Abstract
Voltage-dependent K+ (Kv) channels require lipid phosphates for functioning. The S4 helix, which carries the gating charges in the voltage-sensing domain (VSD), inserts into membranes while being stabilized by a protein-lipid interface in which lipid phosphates play an essential role. To examine the physical basis of the protein-lipid interface in the absence of lipid phosphates, we performed molecular dynamics (MD) simulations of a KvAP S4 variant (S4mut) in bilayers with and without lipid phosphates. We find that in dioleoyltrimethylammoniumpropane (DOTAP) bilayers lacking lipid phosphates, the gating charges are solvated by anionic counterions and, hence, lack the bilayer support provided by phosphate-containing palmitoyloleoylglycerophosphocholine (POPC) bilayers. The result is a water-permeable bilayer with a significantly smaller deformations around the peptide. Together, these results provide an explanation for the non-functionality of VSDs in terms of a destabilizing protein-lipid interface.
Keywords: molecular dynamics simulations, dioleoyltrimethylammoniumpropane (DOTAP), voltage-dependent K+ (Kv) channels, protein-lipid interactions
1. Introduction
The lipid bilayer of the plasma membrane defines the boundary of biological cells and compartmentalizes them by providing a permeability barrier. Membrane proteins embedded in bilayers allow controlled communication with the surrounding environment. The composition of the bilayer is not only important for achieving a proper integration of these membrane proteins, it is also vital for their function. The critical dependence of proteins on the chemical nature of the surrounding lipid bilayer suggests that the two have co-evolved.1 A striking example is the absolute requirement for lipid phosphate groups in the function of voltage-dependent K+ (Kv) channels. 2–4 Molecular dynamics (MD) simulations have identified, and described at atomic detail, a carefully orchestrated protein-lipid interface surrounding the channel gating charges. 5 By removing negatively charged lipid phosphates, the key players in connecting the protein to the bilayer, an entirely new set of protein-lipid interactions is bound to arise. Here, as a first step towards a detailed understanding as to why Kv channels do not function in the absence of lipid phosphates, we examined the interaction of an arginine-rich S4 helix variant with counterions and water molecules in bilayers lacking phosphate groups, by means of MD simulations.
Kv channels help generate action potentials along the cell membrane of excitable neurons by opening and closing their pore domains in response to changes in the transmembrane electrical potential, thereby controlling the flow of ions through the membrane.6–9 Each subunit of these homotetrameric channels contains a voltage-sensing domain (VSD), comprised of transmembrane (TM) helices S1-S4, and a pore domain (PD), which includes TM helices S5-S6. The highly charged, arginine-rich S4 helix of the VSD enables Kv channels to detect and respond to changes in TM potential.10–12 Voltage-induced movements of the VSDs cause the PDs to open or close through mechanical coupling provided by the so-called S4-S5 linker helix. Except for this linkage, structural and functional data13–18 have revealed that the VSDs move at the protein-lipid interface largely independently of the pore domain. However, the energetic penalty for moving charged Arg residues inside the hydrophobic core of a lipid bilayer is expected to be enormous19,20, and this has fueled a debate regarding the nature of the gating mechanism of Kv channels.21 Regardless of the true trajectory of the gating charges within the bilayer, the observation that Kv channels show no activity in dioleoyltrimethylammoniumpropane (DOTAP) bilayers lacking lipid phosphates2 indicates that the interplay between the lipid bilayer and the S4 helix lies at the heart of the gating mechanism.
**The isolated S4 helix has been reported to adopt a TM configuration in lipid bilayers from mixtures of a synthetic S4 peptide and phospholipids.22,23 Also, the propensity for membrane insertion of the S4 helix was addressed in a key experiment showing that a model helix based on the S4 segment (GGPG-LGLFRLVRLLRFLRILLII-GPGG) partitions within the endoplasmic reticulum (ER) membrane with a low apparent free energy penalty.24 A mutated form of S4 (S4mut), in which two Arg residues (R15 and R18) were moved one step closer to the C-terminus, was shown to insert even more efficiently. S4mut was chosen for a computational investigation of the solvation shell that accommodates an S4-like transmembrane helix in lipid bilayer membranes as part of a broader experimental research program.
