Abstract
Double-stranded DNA bacteriophages and their eukaryotic virus counterparts have twelve-fold head-tail connector assemblages embedded at a unique capsid vertex. This vertex is the site of assembly of the DNA packaging motor, and the connector has a central channel through which viral DNA passes during genome packaging and subsequent host infection. Crystal structures of connectors from different phages reveal either disordered residues or structured loops that project into the connector channel. Given the proximity to the translocating DNA substrate, these loops have been proposed to play a role in DNA packaging. Previous models have proposed structural motions in either the packaging ATPase or the connector channel loops as the driving force that translocates the DNA into the prohead. Here we mutate the channel loops of the Bacillus subtilis bacteriophage φ29 connector and show that these loops have no active role in translocation of DNA. Instead, they appear to have an essential function near the end of packaging, acting to retain the packaged DNA in the head in preparation for motor detachment and subsequent tail assembly and virion completion.
Keywords: bacteriophage phi29, DNA packaging, head-tail connector, virus assembly, portal protein
INTRODUCTION
During the assembly of tailed double-stranded DNA (dsDNA) bacteriophages and other dsDNA viruses, the viral chromosome is packaged into a preformed head shell (prohead) by an enzymatic-mechanical packaging motor.1,2 The motor packages the genome into the prohead to near-crystalline density, working against the entropic, electrostatic repulsion and bending energies that resist DNA compaction. In the dsDNA bacteriophages, the packaging motor is comprised of stacked rings of oligomeric components. In general, the catalytic component, a pentameric ATPase ring, binds to the dodecameric head-tail connector ring that is embedded in the portal head vertex to comprise the DNA packaging motor.2 (The head-tail connector is termed portal in several other dsDNA phages). In the Bacillus subtilis phage φ29, an additional ring of a viral-encoded oligomeric RNA (pRNA) bridges the capsid shell protein (gp8), connector (gp10), and the packaging ATPase (gp16) (Fig. 1)3–6. Upon completion of DNA packaging, the packaging ATPase, and in φ29 the pRNA, detach from the connector,1,2,7 temporarily leaving retention of the packaged DNA to the prohead before tail components attach to the connector to complete assembly of an infectious virion.
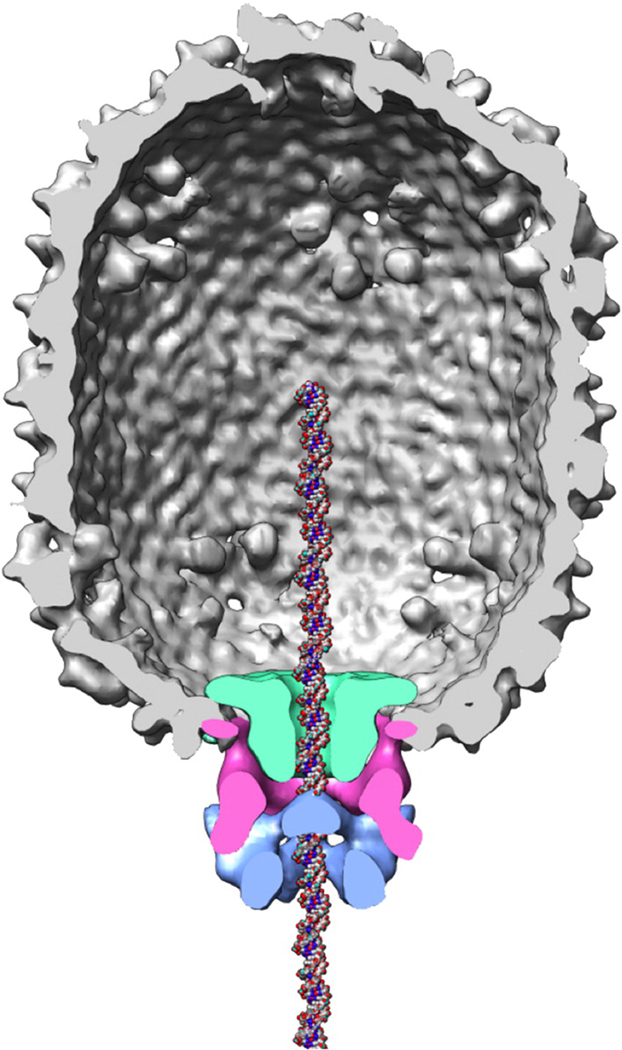
Figure 1.
Model of the φ29 prohead/DNA packaging motor complex. The molecular envelopes of the connector (green), prohead RNA (magenta) and packaging ATPase gp16 (blue) were determined using difference maps of various cryoEM 3-D reconstructions.4 A DNA molecule has been placed in the channel for reference. Figure courtesy of Marc Morais.
A central element of a phage dsDNA packaging motor is the dodecameric connector (Fig. 1). Solutions of connector crystal structures from the podovirus φ293,8 and the siphovirus SPP19 revealed commonalities in both structure and organization. Connectors are roughly cone shaped with a lower, narrow end protruding from the head shell and the central-helical and upper-crown regions embedded in the shell. The SPP1 structure has a set of loops positioned in the central region that protrude into the axial channel of the connector through which the DNA must pass during DNA packaging and ejection.9 This region is not resolved in the φ29 connector crystal structure,3,8 however cryoEM-3D reconstructions of the prohead show density in this region that extends into the channel.3,10 The positioning of these loops within the connector channel places them well within range for contact with the translocating DNA.9 A model has been proposed in which the force against the DNA that drives translocation may in part be generated by conformational changes in the connector that are induced by conformational changes in the ATPase.9 The connector channel loops are thought to play a key role in this mechanism.
