Abstract
trans-4-Hydroxynonenal (HNE) is a peroxidation product of ω-6 polyunsaturated fatty acids. Michael addition of HNE to deoxyguanosine yields four diastereomeric 1,N2-dG adducts. The adduct of (6S,8R,11S) stereochemistry forms inter-strand N2-dG:N2-dG cross-links in the 5′-CpG-3′ sequence. It has been compared with the (6R,8S,11R) adduct, incorporated into 5′-d(GCTAGCXAGTCC)-3′•5′-d(GGACTCGCTAGC)-3′, containing the 5′-CpG-3′ sequence (X = HNE-dG). Both adducts rearrange in DNA to N2-dG aldehydes. These aldehydes exist in equilibrium with diastereomeric cyclic hemiacetals, in which the latter predominate at equilibrium. These cyclic hemiacetals mask the aldehydes, explaining why DNA cross-linking is slow compared to related 1,N2-dG adducts formed by acrolein and crotonaldehyde. Both the (6S,8R,11S) and (6R,8S,11R) cyclic hemiacetals are located within the minor groove. However, the (6S,8R,11S) cyclic hemiacetal orients in the 5′-direction, while the (6R,8S,11R) cyclic hemiacetal orients in the 3′-direction. The conformations of the diastereomeric N2-dG aldehydes, which are the reactive species involved in DNA cross-link formation, have been calculated using molecular mechanics methods. The (6S,8R,11S) aldehyde orients in the 5′-direction, while the (6R,8S,11R) aldehyde orients in the 3′-direction. This suggests a kinetic basis to explain, in part, why the (6S,8R,11S) HNE adduct forms interchain cross-links in the 5′-CpG-3′ sequence, whereas (6R,8S,11R) HNE adduct does not. The presence of these cross-links in vivo is anticipated to interfere with DNA replication and transcription, thereby contributing to the etiology of human disease.
trans-4-Hydroxynonenal (HNE) is produced from the metabolism of membrane lipids [Benedetti et al., 1980]. It is the major peroxidation product of ω-6 polyunsaturated fatty acids in vivo [Esterbauer et al., 1991, Burcham, 1998]. Several routes for the formation of HNE from ω-6 polyunsaturated fatty acids have been described [Lee and Blair, 2000, Schneider et al., 2001, Schneider et al., 2008]. HNE exposures modulate gene expression, cell signaling, cell proliferation, and apoptosis [Parola et al., 1999, Poli and Schaur, 2000, Nakashima et al., 2003, West et al., 2004, West and Marnett, 2005, 2006, Dwivedi et al., 2007]. Human exposures are associated with oxidative stress, and HNE has been implicated in the etiologies of Alzheimer's disease [Sayre et al., 1997], Parkinson's disease [Yoritaka et al., 1996], arteriosclerosis [Napoli et al., 1997], and hepatic ischemia reperfusion injury [Yamagami et al., 2000].
HNE induces the SOS response in Escherichia coli [Benamira and Marnett, 1992]. Chromosomal aberrations are observed upon exposures in a variety of mammalian cells [Esterbauer et al., 1990, Eckl et al., 1993, Karlhuber et al., 1997, Eckl, 2003], including human lymphocytes [Emerit et al., 1991]. HNE is mutagenic in rodent [Cajelli et al., 1987] and human cells [Hussain et al. 2000]. Mammalian genotoxicity depends upon glutathione, which is chemoprotective against the formation of HNE-DNA adducts [Chung et al., 2005, Falletti et al., 2007, Yadav et al., 2008]. Michael addition of the N2-amino group of deoxyguanosine to HNE gives diastereomeric 1,N2-dG adducts (Chart 1) [Winter et al., 1986, Douki et al., 2004, Kowalczyk et al., 2004], which have been detected in cellular DNA [Yi et al., 1997, Chung et al., 2000, Wacker et al., 2000, Wacker et al., 2001, Chung and Zhang, 2002, Liu et al., 2006b, Pan et al., 2006]. These 1,N2-dG adducts bear exocyclic rings through the bonding of guanine N1 and N2 to the HNE moiety; Watson-Crick hydrogen bonding is not possible.
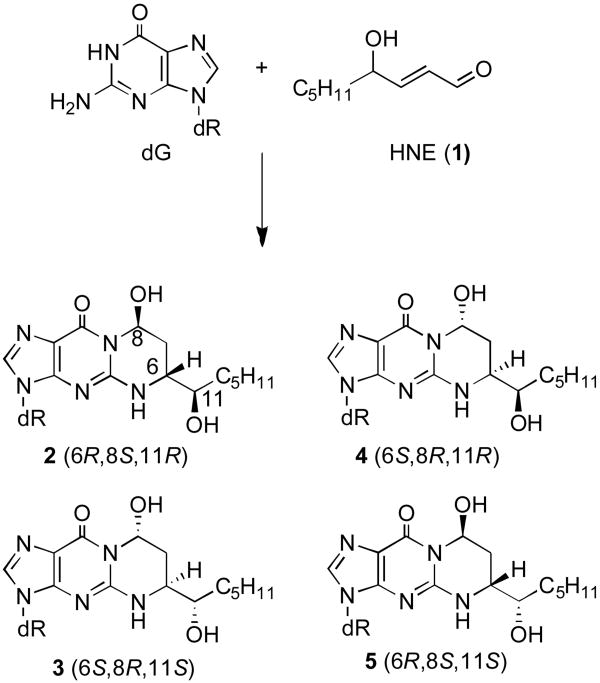
Chart 1.
Formation of exocyclic 1,N2-dG adducts 2-5 by HNE.
Synthesis of Stereospecific HNE-Derived 1,N2-dG Adducts
The stereochemical designations of the 1,N2-dG addition products (2-5) have been established unambiguously through chemical synthesis [Wang and Rizzo, 2001]; the four stereoisomers have been incorporated individually into 5′-d(GCTAGCXAGTCC)-3′•5′-d(GGACTCGCTAGC)-3′, containing the 5′-CpG-3′ sequence, in which X denotes the HNE-dG adduct (Chart 2). The approach involves condensation of stereospecific 4-amino-5-hydroxy-1,2-decane-diols with the corresponding oligodeoxynucleotides containing O6-[(2-trimethylsilyl)-ethyl]-2-fluorohypoxanthine, followed by deprotection and oxidation [Wang et al., 2003].
