphism is not the locus under selection, but rather is in LD with an HA-selected locus in some populations such as the Quechua, but not in other populations. Moreover, one study indicates an I-allele disadvantage with respect to HAPE. This study of Kyrgyz highlanders revealed threefold higher frequency of the I/I genotype in subjects with HAPE (Aldashev et al., 2002). In addition, the highland Kyrgyz actually had lower I-allele frequency (0.56, n = 87) compared with a Bishkek lowland control group, for which the I-allele frequency was 0.65 (n = 276). However, it should be noted that high altitude performance and high altitude disease are two unrelated phenomena that are not mutually exclusive. The I-allele may confer a performance benefit at HA, but it may also render the carrier more susceptible to certain diseases, such as HAPE. This could in fact explain why the I-allele is not found at or near fixation in HA populations if it both grants selective advantage as well as disadvantage.
The phenotypic effects of the ACE I/D polymorphism are still poorly understood, and it is possible (perhaps likely) that the adaptive process involves selection of alleles at multiple loci (i.e., haplotypes) with beneficial effects on the phenotype. Although we did not examine other loci in the ACE gene, such future work will most likely reveal the complex selection history of this gene.
Conclusions
In two groups of Peruvians who differed in altitude at birth and who were tested at 4388-m altitude, this study found that the ACE I/I genotype is associated with higher resting and submaximal exercise 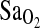 . Moreover, the I-allele effect has little to do with the acclimatization state of subjects. In addition, the ACE genotype was not associated with the isocapnic HVR.
. Moreover, the I-allele effect has little to do with the acclimatization state of subjects. In addition, the ACE genotype was not associated with the isocapnic HVR.
Acknowledgments
We would like to thank the volunteers who gave their time, sweat, and DNA for this research. This work was supported in part by grants from the National Science Foundation BCS-0129377 to T. D. Brutsaert and the National Institutes of Health (HG002154) to M. D. Shriver.
References
- Aldashev A.A. Sarybaev A.S. Sydykov A.S. Kalmyrzaev B.B. Kim E.V. Mamanova L.B. Maripov R. Kojonazarov B.K. Mirrakhimov M.M. Wilkins M.R. Morrell N.W. Characterization of high-altitude pulmonary hypertension in the Kyrgyz: association with angiotensin-converting enzyme genotype. Am. J. Respir. Crit. Care Med. 2002;166:1396–402. doi: 10.1164/rccm.200204-345OC. Epub 2002 Aug 28. [DOI] [PubMed] [Google Scholar]
- Allen A.M. Angiotensin AT1 receptor-mediated excitation of rat carotid body chemoreceptor afferent activity. J. Physiol. 1998;510(Pt. 3):773–781. doi: 10.1111/j.1469-7793.1998.773bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie K.G. Kruglyak L. Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Bartsch P. Maggiorini M. Schobersberger W. Shaw S. Rascher W. Girard J. Weidmann P. Oelz O. Enhanced exercise-induced rise of aldosterone and vasopressin preceding mountain sickness. J. Appl. Physiol. 1991;71:136–143. doi: 10.1152/jappl.1991.71.1.136. [DOI] [PubMed] [Google Scholar]
- Bland J.M. Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- Bonilla C. Shriver M.D. Parra E.J. Jones A. Fernandez J.R. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York City. Hum. Genet. 2004;115:57–68. doi: 10.1007/s00439-004-1125-7. [DOI] [PubMed] [Google Scholar]
-
Brutsaert T. Parra E. Shriver M. Gamboa A. Palacios J. Rivera M. Rodriquez I. Leon-Velarde F. Spanish genetic admixture is associated with larger