Here we focused on dissecting the role of the connector channel loops in φ29 DNA translocation and virion production. We modeled the unresolved loop residues into the φ29 connector crystal structure, identifying likely contacts within each loop, between loops, and with phosphates of the translocating DNA. Residues having possible interactions were mutated, and proheads containing mutant connectors were produced. In vitro DNA packaging assays with mutant particles having modified or deleted channel loops showed that these changes did not significantly impact DNA translocation. However, the packaged heads with mutant connectors did not retain DNA during centrifugation and could not be matured to infectious virions. The inability of the particles with mutant connectors to retain packaged DNA suggests that the channel loops function as a clamp at the end of DNA packaging to prevent the loss of DNA while the motor is replaced by the tail components to complete phage assembly.
RESULTS
Modeling of the channel loops of the connector
To investigate the role of the channel loops in DNA packaging and virion production, disordered residues 229–246 (channel loop), 286–299 (C-terminus), and a segment of B-DNA were modeled into the connector crystal structure (Fig. 2).11 Secondary structure prediction suggested that most of the disordered loop residues participate in a coiled structure, with loop residues 235 to 239 predicted to form a low-confidence-rated α-helix, and C-terminal residues 287 to 299 predicted to form a high-confidence-rated α-helix (data not shown). Based on the secondary structure prediction, the disordered regions of the connector were first built in silico into a single connector protomer using Swiss-PdbViewer 12 and InsightII. The channel loop and the C-terminal helical structures were then optimized using the adopted basis Newton-Raphson (ABNR)13 method while holding the remaining connector coordinates fixed. The modeled regions on the first peptide chain of the dodecamer connector were then replicated to produce an additional eleven copies that were spaced by 30° rotations around the 12-fold axis of the connector. The residues of the disordered regions for the entire connector were then optimized by 100 steps of energy minimization using the ABNR method in CHARMM14 to remove close contacts. Finally, the entire connector was further minimized using the same method with DNA, crystal water and counter ions fixed.
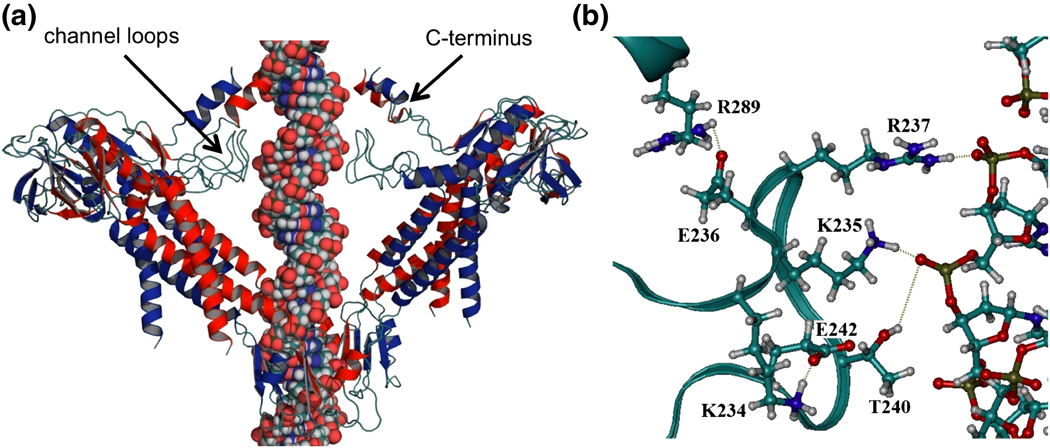
Figure 2.
Modeled structure of the φ29 connector with DNA. (a) The channel loops and C-terminus were modeled into the crystal structure of the connector.11 For clarity, four subunits of the dodecamer are illustrated, including the 1st, 2nd, 8th and 9th monomers. (b) Hydrogen bonds among loop residues (K234-E242), between loop residues (K235, R237, and T240) and DNA, and between the loop and C-terminus (E236-R289).
Modeling of the channel loops suggested the existence of electrostatic interactions between and within connector protomers (Fig. 2). The model predicted that the side chains of the residues K234, K235, and R237 from one protomer interact with the side chain OD2 of D241, the side chain OG1 of T240, and the carbonyl oxygens of M238 and V239 from the adjacent protomer, respectively. Within each protomer, the model predicted that residue K234 interacts with E242, and residue E236 interacts with R289 from the modeled C-terminal region. The putative K234/E242 interaction is supported by strict conservation of these two residues in all φ29 phage relatives (Table 1). Additionally, the channel loops were predicted to interact directly with the double-strand B-DNA in the connector channel (Fig. 2b). Specifically, the residues K235, R237 and T240 from one protomer form hydrogen bonds with the phosphates of one strand of DNA. Due to helical DNA symmetry, the same amino acid residues from the protomer on the opposite side of the connector channel makes similar contacts with the phosphates of the other strand of DNA at the same axial level of the connector (Fig. 2a).
Table 1.