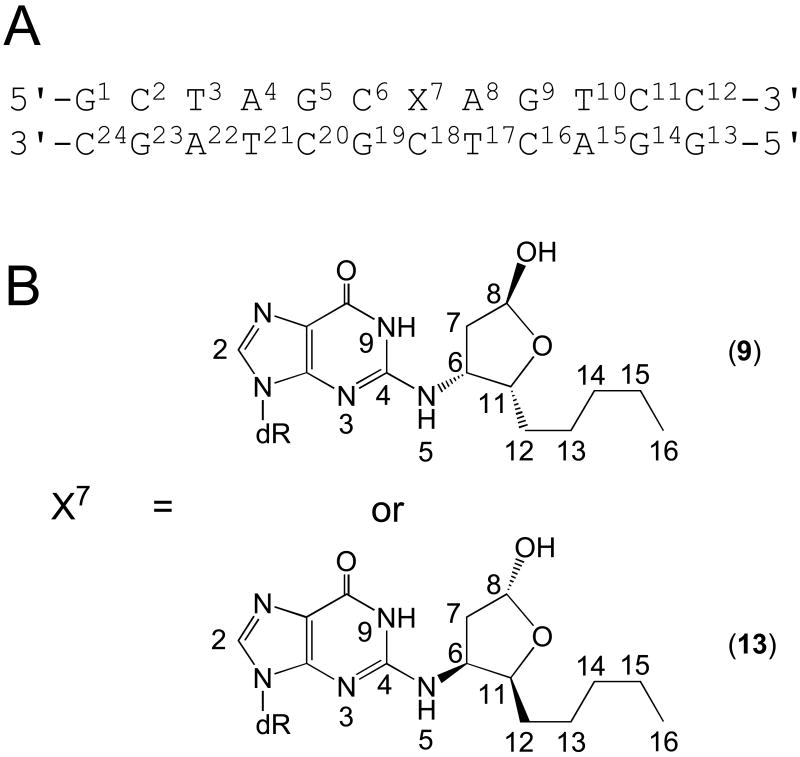
Chart 2.
A. Numbering scheme of the 5′-CpG-3′ duplexes containing stereospecific HNE-dG adducts (X denotes the HNE-derived 1,N2-dG adducts). B. Numbering scheme of the HNE-dG adducts.
Stereospecific Formation of DNA Cross-links
Interest in the cross-linking abilities of the stereoisomers of the HNE-derived 1,N2-dG Michael addition products (2-5) arose from studies of the corresponding 1,N2-dG adducts of acrolein and crotonaldehyde, which formed reversible inter-strand cross-links in this 5′-CpG-3′ sequence, comprised of carbinolamine-type linkages in equilibrium with trace amounts of imines [Stone et al., 2008]. For the crotonaldehyde-derived adduct, the 6R stereoisomer forms cross-links more efficiently than does the 6S stereoisomer [Kozekov et al., 2003]. Of the four HNE-dG adducts (2-5), only stereoisomer 3 possessing (6S,8R,11S) stereochemistry results in inter-strand cross-link formation [Wang et al., 2003]. The (6S,8R,11S) isomer of HNE possesses the same relative stereochemistry as the crotonaldehyde-derived 6R adduct [Kozekov et al., 2003, Stone et al., 2008] (the R vs. S designation at C6 is reversed for the HNE adducts as compared to the crotonaldehyde adducts). The formation of these enal-mediated cross-links is intrinsically slow in vitro, on the order of days for the acrolein-derived adduct and weeks for the crotonaldehyde-derived adduct [Kozekov et al., 2001, Kozekov et al., 2003]. For HNE, cross-link formation requires several months to reach equilibrium at 37 °C, in vitro [Wang et al., 2003]. Nevertheless, as compared to the acrolein- and crotonaldehyde-derived cross-links [Kozekov et al., 2003, Stone et al., 2008], adduct 3 forms high levels of cross-links, suggesting that once formed, the cross-links are stable [Wang et al., 2003].
Ring-Opening to N2-dG Aldehyde Adducts
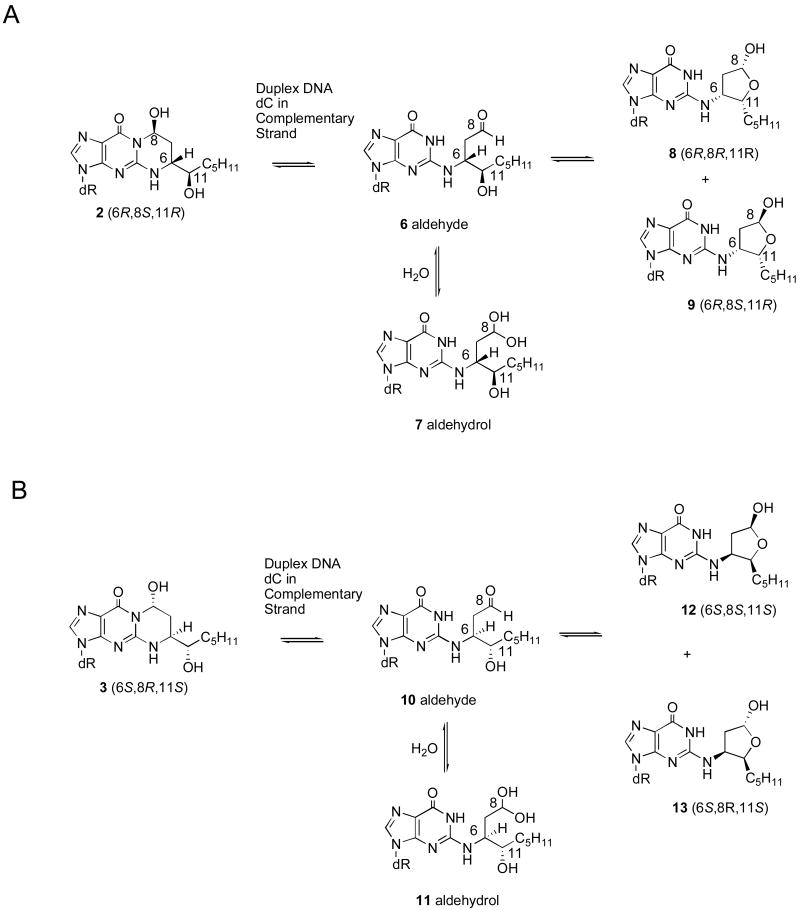
Exocyclic adducts 2 or 3 epimerize at the C8 position; in nucleosides and single-stranded DNA equilibrium favors the ring-closed configuration with the trans arrangement of the C8 hydroxyl group and the C6 alkyl side chain. When either adduct 2 or 3 is placed opposite cytosine in DNA, re-arrangement to aldehydes 6 and 10 is favored (Chart 3) [Huang et al., 2008a]. The observation of the 1H NMR resonance at ∼9.7 ppm indicates the presence of the aldehyde species. Presumably, the ring-opened structures facilitate Watson-Crick hydrogen bonding.
