Abstract
How the pore-forming protein perforin delivers apoptosis-inducing granzymes to the cytosol of target cells is uncertain. Perforin induces a transient Ca2+ flux in the target, which triggers a damaged cell membrane repair process. As a consequence, both perforin and granzymes are endocytosed into enlarged endosomes, called gigantosomes. Here we show that perforin forms pores in the gigantosome membrane, allowing endosomal cargo, including granzymes, to be gradually released. After about 15 minutes, gigantosomes rupture, releasing their remaining content. Thus, perforin delivers granzymes by a two-step process that first involves transient pores in the cell membrane that trigger granzyme and perforin endocytosis and then pore formation in endosomes to trigger cytosolic release.
Keywords: perforin, granzyme, NK cell, cytotoxic T cell, apoptosis
Introduction
Cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells eliminate virus infected or malignantly transformed cells principally by releasing the contents of cytotoxic granules1 into the immune synapse formed with their target cell2–5. The granule serine proteases, known as granzymes (Gzms), induce programmed cell death6–8 after they are delivered into the target cell cytoplasm by the pore-forming granule protein perforin (PFN)9–12. PFN-deficient mice are profoundly immunodeficient. They fail to eliminate many viruses and other intracellular pathogens, spontaneously develop B cell lymphoma and are highly susceptible to carcinogen-induced neoplasia13. Humans bearing genetic mutations that lead to impaired PFN synthesis, function or release develop familial hemophagocytic lymphohistiocytosis14, 15.
The way that PFN delivers Gzms to the cytosol of target cells is not fully understood16. PFN binds in a Ca2+-dependent manner to membranes and multimerizes to form pores. Two models17, both based on the membranolytic properties of PFN18, differ in their predicted site of action. The simplest model is that Gzms are delivered directly to the cytosol by plasma membrane pores19–22. Cells treated with high concentrations of recombinant or purified PFN form pores that are visible by electron microscopy are sufficiently large for Gzms to pass through. However, these high concentrations form stable pores that kill a cell by necrosis, whereas at physiological concentrations that deliver Gzms to induce apoptosis, Gzms are taken up with perforin into endosomes rather than directly to the cytosol, as would be predicted if they entered via plasma membrane pores23, 24. This scenario has prompted a revised model that suggests that perforin acts at the endosomal membrane by damaging its structural integrity (as originally proposed 25, 26), like some bacterial pore-forming proteins, to release Gzms to the cytosol.
At physiologically relevant sublytic concentrations and during killer cell-mediated lysis, PFN perturbs the target plasma membrane (presumably by creating small pores in the target-cell membrane) transiently allowing Ca2+ and small dyes to enter the target cell23. The Ca2+ influx triggers the damaged membrane repair response in which plasma membrane lesions are repaired through calcium-dependent exocytosis of lysosomes and other vesicles27–29. Another recently appreciated aspect of the damaged membrane response is induction of endocytosis to remove the damaged membrane from the cell surface to preserve cell membrane integrity23, 24, 30. Treatment of target cells with PFN and Gzms or killer cells leads to rapid clathrin and dynamin-dependent endocytosis24. Greatly enlarged early endosomal antigen-1 (EEA-1) positive vesicles23, 24, which we have termed “gigantosomes”, that contain PFN and Gzms are formed. When the cellular membrane repair response is inhibited by Ca2+ chelation or inhibitors of endocytosis, treated cells die by necrosis rather than apoptosis, suggesting that activating the membrane repair response is critical for immune-mediated death by apoptosis.
The aim of this study was to investigate how gigantosomes form and how Gzms are released from them. Using live-cell imaging microscopy, we find that gigantosomes formed in target cells by Rab5-dependent homotypic fusion between EEA1-stained early endosomes. However, endosomal fusion was not essential for cell death. Moreover, PFN-induced gigantosomes did not acidify. For the first time, we visualized GzmB and PFN in cells subjected to NK cell attack. Using live cell imaging, we find that Gzm- and PFN-containing gigantosomes formed not only when cells were treated with sublytic concentrations of PFN, but also during cell-mediated cytolysis. PFN-mediated cargo release into the cytosol occurred about 10–15 min after PFN treatment, coincident with PFN multimerization in the gigantosome membrane. Cargo release gradually occurred from discrete locations within the endosomal membrane, followed by endosome rupture and release of their remaining cargo.
Results
GzmB and PFN-mediated apoptosis without gigantosomes
We first verified that large EEA-1+ Lamp1− intracellular vesicles (gigantosomes) containing PFN and Gzms23, 24 formed after sublytic PFN treatment (Supplementary Fig. 1a,b)23, 24. These enlarged endosomes formed by homotypic fusion of early endosomes. (Supplementary Fig. 1c–e). Rab5 is a small GTPase that regulates fusion between endocytic vesicles and early endosomes, as well as the homotypic fusion between early endosomes31, 32, 33. Mutant Rab5(S34N), which has preferential affinity for GDP, acts as a dominant-negative inhibitor of Rab534. Gigantosomes formed within 10 min of sublytic PFN treatment of HeLa cells transfected with monomeric red fluorescent protein (mRFP)-EEA-1 and Rab5(WT), but not when the wild-type protein was replaced with enhanced green fluorescent protein (EGFP)-Rab5(S34N) (Supplementary Fig. 2). Thus gigantosome formation is Rab5-dependent. We next assessed whether gigantosome formation is required for induction of apoptosis by GzmB and PFN. PFN and GzmB-mediated apoptosis, assessed by caspase-3 and cytokeratin 18 cleavage and annexin-V–propidium iodide (PI) staining, was compared in HeLa cells transfected with Rab5(WT) or Rab5(S34N) (Fig. 1 and Supplementary Fig. 3). Apoptosis was similar in untransfected control cells and in cells expressing Rab5(WT) or Rab5(S34N). Thus gigantosome formation is dispensable for GzmB-mediated induction of apoptosis.