Alignment of ϕ29 family connector sequences for unresolved residues in the channel loops of the ϕ29 connector crystal structure. Modeling suggests strictly conserved residues (grey) form an intramolecular contact within a connector protomer that support loop structure.
| ϕ29 | (228) |  |
| PZA | (228) | |
| B103 | (227) | |
| Nf | (227) | |
| GA-1 | (230) | |
| TX1322 | (265) | |
| P1 | (248) | |
| Cp-1 | (258) | |
| asccϕ28 | (215) | |
| CpVI | (215) | |
| Av-1 | (237) | |
| C1 | (243) | |
| 44AHJD | (174) |
DNA packaging activity of connector channel mutants
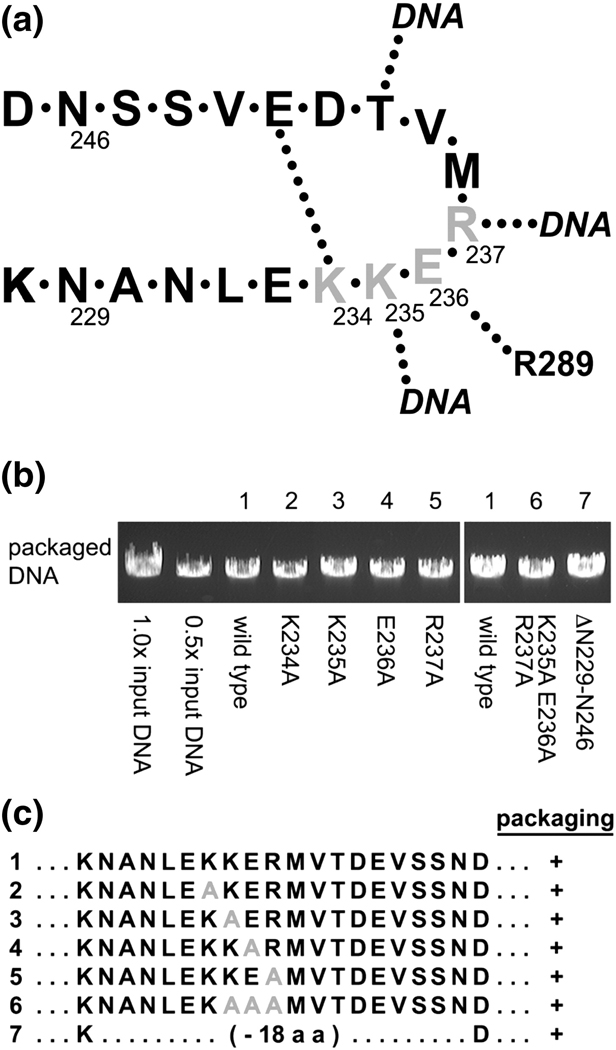
To test whether the channel loops participate in DNA translocation, the cluster of charged residues K234, K235, E236, and R237, predicted to participate in multiple interactions, were mutated to alanines or the entire 18-residue loop (residues 229–246) was deleted in the connector gene of a plasmid expression system for prohead particles (Fig. 3a and c).15 Particles expressed from the plasmid having either single alanine substitutions (K234A, K235A, E236A and R237A), a triple alanine substitution (K235A•E236A•R237A; triple-A), or the loop deletion (DN229-N246) had typical prohead protein compositions as judged by SDS-PAGE (albeit DN229-N246 produced a faster migrating connector band due to the deletion; data not shown).
Figure 3.
In vitro DNA packaging by particles with mutation in the connector channel region (amino acids 229–246). (a) Schematic of the connector channel loop showing contacts within the loops, between loops of connector monomers and with the translocating DNA. (b) DNA packaging assayed by agarose gel electrophoresis. Particles containing wild-type connectors (lane 1) or mutant connectors with alanine substitutions in 234KKER237 (lanes 2–6) or deletion of the entire loop (amino acids 229–246, lane 7) were assayed for biological activity in the in vitro DNA packaging system. DNase treatment was then used to digest the unpackaged DNA and the protected, packaged DNA was extracted and analyzed on a gel. The negative control omitted ATP from the reaction and did not show any protected DNA (data not shown). (c) Summary of mutants assayed. Mutations are highlighted in grey.
Particles were reconstituted with φ29 pRNA and tested for in vitro DNA-gp3 packaging using a DNase-protection assay16 (the φ29 genome has the terminal protein gp3 covalently bound to each 5' end). DNA that has been packaged is protected inside the head from DNase digestion, whereas the unpackaged DNA is degraded. Particles with mutant connectors, including the loop deletion, all displayed similarly high packaging efficiencies compared to particles with wild-type connectors (Fig. 3b). These results demonstrate that the connector channel loops are not essential for DNA translocation.
Conversion of filled heads to infectious phage
During viral assembly, after DNA packaging the DNA-filled head is matured to an infectious virion. At this stage the packaging ATPase, and in φ29 the pRNA, must detach from the head to allow for the assembly of tail components.1,2,7 To test the efficiency of conversion of the mutant DNA-packaged heads into phage, an extract that provided neck and tail components was incubated with the DNA packaging reaction and virus production was assayed (Table 2). Interestingly, all channel loop mutations resulted in severe reductions in phage assembly. Mutation of E236 that is predicted to contact a C-terminus residue outside of the channel loops yielded ~10X reduction in phage production compared to wild-type particles. Mutation of residue K235 that is predicted to contact the DNA had ~100X reduction of phage assembly. The greatest reductions were observed by mutation of the conserved residue K234 that is predicted to make contacts within and between channel loops, and R237 that is expected to interact with the DNA. Both of these mutant particles had assembly levels similar to the background of the extract alone, demonstrating that these mutations essentially abolished phage production, even though they supported in vitro DNA packaging. Similarly, packaged particles with connectors containing the triple-A mutation or the loop deletion also failed to produce virions, yielding virus levels similar to the background of the extract. Clearly the channel loops have a critical function in producing a competent DNA-filled head that can be matured to an infectious virion. Differences among single residue mutants in phage production suggest residue-specific effects.
Table 2.