Chart 3.
Ring-opening chemistry of the HNE-derived exocyclic 1,N2-dG adducts 2 and 3 when placed opposite dC in duplex DNA.
Rearrangement to Cyclic Hemiacetals
In contrast to the acrolein- [de los Santos et al., 2001] and crotonaldehyde-induced [Cho et al., 2006b] 1,N2-dG adducts, at equilibrium in DNA the predominant forms of the ring-opened species arising from HNE-derived adducts 2 or 3 are not the aldehydes 6, 10 or aldehydrols. 1H NMR indicates the presence of only trace amounts of aldehydes 6 and 10. Instead, the NMR and mass spectrometry data indicate that cyclic hemiacetals 8 and 9, or 12 and 13, arising from adducts 2 or 3, respectively (Chart 3) [Huang et al., 2008a]. Starting from adduct 2, 1H NOE studies reveal that cyclic hemiacetal stereoisomer 9 (6R,8S,11R) is the predominant species at equilibrium, and stereoisomer 8 (6R,8R,11R) is the minor species. Likewise, starting from adduct 3, cyclic hemiacetal stereoisomer 13 (6S,8R,11S) is the major species, and stereoisomer 12 (6S,8S,11S) is the minor species. The favored stereochemistry of the cyclic hemiacetal presumably avoids steric repulsion from the large substituents [Huang et al., 2008a]. The formation of cyclic hemiacetals 12 and 13 (Chart 3) [Huang et al., 2008a] provides a plausible explanation as to the slow rate of cross-link formation by adduct 3 in the 5′-CpG-3′ sequence [Wang et al., 2003]. The cyclic hemiacetals 12 and 13 would mask the aldehyde 10 necessary for formation of the 5′-CpG-3′ cross-link.
Stereochemistry Modulates Cross-link Formation in the 5′-CpG-3′ Sequence
The inability of adduct 2 to form cross-links in the 5′-CpG-3′ sequence [Wang et al., 2003], despite the fact that it also undergoes ring-opening when placed opposite dC in DNA [Huang et al., 2008a], involves stereospecific differences in the orientations of cyclic hemiacetals 9 and 13 within the minor groove, which control the orientations of aldehyde 6 as compared to aldehyde 10. NMR studies indicate that the sequential NOE connectivity of both duplexes is complete for both the modified and complementary strands [Huang et al., 2008b]. NOE studies indicate that the tetrahydrofuran ring of stereoisomer 9 is directed in the 3′-direction, while the tetrahydrofuran ring of stereoisomer 13 is directed in the 5′-direction. The aliphatic chain of stereoisomer 9 exhibits NOEs with protons in the 5′-direction and the tetrahydrofuran subunit exhibits NOEs with protons in the 3′-direction. In contrast, the aliphatic chain of stereoisomer 13 exhibits NOEs with protons in the 3′-direction and the tetrahydrofuran subunit exhibits NOEs with protons in the 5′-direction. The presence of the aliphatic chain within the minor groove suggests that the rotation of the HNE-derived cyclic hemiacetals around the X7 N5-X7 C6 bond is restrained. Consequently, to the extent that the cyclic hemiacetals 9 and 13 open to unmask the aldehydes 6 and 10, the latter are anticipated to adopt similar orientations as the respective cyclic hemiacetals. In summary, the (6S,8R,11S) stereoisomer of the 1,N2-dG adduct 3 that exists predominantly as cyclic hemiacetal 13 is positioned to facilitate cross-linking in the 5′-CpG-3′ sequence. In contrast, the (6R,8S,11R) stereoisomer of the 1,N2-dG adduct 2 that exists predominantly as cyclic hemiacetal 9 is not positioned to facilitate cross-link formation. Molecular modeling of the respective aldehydes, 6 and 10, is consistent with this conclusion. Aldehyde 6 orients in the 3′-direction, whereas aldehyde 10 orients in the 5′-direction. Thus, aldehyde 10, arising from 1,N2-dG adduct 3, is predicted to be proximate to C6•G19 base pair, facilitating formation of the cross-link in the 5′-CpG-3′ sequence.
Comparison to Crotonaldehyde-Derived DNA Cross-links
The 6R-configuration of the crotonaldehyde-derived 1,N2-dG adduct produces more DNA cross-links than does the 6S-configuration [Kozekov et al., 2003]. The 6R stereochemistry of the crotonaldehyde-derived adduct corresponds to the (6S,8R,11S) stereochemistry of HNE-derived adduct 3. Thus, the cyclic hemiacetal arising from adduct 3 facilitates inter-strand cross-linking for the same reason that the 6R-crotonaldehyde-derived adduct does [Cho et al., 2006b]: it places the requisite aldehyde in the minor groove proximal to the cross-linking target in the 5′-CpG-3′ sequence. Similarly, for the 6S crotonaldehyde-derived 1,N2-dG adduct, the aldehyde orients towards the A8•T17 base pair, distal to the targeted C•G base pair [Cho et al., 2006a]. In contrast to HNE, crotonaldehyde is small with regard to the width of the minor groove, which allows the aldehydic form of the crotonaldehyde-derived 6S adduct re-orient toward the cross-linking target C6•G19 base pair. This probably explains why <5% cross-link has been observed for the crotonaldehyde-derived 6S adduct [Kozekov et al., 2003]. However, the reduced form of the 6S-crotonaldehyde-derived cross-link is less stable compared to the 6R-crotonaldehyde-derived cross-link [Cho et al., 2007], consistent with modeling studies [Cho et al., 2006b]. Therefore, small amounts of cross-links formed upon reorientation of cyclic hemiacetal 9 would be anticipated to induce a greater destabilization of the DNA, as compared to the crotonaldehyde-derived 6S adduct. Further structural analyses of the cross-link arising from the (6S,8R,11S) HNE-derived adducts 3, currently in progress, are of considerable interest.
Biological Implications
Since (6S,8R,11S) adduct 3 forms inter-strand cross-links in 5′-CpG-3′ DNA sequences in vitro [Wang et al., 2003], it is anticipated that it will also form these cross-links in vivo. Since they occur specifically at 5′-CpG-3′ sequences, only for (6S,8R,11S) HNE adduct 3, and are reversible, they are anticipated to be present at low levels in vivo, challenging the limits of detection by mass spectrometry [Ruan et al., 2006, Stout et al., 2006, Zhang et al., 2006, Goodenough et al., 2007, Zayas et al., 2007].