To gain insights into the protein-lipid interactions that allow a TM configuration of the S4 helix, Freites et al.5 performed an MD simulation of S4 inserted across a phospholipid bilayer. The simulation revealed that the thickness of the bilayer immediately surrounding the helix was significantly reduced to provide a hydrogen-bonded network of lipid phosphates and water molecules to solvate the snorkeling Arg residues. A ~10 Å hydrophobic gap was observed to separate the headgroup-arginine complexes on opposite sides of the bilayer, which suggests that the membrane dielectric barrier is only a fraction of the hydrocarbon core thickness. This structural feature is consistent with experimental 24 and simulation results 25–27 showing that the energetics of arginine-partitioning in a phospholipid bilayer is strongly dependent on the location of the arginine center of charge. The protein-lipid interactions arising in the single S4 helix system are in agreement with those observed in MD simulations of an isolated VSD28,29 and an entire Kv channel 30–32 in bilayers containing lipid phosphates. However, structural studies have revealed that, although the outermost Arg residues are readily accommodated in a protein-lipid interface, Arg residues located deep within the membrane are shielded from a lipid environment 14,17,18. Hence, when drawing an analogy between the protein-lipid interactions of an isolated S4 fragment and those of the entire VSD one needs to consider the caveat that not all Arg residues in the latter are directly exposed to lipids. Nonetheless, an experimental approach combining MD simulations, solid state NMR, and neutron diffraction 33 showed that the interactions between the exposed charged Arg residues of the VSD and the surrounding bilayer head group region, were strong enough to produce a local deformation of the bilayer, ensuring the stabilization of these charges by means of a water defect. Moreover, it has recently been shown that the charge-containing segments of the VSD interact more strongly with the surrounding bilayer than any other segment of the VSD34, indicating a crucial role for the solvation shell surrounding the exposed charged Arg residues.
In this paper, we identify and describe, by means of atomistic MD simulations, the solvation shell surrounding an arginine-rich S4 helix variant in bilayers lacking lipid phosphates. We find that chloride counterions serve as ion-pair partners for the charged Arg residues in the absence of lipid phosphates. The counterion-Arg connection translates into an increased mobility of the Arg residues, because they are no longer anchored to the surrounding lipid bilayer. In addition to the chloride counterions, water molecules are also closely associated with the Arg residues, which can now enter deeply into the hydrophobic core of the bilayer, ultimately forming a continuous water wire leading to a collapse of the bilayer integrity. In phosphate-containing bilayers, on the other hand, the Arg-phosphate interaction induces a deformation of the bilayer as it accommodates itself to anchor the peptide. The nature of the solvation shell in the vicinity of the Arg residues is such that water is prevented from entering the hydrophobic gap, thereby stabilizing protein-lipid complex. As a direct consequence of the failure to form a stable solvation shell around the Arg residues, we observe a significantly smaller deformation of the DOTAP bilayer. Moreover, different rotameric states of the arginines of S4mut were observed to have a strong effect on the secondary structure of the helix.
2. Methods
2.1 Building the systems
We performed 100 ns atomistic MD simulations of lipid bilayers containing S4mut (Figure 1). The simulations were carried out in excess water, which denotes a bilayer containing more water molecules than the minimal amount needed for hydration. For a POPC bilayer, a minimal amount of ~9.5 water molecules per lipid are needed for hydration at 93% relative humidity (Mihailescu et al. unpublished and 35,36) In these simulations, water-lipid ratios of 44.8 and 44.1 were used for POPC and DOTAP, respectively. The sequence of the capped S4mut peptide in our simulations is similar to the wild-type S4:
Figure 1.
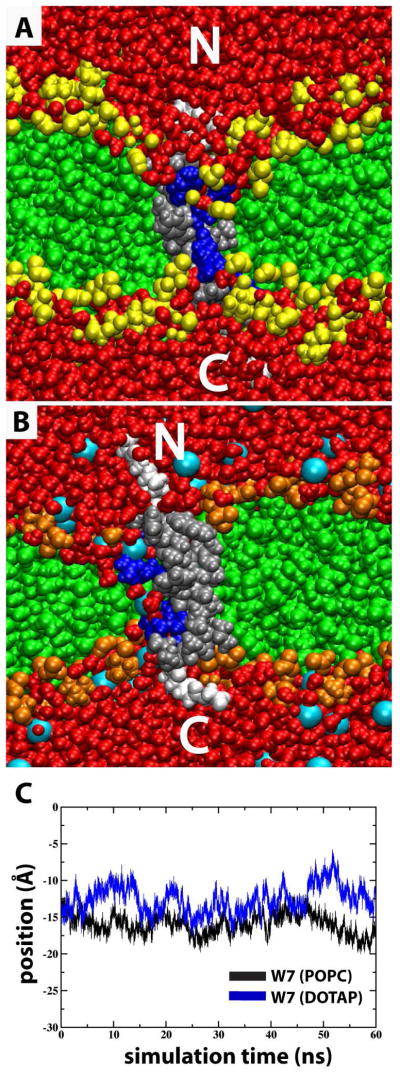
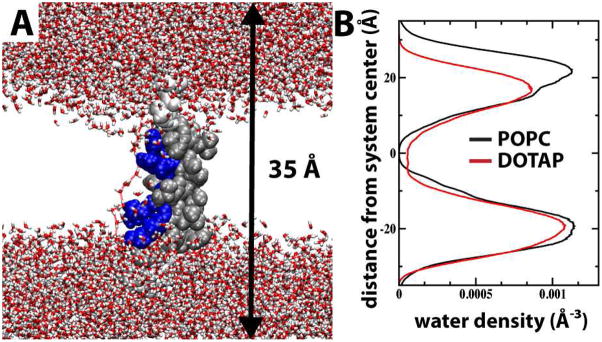
Snapshots of S4mut in POPC and DOTAP bilayers. (A) Snapshot from MD simulation in excess water of GPGG…GGPG-flanked (white) S4mut sequence (silver) carrying four charged Arg residues (blue) residing in a POPC bilayer in which the phosphatidylcholine headgroups are depicted in yellow and the acyl chains in green. (B) Simulation snapshot as in (A), except that the bilayer (DOTAP) lacks lipid phosphates in the head group region (orange). Chloride counter-ions are depicted as cyan spheres. (C) The positional evolution of W7 centers-of-mass (COM) along the bilayer normal (z-dimension) from equilibrated POPC (black) and DOTAP (blue) MD trajectories.
