Abstract
Copper is an essential cofactor for many enzymes but at high concentrations it is toxic for the cell. Copper ion concentrations ≥50 µM inhibited growth of Corynebacterium glutamicum. The transcriptional response to 20 µM Cu2+ was studied using DNA microarrays and revealed 20 genes that showed a ≥ 3-fold increased mRNA level, including cg3281-cg3289. Several genes in this genomic region code for proteins presumably involved in the adaption to copper-induced stress, e. g. a multicopper oxidase (CopO) and a copper-transport ATPase (CopB). In addition, this region includes the copRS genes (previously named cgtRS9) which encode a two-component signal transduction system composed of the histidine kinase CopS and the response regulator CopR. Deletion of the copRS genes increased the sensitivity of C. glutamicum towards copper ions, but not to other heavy metal ions. Using comparative transcriptome analysis of the ΔcopRS mutant and the wild type in combination with electrophoretic mobility shift assays and reporter gene studies the CopR regulon and the DNA-binding motif of CopR were identified. Evidence was obtained that CopR binds only to the intergenic region between cg3285 (copR) and cg3286 in the genome of C. glutamicum and activates expression of the divergently oriented gene clusters cg3285-cg3281 and cg3286-cg3289. Altogether, our data suggest that CopRS is the key regulatory system in C. glutamicum for the extracytoplasmic sensing of elevated copper ion concentrations and for induction of a set of genes capable of diminishing copper stress.
Introduction
Due to its ability to change between the oxidised Cu2+ and reduced Cu+ state, copper has become a versatile cofactor for enzymes involved in electron transport or redox reactions such as cytochrome c oxidases or monooxygenases [1]. However, in high concentrations uncomplexed copper ions can generate reactive oxygen species or lead to sulfhydryl depletion and thereby become toxic for the cell [2]. Hence, the amount of copper ions inside the cell must be tightly regulated to prevent deprivation as well as high, toxic copper concentrations. In prokaryotes several copper resistance systems have been characterised, among the best studied systems being those of Enterococcus hirae for Gram-positive and of Escherichia coli for Gram-negative bacteria (for reviews see [2]–[5]).
In E. hirae the cop operon is mainly responsible for copper homeostasis. It consists of four genes coding for a transcriptional repressor (CopY), a copper chaperon (CopZ) and two copper P-type ATPases (CopA and CopB). In the presence of elevated copper concentrations CopZ donates Cu+ to CopY resulting in a derepression of the cop operon and subsequently in copper export by CopB [2]. In E. coli, copper homeostasis is achieved by the action of the MerR-type regulator CueR in concert with two-component systems, such as the CusRS two-component system [4], [6], [7]. In the presence of elevated copper concentrations, CueR activates the transcription of copA and cueO encoding a P-type ATPase and an oxygen-dependent multicopper oxidase, respectively [8], [9]. CopA is responsible for exporting excess Cu+ from the cytoplasm into the periplasm where it is oxidised to the less toxic Cu2+ by CueO. The two-component system CusRS was found to play a role in copper homeostasis under anoxic conditions. It represents a prototypical two-component system [10] where the membrane-bound sensor kinase CusS monitors the periplasmic copper concentration and autophosphorylates a histidine residue at elevated copper concentrations. The phosphoryl group is then transferred to an aspartate residue of the response regulator CusR, which then activates transcription of the cusRS operon and of the adjacent but divergently oriented cusCFBA operon [11]. The translation products CusCBA (a proton-cation antiporter) and CusF (a copper chaperone) then contribute to copper tolerance under copper stress conditions.
Recently a novel type of copper-sensing transcriptional repressors (CsoR-type) was identified in Mycobacterium tuberculosis [12]. In M. tuberculosis, when there is no copper excess stress, the transcriptional regulator CsoR represses the expression of its own operon (cso operon) which includes a gene coding for the putative copper exporter CtpV [13], [14]. By binding Cu+, CsoR loses its DNA-binding affinity resulting in derepression of the cso operon and export of copper via CtpV.
In the soil bacterium Corynebacterium glutamicum [15], [16], a close relative of M. tuberculosis, the control of copper homeostasis has not been studied yet. C. glutamicum belongs to the Corynebacterium-Mycobacterium-Nocardia group of actinomycetes and serves as a nonpathogenic model organism for studying selected features common to corynebacteria and pathogenic mycobacteria. Additionally, this species is of interest due to its biotechnological importance as a producer of L-glutamate and L-lysine. Recent studies suggested that C. glutamicum possesses four cuproproteins (the cytochrome aa 3 oxidase subunits I and II and two multicopper oxidases), two copper transporters which are likely exporters, and four copper chaperones [1], [17], [18]. Since cytochrome aa 3 oxidase plays a key role in the energy household of C. glutamicum [19], copper homeostasis is probably also important for biotechnological production processes.
Here we investigated copper homeostasis and its regulation in C. glutamicum. We identified the copper excess stimulon and provided evidence that the adaption to excess copper involves the two-component signal transduction system CopRS (previously named CgtRS9, Accession UniProtKB Q6M1P4 and Q8NLH8). We could identify direct target genes of the response regulator CopR and show that these were activated in a copper-dependent manner. The CopR DNA-binding motif was identified and evidence was provided that CopR is active in its phosphorylated state.
Results
Response of C. glutamicum to elevated copper concentrations
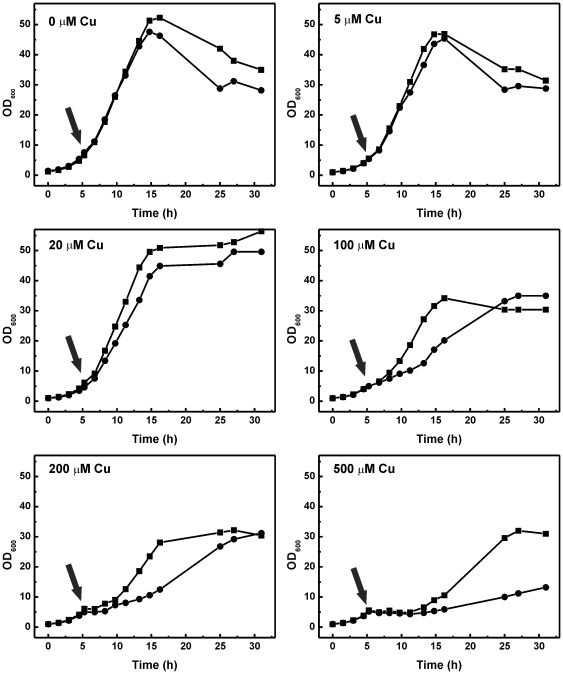
First, the growth of C. glutamicum wild type in the presence of elevated copper ion concentrations was determined. Therefore, cells were grown in CGXII minimal medium (standard copper concentration: 1.25 µM) to an OD600 of 5–6 and then different CuSO4 concentrations (5–500 µM) were added to the cultures (Fig. 1). Whereas the addition of 5 and 20 µM CuSO4 had no effect on the growth of C. glutamicum compared to the control culture (no additional copper), higher CuSO4 concentrations led to reduced growth rates. The addition of 500 µM CuSO4 completely inhibited growth of C. glutamicum, however, after approx. 7 h the culture resumed growth and reached the usual final cell density.
Figure 1. Influence of different copper ion concentrations on the growth of C. glutamicum wild type.
Cells pregrown in CGXII minimal medium with 4% (w/v) glucose were used to inoculate 50 ml of fresh CGXII medium (1.25 µM CuSO4). When the cultures had reached an OD600 of about 5–6, different CuSO4 concentrations were added (indicated by an arrow). The cultures were incubated at 30°C on a rotary shaker at 120 rpm. The growth curves shown here are representative of those from three independent growth experiments with comparable results.