The genotoxic consequences arising from low levels of these cross-links may be of considerable significance. Human inter-strand cross-link repair seems to require the cooperation of multiple proteins belonging to different pathways, including nucleotide excision repair (NER), homologous recombination (HR), trans-lesion synthesis (TLS), double-strand break (DSB) repair, and the Fanconi anemia (FA) pathway [Kennedy and D'Andrea, 2005, Niedernhofer et al., 2005, Nojima et al., 2005, Mirchandani and D'Andrea, 2006, Noll et al., 2006, Patel and Joenje, 2007]. One HR-independent model for inter-strand cross-link repair utilizes endonucleases for strand incision surrounding the cross-link on one of the two DNA strands and trans-lesion polymerases for gap-filling replication past the cross-link site on the other strand [Wang et al., 2001, Zheng et al., 2003, Richards et al., 2005, Liu et al., 2006a, Sarkar et al., 2006, Shen et al., 2006]. In this repair model, the dually incised strand possesses sufficient mobility that a bypass DNA polymerase can strand displace the nucleotide patch that is 5′ to the lesion, then replicate past the ICL site to complete the repair gap-filling synthesis.
Because enal-mediated inter-strand cross-links are reversible, most studies to date have utilized saturated inter-strand N2-dG•N2-dG propano cross-links as models to address molecular mechanisms of repair. The saturated cross-link has been used to investigate processing by the XPF/ERCC1 heterodimer; the results suggest a role for XPF/ERCC1 in the processing of a double-strand break that could be created when the cross-link encounters the replication fork [Mu et al., 2000]. In E. coli, a mechanism has been proposed in which repair is initiated by NER followed by trans-lesion DNA synthesis (TLS) and completed through another round of NER [Kumari et al., 2008]. Thus, pol IV catalyzes TLS when the nucleotides that are 5′ to the cross-link are removed. The efficiency of TLS is further increased when the nucleotides 3′ to the cross-linked site are also removed. Moriya and co-workers have examined the repair of crotonaldehyde-derived N2-dG•N2-dG inter-strand cross-links following replication of site-specifically modified vectors in E. coli and mammalian cells [Liu et al., 2006a]. Their results suggest that the native cross-link partially reverts, but are consistent with earlier reports that NER is essential for inter-strand cross-link repair in E. coli [Cole, 1973, Berardini et al., 1997]. In human XPA cells, the reduced cross-link is removed, suggesting a repair pathway unique to higher eukaryotes that does not require damage recognition by NER [Liu et al., 2006a]. Minko et al. [Minko et al., 2008] have reported that a vector containing a model of the incised product following dual incision around the saturated N2-dG•N2-dG propano cross-link is replicated in mammalian cells. Human polymerase κ catalyzes accurate incorporation opposite this cross-link and also replicates beyond the lesion. The reversibility of these HNE-derived inter-strand cross-links, as noted by Liu et al. [Liu et al., 2006a] might reduce their ability to block DNA processing, in vivo. Cross-link reversion would be anticipated to target removal of the resulting bulky N2-dG adducts by nucleotide excision repair [Chung et al., 2003, Feng et al., 2003, Choudhury et al., 2004].
Site-specific mutagenesis in the mammalian COS-7 system shows that stereoisomers 2 and 3 of the HNE-derived 1,N2-dG adduct induce low levels of G→T transversions and G→A transitions, whereas stereoisomers 4 and 5 are inactive [Fernandes et al., 2003]. The re-arrangement of these adducts into the cyclic hemiacetals 8 and 9, and 12 and 13, respectively, provides a potential explanation for the low levels of mutations induced by adducts 2 and 3 in the COS-7 system. The cyclic hemiacetals are anticipated to facilitate Watson-Crick hydrogen bonding during replication bypass [Fernandes et al., 2003]. Similar explanations have been advanced to explain low levels of mutations induced by acrolein- [VanderVeen et al., 2001, Yang et al., 2001] and crotonaldehyde-induced 1,N2-dG adducts [Fernandes et al., 2005]. While modestly higher levels of mutations are observed for the crotonaldehyde-derived adducts [Stein et al., 2006], these probably correlate with modestly higher levels of the intact 1,N2-dG products in DNA [Cho et al., 2006b]. This correlates with the observation that significantly higher levels of G→T mutations are associated with the ring-closed 1,N2-dG adducts [Xing et al., 2007]. The chemically stable 1,N2-propano-dG (PdG) adduct exhibits significant mutagenicity [Moriya et al., 1994, Moriya et al., 1999]. Incorporation of PdG into DNA precludes Watson-Crick hydrogen bonding and results in structural [Kouchakdjian et al., 1989, Kouchakdjian et al., 1990, Singh et al., 1993, Weisenseel et al., 2002] and thermodynamic [Plum et al., 1992] perturbations.
Consistent with these observations, HNE causes G→T transversions at codon 249 of p53 in lymphoblastoid cells [Hussain et al., 2000], and HNE adducts preferentially form with dG in codon 249 in the p53 gene [Hu et al., 2002]. The mutational spectrum induced by HNE-dG adducts in the supF gene of shuttle vector pSP189 replicated in human cells also shows primarily G→T transversions, accompanied by G→A transitions [Feng et al., 2003]. On the other hand, the mutational spectrum induced by HNE in the lacZ gene of the single-stranded M13 phage transfected into wild type Escherichia coli reveals recombination events, C→T transitions, and lesser amounts of G→C and A→C transversions, and frameshift mutations [Kowalczyk et al., 2004].
The acrolein-derived 1,N2-dG adduct provides a block to replicative mammalian DNA polymerases, pol δ and pol ε [Kanuri et al., 2002]. Consequently, it seems that replicative polymerases will also be blocked by the larger HNE-derived 1,N2-dG adducts. In contrast, the sequential action of human pols ι and κ, Y-family polymerases facilitates error-free bypass of the (6S,8R,11R) and (6S,8R,11S) diastereomers of the 1,N2-dG HNE adduct [Wolfle et al., 2006]. In this case, pol ι inserts dCTP and to a lesser extent dTTP opposite the HNE adduct but is unable to further elongate the primer. Further extension is observed in the presence of pol κ, which elongates from a primer terminus C opposite the 1,N2-dG HNE adducts more efficiently than when T is opposite the adducts [Wolfle et al., 2006].