Conversion of DNA-filled heads to phage. After in vitro DNA packaging, an extract that provided neck and tails components was added to the reaction to complete viral assembly and the formation of phage measured by plaque-forming units (pfu).
| Mutuant | pfu/ml |
|---|---|
| wild type | 1.8 ± 0.3 × 1011 |
| K234A | 1.1 ± 0.5 × 108 |
| K235A | 2.6 ± 0.5 × 109 |
| E236A | 2.4 ± 0.6 × 1010 |
| R237A | 9.8 ± 3.9 × 107 |
| extract | 9.2 ± 6.1 × 107 |
| wild type | 9.0 ± 0.3 × 1010 |
| K235A E236A R237A | 6.4 ± 2.8 × 107 |
| ΔN229-N246 | 6.7 ± 0.8 × 107 |
| Extract* | 8.0 × 107 |
Mean and standard deviation from three experiments (except where noted by * where n=1).
Stability of DNA-filled heads
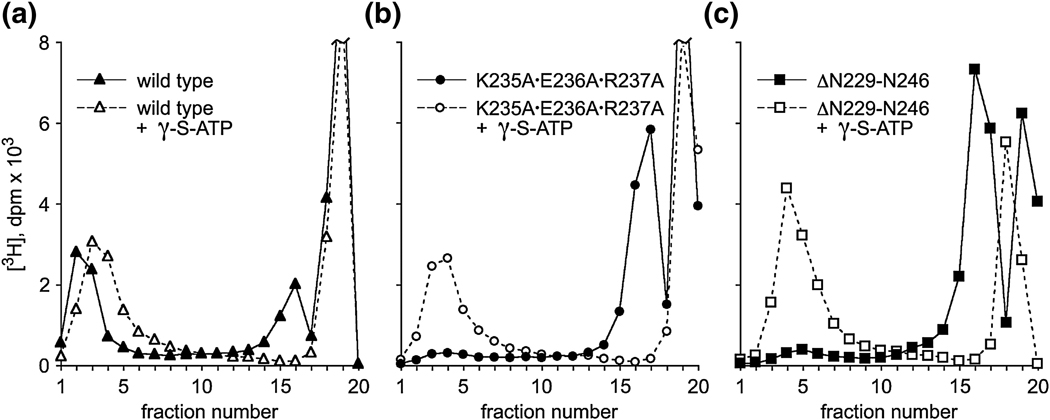
Since the channel loop mutants are compromised in the ability to convert to virions, packaging was further assessed by sucrose density gradient centrifugation,17–18 which assays both packaging efficiency and stability of the DNA-filled head. DNA-filled particles are fast-sedimenting, whereas the DNase-digested unpackaged DNA is found at the top of the gradient. Proheads containing the wild-type, the triple-A, or the loop deletion connector were tested for in vitro packaging using [3H]DNA-gp3. A portion of each reaction was assessed in an agarose gel to confirm packaging efficiency, while the remainder was centrifuged through a sucrose density gradient to assess stability of the packaged DNA. All particles had similar packaging efficiencies (as those shown in Fig. 3; data not shown). However, in gradient centrifugation only particles with wild-type channel loops retained their packaged DNA, demonstrated by the appearance of [3H]DNA at the position of filled heads (fractions 2–3 in Fig. 4a, filled symbol). Both the triple-A and deletion mutant particles yielded little radiolabel at the filled head position (Fig. 4b and c, filled symbol), and instead showed a large peak near the top of the gradient (fraction 16–17), the position of DNA that has been lost from packaged heads during centrifugation.19 The packaged particles with single alanine substitutions were also unstable during gradient centrifugation (data not shown). Thus, it appears that although particles with mutant connectors packaged DNA-gp3 efficiently, they were unable to retain DNA under the conditions of gradient centrifugation. Since the residues K234 and E236 are predicted to make intramolecular contacts, loss of DNA from these mutant particles suggests that these residues are important in maintaining loop structure.
Figure 4.
DNA packaging assayed by sucrose gradient centrifugation. Wild-type particles (a, filled symbol) or particles containing the mutation triple-A (b) or the loop deletion (c) were added to the packaging system containing [3H]DNA. After incubation, the unpackaged DNA was digested with DNase, and the reaction assayed for packaging by sedimentation in a 5–20% sucrose gradient. Sedimentation is from right to left. Filled heads sediment at fraction 2–4 and unpackaged DNA is found at fraction 19. The peak at fractions 16–17 is from DNA that escaped from the packaged head early in centrifugation. The open symbols represent the addition of γ-S-ATP in the gradients and in the reaction to stall the packaging motor prior to centrifugation.
The production of DNA-filled, yet defective, heads in the channel loop mutants suggests failure at a late stage of packaging. This assembly defect may by due to packaging that is incomplete or could represent an aberrant, off-pathway event. It has been shown that the motor ATPase engages the DNA in a nucleotide-dependent manner during packaging.17 If the packaging process is not fully complete, but on-pathway, it should be possible to engage the ATPase component of the motor to stabilize the packaged DNA. Therefore, γ-S-ATP was added at the end of the packaging reaction and to the gradients to maintain potential ATPase-DNA contacts.17 The filled heads produced from particles with wild-type connectors were stable during centrifugation irrespective of the presence of ATP analog (Fig. 4a). In contrast, while the triple-A and deletion mutants normally lose their DNA during centrifugation (Fig. 4b and c, filled symbol), γ-S-ATP stabilized the DNA-packaged head as evidenced by the disappearance of radiolabel in peak 17 that coincided with the appearance of radiolabel in the filled head position (Fig. 4b and c, open symbol). This result showed that there is a transition near the end of packaging that renders the wild-type DNA-filled head stable independent of the ATPase component of the motor. The fact that the packaged DNA in the mutant particles could only be retained by engaging the ATPase portion of the motor showed a failure to undergo this transition, and that part of the DNA is still in contact with the ATPase.