In order to identify genes that were differentially expressed in the presence of elevated copper ion concentrations in the medium, DNA microarray experiments were performed. C. glutamicum wild type cells were pre-cultivated in CGXII minimal medium overnight and then used to inoculate fresh standard CGXII medium (containing 1.25 µM CuSO4) or CGXII medium containing 21.25 µM CuSO4. After the cultures had reached an OD600 of 5–6, the cells were harvested and used for RNA preparation. Altogether 26 genes showed a more than threefold changed mRNA level in at least two of four independent biological replicates (P-value≤0.05). Six of these genes, all of which encode proteins of unknown function, showed a decreased mRNA level and 20 genes an increased mRNA level in the presence of 21.25 µM CuSO4 (Table 1). The latter group included two genes coding for the transcriptional regulators ArsR1 and CopR (previously named CgtR9). ArsR1 belongs to the SmtB/ArsT family of metal-sensing transcriptional repressors and regulates the ars operons (cg0318-cg0319 and cg1705-cg1707) in an arsenic-dependent manner [20]. In the presence of As3+, a derepression of the ars operons occurs which leads to increased tolerance to elevated arsenic concentrations. Despite the increased mRNA level of arsR1 the expression level of the ars operons was not altered, indicating that an elevated copper ion concentration does not cause a dissociation of ArsR1 from its target promoters and hence no derepression of the target genes. CopR is a response regulator and part of the CopR-CopS two-component system [21]. The copRS genes (cg3285 and cg3284, respectively) as well as the up- and downstream genes (cg3286-cg3289 and cg3283-cg3281) exhibited highly increased mRNA levels (Table 1), indicating that this gene region is particularly important for the adaption to copper excess conditions. This assumption is supported by the fact that several of these genes code for proteins that are obviously linked to copper homeostasis, such as a putative copper-transporting ATPase (CopB, Cg3281; Accession UniProtKB Q8NLI0) and a secreted multicopper oxidase (CopO, Cg3287; Accession UniProtKB Q8NLH5). Since genes encoding transcriptional regulators are often localised in the immediate vicinity of their target genes, the CopRS two-component system might be responsible for the altered expression of cg3281-cg3289.
Table 1. Transcriptome comparison of C. glutamicum ATCC 13032 cultivated in CGXII minimal medium supplemented with 21.25 µM CuSO4 (Cu↑) and in standard CGXII medium with 1.25 µM CuSO4 (Cus) using DNA microarrays.
| cg no. | NCgl no. | Gene | Known or predicted function of gene product | Cu↑/Cus |
| Copper-related proteins | ||||
| cg3281 | NCgl2859 | copB | probable cation-transporting ATPasetransmembrane protein | 11.22 |
| cg3282 | NCgl2860 | heavy metal binding transport protein | 14.91 | |
| cg3283 | protein of unknown function | 12.36 | ||
| cg3284 | NCgl2862 | copS | two component sensor kinase | 545.93 |
| cg3285 | NCgl2863 | copR | two component response regulator | 38.68 |
| cg3286 | NCgl2864 | secreted protein of unknown function | 52.96 | |
| cg3287 | NCgl2865 | copO | secreted multicopper oxidase | 267.14 |
| cg3288 | protein of unknown function | 422.20 | ||
| cg3289 | NCgl2866 | tlpA | thioredoxin-like protein | 235.59 |
| Proteins involved in heme biosynthesis and cytochrome c maturation | ||||
| cg0518 | NCgl0422 | hemL | glutamate-1-semialdehyde-2,1-aminomutase | 3.63 |
| cg0519 | NCgl0423 | putative phosphoglycerate mutase | 4.13 | |
| cg0520 | NCgl0424 | periplasmic thioredoxin | 4.62 | |
| cg0522 | NCgl0425 | ccsA | cytochrome c biogenesis protein membrane protein | 3.61 |
| cg0524 | NCgl0427 | ccsB | cytochrome c assembly membrane protein | 3.65 |
| Other proteins | ||||
| cg0915 | NCgl0769 | ftsX | putative cell division protein | 3.79 |
| cg1109 | NCgl0933 | porB | anion-specific porin precursor | 3.61 |
| cg1704 | arsR1 | ArsR-type transcriptional regulator | 6.02 | |
| Proteins of unknown function | ||||
| cg0905 | NCgl0760 | secreted protein of unknown function | 0.19 | |
| cg1514 | NCgl1289 | protein of unknown function | 0.32 | |
| cg1918 | NCgl1635 | secreted protein of unknown function | 0.32 | |
| cg2058 | protein of unknown function | 0.25 | ||
| cg2348 | NCgl2059 | lipoprotein of unknown function | 4.55 | |
| cg2799 | NCgl2452 | secreted protein of unknown function | 3.88 | |
| cg3343 | NCgl2912 | secreted membrane protein of unknown function | 0.31 | |
| cg3344 | NCgl2913 | protein of unknown function | 52.92 | |
| cg4005 | NCgl1288 | lipoprotein of unknown function | 0.22 | |
The mRNA ratios shown represent mean values from four independent DNA microarray experiments starting from independent cultures. The wild type was cultivated in CGXII minimal medium with 4% (w/v) glucose with or without additional 20 µM CuSO4 and mRNA was prepared from cells in the exponential growth phase. The table includes those genes which showed an at least threefold changed mRNA level (increased or decreased) in at least two of the four replicates with a P-value of ≤0.05.
In summary, the transcriptome data led to the identification of the copper excess stimulon and provided evidence for the involvement of CopR in the regulation of copper homeostasis in C. glutamicum. In the remaining part of this study the role of the CopRS two-component system in copper homeostasis was investigated.
Influence of CopRS on the resistance to heavy metals
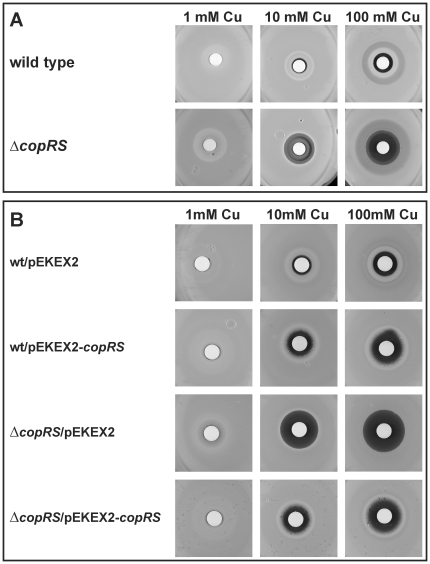
In order to verify the involvement of the CopRS two-component system in copper homoeostasis, the resistance of a copRS deletion mutant [21] to copper ions and other heavy metal salts was compared to the wild type using agar diffusion assays. An increased susceptibility was only observed for copper ions (Fig. 2A) but not for all other heavy metal ions tested (nickel, manganese, zinc, silver, cobalt, lead or cadmium; data not shown). Cultivation of ΔcopRS and C. glutamicum wild type in CGXII minimal medium to which different CuSO4 concentrations (5–500 µM) were added when the cultures had reached an OD600 of 5–6 corroborated the reduced resistance to copper ions of the mutant strain (Fig. 3). The phenotype could be complemented by plasmid-borne copRS expression (Fig. 2B). These results confirmed the assumption that CopRS is involved in copper homeostasis of C. glutamicum.
Figure 2. Agar diffusion assays showing growth inhibition of different C. glutamicum strains by copper ions.
(A) Comparison of the copper sensitivity of C. glutamicum wild type and the ΔcopRS deletion mutant. The inhibition zone (black halo) increased with higher copper concentrations and was larger for the deletion mutant. (B) The copRS deletion can be complemented using the plasmid encoded copRS genes. For experimental details see Material and Methods.
Figure 3. Influence of increasing copper ion concentrations on growth of C. glutamicum wild type (▪) and C. glutamicum ΔcopRS (•).
For experimental details see legend to Fig. 1. The time points of CuSO4 addition are indicated by arrows.