Summary
The (6R,8S,11R) and (6S,8R,11S) HNE-derived 1,N2-dG adducts 2 and 3 have been examined in an oligodeoxynucleotide containing the 5′-CpG-3′ sequence in which adduct 3, but not adduct 2, forms inter-strand cross-links. At equilibrium the predominant forms of the ring-opened species arising from adducts 2 or 3 are cyclic hemiacetals 8 and 9, or 12 and 13 (Chart 3) [Huang et al., 2008a]. Starting from adduct 2, cyclic hemiacetal stereoisomer 9 (6R,8S,11R) is the major species. Starting from adduct 3, cyclic hemiacetal stereoisomer 13 (6S,8R,11S) is the major species. The orientations of the cyclic hemiacetal groups within the minor groove differ. The tetrahydrofuran ring of cyclic hemiacetal 13, arising from adduct 3, orients in the 5′-direction toward base pair C6•G19, while the tetrahydrofuran ring of cyclic hemiacetal 9, arising from adduct 2 with (6R,8S,11R) stereochemistry, orients in the 3′-direction toward base pair A8•T17. Thus, adduct 3 with (6S,8R,11S) stereochemistry facilitates formation of inter-strand cross-links, whereas adduct 2 with (6R,8S,11R) stereochemistry, does not form inter-strand cross-links. Detailed structural studies of the cross-links are currently in progress.
Figure 1.
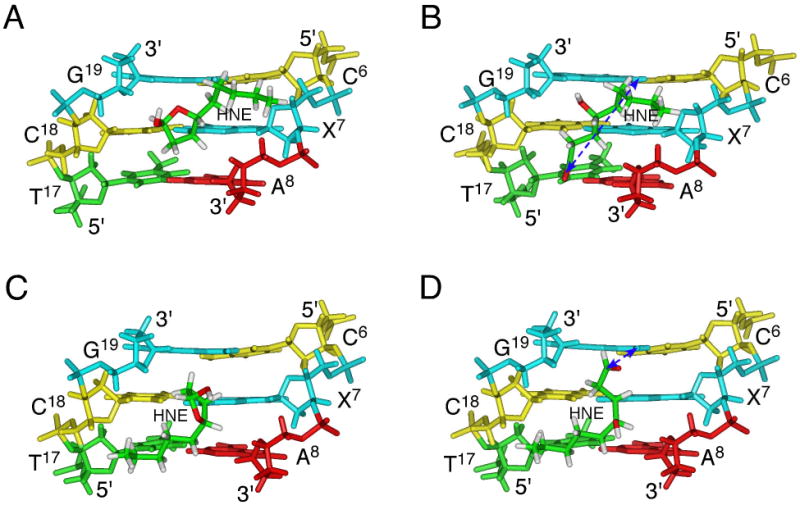
The adducted regions of the oligodeoxynucleotide duplexes containing the 5′-CpX-3′ sequence, viewed from the minor grooves. A. Average refined structure emergent from rMD calculations of the duplex containing cyclic hemiacetal 8. B. Predicted structure, obtained by molecular mechanics calculations, of the duplex containing aldehyde 6. The dashed arrows indicate the spatial relationship between the reactive aldehyde carbon and the exocyclic amino nitrogen of cross-linking target G19 (7.1 Å). C. Average refined structure emergent from rMD calculations of the duplex containing cyclic hemiacetal 10. D. Predicted structure, obtained by molecular mechanics calculations, of the duplex containing aldehyde 7. The cyan sticks represent nucleotides. The blue sticks represent the two amino nitrogens of X7 and G19. The white, green, and red sticks represent hydrogens, carbons, and oxygens of the HNE moiety. The dashed arrows indicate the spatial relationship between the reactive aldehyde carbon and the exocyclic amino nitrogen of cross-linking target G19 (4.4 Å). Adopted with permission from Huang et al., Biochemistry 2008 47: 11457-11472. Copyright 2008 American Chemical Society.
Figure 2.
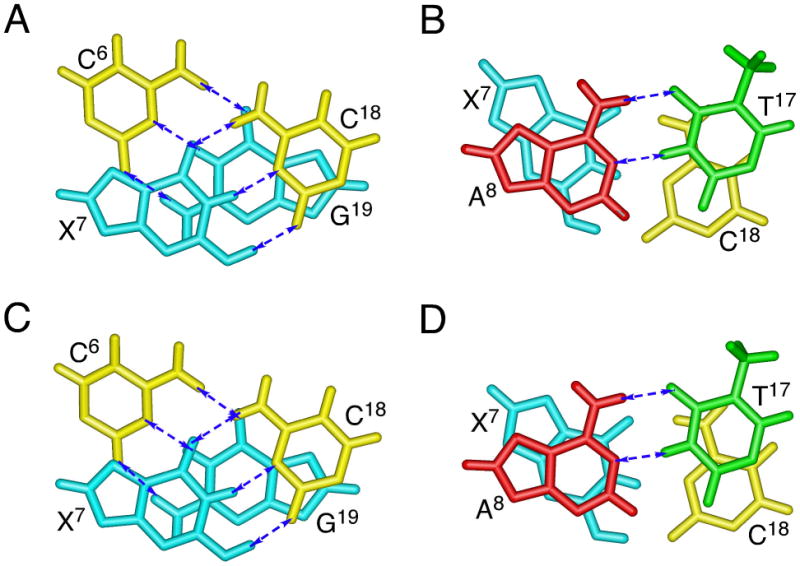
Base stacking of the adduct region for oligodeoxynucleotide duplexes containing the 5′-CpX-3′ sequence. A. The duplex containing cyclic hemiacetal 8. Stacking of base pair C6•G19 above base pair X7•C18. B. The duplex containing cyclic hemiacetal 8. Stacking of base pair X7•C18 above base pair A8•T17. C. The duplex containing cyclic hemiacetal 10. Stacking of base pair C6•G19 above base pair X7•C18. D. The duplex containing cyclic hemiacetal 10. Stacking of base pair X7•C18 above base pair A8•T17. For both duplexes containing either cyclic hemiacetals 8 or 10, base pairs C6•G19, X7•C18, and A8•T17 adopt Watson-Crick pairing.