Termination of packaging
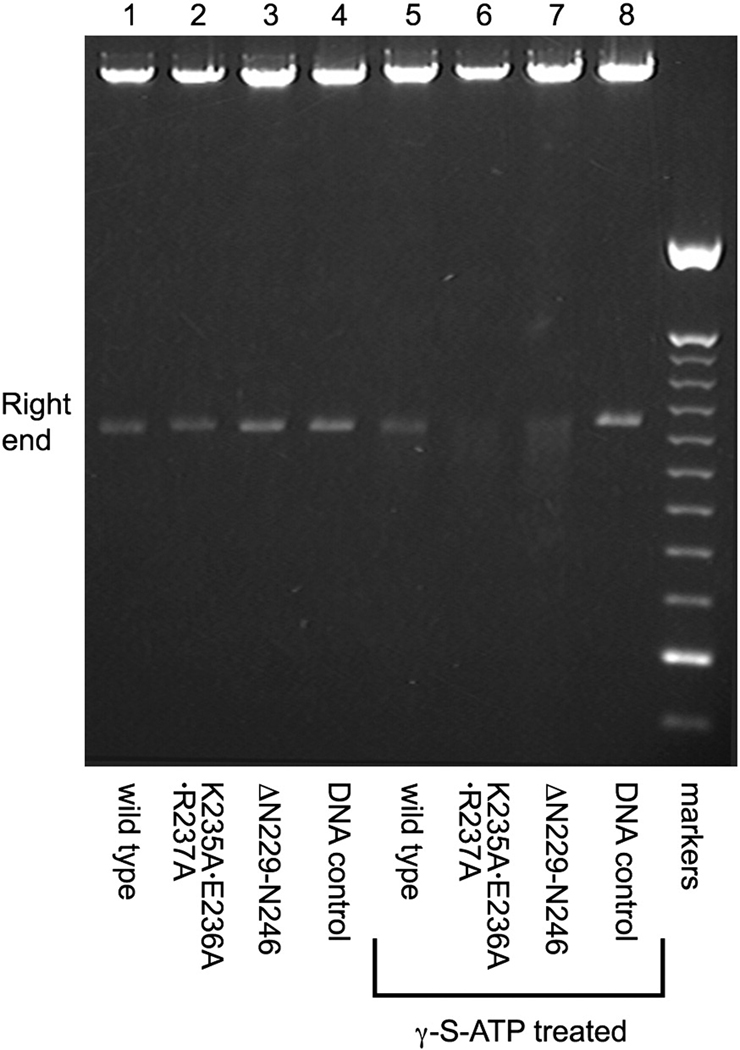
Given that connector-loop mutants appear defective in retaining full-length DNA, we sought to determine whether the last portion of DNA is fully packaged or protrudes from the packaging motor. Particles were packaged in vitro, treated with DNase, and the packaged DNA extracted to analyze the integrity of the right end of DNA-gp3, the last part portion to enter the capsid.19 γ-S-ATP was added to a portion of the reaction to lock contacts between the ATPase and DNA at the end of the in vitro packaging assay. Packaged DNA-gp3 was extracted from the particles, and the DNA digested with ApaLI to assay the last 7% of the packaging process. In the absence of γ-S-ATP treatment, packaged DNA extracted from wild-type and mutant particles yielded the expected 1149bp right-end ApaL1 fragment, indicative of complete DNase protection (Fig. 5). In contrast, in the γ-S-ATP treated samples, only the proheads with wild-type connectors yielded this size right-end fragment. The packaged DNA extracted from both the triple-A and the deletion mutant particles yielded heterogeneous right-end fragments that were several hundred base pairs shorter than the 1149bp fragment length (Fig. 5). Thus, although the mutant particles appeared to complete packaging in the absence of γ-S-ATP, this likely reflects that in the packaging reaction the ATPase is repackaging the last, unstably packaged piece of DNA, thereby protecting it from DNase digestion. Addition of the nucleotide analogue both blocks repackaging of the slipping DNA and stabilizes the DNA-ATPase contact, thus leaving the end of the DNA exposed and sensitive to DNase digestion. These results indicate that the connector channel loops likely interact with the right-end portion of DNA-gp3, stabilizing the packaged DNA in an ATPase-independent manner.
Figure 5.
Determination of the completion of DNA packaging. Particles containing wild-type (lanes 1 and 5), the triple-A mutation (lanes 2 and 6) or loop deletion of (lanes 3 and 7) were added to the packaging system. After 15 min. incubation, the reaction was split and γ-S-ATP added to one part to stall the motor. DNase was then added to digest the unpackaged DNA, and the packaged DNA was extracted and cut with ApaLI. Lanes 1–3 were derived from the standard packaging samples (no γ-S-ATP addition). Lanes 5–7 were derived from the portion of the reaction that was stalled with γ-S-ATP. Lane 4 and 8 were control digests of φ29 DNA alone treated with ApaLI ; markers=100 bp DNA ladder.