Comparison of the expression profiles of the ΔcopRS mutant and the wild type
To identify the regulon of the response regulator CopR, the transcriptome of the ΔcopRS deletion mutant was compared to that of the wild type using DNA microarrays. For cells grown in standard CGXII medium (1.25 µM CuSO4), no significant gene expression differences were observed, indicating that the CopRS two-component system is not active under this condition (data not shown). However, in the presence of elevated copper ion concentrations (21.25 µM), the mRNA level of 43 genes was changed more than threefold (Table 2) in the ΔcopRS mutant, showing that CopRS is active in the presence of elevated copper concentrations and further supporting our hypothesis that CopRS is involved in copper-induced stress regulation. Besides copS and copR, the strongest downregulation was observed for the gene cluster cg3286-cg3289 (50- to 100-fold decreased mRNA level in the mutant strain), which is located directly upstream of copRS in reverse orientation and belongs to the copper excess stimulon. The genes encode a secreted protein of unknown function (Cg3286), a putative secreted multicopper oxidase (CopO, Cg3287), a protein of unknown function (Cg3288), and a thioredoxin-like protein (TlpA, Cg3289). In addition, several genes which encode transport systems were downregulated in the mutant (Table 2). Except for the glutamate uptake system (GluABCD), none of these transport systems has been characterised in detail yet, but it was supposed that Cg0507 and Cg0508, Cg0924-Cg0927, and Cg3434 are involved in iron uptake [22], [23]. The genes cg3281-cg3283, which were also part of the copper excess stimulon, showed a 1.6-2.3-fold (P-value≤0.05) decreased mRNA level in the ΔcopRS mutant, as well (data not shown).
Table 2. Transcriptome comparison of C. glutamicum wild type (wt) and the ΔcopRS deletion mutant after cultivation in CGXII minimal medium supplemented with 21.25 µM CuSO4 (Cu↑) using DNA microarrays.
| cg no. | NCgl no. | Gene | Known or predicted function of gene product | ΔcopRS Cu↑/wt Cu↑ |
| Copper-related proteins | ||||
| cg3284 | NCgl2862 | copS | two component sensor kinase | 0.00 |
| cg3285† | NCgl2863 | copR | two component response regulator | 0.00 |
| cg3286† | NCgl2864 | secreted protein of unknown function | 0.01 | |
| cg3287† | NCgl2865 | copO | secreted multicopper oxidase | 0.01 |
| cg3288 | protein of unknown function | 0.01 | ||
| cg3289† | NCgl2866 | tlpA | thioredoxin-like protein | 0.02 |
| Transporter or transport-related proteins | ||||
| cg0507 | NCgl0412 | ABC-type transporter, permease component | 0.30 | |
| cg0508† | NCgl0413 | secreted substrate-binding lipoprotein | 0.29 | |
| cg0622 | NCgl0510 | ABC-type cobalt transport system, ATPase component | 0.24 | |
| cg0623 | NCgl0511 | ABC-type cobalt transport system, permease component | 0.26 | |
| cg0624† | NCgl0512 | secreted oxidoreductase; protein of unknown function | 0.25 | |
| cg0924† | NCgl0776 | secreted siderophore-binding lipoprotein | 0.20 | |
| cg0926† | NCgl0777 | siderophore ABC transporter, permease protein | 0.30 | |
| cg0927 | NCgl0778 | siderophore ABC transporter, permease protein | 0.30 | |
| cg2136† | NCgl1875 | gluA | glutamate uptake system ATP-binding protein | 0.38 |
| cg2137 | NCgl1876 | gluB | secreted glutamate binding protein | 0.37 |
| cg2138 | NCgl1877 | gluC | glutamate permease | 0.39 |
| cg2181† | NCgl1915 | ABC-type peptide transport system, secreted component | 0.24 | |
| cg2182 | NCgl1916 | ABC-type peptide transport system, permease component | 0.22 | |
| cg2183 | NCgl1917 | ABC-type peptide transport system, permease component | 0.20 | |
| cg2184 | NCgl1918 | ABC-type peptide transport system, ATPase component | 0.19 | |
| cg2610† | NCgl2294 | ABC-type transport system, secreted component | 0.34 | |
| cg3320† | NCgl2891 | ABC-type transporter, permease component | 0.20 | |
| cg3404† | NCgl2970 | secreted siderophore-binding lipoprotein | 0.26 | |
| Other | ||||
| cg0414† | NCgl0337 | wzz | cell surface polysaccharide biosynthesis/chain length determinant protein | 0.25 |
| cg0424† | NCgl0347 | putative glycosyltransferase | 0.09 | |
| cg0797 | NCgl0665 | prpB1 | methylisocitric acid lyase | 3.16 |
| cg0998 | NCgl0841 | trypsin-like serine protease | 3.53 | |
| cg1055 | NCgl0888 | menG | ribonuclease activity regulator protein RraA | 3.24 |
| cg1290 | NCgl1094 | metE | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 0.29 |
| cg1487 | NCgl1262 | leuC | isopropylmalate isomerase, large subunit | 0.37 |
| cg2925 | NCgl2553 | ptsS | enzyme II sucrose protein | 0.38 |
| cg3226† | NCgl2816 | putative L-lactate permease | 0.02 | |
| Proteins of unknown function | ||||
| cg0077† | NCgl0057 | hypothetical protein | 11.67 | |
| cg0078 | NCgl0058 | hypothetical protein | 10.02 | |
| cg0416 | NCgl0339 | secreted protein | 0.34 | |
| cg0625 | NCgl0513 | secreted protein | 0.29 | |
| cg1326† | NCgl1126 | hypothetical protein | 0.31 | |
| cg2799† | NCgl2452 | secreted protein | 3.29 | |
| cg3009 | hypothetical protein | 0.27 | ||
| cg3213 | NCgl2805 | secreted protein | 3.71 | |
| cg3377† | NCgl2945 | hypothetical protein | 8.76 | |
| cg3378† | NCgl2946 | hypothetical protein | 11.29 | |
The mRNA ratios shown represent mean values from five independent DNA microarray experiments starting from independent cultures. The strains were cultivated in CGXII minimal medium with 4% (w/v) glucose with additional 20 µM CuSO4 and mRNA was prepared from cells in the exponential growth phase. The table includes those genes which showed an at least threefold changed mRNA level (increased or decreased) in at least three of the five replicates with a P-value of ≤0.05. The putative promoter regions of the genes indicated with a † were chosen for the EMSAs (see Fig. S1).
Search for direct target genes of CopR
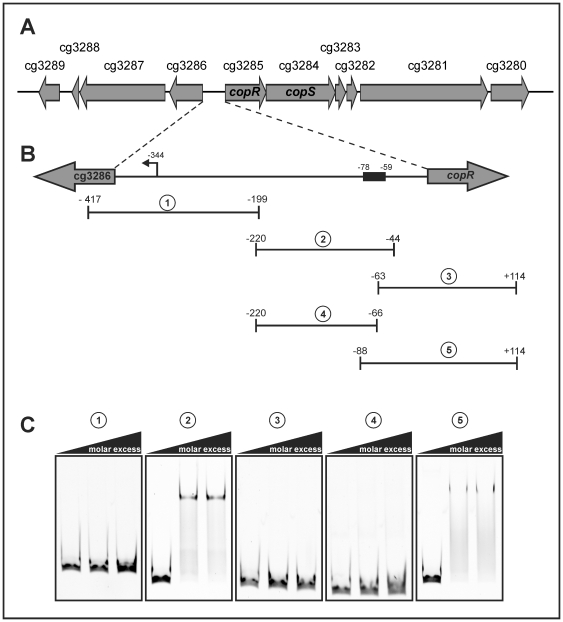
In order to test which of the genes with an altered expression in the ΔcopRS mutant are direct target genes of the response regulator CopR, electrophoretic mobility shift assays (EMSAs) were performed with purified CopR and the relevant promoter regions. As phosphorylated CopR protein was found to have a higher binding affinity compared to unphosphorylated CopR (see below), all gel shift experiments were performed with CopR which had been preincubated with the low molecular weight phosphoryl donor acetylphosphate. In a first set of experiments, 21 promoter regions of putative CopR target genes (see Table 2, indicated with †) and as a negative control the promoter region of an arbitrarily chosen gene (cg1665) were tested. As shown in Fig. S1, binding of CopR was only observed with the DNA fragment containing the promoter region of copR (229 bp, +9 to −220 corresponding to the copR translational start site). A complete retardation was observed in the presence of a 25-fold molar excess of phosphorylated CopR. Since the putative binding site of CopR is located in the intergenic region of copR and cg3286, which are orientated divergently (Fig. 4A), not only the cg3285-cg3281 genes, but also the cg3286-cg3289 genes might be regulated by CopR.