| S4 | GGPG–LGLFRLVRLLRFLRILLII–GPGG |
| S4mut | GGPG–LGWFRLVRLLFRLIRLLII–GPGG |
The mutated arginine residues are underlined. In addition, S4mut carries an L7W mutation to facilitate optical detection in ongoing experiments. Hessa et al. found that peptides carrying Trp and Leu at near-terminal equivalent positions inserted equally well into a biological membrane.37 Furthermore, Nishizawa et al.38 could not differentiate the structural dynamics displayed by S4 helices carrying either F8W, F8L, or F8V mutations when simulating the behavior of S4-related peptides at the membrane-water interface. The W7L mutation is, therefore, not likely to alter significantly the dynamics displayed by this residue. Indeed, the evolution of the W7 centers-of-mass in both the POPC and DOTAP simulations indicate no tendency to drift along the bilayer normal (Figure 1C).
To investigate whether two different sidechain configurations, placing one of the S4mut charges at different positions along the bilayer normal, affected the conformational stability of the peptide in the bilayer, we constructed two rotameric states of S4mut, one in which R16 is “up”, towards the C- or “down” towards the N-terminus (Figure 2A and 2B). We refer to these configurations as 2-up-2-down and 1-up-3-down, respectively. The 1-up-3-down configuration of S4mut was built as a perfect α-helix using the psfgen plugin of the VMD 1.8.7 software package,39 and the 2-up-2-down configuration was constructed by exploring further rotameric configurations of Arg 16 using the software COOT.40 Both configurations of the S4mut helix were then inserted into two separate pre-equilibrated POPC lipid bilayers by aligning the principal axes of the helix to the bilayer normal, and then making the peptide centers-of-mass coincident with the bilayer center-of-mass. Each system was composed of 268 lipid molecules, 12,107 water molecules, 4 counter ions, adding up to a total of 72,689 atoms and a water:lipid ratio of 45.2.
Figure 2.
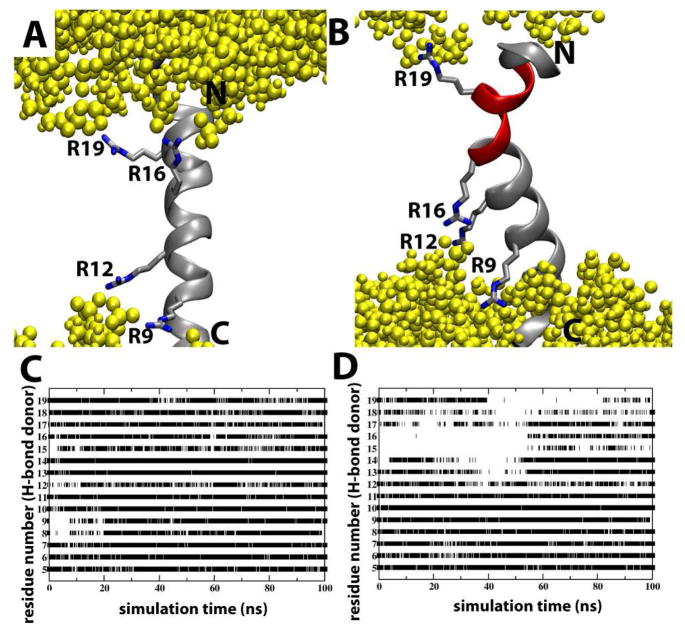
The effect of Arg rotamers on S4mut conformation in POPC bilayers. (A) A representative simulation snapshot of the S4mut helix with a 2-up-2-down distribution of its Arg residues (R9, R12, R16 and R19). Phospholipid headgroups are colored yellow. (B) A representative simulation snapshot of the S4mut helix with a 1-up-3-down distribution of its Arg residues (R9, R12, R16 and R19). The snapshot shows the severely kinked region (red) of S4mut. (C) The presence of backbone H-bonds in S4mut from an H-bond donor at position i to an H-bond acceptor at position i+4 for the 1-up-3-down Arg distribution and (D) the 2-up-2-down Arg distribution. The flanking -GPGG- regions were left out of this analysis. The criteria for H-bonds in this analysis were defined by cut-off values; 3.5 Å for the distance and 40 degrees for the angle.