DISCUSSION
Various models for dsDNA viral packaging have been proposed in which different motor components are responsible for the catalytic and mechanical events during packaging (reviewed in ref. 2). In all models, the ATPase component of the motor plays a central role, mediating the nucleotide binding and hydrolysis that provides the input energy to drive the motor. One class of models proposes that the force that moves the DNA is generated directly by the ATPase, where ATP binding and hydrolysis produce the conformational changes needed for DNA translocation.20,21 A second class of models proposes that mechanical events occur in both the ATPase and connector components, where conformational changes in the ATPase produce conformational changes in the connector, resulting in the channel loops driving the DNA into the head.9
Here we examine the role of the φ29 connector channel loops, which include residues 229–246 that are not resolved in the crystal structure,3,8 in φ29 DNA packaging and virion production. Simpson et al.3 suggested that the charged residues in the loops might interact with DNA, and Guasch et al.8 speculated that the loops might close the channel after DNA packaging. We show that the entire channel loop can be deleted (DN229-N246) with little effect on the observed efficiency of DNA packaging (Fig. 3), suggesting that the loops are not essential for translocation, i.e., force generation by the motor. However, the loops are crucial for some aspect of the DNA packaging process. Whereas wild-type DNA-filled heads can be successfully converted to virions by providing tail components, the DNA-filled heads produced by channel-mutant particles were severely defective in phage assembly (Table 2). Sedimentation analysis showed that the DNA-filled heads produced by the channel mutants are incapable of retaining the full DNA complement, losing their DNA during centrifugation (Fig. 4b and c). Examination of the packaged DNA from mutant particles showed that the right-end of the DNA-gp3, the last portion to be packaged, is unable to be retained by the prohead (Fig. 5). Thus, in the channel loop mutants a mechanism for stabilizing the packaged DNA is missing.
Rather than an active force-generating role, we conclude that one function of the channel loops is to serve as a clamp to retain the pressurized DNA in the packaged head. At the end of packaging, the connector channel loops would serve to restrain the DNA while the pRNA and ATPase detach from the head and the tail proteins assemble onto the head to complete viral assembly. Here we show that in the normal packaging process a transition occurs near the end of packaging in which the DNA-filled heads become stable independent of the ATPase component of the motor (Figs. 4a and 5). In contrast, the DNA-filled heads produced by channel loop mutants cannot make this transition; the packaged DNA can only be stabilized by retaining a nucleotide-dependent ATPase/DNA contact (Fig. 4b and c). This precludes the successful retention of DNA when the ATPase detaches for tail assembly, and therefore explains the observed reduction in phage production. The clamping function that the channel loops provide must be provisional, as the DNA must exit the head via the connector channel during DNA ejection. In fact, cryoEM-3D reconstruction of the mature φ29 virion shows DNA filling the tail tube, suggesting the release of the clamp function during tail assembly.22
The connector protein has been extensively analyzed in phage SPP1 (termed portal protein in SPP1). The channel (tunnel) loops are clearly resolved in the SPP1 connector (portal) crystal structure, and fitting the crystal structure into the cryoEM density map of the connector suggests that the channel loops would be in close proximity to the translocating DNA.9 While the φ29 channel loop contains multiple charged residues (Table 1), the SPP1 sequence (345QAVDNSPETIGGGAT359) is predominantly charge-neutral and has been suggested to interact with the DNA through non-ionic interactions.9 However, it is likely that the φ29 and SPP1 channel loops play similar roles given the overall similarity of connector architecture and the nearly identical positioning of the loops in these structures.9 In SPP1, single residue substitutions in the channel loops disrupted full-length DNA packaging,23–25 consistent with the observed inability to retain the full-length genome in φ29 channel loop mutants. At the completion of packaging, the SPP1 packaging terminase complex (including the ATPase) detaches from the connector and is replaced by the head completion proteins gp15 and gp16 that ultimately retain DNA in the packaged head.26,27 Therefore, similar to φ29, there is a need for DNA retention while this transition is occurring. Mutation in the SPP1 completion proteins results in loss of packaged DNA,26,27 suggesting an eventual failure of this clamping mechanism in the absence of the next step of virion assembly.
Studies have also suggested a role for the channel loops in communication/coordination in the packaging motor. In SPP1, mutagenesis,23–25 cross-linking,28 and structural9 studies of the connector suggest the channel loops function in coordination with the packaging ATPase to translocate the DNA into the prohead. Mutations in the connector affect ATP hydrolysis by the packaging ATPase25 and some mutations result in shorter than normal lengths of DNA being packaged,23,24,29 supporting the existence of “cross-talk” between the connector protein and the packaging ATPase. This cross-talk, mediated by movement of protein helices in the connector, are proposed to provide coordination between ATP hydrolysis by the packaging ATPase and a wave-like motion by the channel loops.9,28
While our data does not support a model where the channel loops are the main force-generating component of the motor, it does not rule out a role for coordination/communication in the motor. It is possible that at the end of packaging a signal is exchanged between the ATPase and connector to prepare the ATPase for detachment and engage the channel loops for DNA retention. Such a signaling may be disrupted in the channel loop mutants resulting in an ATPase that is still in packaging mode. Additionally, high-resolution laser tweezers studies on φ29 DNA packaging revealed a highly coordinated motor with a two-phase mode of operation: the motor “dwells” while multiple ATPase subunits bind ATP, followed by translocation "bursts" of 10bp that are comprised of four 2.5 bp sub-steps.30 Whether the channel loops contribute to this coordination during packaging remains to be determined. Future high-resolution laser tweezers studies will allow further dissection of the role of the channel loops in packaging. Studies with channel loops mutants will reveal if the dwell/burst pattern of motor operation is affected and if motor behavior near the end of packaging is altered by defective connector channel loops.