Figure 4. Search for the CopR-binding motif within the cg3286-copR intergenic region.
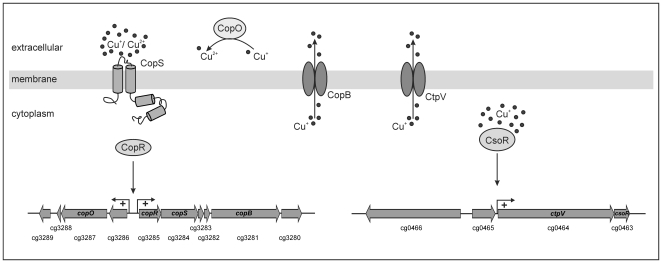
(A) Scheme of the genome region covering cg3280-cg3289 based on the CoryneRegNet annotation (http://coryneregnet.cebitec.uni-bielefeld.de). The genes code for a secreted protein of unknown function (cg3280), a probable cation-transporting ATPase transmembrane protein (copB, cg3281), a heavy metal binding transport protein (cg3282), a protein of unknown function (cg3283), a secreted protein of unknown function (cg3286), a secreted multicopper oxidase (copO, cg3287), a protein of unknown function (cg3288) and a thioredoxin-like protein (tlpA, cg3289). (B) Enlargement of the cg3286-copR intergenic region and position of DNA fragments 1-5 used for EMSAs. The numbers above the DNA fragments indicate the bp distance of their ends to the translation start site of copR. (C) EMSAs with fragments 1–5 (100 nM, 155–219 bp in length) and phosphorylated CopR. The molar excesses of the CopR used were 0-, 25-, 50-fold.
Influence of phosphorylation on CopR binding affinity
To analyse the influence of phosphorylation on the DNA-binding properties of the response regulator CopR, the apparent Kd value of unphosphorylated CopR and of CopR that had been preincubated with acetylphosphate were determined using a 5′-Cy3-labelled DNA fragment (90 bp) that covers the DNA region from −44 to −110 upstream of the copR translational start site (Fig. S2). For CopR preincubated with acetylphosphate an apparent Kd value of 0.4 µM was calculated (CopR concentration required for binding half of the DNA). Unphosphorylated CopR exhibited a significantly lower affinity and showed an apparent Kd value of 2.3 µM. These results indicate that CopR is phosphorylated by acetylphosphate and that phosphorylation increases the DNA-binding affinity.
Identification of the CopR binding site
In order to identify the CopR binding motif, EMSAs with subfragments of the intergenic region between cg3286 and copR were performed. The subfragments were incubated with phosphorylated CopR at two different molar excesses (25- and 50-fold) and separated by 10% native PAGE. As shown in Fig. 4B and C, binding was observed for fragments 2 and 5, but not for fragments 1, 3, and 4. Consequently, a CopR binding site must be localised between bp −59 and −78 with respect to the copR translational start site. This corresponds to position −266 and −285 with respect to the cg3286 transcriptional start site, which had been mapped by primer extension analysis 59 bp upstream of the cg3286 start codon (Fig. S3). The determination of the copR transcriptional start site failed so far although different methods were tested.
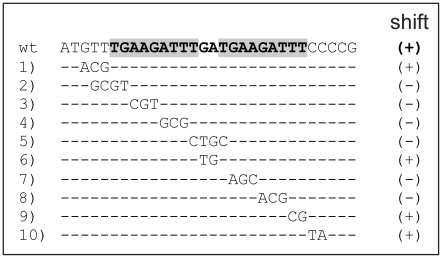
Analysis of the identified DNA region with the motif alignment and search tool MAST (http://meme.sdsc.edu/meme4_4_0/cgi-bin/mast.cgi) revealed the presence of a perfect direct repeat separated by a two base pair linker (TGAAGATTTgaTGAAGATTT). The relevance of this direct repeat for CopR binding was tested by EMSAs with DNA fragments in which two to four nucleotides of the motif or of the surrounding sequence were exchanged (Fig. 5). Mutations in the motifs' flanking sequence or in the linker did not affect CopR binding, whereas mutations of three base pairs within the binding motif led to a complete inhibition of CopR binding. These results confirmed that the predicted DNA motif is relevant for CopR binding.
Figure 5. Mutational analysis of the putative CopR binding site within the cg3286-copR intergenic region.
To determine the importance of the predicted binding motif (sequence in the grey boxes) for CopR binding, fragments with different mutations were tested in EMSAs with phosphorylated CopR. According to the results of the shifts, the fragments were classified into two categories: +, mutated fragment shifted like the wild type fragment; -, the fragment was not shifted.
To identify further CopR target genes, the binding motif TGAAGATTTnnTGAAGATTT was used to search for similar sequences in the whole C. glutamicum genome using the ERGO™ bioinformatics suite (Integrated Genomics, Illinois, USA) allowing four mutations, no deletions and no insertions. 46 hits were found, but only in six cases the putative CopR binding site was located in intergenic regions up to 200 bp upstream of the start codon of the neighbouring gene (cg1336, cg2976, cg3187, cg3337, cg3357 and cg0414). Only one of these genes (cg0414) showed an altered mRNA level in the DNA microarrays comparing ΔcopRS with the wild type (Table 2). However, no binding of CopR to the promoter region of cg0414 was observed in EMSAs (see Fig. S1), indicating that cg0414 is not a direct target gene of CopR.
CopR regulation of the copR and cg3286 gene
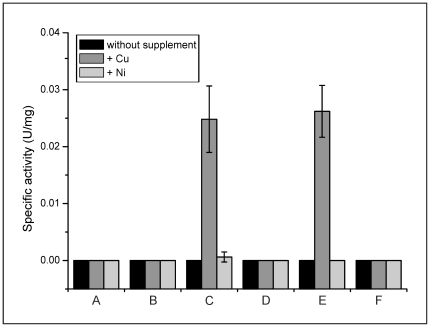
The data reported above showed that CopR binds to the intergenic region between cg3286 and copR, but it was still unclear if CopR activates expression of only the copR promoter, of only the cg3286 promoter or of both promoters. To answer this question, reporter gene assays were performed. The plasmids pET2-IGR and pET2-IGR_inverse, which contain the cg3286-copR intergenic region in both orientations upstream of a promoterless chloramphenicol acetyltransferase (cat) gene, were transferred into C. glutamicum wild type and the ΔcopRS mutant. The resulting strains and the control strains carrying the vector pET2 were cultivated in CGXII minimal medium with or without the addition of 20 µM CuSO4 or 20 µM NiSO4, respectively. When cultivated in standard CGXII medium with 1.25 µM CuSO4 (Fig. 6), none of the strains exhibited Cat activity. Same results were observed when NiSO4 was added (Fig. 6). Thus, CopR did not activate transcription of the reporter gene under these conditions. When the medium was supplemented with 20 µM CuSO4 (Fig. 6), Cat activity was measurable for the wild type strain harbouring the plasmid pET2-IGR or pET2-IGR_inverse, but not for the ΔcopRS mutant harbouring one of these plasmids. Also the control strains carrying pET2 had no measurable Cat activity. These results showed that both the cg3286 promoter and the copR promoter are activated by CopRS in a strictly copper-dependent manner.