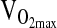 decrement from sea level to 4.338 m in Peruvan Quechua. J. Appl. Physiol. 2003;95:519–528. doi: 10.1152/japplphysiol.01088.2002. [DOI] [PubMed] [Google Scholar]
decrement from sea level to 4.338 m in Peruvan Quechua. J. Appl. Physiol. 2003;95:519–528. doi: 10.1152/japplphysiol.01088.2002. [DOI] [PubMed] [Google Scholar] - Brutsaert T. Parra E. Shriver M. Gamboa A. Palacios J. Rivera M. Rodriquez I. Leon-Velarde F. Effects of birth place and individual admixture on lung volume and exercise phenotypes of Peruvian Quechua. Am. J. Phys. Anthropol. 2004;123:390–398. doi: 10.1002/ajpa.10319. [DOI] [PubMed] [Google Scholar]
- Brutsaert T.D. Parra E.J. What makes a champion? Explaining variation in human athletic performance. Respir. Physiol. Neurobiol. 2006;151:109–123. doi: 10.1016/j.resp.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Brutsaert T.D. Parra E.J. Shriver M.D. Gamboa A. Rivera M. Leon-Velarde F. Ancestry explains the blunted ventilatory response to sustained hypoxia and lower exercise ventilation of Quechua altitude natives. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;289:225–234. doi: 10.1152/ajpregu.00105.2005. [DOI] [PubMed] [Google Scholar]
- Castellano M. Glorioso N. Cusi D. Sarzani R. Fabris B. Opocher G. Zoccali C. Golin R. Veglio F. Volpe M. Mantero F. Fallo F. Rossi G.P. Barlassina C. Tizzoni L. Filigheddu F. Giacche M. Rossi F. Genetic polymorphism of the renin-angiotensin-aldosterone system and arterial hypertension in the Italian population: the GENIPER Project. J. Hypertens. 2003;21:1853–1860. doi: 10.1097/00004872-200310000-00012. [DOI] [PubMed] [Google Scholar]
- Costerousse O. Allegrini J. Lopez M. Alhenc-Gelas F. Angiotensin I-converting enzyme in human circulating mononuclear cells: genetic polymorphism of expression in T-lymphocytes. Biochem. J. 1993;290(Pt. 1):33–40. doi: 10.1042/bj2900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danser A.H. Schalekamp M.A. Bax W.A. van den Brink A.M. Saxena P.R. Riegger G.A. Schunkert H. Angiotensin-converting enzyme in the human heart. Effect of the deletion/insertion polymorphism. Circulation. 1995;92:1387–1388. doi: 10.1161/01.cir.92.6.1387. [DOI] [PubMed] [Google Scholar]
- Dempsey J.A., editor; Forster H.V., editor; Ainsworth D.M., editor. Regulation of hyperpnea, hyperventilation, and respiratory muscle recruitment during exercise. Lung Biology in Health and Disease: Regulation of Breathing. Marcel Dekker; New York: 1995. [Google Scholar]
- Durnin J.V. Womersley J. Body fat assessed from total body density and its estimation from skinfold thickness: measurements on 481 men and women aged from 16 to 72 years. Br. J. Nutr. 1974;32:77–97. doi: 10.1079/bjn19740060. [DOI] [PubMed] [Google Scholar]
- Evans A.E. Poirier O. Kee F. Lecerf L. McCrum E. Falconer T. Crane J. O'Rourke D.F. Cambien F. Polymorphisms of the angiotensin-converting-enzyme gene in subjects who die from coronary heart disease. Q. J. Med. 1994;87:211–214. [PubMed] [Google Scholar]
- Gesang L. Liu G. Cen W. Qiu C. Zhuoma C. Zhuang L. Ren D. Pincuo Z. Chan Y. Angiotensin-converting enzyme gene polymorphism and its association with essential hypertension in a Tibetan population. Hypertens Res. 2002;25:481–485. doi: 10.1291/hypres.25.481. [DOI] [PubMed] [Google Scholar]
- Hackett P.H. Rennie D. Hofmeister S.E. Grover R.F. Grover E.B. Reeves J.T. Fluid retention and relative hypoventilation in acute mountain sickness. Respiration. 1982;43:321–329. doi: 10.1159/000194501. [DOI] [PubMed] [Google Scholar]
- Hagberg J.M. McCole S.D. Brown M.D. Ferrell R.E. Wilund K.R. Huberty A. Douglass L.W. Moore G.E. ACE insertion/deletion polymorphism and submaximal exercise hemodynamics in postmenopausal women. J. Appl. Physiol. 2002;92:1083–1088. doi: 10.1152/japplphysiol.00135.2001. [DOI] [PubMed] [Google Scholar]