MATERIALS AND METHODS
DNA and disordered connector residues modeling
The starting structure for connector modeling was the PDB map 1IJG.11 The initial structure of B-DNA was built using QUANTA, which was then manually moved to the connector channel. After coordinate manipulation, the center of mass and 10-fold symmetry axis of DNA was overlapped with the center of mass and 12-fold symmetry axis of the connector. The disordered regions on the connector were built using Swiss-PdbViewer12 and InsightII for one subunit according to secondary structure prediction with PSIPRED server, then the adopted basis Newton-Raphson (ABNR) method13 was used to minimize the built regions with the rest of the connector fixed. The reconstructed regions were then made 11 copies with 30° rotation away from each other with respect to the central 12-fold axis. Each subunit of the connector includes three histidines; two of them (His26 and His188) were protonated and the other one (His58) takes the neutral form HSE based on the surrounding interactions. All of the lysines and arginines were protonated, and the glutamates and aspartates unprotonated. In addition, sodium ions were added to neutralize the charge of connector. The crystal water molecules from the X-ray structure were kept in the system but those that overlapped with the modeled residues were deleted.
The modeled residues for the whole connector were first minimized for 100 steps using the ABNR method with the rest of the system fixed. The whole connector was then minimized for 100 steps using the same method with the DNA, crystal water and counter ions fixed. In all of the minimizations, we applied RDIE method, fswitch and shift functions were used for smoothing the electrostatic and vdW interactions at cutoff distance; a cutoff of 9.0 Å was used for calculation of nonbonded interactions and 10.0 Å was used for generating the nonbonded interaction list, which was updated every 25 steps.
Connector sequence alignment
Head-tail connector sequences for φ29 Bacillus relatives and φ29-like relatives were identified by BLAST31 using the φ29 connector protein sequence and aligned with T-coffee.32 Proteins in the alignment were the Bacillus phage φ29 upper collar (connector) (UniProtKB/Swiss-Prot accession no. P04332.1), Bacillus phage PZA upper collar protein (NCBI accession no. NP_040728.1), Bacillus phage B103 upper collar protein gene 10 (NCBI accession no. NP_690644.1), Bacillus phage GA-1 head-tail connector gene 10 (GenBank accession no. CAC21531.1) Bacillus phage Nf upper collar connector (GenBank accession no. ACH57078.1), Staphylococcus phage phiP68 upper collar protein orf19 (NCBI accession no. NP_817335.1), Streptococcus phage C1 head-tail connector protein orf15 (NCBI accession no. NP_852021.1), Streptococcus phage Cp-1 connector protein orf10 (GenBank accession no. CAA87731.1), Mycoplasma virus P1 P38.4 protein (NCBI accession no. NP_064644.1), Actinomyces phage Av-1 upper collar orf9 (NCBI accession no. NC_009643.1), Enterococcus faecalis TX1322 contig00097 upper collar protein (NCBI accession no. ZP_04435956.1), Clostridium phage CpV1 connector protein (GenBank accession no. ADR30486.1), and Lactococcus phage asccphi28 upper collar orf21(NCBI accession no. NC_010363.1).
Creation of connector channel loop mutants
Alanine substitutions of channel loop residues were introduced into the E. coli prohead expression plasmid pAR7-8-8.5-1015 by first mutating the L232 and T240 codons of gene 10 (connector) in the plasmid by site directed mutagenesis33 to create unique XhoI and BstEII sites, respectively. Complementary oligonucleotides (Sigma-Genosys) that contained the desired alanine substitutions (K234A, K235A, E236A, R237A, and K235A•E236A•R237A) and overhanging XhoI and BstEII sites were then ligated into the double-digested plasmid. The channel loop deletion mutant DN229-N246 was made by inverse PCR using the pAR7-8-8.5-10 template.34 The resulting plasmids were transformed into E. coli HMS174(DE3) and ApR-transformants were selected on plates containing 100 µg/ml ampicillin. The plasmid genes that encode gp7 (scaffold), gp8 (shell protein) and the mutagenized gp10 (connector) were sequenced to verify the maintenance of wild-type sequence for the scaffold and shell genes and the presence of the desired mutation in the connector gene.
Protein expression and purification of proheads
φ29 wild-type and connector channel loop mutant particles were expressed in E. coli from the pAR7-8-8.5-10 plasmid15 and were isolated and purified as previously described.35 Briefly, after induction cells were concentrated and lysed in TES buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA and 100 mM NaCl) containing 10 mg/ml lysozyme for 20 min at 37°C. Cells were lysed in the presence of 0.2% sodium deoxycholate, 3mM MgCl2 and 1.5µg/ml DNase I (final concentrations) and the lysate was incubated for an additional 20 min at 37°C. The prohead particles were isolated in a 10 to 40% (w/v) linear sucrose density gradients containing TMS buffer (50 mM Tris-HCl [pH 7.8], 10 mM MgCl2, and 100 mM NaCl) and 2 mM dithiothreitol (DTT) in a Beckman SW28 rotor at 15K rpm for 16.5 hr. The particles were pelleted in a Beckman Ti-50.2 rotor at 35K rpm for 5 hr, resuspended in TMS buffer containing 2 mM DTT, further purified in a 0.42% (w/v) CsCl isopycnic density gradient, and concentrated in a Centricon YM-30 (Amicon) in TMS buffer containing 2mM DTT.
Production of packaging components and the in vitro DNA-gp3 packaging assay
The wild-type and mutant proheads were produced as described above. The packaging ATPase gp16 was expressed from the plasmid pSACB-gp16 in Bacillus subtilis and purified as described previously.36 DNA-gp3 and [3H]DNA-gp3 were purified from phage on CsCl density gradients as described previously.16 120nt pRNA was produced by in vitro transcription and purified by denaturing urea polyacrylamide gel electrophoresis as described previously.36
In vitro DNA-gp3 packaging was performed as described previously.16,36 Briefly, purified φ29 wild-type and mutant prohead particles (83nM) were reconstituted with 120nt pRNA (830nM) in 0.5X TMS for 10 min at ambient temperature in a final volume of 10µl. For in vitro DNA packaging, reconstituted particles (8.3nM) are incubated with DNA-gp3 (4.2nM), the packaging ATPase gp16 (100–125nM), and 500 µM ATP in 0.5X TMS for 10 min at ambient temperature in a final volume of 20µl. The unpackaged DNA-gp3 was then digested with DNase I (1 µg/ml) for 10 min at ambient temperature, and the packaged, protected DNA-gp3 was extracted from the particles with 25 mM EDTA and 500 µg/ml Proteinase K for 30 min at 65°C. The packaged DNA was then analyzed by agarose gel (0.8%) electrophoresis.