Figure 6. Specific Cat activities of C. glutamicum wild type (wt) and the ΔcopRS mutant.
Both strains carrying either the pET2, the pET2-IGR (copR promoter) or the pET2-IGR_inverse (cg3286 promoter) vector. Cat activities were measured after growth in CGXII medium containing either 1.25 µM CuSO4 (black bars), 20 µM additional CuSO4 (dark grey bars) or 20 µM additional NiSO4 (light grey bars). A, wt/pET2; B, ΔcopRS/pET2; C, wt/pET2-IGR (copR promoter); D, ΔcopRS/pET2-IGR; E, wt/pET2-IGR_inverse (cg3286 promoter); F, ΔcopRS/pET2-IGR_inverse. The values represent averages and standard deviations of three biological replicates.
Discussion
Copper-dependent organisms have evolved sophisticated regulatory mechanisms to ensure a sufficient supply of copper ions on the one hand and a resistance against high concentrations on the other. To enable the cell to react to copper-induced stress, several copper ion sensors were evolved like CueR of E. coli, CopY of E. hirae and CsoR of M. tuberculosis [2], [4], [12], [24]. Besides these cytoplasmic one-component regulators, two-component regulatory systems were identified which are involved in the sensing of elevated copper ion concentrations in the periplasm. Examples are the CusRS system of E. coli [7], [11] and the CinSR system of Pseudomonas putida [25].
In the present work we studied the response of C. glutamicum to elevated copper ion concentrations and identified a novel copper-responsive two-component system, CopRS, which is the key regulatory system for copper ion resistance in this bacterium. The sensor kinase CopS shows the typical domain structure of two-component sensor kinases. The intracellular N-terminus is followed by two predicted transmembrane helices (TMHHM Server: http://www.cbs.dtu.dk/services/TMHMM/) connected via a short periplasmic loop (31 aa). The cytoplasmatic C-terminus harbours a HAMP domain, a HisKA domain and a HATPase_c domain (SMART: http://smart.embl-heidelberg.de/). The sensor kinase CopS of C. glutamicum shows 21% and 22% sequence identity to CusS of E. coli and CinS of P. putida, respectively. As expected, the C-terminal catalytic domains are highly similar but the periplasmic loop of C. glutamicum CopS differs from the others since it is much shorter. Within the periplasmic loop of CinS two histidine residues (H37, H147) were identified as potential copper binding site [25]. These two histidine residues are also conserved in CusS but not in CopS. Nevertheless CopS harbours three histidine (H39, H41, H56) and two methionine residues (M42, M44) within its periplasmic loop that might play a role in copper recognition/binding. The response regulator CopR exhibits 33% and 34% sequence identity to E. coli CusR and P. putida CinR, respectively.
C. glutamicum wild type grows unaffectedly up to 20 µM CuSO4 (Fig. 1) which is similar to the copper tolerance of M. tuberculosis, which grows normally up to 50 µM copper [14] and to that of E. coli [7], while it is much lower than that of bacteria like E. hirae and Pseudomonas aeruginosa, which can grow at copper concentrations up to approx. 8 mM [2], [26].
DNA microarray analyses indicated the involvement of the two-component system CopRS in copper homeostasis (Table 1). This was confirmed by copper sensitivity tests on agar plates and by growth experiments in the presence of different copper concentrations (Fig. 2 and 3). Our approaches revealed two gene clusters regulated by CopR, cg3286-cg3289 and cg3285-cg3281 with cg3285 and cg3284 coding for CopR and CopS, respectively. The translation products of these genes possibly play a role in copper resistance. For example the multicopper oxidase CopO (Cg3287) can detoxify Cu+ by converting it to the less toxic Cu2+ and by binding free Cu ions while the putative copper transporting P-type ATPase CopB (Cg3281) most likely functions as a copper export pump.
The determined apparent Kd value of 0.4 µM for the phosphorylated form of CopR is in the same range as described for other response regulators [27]. The binding site (TGAAGATTTnnTGAAGATTT) of CopR within the cg3286-copR intergenic region (Fig. 4) seems to be the only one within the entire genome of C. glutamicum. This situation is not unusual for copper regulatory systems, as e.g. the E. coli CusRS system also only activates the transcription of its own genes and the divergently orientated cusCFB operon [6]. Binding of CopR to this single binding site results in transcriptional activation of both divergently located gene regions (copR-cg3281 and cg3286-cg3289). Further the activation is depended on the existence of the CopRS system and this system by itself is only active in the presence of elevated copper ion concentrations (Fig. 6). Since the binding motif is rather far upstream of the transcriptional start site of cg3286 (−266 to −285 bp) the question arises how these genes can be under the control of CopR.
The cop gene region (cg3281-cg3289) exhibits an extraordinary high G+C content in comparison to the rest of the genome and is highly conserved in Corynebacterium diphtheriae (more than 95% identical) [15], [16]. This high level of sequence conservation points to a recent horizontal gene transfer between these two species possibly mediated by the natural Corynebacterium plasmid pLEW279b. This plasmid contains a gene region with 97% nucleotide similarity to the cop gene region of C. glutamicum wild type. It was shown that the plasmid sequence is a mosaic of genes and accessory elements acquired from related Gram-positive sources [28].
In C. diphtheriae a gene encoding a small protein of about 9 kDa was proposed to be located between the homologues of cg3286 and copR. In C. glutamicum the nucleotide sequence of this gene can also be found within the intergenic region of cg3286 and copR extending from position −108 to −334 bp with respect to the copR translational start site and orientated divergently to copR. The translational start point of the hypothetical gene would be located 30 bp upstream of the CopR binding site and might form an operon structure with the following cg3286 gene. However, so far no evidence is available that this putative gene is expressed in C. diphtheriae or C. glutamicum. Another explanation for the long distance between the CopR binding site and the cg3286 transcriptional start site could be the existence of a regulatory RNA element within the intergenic region, but inspection of this region with the software tool RNAstructure [29] revealed no significant RNA structure. Further experiments are on the way to assign a function to this DNA region.
The copper stimulon not only consists of the cop region (cg3281-cg3289), but also of genes coding for proteins involved in heme biosynthesis and cytochrome c maturation (Table 1). It can be assumed that excess of copper stimulates synthesis of heme and cytochrome c since these are also cofactors of the respiratory chain protein complexes as well as copper. A link between iron and copper was previously also shown for E. coli [30], P. aeruginosa [26], [31], yeasts [32] and mammals [33]. The existence of this mechanistic link in C. glutamicum is further underlined by the finding that the putative copper transporter CtpV (Cg0464; Accession UniProtKB Q6M7X6) belongs to the regulon of the master regulator of iron homeostasis, DtxR [34]. The ctpV gene together with the downstream located gene csoR were highly upregulated in the presence of elevated copper ion concentrations (1091-fold (P-value of 0.07) and 9-fold (P-value of 0.06) changed mRNA level, respectively) in C. glutamicum wild type. csoR codes for a homologue of the transcriptional regulator CsoR from M. tuberculosis (27% identity) which was shown to be involved in the copper stress response [12] by binding intracellular Cu+ followed by derepression of the ctpV gene coding for a putative copper-exporting P-type ATPase [13]. Therefore, it is reasonable to assume that CsoR (Accession UniProtKB Q8NTC2) of C. glutamicum also functions as a cytoplasmatic copper sensor and regulates expression of ctpV. Studies are underway to confirm this hypothesis. Taken together, our work suggests that C. glutamicum possesses both an intracellular copper sensor, i. e. CsoR, and the extracellular copper sensor system CopRS.