- Hanis C.L. Chakraborty R. Ferrell R.E. Schull W.J. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am. J. Phys. Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- Hoggart C.J. Parra E.J. Shriver M.D. Bonilla C. Kittles R.A. Clayton D.G. McKeigue P.M. Control of confounding of genetic associations in stratified populations. Am. J. Hum. Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajantie E. Rautanen A. Kere J. Andersson S. Yliharsila H. Osmond C. Barker D.J. Forsen T. Eriksson J. The effects of the ACE gene insertion/deletion polymorphism on glucose tolerance and insulin secretion in elderly people are modified by birth weight. J. Clin. Endocrinol. Metab. 2004;89:5738–5741. doi: 10.1210/jc.2004-0492. [DOI] [PubMed] [Google Scholar]
- Kiely D.G. Cargill R.I. Lipworth B.J. Acute hypoxic pulmonary vasoconstriction in man is attenuated by type I angiotensin II receptor blockade. Cardiovasc Res. 1995;30:875–880. [PubMed] [Google Scholar]
- Kittles R.A. Chen W. Panguluri R.K. Ahaghotu C. Jackson A. Adebamowo C.A. Griffin R. Williams T. Ukoli F. Adams-Campbell L. Kwagyan J. Isaacs W. Freeman V. Dunston G.M. CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum. Genet. 2002;110:553–560. doi: 10.1007/s00439-002-0731-5. [DOI] [PubMed] [Google Scholar]
- Milledge J.S. Catley D.M. Williams E.S. Withey W.R. Minty B.D. Effect of prolonged exercise at altitude on the renin–aldosterone system. J. Appl. Physiol. 1983;55:413–418. doi: 10.1152/jappl.1983.55.2.413. [DOI] [PubMed] [Google Scholar]
- Montgomery H.E. Marshall R. Hemingway H. Myerson S. Clarkson P. Dollery C. Hayward M. Holliman D.E. Jubb M. World M. Thomas E.L. Brynes A.E. Saeed N. Barnard M. Bell J.D. Prasad K. Rayson M. Talmud P.J. Humphries S.E. Human gene for physical performance. Nature. 1998;393:221–222. doi: 10.1038/30374. [DOI] [PubMed] [Google Scholar]
- Mortimer H. Patel S. Peacock A.J. The genetic basis of high-altitude pulmonary oedema. Pharmacol. Ther. 2004;101:183–192. doi: 10.1016/j.pharmthera.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Mou X.B. Howard L.S. Robbins P.A. A protocol for determining the shape of the ventilatory response to hypoxia in humans. Respir. Physiol. 1995;101:139–143. doi: 10.1016/0034-5687(95)00027-b. [DOI] [PubMed] [Google Scholar]
- Muller D.N. Bohlender J. Hilgers K.F. Dragun D. Costerousse O. Menard J. Luft F.C. Vascular angiotensin-converting enzyme expression regulates local angiotensin II. Hypertension. 1997;29:98–104. doi: 10.1161/01.hyp.29.1.98. [DOI] [PubMed] [Google Scholar]
- Patel S. Woods D.R. Macleod N.J. Brown A. Patel K.R. Montgomery H.E. Peacock A.J. Angiotensin-converting enzyme genotype and the ventilatory response to exertional hypoxia. Eur. Respir. J. 2003;22:755–760. doi: 10.1183/09031936.03.00086402. [DOI] [PubMed] [Google Scholar]
- Paton J.F. Kasparov S. Differential effects of angiotensin II on cardiorespiratory reflexes mediated by nucleus tractus solitarii—a microinjection study in the rat. J. Physiol. 1999;521(Pt. 1):213–225. doi: 10.1111/j.1469-7793.1999.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff C.L. Parra E.J. Bonilla C. Hiester K. McKeigue P.M. Kamboh M.I. Hutchinson R.G. Ferrell R.E. Boerwinkle E. Shriver M.D. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am. J. Hum. Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadar Pasha M.A. Khan A.P. Kumar R. Grover S.K. Ram R.B. Norboo T. Srivastava K.K. Selvamurthy W. Brahmachari S.K. Angiotensin converting enzyme insertion allele in relation to high altitude adaptation. Ann. Hum. Genet. 2001;65:531–536. doi: 10.1017/S0003480001008879. [DOI] [PubMed] [Google Scholar]