The DOTAP chloride salt has been observed to form stable bilayers at different levels of hydration, as we have previously reported in a concerted neutron diffraction and MD simulation study on the structure of pure DOTAP bilayers,41 the details of which will be provided in a manuscript that is in preparation. In the present study, S4mut was inserted into a DOTAP bilayer in the 2-up-2-down configuration. The numbers of lipids and water molecules of the DOTAP bilayer system were chosen to have a water:lipid ratio similar to that of the POPC system (44.1). The final DOTAP system contained 284 lipid molecules, 12,515 water molecules, 4 chloride counter ions for the protein and 284 chloride counter ions for the lipid bilayer, adding up to a total of 74,347 atoms.
2.2 MD simulations
Each system was relaxed using a 2500-step conjugate-gradient energy minimization followed by eight 100-ps simulation runs at constant temperature (300 K) and volume during which the peptide was progressively released from its initial configuration using harmonic restraints. The subsequent production simulations were carried out for 100 ns at constant temperature (300 K) and constant pressure (1 atm). All molecular dynamics simulations were run with the NAMD 2.7 software package.42,43 The CHARMM22 and CHARMM36 force fields 44,45 were used for the peptide and lipids, respectively, and the TIP3P model43 was used for the water molecules. A time-step of 1 fs was used to integrate the equations of motion and a reversible multiple time-step algorithm46 of 4 fs was used for the electrostatic forces and 2 fs for short-range, nonbonded forces. The smooth particle mesh Ewald method47,48 was used to calculate electrostatic interactions. The short-range interactions were cut off at 12 Å. All bond lengths involving hydrogen atoms were held fixed using the SHAKE49 and SETTLE50 algorithms. A Langevin dynamics scheme was used for thermostatting. Nosé-Hoover-Langevin pistons were used for pressure control.51,52 Molecular graphics and simulation analyses were generated with the VMD 1.8.7 software package.39
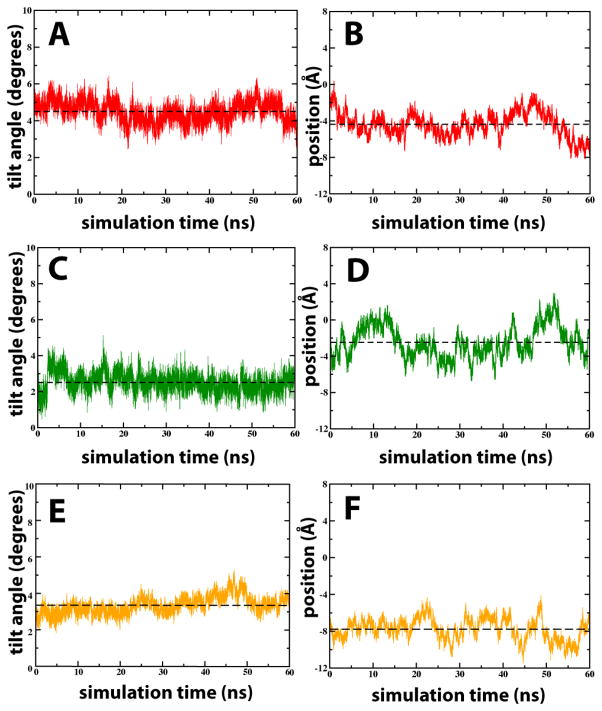
The evolution of the peptide tilt angle with respect to the bilayer normal, as well as the position of the peptide center-of-mass along the bilayer normal relative to the bilayer, indicated that all three systems (1-up-3-down and 2-up-2-down distribution of S4mut in POPC, and the 2-up-2-down distribution in DOTAP) were equilibrated after 40 ns (Figure 3). All subsequent analyses were, therefore, based on the last 60 ns of each trajectory.
Figure 3.
Trajectories of transmembrane tilt angles and positions of different S4mut configurations in POPC and DOTAP bilayers. (A) and (B) show the tilt angles and positions, respectively for the 2-up-2-down Arg configuration of S4mut in POPC. (C) and (D) The same as (A) and (B) except the lipid is DOTAP. (D) and (E) show the tilt angles and positions, respectively for the 1-up-3-down Arg configuration of S4mut in POPC. Tilt angles refer to the angle (in degrees) between the peptide and the bilayer normal. The position refers to the S4mut center-of-mass (COM) along the bilayer normal (z-dimension).