Based on these data we can propose a model of the dual copper resistance mechanism in C. glutamicum (Fig. 7). Under aerobic conditions copper is present in the periplasmic space in the Cu+ and Cu2+ state [35]. The two-component system CopRS recognises high extracellular copper concentrations followed by transcriptional activation of the two putative operons cg3286-cg3289 and copR-cg3281 containing genes encoding copper resistance proteins, e.g. the putative multicopper oxidase CopO and the putative copper export ATPase CopB. As a result CopB exports excess of Cu+ from the cytoplasm to the ‘periplasm’. In the ‘periplasm’ CopO can detoxify Cu+ by oxidising it to Cu2+ which is less toxic and less able to diffuse through the cytoplasmic membrane [7] and by sequestering free Cu ions. The Cu-specific regulator CsoR senses high intracellular copper concentrations and activates (or derepresses) transcription of the copper export ATPase CtpV which also might be responsible for exporting excess copper from the cytoplasm.
Figure 7. Model of copper excess response in C. glutamicum.
The CopS sensor kinase recognises high extracellular copper concentrations followed by autophosphorylation and phosphotransfer to the response regulator CopR. Phosphorylated CopR binds to the direct repeat (TGAAGATTTnnTGAAGATTT) within the cg3286-copR intergenic region. This results in a transcriptional activation of both putative operons (cg3286-cg3289 and copR-cg3281) containing genes encoding copper resistance proteins, e.g. a putative multicopper oxidase (CopO) and a copper export ATPase (CopB). CopO can detoxify Cu+ by converting it to the less toxic Cu2+ and by binding free Cu ions. CopB is a cation ATPase and likely functions as a copper export pump. The Cu-specific regulator CsoR senses high intracellular copper concentrations and activates (or derepresses) the transcription of the copper export ATPase CtpV which is part of the copper detoxification process.
Materials and Methods
Bacterial strains, media, and growth conditions
Bacterial strains and plasmids used or constructed in the course of this work are listed in Table 3, oligonucleotides in supplementary Table S1. C. glutamicum was routinely cultivated aerobically in 500-ml shaking flasks with 50 ml CGXII minimal medium [36] containing 4% (w/v) glucose as carbon and energy source on a rotary shaker (120 rpm) at 30°C. For strain construction and maintenance, BHIS agar plates (BHI agar (Difco, Detroit, USA) with 0.5 M sorbitol) were used. E. coli DH5α was grown aerobically on a rotary shaker (120 rpm) at 37°C in LB medium or on LB agar plates (LB medium with 1.5% (w/v) agar). If appropriate, kanamycin was added to final concentrations of 25 µg/ml (C. glutamicum) or 50 µg/ml (E. coli).
Table 3. Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
| Strains | ||
| C. glutamicum ATCC13032 | Biotin-auxotrophic wild type strain | [51] |
| C. glutamicum ΔcopRS | Derivative of ATCC13032 with an in-frame deletion of the copRS genes | [21] |
| E. coli DH5α | F- φ80dlacΔ (lacZ)M15 Δ (lacZYA-argF) U169 endA1 recA1 hsdR17 (rK -, mK +) deoR thi-1 phoA supE44 λ- gyrA96 relA1 | Invitrogen |
| E. coli BL21(DE3) | ompT hsdS B (rB -mB -) gal dcm (DE3) | [52] |
| Plasmids | ||
| pET28b | Kanr; vector for overexpression of genes in E. coli, adding an N-terminal hexahistidine affinity tag to the synthesised protein (pBR322 oriVE.c. PT7 lacI) | Novagen, Merck KGaA |
| pET28b-NHis6-CopR | Kanr; pET28b derivative for overproduction of CopR with an N-terminal hexahistidine tag | This work |
| pEKEx2 | Kanr; C. glutamicum/E. coli shuttle vector for regulated gene expression (Ptac, lacI Q, pBL1 oriVC. g ., pUC18 oriVE. c.) | [39] |
| pEKEx2-copRS | Kanr; pEKEx2 derivative containing the copRS genes from C. glutamicum under control of the tac promoter | This work |
| pET2 | Kanr; promoter-probe vector | [40] |
| pET2-IGR | Kanr; pET2 with the whole intergenic region of cg3286 and copR (402 bp), orientated cg3286 to copR | This work |
| pET2-IGR_inverse | Kanr; pET2 with the whole intergenic region of cg3286 and copR (402 bp), orientated copR to cg3286 | This work |
Agar diffusion assay
To compare the resistance of C. glutamicum to heavy metal ions, strains were grown in CGXII minimal medium to an OD600 of 6. The resulting cells were diluted to an OD600 of 0.05 in CGXII soft agar (CGXII medium with 0.75% (w/v) agar) and 4 ml thereof were poured onto a CGXII agar plate. Once the agar became solid, a glass fibre filter disc (10 mm diameter, 2 µm pore size; Millipore) was placed onto the plate and 70 µl of a heavy metal ion solution (different concentrations of CuSO4, NiSO4, MnCl2, ZnCl2, AgNO3, CoCl2, PbCl2 or CdCl2) were spotted onto the paper disc. The plates were incubated at 30°C and the zones of inhibition were determined after 48 h. If appropriate, kanamycin (25 mg/ml) and IPTG (1 mM) were added to the media.
Recombinant DNA work
The enzymes for recombinant DNA work were obtained from Roche Diagnostics or New England Biolabs. All oligonucleotides were synthesised by Eurofins MWG Operon (Table S1). Routine methods like PCR, restriction or ligation were carried out according to standard protocols [37]. Plasmids were isolated from E. coli with the QIAprepspin miniprep kit (Qiagen). E. coli was transformed by the RbCl method [38]. All constructs described below were confirmed via DNA sequencing performed by LGC genomics.
Construction of expression plasmids
For IPTG-inducible expression of copRS, the coding region of these genes including 9 bp upstream of the copR start codon plus an artificial ribosomal binding site (AAGGAGA) was amplified using chromosomal DNA of C. glutamicum ATCC 13032 as template, the oligonucleotide pair copRS_SalI_fw/copRS_EcoRI_rv and the Expand High Fidelity PCR kit (Roche Diagnostics). The PCR product was digested with SalI and EcoRI and cloned into pEKEx2 [39] cut with the same enzymes.
For overproduction and purification of CopR with an amino-terminal histidine tag, the copR coding region was amplified using oligonucleotides that introduce an NdeI restriction site (copR_NdeI_fw) and a XhoI restriction site after the stop codon (copR_XhoI_rv). The purified 740 bp PCR product was digested with NdeI and XhoI and cloned into the expression vector pET28b, resulting in plasmid pET28b-NHis6-CopR. The CopR protein encoded by this plasmid (260 amino acids, 28.8 kDa) contains 20 additional amino acids (MGSSHHHHHHSSGLVPRGSH) at the N-terminus including a hexahistidine tag and a thrombin cleavage site.
Construction of reporter gene plasmids and measurement of chloramphenicol acetyltransferase activity
The whole intergenic region, meaning the region between the start codons of the genes cg3286 and copR (402 bp), was amplified with the primer pairs PstI_fw IGR-cg3286_copR/BamHI_rv IGR-cg3286_copR and BamHI_fw IGR-cg3286_copR/PstI_rv IGR-cg3286_copR, respectively, and cloned into the corynebacterial promoter-probe vector pET2 [40]. The resulting plasmids pET2-IGR and pET2-IGR_inverse contained the intergenic region upstream of the promoterless cat gene and were transferred into C. glutamicum wild type and the ΔcopRS mutant by electroporation. The promoter activities were determined by measuring the chloramphenicol acetyltransferase (CAT) activity via the formation of 5-thio-2-nitrobenzoate photometrically at 412 nm and 30°C [41]–[43].