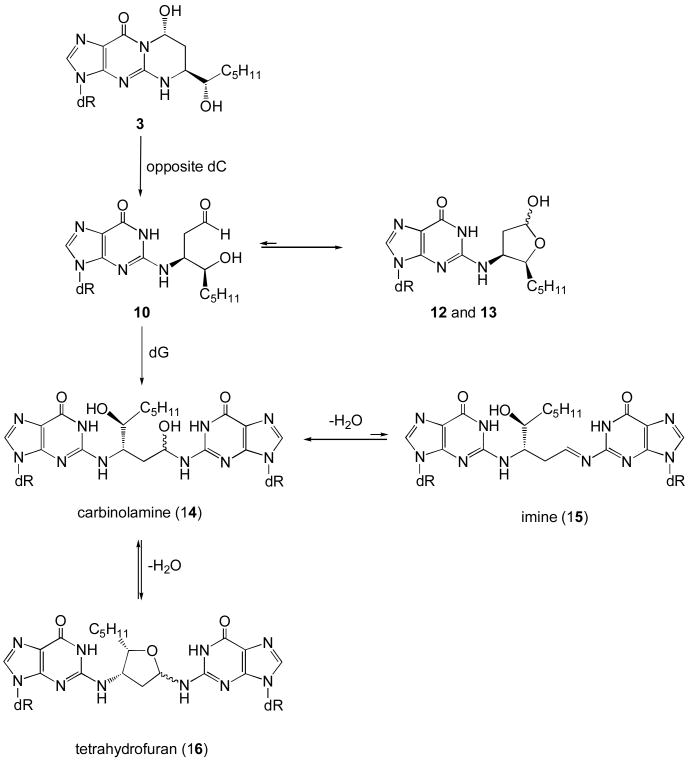
Chart 4. Formation of the inter-strand cross-link by HNE derived (6S,8R,11S) 1,N2-dG adduct.
Acknowledgments
Dr. Markus Voehler assisted with NMR experiments. Drs. Constance M. Harris and Thomas M. Harris provided thoughtful comments regarding the chemistry of these adducts. Dr. Irina G. Minko consulted on the biology of HNE. This work was supported by NIH Grant ES05355 (I.D.K., R.S.L., C.J.R. and M.P.S.). The Vanderbilt University Center in Molecular Toxicology is supported by NIH grant P30 ES00267. Vanderbilt University and NIH Grant RR05805 provided additional funding for NMR instrumentation.
Abbreviations
- HNE
trans-4-hydroxynonenal
- HNE-dG
trans-4-hydroxynonenal derived deoxyguanosine adduct
- NOESY
nuclear Overhauser effect spectroscopy
- NOE
nuclear Overhauser effect
- pol
DNA polymerase
References
- Benamira M, Marnett LJ. The lipid peroxidation product 4-hydroxynonenal is a potent inducer of the SOS response. Mutat Res. 1992;293:1–10. doi: 10.1016/0921-8777(92)90002-k. [DOI] [PubMed] [Google Scholar]
- Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim Biophys Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- Berardini M, Mackay W, Loechler EL. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry. 1997;36:3506–3513. doi: 10.1021/bi962778w. [DOI] [PubMed] [Google Scholar]
- Burcham PC. Genotoxic lipid peroxidation products: Their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis. 1998;13:287–305. doi: 10.1093/mutage/13.3.287. [DOI] [PubMed] [Google Scholar]
- Cajelli E, Ferraris A, Brambilla G. Mutagenicity of 4-hydroxynonenal in V79 Chinese hamster cells. Mutat Res. 1987;190:169–171. doi: 10.1016/0165-7992(87)90050-9. [DOI] [PubMed] [Google Scholar]
- Cho YJ, Kozekov ID, Harris TM, Rizzo CJ, Stone MP. Stereochemistry modulates the stability of reduced interstrand cross-links arising from R- and S-α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine in the 5′-CpG-3′ DNA sequence. Biochemistry. 2007;46:2608–2621. doi: 10.1021/bi061381h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YJ, Wang H, Kozekov ID, Kozekova A, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Orientation of the crotonaldehyde-derived N2-[3-Oxo-1S-methyl-propyl]-dG DNA adduct hinders interstrand cross-link formation in the 5′-CpG-3′ sequence. Chem Res Toxicol. 2006a;19:1019–1029. doi: 10.1021/tx0600604. [DOI] [PubMed] [Google Scholar]
- Cho YJ, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem Res Toxicol. 2006b;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Pan J, Amin S, Chung FL, Roy R. Repair kinetics of trans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 2004;43:7514–7521. doi: 10.1021/bi049877r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung FL, Komninou D, Zhang L, Nath R, Pan J, Amin S, Richie J. Glutathione depletion enhances the formation of endogenous cyclic DNA adducts derived from t-4-hydroxy-2-nonenal in rat liver. Chem Res Toxicol. 2005;18:24–27. doi: 10.1021/tx049728+. [DOI] [PubMed] [Google Scholar]
- Chung FL, Nath RG, Ocando J, Nishikawa A, Zhang L. Deoxyguanosine adducts of t-4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: Detection and potential sources. Cancer Res. 2000;60:1507–1511. [PubMed] [Google Scholar]
- Chung FL, Pan J, Choudhury S, Roy R, Hu W, Tang MS. Formation of trans-4-hydroxy-2-nonenal- and other enal-derived cyclic DNA adducts from ω-3 and ω-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutat Res. 2003;531:25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Chung FL, Zhang L. Deoxyguanosine adducts of tert-4-hydroxy-2-nonenal as markers of endogenous DNA lesions. Methods Enzymol. 2002;353:523–536. doi: 10.1016/s0076-6879(02)53074-3. [DOI] [PubMed] [Google Scholar]
- Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: Sequential excision and recombination. Proc Natl Acad Sci USA. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2′-deoxyguanosine. J Biol Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- Douki T, Odin F, Caillat S, Favier A, Cadet J. Predominance of the 1,N2-propano 2′-deoxyguanosine adduct among 4-hydroxy-2-nonenal-induced DNA lesions. Free Radic Biol Med. 2004;37:62–70. doi: 10.1016/j.freeradbiomed.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Dwivedi S, Sharma A, Patrick B, Sharma R, Awasthi YC. Role of 4-hydroxynonenal and its metabolites in signaling. Redox Rep. 2007;12:4–10. doi: 10.1179/135100007X162211. [DOI] [PubMed] [Google Scholar]
- Eckl PM. Genotoxicity of HNE. Mol Aspects Med. 2003;24:161–165. doi: 10.1016/s0098-2997(03)00010-4. [DOI] [PubMed] [Google Scholar]
- Eckl PM, Ortner A, Esterbauer H. Genotoxic properties of 4-hydroxyalkenals and analogous aldehydes. Mutat Res. 1993;290:183–192. doi: 10.1016/0027-5107(93)90158-c. [DOI] [PubMed] [Google Scholar]
- Emerit I, Khan SH, Esterbauer H. Hydroxynonenal, a component of clastogenic factors? Free Radic Biol Med. 1991;10:371–377. doi: 10.1016/0891-5849(91)90045-5. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Eckl P, Ortner A. Possible mutagens derived from lipids and lipid precursors. Mutat Res. 1990;238:223–233. doi: 10.1016/0165-1110(90)90014-3. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- Falletti O, Cadet J, Favier A, Douki T. Trapping of 4-hydroxynonenal by glutathione efficiently prevents formation of DNA adducts in human cells. Free Radic Biol Med. 2007;42:1258–1269. doi: 10.1016/j.freeradbiomed.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, Amin S, Tang MS. Mutational spectrum and genotoxicity of the major lipid peroxidation product, trans-4-hydroxy-2-nonenal, induced DNA adducts in nucleotide excision repair-proficient and -deficient human cells. Biochemistry. 2003;42:7848–7854. doi: 10.1021/bi034431g. [DOI] [PubMed] [Google Scholar]
- Fernandes PH, Kanuri M, Nechev LV, Harris TM, Lloyd RS. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ Mol Mutagen. 2005;45:455–459. doi: 10.1002/em.20117. [DOI] [PubMed] [Google Scholar]