To assess DNA-gp3 packaging by sucrose gradient density centrifugation, [3H]DNA-gp3 was used as the packaging substrate. After 15 min incubation, a portion of each in vitro packaging reaction was removed and γ-S-ATP added to 250µM final concentration. To both portions of the sample, DNase I was then added to 1µg/ml and the reaction incubated for 5 min. Each reaction was then split for agarose gel analysis and gradient centrifugation. For agarose gel analysis, the samples were treated with Proteinase K/EDTA as indicated above and analyzed for packaging efficiency on an agarose gel. For gradient analysis, 10µl of the reaction was removed and diluted into 125µl of 0.5X TMS. Samples were then layered on top of a 5 to 20% (w/v) linear sucrose gradient in 0.5X TMS or 0.5X TMS with 1µM γ-S-ATP and centrifuged in a Beckman SW55 rotor at 35,000 rpm for 33 min. Gradients were fractionated from the bottom, and 16 drop fractions collected and assayed for [3H] by liquid scintillation counting.
Size determination of the right end DNA-gp3 ApaL1 restriction fragment
To determine whether the entire genome has been packaged with mutant particles, after 15 min incubation for in vitro DNA packaging, γ-S-ATP was added to 250µM final concentration to the reaction and incubated for 1 min. The reactions were then treated with DNase I, followed by Proteinase K/EDTA as described above. DNA was extracted from the packaged particles by phenol/chloroform extraction and precipitated with ethanol. The DNA pellet was resuspended in restriction buffer and digested with ApaL1 (10u) at 37°C for 60 min. The 1.1kbp DNA-gp3 right end fragment was analyzed for size on a 0.8% agarose gel.
Conversion of filled heads to phage
To assay for the ability to convert the filled heads into infectious phage, an extract providing tail components was made by infection of Bacillus subtilis SpoA12 (sup-) with the φ29 mutant sus7(614)sus8(769)sus14(1241) that is deficient in the scaffold and shell proteins needed for prohead production.18 After in vitro packaging (described above), an equal volume of this extract was added to the packaging reaction and incubated for 60 min at ambient temperature. The mixture was then plated on Bacillus subtilis su+44 to measure plaque-forming units.
ACKNOWLEDGEMENTS
We thank Marc Morais for providing Figure 1 and Dwight Anderson for helpful discussion. This research was supported by NIH grants DE-003606, GM-059604 to S.G. and GM-046736 to J.G.
REFERENCES
- 1.Jardine PJ, Anderson D. DNA packaging in double-stranded DNA bacteriophages. Chapter 6. In: Calendar R, editor. The Bacteriophages. Oxford Press; 2006. pp. 49–65. [Google Scholar]
- 2.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 3.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage φ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage φ29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage φ29 DNA in vitro. Mol. Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 6.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage φ29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 7.Guo P, Erickson S, Anderson DL. A small viral RNA is required for in vitro packaging of bacteriophage φ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 8.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 9.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol. Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head-tail connector of bacteriophage φ29. Acta Cryst. 2001;D57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- 12.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 13.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 14.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, A. R. Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo PX, Erickson S, Xu W, Olson N, Baker TS, Anderson D. Regulation of the phage phi 29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 17.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 18.Guo P, Grimes S, Anderson DL. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage φ29. Proc. Natl. Acad. Sci. USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjornsti MA, Reilly BE, Anderson DL. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA-gp3. J. Virol. 1983;45:383–396. doi: 10.1128/jvi.45.1.383-396.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage phi29. Structure. 2008;16:935–943. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004;322:253–263. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Isidro A, Santos MA, Henriques AO, Tavares P. The high-resolution functional map of bacteriophage SPP1 portal protein. Mol. Microbiol. 2004;51:949–962. doi: 10.1046/j.1365-2958.2003.03880.x. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 2006;281:21914–21923. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 26.Becker B, de la Fuente N, Gassel M, Günther D, Tavares P, Lurz R, Trautner TA, Alonso JC. Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 1997;268:822–839. doi: 10.1006/jmbi.1997.0997. [DOI] [PubMed] [Google Scholar]
- 27.Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J. Biol. Chem. 2007;282:18907–18913. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- 29.Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 1992;225:81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- 30.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Poirot O, Suhre K, Abergel C, O'Toole E, Notredame C. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucl. Acids Res. 2004;32:W37–W40. doi: 10.1093/nar/gkh382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Wilkinson MF. Site-directed mutagenesis of large (13-kb) plasmids in a single-PCR procedure. BioTechniques. 2000;29:976–978. doi: 10.2144/00295bm09. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Wilkinson MF. Deletion mutagenesis of large (12-kb) plasmids by a one-step PCR protocol. BioTechniques. 2001;31:722–724. doi: 10.2144/01314bm01. [DOI] [PubMed] [Google Scholar]
- 35.Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine scanning and Fe-BABE probing of the bacteriophage φ29 prohead RNA-connector interaction. J. Mol. Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Morais MC, Anderson DL, Jardine PJ, Grimes S. Role of the CCA bulge of prohead RNA of bacteriophage φ29 in DNA packaging. J. Mol. Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]