DNA microarray analysis
For RNA preparation, C. glutamicum strains were cultivated overnight in CGXII minimal medium containing 4% (w/v) glucose. Cells from these precultures were washed in 0.9% (w/v) NaCl and used for inoculation of CGXII minimal medium containing 4% (w/v) glucose with or without the addition of 20 µM CuSO4. At an OD600 of 5–6, 20 ml of the cultures were poured into ice-containing tubes precooled to −20°C and cells were harvested by centrifugation (3 min, 4200 x g, 4°C). The cell pellet was directly used for RNA isolation as described before [44]. All DNA microarray analyses were performed with custom-made DNA microarrays based on 70-mer oligonucleotides obtained from Operon Biotechnologies. The comparisons were performed from four or five independent biological replicates. The experimental details for handling of these microarrays were described before [45]. We defined the experimental data as significant if the mRNA level showed an at least threefold change in at least two of four (Table 1) or three of five (Table 2) replicates and the P-value was ≤0.05. Processed and normalized data as well as experimental details conformed to the MIAME guidelines [46] were stored in the in-house microarray database [47] for further analysis and in the Gene Expression Omnibus (GEO) repository under the accession number GSE27510.
Primer extension analysis
Nonradioactive primer extension analyses of cg3286 were performed as described previously [48], [49] using the IRD800-labeled oligonucleotides PE_cg3286_30 and PE_cg3286_80 (Table S1) and 10 µg RNA from C. glutamicum wild type or strain ΔcopRS as template. The length of the primer extension product was determined by running the four lanes of a DNA-sequencing reaction mixture set up using the same oligonucleotide as that used for the reverse transcription alongside the primer extension product. The template for DNA sequencing covered the whole promoter region and was obtained by PCR using chromosomal DNA of C. glutamicum ATCC13032 and the oligonucleotide pair cg3286_fw/cg3286_rv (Table S1).
Overproduction and purification of CopR
The response regulator CopR was overproduced in E. coli BL21(DE3) using the expression plasmid pET28b-NHis6-CopR. Cells were grown at 37°C and 120 rpm in 500 ml LB medium with 50 µg/ml kanamycin to an OD600 of ∼0.6 before adding 1 mM isopropyl β–D-thiogalactoside (IPTG). After cultivation for another 4 hours, cells were harvested by centrifugation at 4°C and stored at −20°C. NHis6-CopR was purified by Ni2+-chelate affinity chromatography followed by buffer exchange against bandshift buffer (50 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 10% (v/v) glycerol, pH 7.5) as described for the response regulator CitB previously [44]. Protein concentrations were determined with the Bradford protein assay (Uptima) using bovine serum albumin (BSA) as standard.
Electrophoretic mobility shift assays (EMSAs)
For testing the binding of CopR to putative target promoters, purified CopR protein (0–18 µM) was mixed with 100 ng DNA fragments (30–229 bp, final concentration 25–255 nM) in a total volume of 20 µl. The binding buffer contained 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA and 10% (v/v) glycerol. NHis6-CopR, either in the unphosphorylated state or after phosphorylation with 50 mM acetylphosphate, was mixed with the DNA fragment, incubated at room temperature for 30 min and then loaded onto a 10% (DNA fragments >100 bp) or 15% (DNA fragments <100 bp) native polyacrylamide gel. For phosphorylation of NHis6-CopR, the protein was incubated for 60 min with 50 mM acetylphosphate before addition of the DNA fragments. Electrophoresis and DNA detection with SybrGreen I were performed as described before [50]. The DNA fragments were generated by PCR or by primer annealing for insertion of base pair mutations, purified with the PCR purification Kit (Qiagen) and eluted in double-distilled H2O. For determination of the apparent Kd value, the DNA fragment (90 bp) was amplified using an oligonucleotide labelled with the fluorescent dye Cy3 (Eurofins MWG Operon). Purified and phosphorylated CopR was mixed with the DNA fragment (8 nM), incubated at room temperature for 30 min and then loaded onto a 15% native polyacrylamide gel. Electrophoresis was performed as described before [50]. The DNA was detected using a fluorescence scanner (Typhoon Trio, GE Healthcare) at an excitation of 532 nm and an emission of 580 nm.
Supporting Information
EMSAs for testing the binding of CopR. The binding of CopR to the promoter regions of putative target genes referring to Table 2 was analysed. Fragment cg1665 served as negative control. DNA fragments (100 ng, 157–263 bp) were incubated for 30 min at room temperature with phosphorylated CopR at different molar excesses (0- to 100-fold, see legend). For experimental details see Material and Methods.
(TIF)
Electrophoretic mobility shift assays and calculated plots for the determination of the apparent Kd values. The Kd values of unphosphorylated CopR (A) and phosphorylated CopR (B) binding to the copR promoter region were determined. The 5′-Cy3-labelled DNA probes (8 nM) were incubated with various amounts of phosphorylated (0–500 nM) and unphosphorylated (0–5000 nM) CopR, respectively. Free and CopR-bound DNA were separated by electrophoresis using a 15% native polyacylamide gel and detected using a fluorescence scanner. The amount of free and protein-bound DNA was quantified using ImageQuant™ TL (GE Healthcare). The ratios of the amount of bound to total DNA were calculated and plotted against the protein concentration in order to determine the Kd values.
(TIF)
Primer extension analysis of the gene cg3286. The analysis was performed using the oligonucleotide PE_cg3286_30 and 10 µg of total RNA from wild type and the ΔcopRS mutant. The transcriptional start site is indicated by an asterisk. The strains were grown in CGXII medium with 4% (w/v) glucose supplemented with 20 µM CuSO4 and harvested in the exponential growth phase for RNA isolation.
(TIF)
Oligonucleotides used in this study.
(DOC)
Acknowledgments
We thank Julia Frunzke for critically reading the manuscript and for her helpful feedback.
Footnotes
Competing Interests: All authors are affiliated to the “Forschungszentrum Jülich GmbH”. This does not alter the authors' adherence to all PloS ONE policies on sharing data and materials.
Funding: The authors have no support or funding to report.