3. Results and Discussion
3.1 Integrity of the lipid bilayer
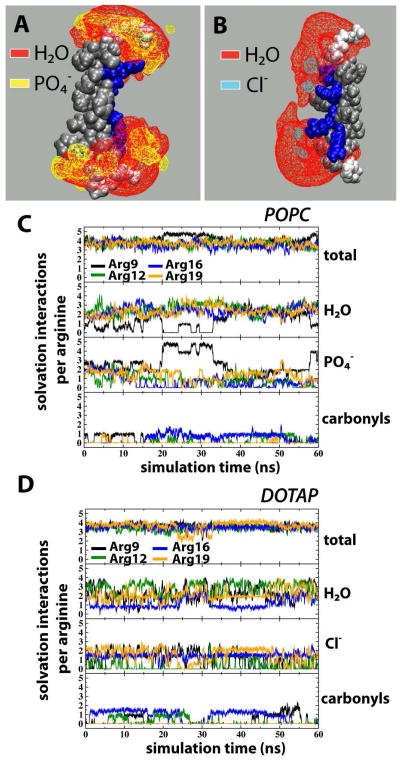
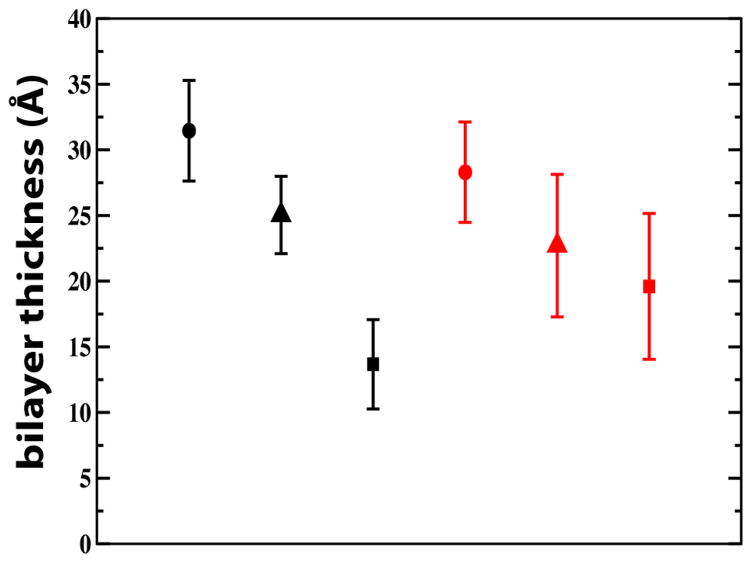
In a palmitoyloleoylglycerophosphocholine (POPC) bilayer, the 2-up-2-down S4mut helix adopted a TM configuration and the simulation revealed an H-bonding pattern where lipid phosphates, water molecules and, to a lesser extent, lipid carbonyls alternated in solvating the Arg residues (Figure 4A and 4C), in a similar fashion to that observed in an earlier simulation of S4.5 Although the maximum number of H-bonding partners to an Arg residue is five, the average orientation of the Arg sidechain was such that four H-bonding partners are preferred to avoid steric clashes. This is reflected in the simulations; the average total number of solvation interactions per Arg residue was around four, with an occasional jump to the maximum of five H-bond partners for Arg9 (Figure 4C). This shows that the Arg residues were essentially fully H bonded throughout the simulation. As a means of observing distortions in the lipid bilayer, we measured the bilayer thickness, defined as the distance between carbonyl carbons in each leaflet. The calculated bilayer thickness for the entire POPC bilayer is 31.5 ± 3.8 Å (Figure 5), whereas close to the peptide, we observed significant distortions of the bilayer. The thickness of the 2nd and 1st coordination shells, centered at 7 Å and 4 Å from the center of the peptide, decreased to 25.0 ± 3.0 Å and 13.7 ± 3.4 Å, respectively. Hence, in order for the structural H-bonding framework to become established around S4mut, the thickness of the lipid bilayer close vicinity of the TM helix is reduced by 6.5 Å and 17.8 Å for the 2nd and 1st coordination shells respectively, thereby bringing the bilayer polar region into close contact with the snorkeling arginines.
Figure 4.
Isosurfaces and solvation-interaction counts for S4mut (2-up-2-down) in POPC and DOTAP bilayers. (A) Isosurfaces of lipid phosphates (yellow) and water molecules (red) based on 60 ns equilibrated simulation of S4mut in POPC. (B) Isosurfaces of water molecules (red) and chloride counterions (cyan) based on 60 ns equilibrated simulation of S4mut in DOTAP. For clarity, the isosurfaces are depicted at 10% occupancy. (C) The total number of solvation interactions (first panel), the number of water H-bonds (second panel), the number of lipid phosphate interactions (third panel), and the number of H-bonds to the lipid carbonyl oxygens (fourth panel), formed by the S4mut helix in POPC. (D) The total number of solvation interactions (upper panel), the number of water H-bonds (second panel), the chloride ion interactions (third panel), and the number of H-bonds to the lipid carbonyl oxygens (fourth panel), formed by S4mut helix in DOTAP. The criteria for the presence of a solvation interaction were defined by cut-off values; 3.5 Å for the distance and 40 degrees for the angle.