- Qi Y. Niu W. Zhu T. Zhou W. Qiu C. Synergistic effect of the genetic polymorphisms of the renin–angiotensin–aldosterone system on high-altitude pulmonary edema: a study from Qinghai–Tibet altitude. Eur. J. Epidemiol. 2007 doi: 10.1007/s10654-007-9208-0. [DOI] [PubMed] [Google Scholar]
- Rankinen T. Perusse L. Gagnon J. Chagnon Y.C. Leon A.S. Skinner J.S. Wilmore J.H. Rao D.C. Bouchard C. Angiotensin–converting enzyme ID polymorphism and fitness phenotype in the HERITAGE Family Study. J. Appl. Physiol. 2000;88:1029–1035. doi: 10.1152/jappl.2000.88.3.1029. [DOI] [PubMed] [Google Scholar]
- Rigat B. Hubert C. Alhenc-Gelas F. Cambien F. Corvol P. Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert J.L. Devine D.V. Monsalve M.V. Hochachka P.W. Angiotensin-converting enzyme (ACE) alleles in the Quechua, a high altitude South American native population. Ann. Hum. Biol. 1999;26:375–380. doi: 10.1080/030144699282688. [DOI] [PubMed] [Google Scholar]
- Rupert J.L. Koehle M.S. Evidence for a genetic basis for altitude-related illness. High Alt. Med. Biol. 2006;7:150–167. doi: 10.1089/ham.2006.7.150. [DOI] [PubMed] [Google Scholar]
- Ryu S.K. Cho E.Y. Park H.Y. Im E.K. Jang Y.S. Shin G.J. Shim W.H. Cho S.Y. Renin–angiotensin–aldosterone system (RAAS) gene polymorphism as a risk factor of coronary in-stent restenosis. Yonsei Med. J. 2002;43:461–472. doi: 10.3349/ymj.2002.43.4.461. [DOI] [PubMed] [Google Scholar]
- Severinghaus J.W. Simple, accurate equations for human blood O2 dissociation computations. J. Appl. Physiol. 1979;46:599–602. doi: 10.1152/jappl.1979.46.3.599. [DOI] [PubMed] [Google Scholar]
- Shriver M.D. Parra E.J. Dios S. Bonilla C. Norton H. Jovel C. Pfaff C. Jones C. Massac A. Cameron N. Baron A. Jackson T. Argyropoulos G. Jin L. Hoggart C.J. McKeigue P.M. Kittles R.A. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum. Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- Siri W.E. Body composition from fluid spaces and density: analysis of methods. In: Brozek J., editor; Henschel A., editor. Techniques for Measuring Body Composition. National Academy of Sciences, National Research Council; Washington, DC: 1961. pp. 223–224. [Google Scholar]
- Swenson E.R. ACE inhibitors and high altitude. High Alt. Med. Biol. 2004;5:92–94. doi: 10.1089/152702904322963762. [DOI] [PubMed] [Google Scholar]
- Tsianos G. Eleftheriou K.I. Hawe E. Woolrich L. Watt M. Watt I. Peacock A. Montgomery H. Grant S. Performance at altitude and angiotensin I-converting enzyme genotype. Eur. J. Appl. Physiol. 2005;93:630–633. doi: 10.1007/s00421-004-1284-1. [DOI] [PubMed] [Google Scholar]
- Wang B. Jin F. Yang Z. Lu Z. Kan R. Li S. Zheng C. Wang L. The insertion polymorphism in angiotensin-converting enzyme gene associated with the APOE epsilon 4 allele increases the risk of late-onset Alzheimer disease. J. Mol. Neurosci. 2006;30:267–271. doi: 10.1385/JMN:30:3:267. [DOI] [PubMed] [Google Scholar]
- Woods D.R. Pollard A.J. Collier D.J. Jamshidi Y. Vassiliou V. Hawe E. Humphries S.E. Montgomery H.E. Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene and arterial oxygen saturation at high altitude. Am. J. Respir. Crit. Care Med. 2002;166:362–366. doi: 10.1164/rccm.2103060. [DOI] [PubMed] [Google Scholar]
- Zhang S. Robbins P.A. Methodological and physiological variability within the ventilatory response to hypoxia in humans. J. Appl. Physiol. 2000;88:1924–1932. doi: 10.1152/jappl.2000.88.5.1924. [DOI] [PubMed] [Google Scholar]
- Zhu X. Bouzekri N. Southam L. Cooper R.S. Adeyemo A. McKenzie C.A. Luke A. Chen G. Elston R.C. Ward R. Linkage and association analysis of angiotensin I-converting enzyme (ACE)-gene polymorphisms with ACE concentration and blood pressure. Am. J. Hum. Genet. 2001;68:1139–1148. doi: 10.1086/320104. [DOI] [PMC free article] [PubMed] [Google Scholar]