References
- 1.Ridge PG, Zhang Y, Gladyshev VN. Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One. 2008;3:e1378. doi: 10.1371/journal.pone.0001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev. 2003;27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 3.Solioz M, Abicht HK, Mermod M, Mancini S. Response of gram-positive bacteria to copper stress. J Biol Inorg Chem. 2010;15:3–14. doi: 10.1007/s00775-009-0588-3. [DOI] [PubMed] [Google Scholar]
- 4.Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 5.Osman D, Cavet JS. Copper homeostasis in bacteria. Adv Appl Microbiol. 2008;65:217–247. doi: 10.1016/S0065-2164(08)00608-4. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005;56:215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- 7.Outten FW, Huffman DL, Hale JA, O'Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 8.Fan B, Grass G, Rensing C, Rosen BP. Escherichia coli CopA N-terminal Cys(X)(2)Cys motifs are not required for copper resistance or transport. Biochem Biophys Res Commun. 2001;286:414–418. doi: 10.1006/bbrc.2001.5367. [DOI] [PubMed] [Google Scholar]
- 9.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci U S A. 2000;97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 11.Munson GP, Lam DL, Outten FW, O'Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182:5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 13.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J Bacteriol. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggeling L, Bott M. CRC Press; 2005. Handbook Of Corynebacterium glutamicum. [Google Scholar]
- 16.Burkovski A. Genomics and Molecular Biology. Norfolk, United Kingdom: Caister Academic Press; 2008. Corynebacteria. [Google Scholar]
- 17.Niebisch A, Bott M. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch Microbiol. 2001;175:282–294. doi: 10.1007/s002030100262. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Gladyshev VN. General trends in trace element utilization revealed by comparative genomic analyses of Co, Cu, Mo, Ni, and Se. J Biol Chem. 2010;285:3393–3405. doi: 10.1074/jbc.M109.071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bott M, Niebisch A. The respiratory chain of Corynebacterium glutamicum. J Biotechnol. 2003;104:129–153. doi: 10.1016/s0168-1656(03)00144-5. [DOI] [PubMed] [Google Scholar]
- 20.Ordonez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, et al. Evolution of metal(loid) binding sites in transcriptional regulators. J Biol Chem. 2008;283:25706–25714. doi: 10.1074/jbc.M803209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocan M, Schaffer S, Ishige T, Sorger-Herrmann U, Wendisch VF, et al. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J Bacteriol. 2006;188:724–732. doi: 10.1128/JB.188.2.724-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kronemeyer W, Peekhaus N, Kramer R, Sahm H, Eggeling L. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J Bacteriol. 1995;177:1152–1158. doi: 10.1128/jb.177.5.1152-1158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frunzke J, Bott M. Chapter 11: Regulation of iron homeostasis in Corynebacterium glutamicum. In: Burkovski A, editor. Corynebacteria: Genomics and Molecular Biology. Norfolk, United Kingdom: Caister Academic Press; 2008. [Google Scholar]
- 24.Stoyanov JV, Magnani D, Solioz M. Measurement of cytoplasmic copper, silver, and gold with a lux biosensor shows copper and silver, but not gold, efflux by the CopA ATPase of Escherichia coli. FEBS Lett. 2003;546:391–394. doi: 10.1016/s0014-5793(03)00640-9. [DOI] [PubMed] [Google Scholar]
- 25.Quaranta D, McEvoy MM, Rensing C. Site-directed mutagenesis identifies a molecular switch involved in copper sensing by the histidine kinase CinS in Pseudomonas putida KT2440. J Bacteriol. 2009;191:5304–5311. doi: 10.1128/JB.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, et al. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol. 2006;188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janausch IG, Garcia-Moreno I, Lehnen D, Zeuner Y, Unden G. Phosphorylation and DNA binding of the regulator DcuR of the fumarate-responsive two-component system DcuSR of Escherichia coli. Microbiology. 2004;150:877–883. doi: 10.1099/mic.0.26900-0. [DOI] [PubMed] [Google Scholar]
- 28.Williams LE, Detter C, Barry K, Lapidus A, Summers AO. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Applied and Environmental Microbiology. 2006;72:4899–4906. doi: 10.1128/AEM.00354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kershaw CJ, Brown NL, Constantinidou C, Patel MD, Hobman JL. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology. 2005;151:1187–1198. doi: 10.1099/mic.0.27650-0. [DOI] [PubMed] [Google Scholar]
- 31.Frangipani E, Slaveykova VI, Reimmann C, Haas D. Adaptation of aerobically growing Pseudomonas aeruginosa to copper starvation. J Bacteriol. 2008;190:6706–6717. doi: 10.1128/JB.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Bakel H, Strengman E, Wijmenga C, Holstege FC. Gene expression profiling and phenotype analyses of S. cerevisiae in response to changing copper reveals six genes with new roles in copper and iron metabolism. Physiol Genomics. 2005;22:356–367. doi: 10.1152/physiolgenomics.00055.2005. [DOI] [PubMed] [Google Scholar]
- 33.Winzerling JJ, Law JH. Comparative nutrition of iron and copper. Annu Rev Nutr. 1997;17:501–526. doi: 10.1146/annurev.nutr.17.1.501. [DOI] [PubMed] [Google Scholar]
- 34.Wennerhold J, Bott M. The DtxR regulon of Corynebacterium glutamicum. J Bacteriol. 2006;188:2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim EH, Rensing C, McEvoy MM. Chaperone-mediated copper handling in the periplasm. Nat Prod Rep. 2010;27:711–719. doi: 10.1039/b906681k. [DOI] [PubMed] [Google Scholar]
- 36.Keilhauer C, Eggeling L, Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, MacCallum P, Russell D. A Laboratory Manual. 3rd ed. Ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning. [Google Scholar]
- 38.Hanahan D. Techniques of Transformation of E. coli. In: Glover DM, editor. DNA-Cloning. IRL-Press; 1985. pp. 109–135. [Google Scholar]
- 39.Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene. 1991;102:93–98. doi: 10.1016/0378-1119(91)90545-m. [DOI] [PubMed] [Google Scholar]
- 40.Vasicova P, Abrhamova Z, Nesvera J, Patek M, Sahm H, et al. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnology Techniques. 1998;12:743–746. [Google Scholar]
- 41.Engels V, Wendisch VF. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J Bacteriol. 2007;189:2955–2966. doi: 10.1128/JB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Netzer R, Krause M, Rittmann D, Peters-Wendisch PG, Eggeling L, et al. Roles of pyruvate kinase and malic enzyme in Corynebacterium glutamicum for growth on carbon sources requiring gluconeogenesis. Arch Microbiol. 2004;182:354–363. doi: 10.1007/s00203-004-0710-4. [DOI] [PubMed] [Google Scholar]
- 43.Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, et al. Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol. 2003;104:99–122. doi: 10.1016/s0168-1656(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 44.Brocker M, Schaffer S, Mack C, Bott M. Citrate utilization by Corynebacterium glutamicum is controlled by the CitAB two-component system through positive regulation of the citrate transport genes citH and tctCBA. J Bacteriol. 2009;191:3869–3880. doi: 10.1128/JB.00113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frunzke J, Engels V, Hasenbein S, Gatgens C, Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol. 2008;67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 47.Polen T, Wendisch VF. Genomewide expression analysis in amino acid-producing bacteria using DNA microarrays. Appl Biochem Biotechnol. 2004;118:215–232. doi: 10.1385/abab:118:1-3:215. [DOI] [PubMed] [Google Scholar]
- 48.Engels S, Ludwig C, Schweitzer J-E, Mack C, Bott M, et al. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Molecular Microbiology. 2005;57:576–591. doi: 10.1111/j.1365-2958.2005.04710.x. [DOI] [PubMed] [Google Scholar]
- 49.Engels S, Schweitzer JE, Ludwig C, Bott M, Schaffer S. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor sigmaH. Mol Microbiol. 2004;52:285–302. doi: 10.1111/j.1365-2958.2003.03979.x. [DOI] [PubMed] [Google Scholar]
- 50.Wennerhold J, Krug A, Bott M. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem. 2005;280:40500–40508. doi: 10.1074/jbc.M508693200. [DOI] [PubMed] [Google Scholar]
- 51.Abe S, Takayama K, Kinoshita S. Taxonomical studies on glutamic acid producing bacteria. J Gen Appl Microbiol. 1967;13:279–301. [Google Scholar]
- 52.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EMSAs for testing the binding of CopR. The binding of CopR to the promoter regions of putative target genes referring to Table 2 was analysed. Fragment cg1665 served as negative control. DNA fragments (100 ng, 157–263 bp) were incubated for 30 min at room temperature with phosphorylated CopR at different molar excesses (0- to 100-fold, see legend). For experimental details see Material and Methods.
(TIF)
Electrophoretic mobility shift assays and calculated plots for the determination of the apparent Kd values. The Kd values of unphosphorylated CopR (A) and phosphorylated CopR (B) binding to the copR promoter region were determined. The 5′-Cy3-labelled DNA probes (8 nM) were incubated with various amounts of phosphorylated (0–500 nM) and unphosphorylated (0–5000 nM) CopR, respectively. Free and CopR-bound DNA were separated by electrophoresis using a 15% native polyacylamide gel and detected using a fluorescence scanner. The amount of free and protein-bound DNA was quantified using ImageQuant™ TL (GE Healthcare). The ratios of the amount of bound to total DNA were calculated and plotted against the protein concentration in order to determine the Kd values.
(TIF)
Primer extension analysis of the gene cg3286. The analysis was performed using the oligonucleotide PE_cg3286_30 and 10 µg of total RNA from wild type and the ΔcopRS mutant. The transcriptional start site is indicated by an asterisk. The strains were grown in CGXII medium with 4% (w/v) glucose supplemented with 20 µM CuSO4 and harvested in the exponential growth phase for RNA isolation.
(TIF)
Oligonucleotides used in this study.
(DOC)