Figure 5.
Bilayer thickness measurements for POPC (black symbols) and DOTAP bilayers (red symbols) at various distances from S4mut (2-up-2-down). Measurements represent transbilayer carbonyl-to-carbonyl distances. At large distances from the peptide, the thicknesses are 31.5 ± 3.8 Å for POPC and 28.3 ± 3.8 Å for DOTAP (solid circles). For the second coordination shell, the thicknesses are 25.0 ± 3.0 Å and 22.7 ± 5.4 Å for POPC and DOTAP, respectively (triangles). For the first coordination shell, the distances decrease to 13.7 ± 3.4 Å for POPC and 19.6 ± 5.6 Å for POPC and DOTAP, respectively (solid squares).
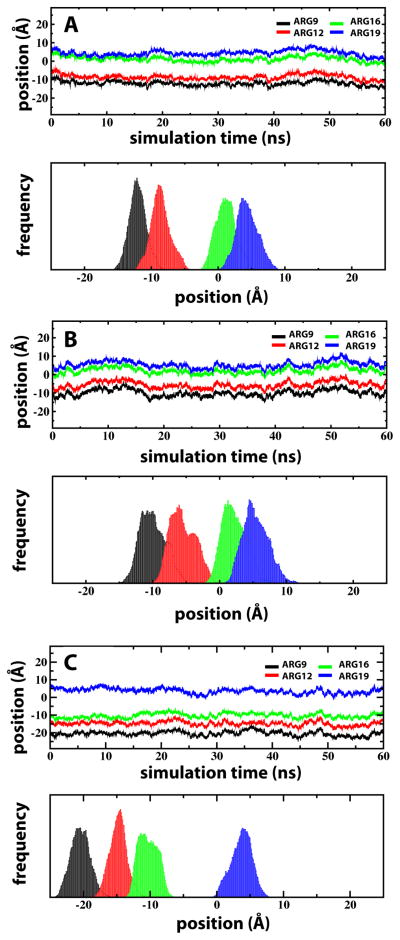
The configuration of S4mut in POPC contrasts sharply to that obtained when solvating the peptide in the cationic lipid DOTAP, which lacks phosphate groups. Whereas well-defined positions of the Arg side chains were obtained for S4mut in POPC, the centers-of-mass of the Arg side chains in DOTAP were widely distributed (Figure 6). These results are consistent with free energy profiles of an isolated arginine side chain displaying relatively high energetics at the DOTAP bilayer interface and more favorable energetics at the same position in bilayers containing lipid phosphates. 25–27,53
Figure 6.
Transbilayer distributions of Arg residues for S4mut in POPC and DOTAP bilayers. The equilibrated positions of the Arg residues are represented by the center-of-mass (COM) trajectories along the bilayer normal (upper panels) and their corresponding distribution histograms (lower panels). In all cases of R9 (black), R12 (red), R16 (green) and R19 (blue) the COM were taken to be the position of the guanidinium carbon atom. (A) The Arg residues (2-up-2-down) in S4mut in POPC. (B) The Arg residues (2-up-2-down) in DOTAP. (C) Arg residues (1-up-3-down configuration) for S4mut in POPC.
The lack of lipid phosphate groups in the DOTAP bilayer resulted in charge-pairing between the Arg residues and chloride counter-ions (Figure 4D). We observed no significant difference in the number of Arg - carbonyl interactions between the POPC and DOTAP systems and, hence, the Arg residues did not seem to use the lipid carbonyl oxygens as additional anchor points to a greater extent when lipid phosphates were not present. Importantly, by interacting electrostatically with the mobile DOTAP chloride counter-ions, the Arg residues are not anchored to the polar region of the DOTAP bilayer (Figure 4B). The resulting increase in conformational flexibility of the Arg side chains affects the water distribution. In POPC, the water molecules are distributed around the snorkeling Arg residues and the hydrophobic gap between the two solvation shells is maintained (Figure 4A). The dynamical feature of this structural framework is illustrated by the number density plot in Figure 7B, which shows the water profile along the normal of the POPC bilayer. The symmetrically distributed peak regions of the water profile show the preference of water molecules for the polar headgroups of the lipid bilayer. The Gaussian character of the water distribution indicates that water penetrates quite deep into the bilayer, although it never enters the hydrophobic gap of the bilayer. In DOTAP, on the other hand, the hydrophobic gap collapses, because water molecules are being dragged through the bilayer by the unanchored Arg residues (Figure 7B). In several instances during the S4mut-DOTAP simulation, we observed the formation of a water wire stretching across the entire bilayer. The occurrence of such a water wire was strictly confined to the immediate vicinity of the peptide at all times during the simulations (Figure 7A). At no instance during the simulation did we observe a chloride ion crossing over the bilayer. Although ions do enter the hydrophobic core in the neighborhood of S4mut, the charge-charge interactions to the arginines prevent any flux of ions over the bilayer. Moreover, because the phosphate-Arg solvation shells were disrupted, bilayer distortion in the vicinity of S4mut was reduced. The average thickness of the DOTAP bilayer was found to be 28.3 ± 3.8 Å, while the 2nd and 1st coordination shell around the peptide were 22.7 ± 5.4 Å and 19.6 ± 5.6 Å, respectively (Figure 5). This amounts to a distortion of the DOTAP bilayer in the vicinity of the S4mut peptide of ~5.6–8.7 Å, which is significantly less than the distortion of the POPC system.