- Fernandes PH, Wang H, Rizzo CJ, Lloyd RS. Site-specific mutagenicity of stereochemically defined 1,N2-deoxyguanosine adducts of trans-4-hydroxynonenal in mammalian cells. Environ Mol Mutagen. 2003;42:68–74. doi: 10.1002/em.10174. [DOI] [PubMed] [Google Scholar]
- Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2′-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Feng Z, Eveleigh J, Iyer G, Pan J, Amin S, Chung FL, Tang MS. The major lipid peroxidation product, trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at codon 249 of human p53 gene, a unique mutational hotspot in hepatocellular carcinoma. Carcinogenesis. 2002;23:1781–1789. doi: 10.1093/carcin/23.11.1781. [DOI] [PubMed] [Google Scholar]
- Huang H, Wang H, Qi N, Kozekova A, Rizzo CJ, Stone MP. Rearrangement of the (6S,8R,11S) and (6R,8S,11R) exocyclic 1,N2-deoxyguanosine adducts of trans-4-hydroxynonenal to N2-deoxyguanosine cyclic hemiacetal adducts when placed complementary to cytosine in duplex DNA. J Am Chem Soc. 2008a;130:10898–10906. doi: 10.1021/ja801824b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Wang H, Qi N, Lloyd RS, Rizzo CJ, Stone MP. The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2′-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5′-CpG-3′ sequence. Biochemistry. 2008b;47:11457–11472. doi: 10.1021/bi8011143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C, Hohler T, Galle PR, Phillips DH, Markin R, Marrogi AJ, Harris CC. Increased p53 mutation load in nontumorous human liver of Wilson disease and hemochromatosis: Oxyradical overload diseases. Proc Natl Acad Sci USA. 2000;97:12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlhuber GM, Bauer HC, Eckl PM. Cytotoxic and genotoxic effects of 4-hydroxynonenal in cerebral endothelial cells. Mutat Res. 1997;381:209–216. doi: 10.1016/s0027-5107(97)00170-x. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: New faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M, Eisenberg M, Live D, Marinelli E, Grollman AP, Patel DJ. NMR studies of an exocyclic 1,N2-propanodeoxyguanosine adduct (X) located opposite deoxyadenosine (A) in DNA duplexes at basic pH: Simultaneous partial intercalation of X and A between stacked bases. Biochemistry. 1990;29:4456–4465. doi: 10.1021/bi00470a028. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M, Marinelli E, Gao X, Johnson F, Grollman A, Patel D. NMR studies of exocyclic 1,N2-propanodeoxyguanosine adducts (X) opposite purines in DNA duplexes: Protonated X(syn):A(anti) pairing (acidic pH) and X(syn):G(anti) pairing (neutral pH) at the lesion site. Biochemistry. 1989;28:5647–5657. doi: 10.1021/bi00439a047. [DOI] [PubMed] [Google Scholar]
- Kowalczyk P, Ciesla JM, Komisarski M, Kusmierek JT, Tudek B. Long-chain adducts of trans-4-hydroxy-2-nonenal to DNA bases cause recombination, base substitutions and frameshift mutations in M13 phage. Mutat Res. 2004;550:33–48. doi: 10.1016/j.mrfmmm.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Interchain cross-linking of DNA mediated by the principal adduct of acrolein. Chem Res Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J Biol Chem. 2008;283:27433–27437. doi: 10.1074/jbc.M801237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Blair IA. Characterization of 4-oxo-2-nonenal as a novel product of lipid peroxidation. Chem Res Toxicol. 2000;13:698–702. doi: 10.1021/tx000101a. [DOI] [PubMed] [Google Scholar]
- Liu X, Lao Y, Yang IY, Hecht SS, Moriya M. Replication-coupled repair of crotonaldehyde/acetaldehyde-induced guanine-guanine interstrand cross-links and their mutagenicity. Biochemistry. 2006a;45:12898–12905. doi: 10.1021/bi060792v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lovell MA, Lynn BC. Detection and quantification of endogenous cyclic DNA adducts derived from trans-4-hydroxy-2-nonenal in human brain tissue by isotope dilution capillary liquid chromatography nanoelectrospray tandem mass spectrometry. Chem Res Toxicol. 2006b;19:710–718. doi: 10.1021/tx0502903. [DOI] [PubMed] [Google Scholar]
- Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase κ in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani KD, D'Andrea AD. The Fanconi anemia/BRCA pathway: A coordinator of cross-link repair. Exp Cell Res. 2006;312:2647–2653. doi: 10.1016/j.yexcr.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Moriya M, Pandya GA, Johnson F, Grollman AP. Cellular response to exocyclic DNA adducts. IARC Sci Publ. 1999;150:263–270. [PubMed] [Google Scholar]
- Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: Marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima I, Liu W, Akhand AA, Takeda K, Kawamoto Y, Kato M, Suzuki H. 4-hydroxynonenal triggers multistep signal transduction cascades for suppression of cellular functions. Mol Aspects Med. 2003;24:231–238. doi: 10.1016/s0098-2997(03)00018-9. [DOI] [PubMed] [Google Scholar]
- Napoli C, D'Armiento FP, Mancini FP, Postiglione A, Witztum JL, Palumbo G, Palinski W. Fatty streak formation occurs in human fetal aortas and is greatly enhanced by maternal hypercholesterolemia. Intimal accumulation of low density lipoprotein and its oxidation precede monocyte recruitment into early atherosclerotic lesions. J Clin Invest. 1997;100:2680–2690. doi: 10.1172/JCI119813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Davis W, Trushin N, Amin S, Nath RG, Salem N, Jr, Chung FL. A solid-phase extraction/high-performance liquid chromatography-based 32P-postlabeling method for detection of cyclic 1,N2-propanodeoxyguanosine adducts derived from enals. Anal Biochem. 2006;348:15–23. doi: 10.1016/j.ab.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Parola M, Bellomo G, Robino G, Barrera G, Dianzani MU. 4-Hydroxynonenal as a biological signal: Molecular basis and pathophysiological implications. Antioxid Redox Signal. 1999;1:255–284. doi: 10.1089/ars.1999.1.3-255. [DOI] [PubMed] [Google Scholar]
- Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of an exocyclic guanine adduct on the thermal stability, conformation, and melting thermodynamics of a DNA duplex. Biochemistry. 1992;31:12096–12102. doi: 10.1021/bi00163a019. [DOI] [PubMed] [Google Scholar]