Figure 7.
Formation of water wires in DOTAP bilayers containing S4mut (2-up-2-down). (A) A snapshot from the DOTAP simulation showing the formation of a water wire that extends from one side of the bilayer to the other. Connecting water molecules were defined by cut-of-values; 3.5 Å for the distance and 40 degrees for the angle. (B) The number densities of water in the 1st and 2nd solvation shells around the Arg residues in POPC (black line) and DOTAP (red line). Notice the absence waters in the center of the POPC bilayer due to the so-called hydrophobic gap (see text).
Comparing the simulations of S4mut in POPC and DOTAP, the nature of the solvation-shell in the protein-lipid interface emerges as the most prominent discriminatory feature. Bilayers lacking lipid phosphates are not capable of charge pairing to the Arg residues of the S4mut helix, and as a consequence the bilayer deformation around the peptide is significantly smaller in the DOTAP bilayer. The protein-lipid interactions in these simulations provide an atomistic view of the lipid phosphate requirement by Kv channels.
3.2 Anchoring affects helical conformation
The anchoring by lipid phosphates in the headgroup region of the bilayer also has a strong influence on the helical conformation of the peptide. This becomes clear by comparing the two rotameric states of S4mut in POPC bilayers. When R16 was pointing up in the 2-up-2-down configuration, the integrity of the helix was maintained throughout the simulation (Figure 2A). Although the 1-up-3-down rotamer distribution (R16 pointing down) was seen earlier to generate a conformationally stable S4 helix in POPC5, our 100 ns MD simulation of S4mut with the same rotamer distribution in POPC produced a strongly kinked peptide, suggesting a conformationally unstable state (Figure 2B). A similar behavior, where the rotameric state of a charged sidechain dictates the secondary structure of a TM segment, was observed in simulations of the membrane-spanning domain (MSD) of the HIV-1 glycoprotein gp41.54 Furthermore, Nishizawa et al. reported highly tilted helix orientations in MD simulations where the S4 helix was placed at different initial depths in a dipalmitoylphosphatidylcholine (DPPC) bilayer.38 Interestingly, in this latter work the rotameric distribution of the Arg residues is either in a 1-up-3-down or a 3-up-1-down configuration, thereby confirming our observation that strain is imposed on the helix when the arginines are not in a 2-up-2-down configuration. The movement of the sidechain of either R12 or R16, in the 3-up-1-down and 1-up-3-down configurations, respectively, toward the edge of the bilayer reduces the energetic penalty of charge burial in the non-polar interior of the membrane, and results in a conformationally unstable state. The structural consequences seem to be either a pronounced helical tilt38 or, as seen in our simulations, a strong kink in the helix that disturbs the normal α-helical H-bonding pattern in the upper part of the helix (Figure 2D).
Because switching through different rotameric states of charged residues within the lipid bilayer involves the crossing of high energy barriers,26 the most energetically favorable set of sidechain configurations is maintained throughout a simulation. This analysis prompted us to use the 2-up-2-down configuration of S4mut in the simulation of the DOTAP bilayer.
4. Summary
Taken together, our simulations show that in bilayers lacking lipid phosphates, the charged Arg residues of S4mut are solvated by chloride counterions, water molecules and, to a much lesser degree, the carbonyl groups of the surrounding lipids. In such a protein-lipid interface, the Arg residues lack anchor points to the lipid bilayer and, hence, display an increased structural flexibility along the bilayer normal. As a direct consequence of the nature of this protein-lipid interface, we make two observations that might explain the non-functionality of Kv channels in DOTAP bilayers. First, because of the lack of lipid anchors, Arg residues, solvated by chloride counterions and water molecules, move freely into the bilayer core and ultimately disrupt the integrity of the hydrophobic gap. Second, the absence of strong lipid connections leads to a significantly less distorted DOTAP bilayer around the charged Arg residues. Moreover, this set of simulations show that phosphate-Arg interactions have a strong influence on the secondary structure of the TM segment.
Acknowledgments
We thank Joseph Farran of the MPC computing center (University of California, Irvine) for outstanding technical support and Harindar Keer for providing the initial configuration of the DOTAP bilayer. This work was supported by grants from the National Institutes of Health (GM74637 to SHW and GM86685 to SHW and DJT) and the National Science Foundation (CHE-0750175 to DJT), and in part by the National Science Foundation through TeraGrid resources provided by the Texas Advanced Computing Center at the University of Texas at Austin.
References
- 1.Lee AG. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt D, Jiang Q-X, MacKinnon R. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 3.Ramu Y, Xu Y, Lu Z. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Ramu Y, Lu Z. Nature. 2008;451:826–830. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freites JA, Tobias DJ, von Heijne G, White SH. Proc Natl Acad Sci USA. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgkin AL, Huxley AF. J Physiol (Lond) 1952;116:449–472. doi: 10.1113/jphysiol.1952.sp004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkin AL, Huxley AF. J Physiol (Lond) 1952;116:473–496. doi: 10.1113/jphysiol.1952.sp004718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodgkin AL, Huxley AF. J Physiol (Lond) 1952;116:497–506. doi: 10.1113/jphysiol.1952.sp004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodgkin AL, Huxley AF. J Physiol (Lond) 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal SK, MacKinnon R. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 12.Swartz KJ. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang YX, Ruta V, Chen JY, Lee A, MacKinnon R. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 14.Cuello LG, Cortes DM, Perozo E. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 15.Ruta V, Chen J, MacKinnon R. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 16.Alabi AR, Bahamonde MI, Jung HJ, Kin JI, Swartz KJ. Nature. 2007;450:370–376. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long SB, Tao X, Campbell EB, Mackinnon R. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 18.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsegian A. Nature. 1969;221:844–846. doi: 10.1038/221844a0. [DOI] [PubMed] [Google Scholar]
- 20.Grabe M, Lecar H, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2004;101:17640–17645. doi: 10.1073/pnas.0408116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tombola F, Pathak MM, Isacoff EY. Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 22.Castro-Román F, Fernandez-Vidal M, Mihailescu M, White SH. Biophys J. 2007;92:294a. [Google Scholar]
- 23.Doherty T, Su Y, Hong M. J Mol Biol. 2010;401:642–652. doi: 10.1016/j.jmb.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hessa T, White SH, von Heijne G. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 25.MacCallum JL, Bennett WFD, Tieleman DP. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorairaj S, Allen TW. Proc Natl Acad Sci USA. 2007;104:4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schow EV, Freites JA, Cheng P, Bernsel A, von Heijne G, White SH, Tobias DJ. J Membr Biol. 2011;239:35–48. doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freites JA, Tobias DJ, White SH. Biophys J. 2006;91:L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sands ZA, Sansom MSP. Structure. 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Treptow W, Tarek M. Biophys J. 2006;90:L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jogini V, Roux B. Biophys J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjelkmar P, Niemelä PS, Vattulainen I, Lindahl E. PLoS Comp Biol. 2009;5:1–14. doi: 10.1371/journal.pcbi.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krepkiy D, Mihailescu M, Freites JA, Schow EV, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butterwick JA, MacKinnon R. J Mol Biol. 2010;403:591–606. doi: 10.1016/j.jmb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hristova K, White SH. Biophys J. 1998;74:2419–2433. doi: 10.1016/S0006-3495(98)77950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIntosh TJ, Magid AD, Simon SA. Biophys J. 1989;55:897–904. doi: 10.1016/S0006-3495(89)82888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson IM, White SH, von Heijne G. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 38.Nishizawa M, Nishizawa K. Biophys J. 2008;95:1729–1744. doi: 10.1529/biophysj.108.130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke W, Schulten K. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Keer HS, Freites JA, Mihailescu E, White SH, Tobias DJ. Biophys J. 2010;98(Supplement 1):492a. [Google Scholar]
- 42.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comput Phys. 1999;151:283–312. [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 44.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, Mackerell AD, Pastor RW. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grubmüller H, Heller H, Windemuth A, Schulten K. Mol Simul. 1991;6:121–142. [Google Scholar]
- 47.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 48.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 49.Ryckaert J-P, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 50.Miyamoto S, Kollman P. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 51.Martyna GJ, Tobias DJ, Klein ML. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 52.Feller SE, Zhang Y, Pastor RW, Brooks BR. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 53.Johansson AC, Lindahl E. J Chem Phys. 2009;130:185101–185101 . 185101–185108. doi: 10.1063/1.3129863. [DOI] [PubMed] [Google Scholar]
- 54.Gangupomu VK, Abrams CF. Biophys J. 99:3438–3444. doi: 10.1016/j.bpj.2010.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]