- Poli G, Schaur RJ. 4-Hydroxynonenal in the pathomechanisms of oxidative stress. IUBMB Life. 2000;50:315–321. doi: 10.1080/713803726. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu ST, Majumdar A, Liu JL, Nairn RS, Bernier M, Maher V, Seidman MM. Triplex targeted genomic crosslinks enter separable deletion and base substitution pathways. Nucleic Acids Res. 2005;33:5382–5393. doi: 10.1093/nar/gki851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Kim HY, Jiang H, Penning TM, Harvey RG, Blair IA. Quantification of benzo[a]pyrene diol epoxide DNA-adducts by stable isotope dilution liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1369–1380. doi: 10.1002/rcm.2457. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase ζ. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre LM, Zelasko DA, Harris PL, Perry G, Salomon RG, Smith MA. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- Schneider C, Porter NA, Brash AR. Routes to 4-hydroxynonenal: Fundamental issues in the mechanisms of lipid peroxidation. J Biol Chem. 2008;283:15539–15543. doi: 10.1074/jbc.R800001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Tallman KA, Porter NA, Brash AR. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J Biol Chem. 2001;276:20831–20838. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- Shen X, Jun S, O'Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J Biol Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- Singh US, Moe JG, Reddy GR, Weisenseel JP, Marnett LJ, Stone MP. 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of Salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem Res Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- Stein S, Lao Y, Yang IY, Hecht SS, Moriya M. Genotoxicity of acetaldehyde- and crotonaldehyde-induced 1,N2-propanodeoxyguanosine DNA adducts in human cells. Mutat Res. 2006;608:1–7. doi: 10.1016/j.mrgentox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout MD, Jeong YC, Boysen G, Li Y, Sangaiah R, Ball LM, Gold A, Swenberg JA. LC/MS/MS method for the quantitation of trans-2-hexenal-derived exocyclic 1,N2-propanodeoxyguanosine in DNA. Chem Res Toxicol. 2006;19:563–570. doi: 10.1021/tx050346t. [DOI] [PubMed] [Google Scholar]
- VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J Biol Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- Wacker M, Schuler D, Wanek P, Eder E. Development of a 32P-postlabeling method for the detection of 1,N2-propanodeoxyguanosine adducts of trans-4-hydroxy-2-nonenal in vivo. Chem Res Toxicol. 2000;13:1165–1173. doi: 10.1021/tx000058r. [DOI] [PubMed] [Google Scholar]
- Wacker M, Wanek P, Eder E. Detection of 1,N2-propanodeoxyguanosine adducts of trans-4-hydroxy-2-nonenal after gavage of trans-4-hydroxy-2-nonenal or induction of lipid peroxidation with carbon tetrachloride in F344 rats. Chem Biol Interact. 2001;137:269–283. doi: 10.1016/s0009-2797(01)00259-9. [DOI] [PubMed] [Google Scholar]
- Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J Am Chem Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenseel JP, Reddy GR, Marnett LJ, Stone MP. Structure of an oligodeoxynucleotide containing a 1,N2-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2. Chem Res Toxicol. 2002;15:127–139. doi: 10.1021/tx0101090. [DOI] [PubMed] [Google Scholar]
- West JD, Ji C, Duncan ST, Amarnath V, Schneider C, Rizzo CJ, Brash AR, Marnett LJ. Induction of apoptosis in colorectal carcinoma cells treated with 4-hydroxy-2-nonenal and structurally related aldehydic products of lipid peroxidation. Chem Res Toxicol. 2004;17:453–462. doi: 10.1021/tx034248o. [DOI] [PubMed] [Google Scholar]
- West JD, Marnett LJ. Alterations in gene expression induced by the lipid peroxidation product, 4-hydroxy-2-nonenal. Chem Res Toxicol. 2005;18:1642–1653. doi: 10.1021/tx050211n. [DOI] [PubMed] [Google Scholar]
- West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- Winter CK, Segall HJ, Haddon WF. Formation of cyclic adducts of deoxyguanosine with the aldehydes trans-4-hydroxy-2-hexenal and trans-4-hydroxy-2-nonenal in vitro. Cancer Res. 1986;46:5682–5686. [PubMed] [Google Scholar]
- Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2006;26:381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing DX, Sun LX, Cukier RI, Bu YX. Theoretical prediction of the p53 gene mutagenic mechanism induced by trans-4-hydroxy-2-nonenal. J Phys Chem B. 2007;111:5362–5371. doi: 10.1021/jp0673922. [DOI] [PubMed] [Google Scholar]
- Yadav UC, Ramana KV, Awasthi YC, Srivastava SK. Glutathione level regulates HNE-induced genotoxicity in human erythroleukemia cells. Toxicol Appl Pharmacol. 2008;227:257–264. doi: 10.1016/j.taap.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami K, Yamamoto Y, Kume M, Ishikawa Y, Yamaoka Y, Hiai H, Toyokuni S. Formation of 8-hydroxy-2′-deoxyguanosine and 4-hydroxy-2-nonenal-modified proteins in rat liver after ischemia-reperfusion: Distinct localization of the two oxidatively modified products. Antioxid Redox Signal. 2000;2:127–136. doi: 10.1089/ars.2000.2.1-127. [DOI] [PubMed] [Google Scholar]
- Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J Biol Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- Yi P, Zhan D, Samokyszyn VM, Doerge DR, Fu PP. Synthesis and 32P-postlabeling/high-performance liquid chromatography separation of diastereomeric 1,N2-(1,3-propano)-2′-deoxyguanosine 3′-phosphate adducts formed from 4-hydroxy-2-nonenal. Chem Res Toxicol. 1997;10:1259–1265. doi: 10.1021/tx970100r. [DOI] [PubMed] [Google Scholar]
- Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci USA. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas B, Stillwell SW, Wishnok JS, Trudel LJ, Skipper P, Yu MC, Tannenbaum SR, Wogan GN. Detection and quantification of 4-ABP adducts in DNA from bladder cancer patients. Carcinogenesis. 2007;28:342–349. doi: 10.1093/carcin/bgl142. [DOI] [PubMed] [Google Scholar]
- Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1,N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem Res Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase η-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]