Figure 1. Inhibition of gigantosome formation does not impair GzmB-induced apoptosis.
(a–b) HeLa cells transfected with EGFP-Rab5(WT) or EGFP-Rab5(S34N) dominant negative mutant (a, top row) were treated with buffer or sublytic rat PFN ± 100 nM native human GzmB, and apoptosis in EGFP+ cells was measured 2 h later by labeling with M30 mAb (which recognize a cytokeratin-18 epitope, revealed after caspase cleavage). Representative flow cytometry histograms (a) (MFI, mean fluorescence intensity) and mean ± s.d. of percentage of M30+ cells from three independent experiments (b) are shown. P values were determined by unpaired two-tailed student’s t-test. There was no significant (NS) difference in GzmB-mediated apoptosis in Rab5(S34N)-transfected cells relative to Rab5(WT)-transfected cells. (c) Analysis of procaspase-3 activation by immunoblot in HeLa cells transfected with EGFP-Rab5(WT) or EGFP-Rab5(S34N) and treated with buffer or sublytic rat PFN ± 50 nM native human GzmB for 30 min. Actin was a loading control. Data are representative of two independent experiments.
PFN inhibits early endosome acidification
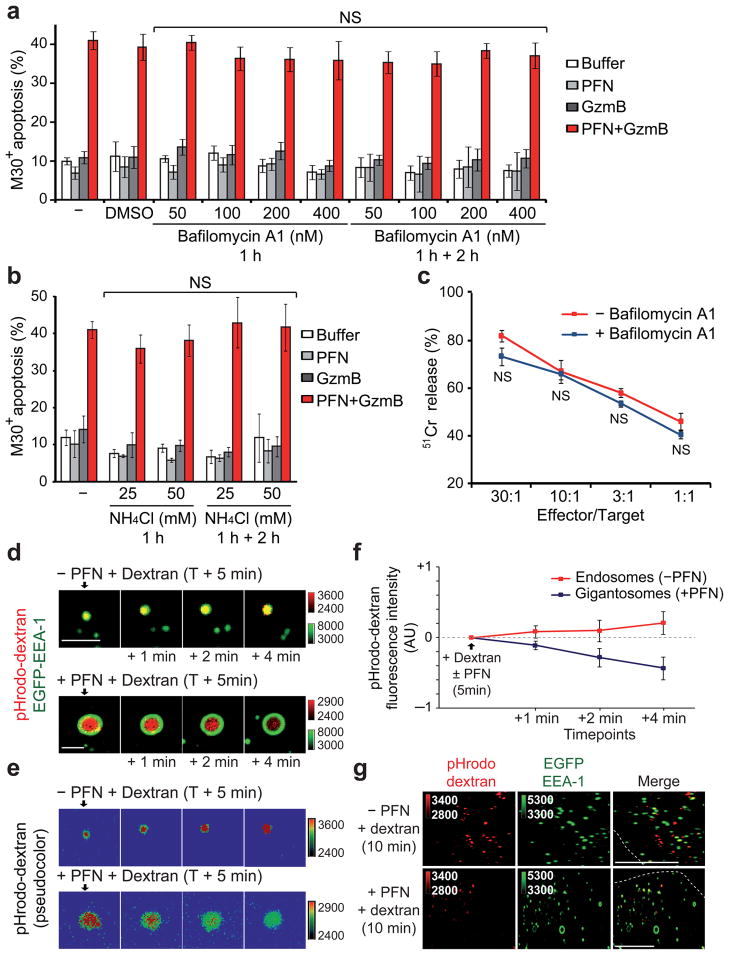
Gzms need to be released into the target cell cytosol to trigger apoptosis23, 24. We hypothesized that Gzms are released when PFN forms endosomal membrane pores. However, early endosomes normally rapidly acidify through the actions of the vacuolar ATPase35, and PFN pore formation is severely compromised at pH<6.5 (36 and data not shown). We therefore predicted that PFN might interfere with endosomal acidification. We first assessed whether PFN-mediated delivery of GzmB and apoptosis induction requires endosomal acidification by treating target cells with Bafilomycin A1, an inhibitor of the vacuolar-type H+-ATPase37, 38 (Fig. 2a) or with ammonium chloride, a weak base that increases endosomal pH by unidirectional diffusion into endosomes39 (Fig. 2b). PFN and GzmB-mediated apoptosis was not altered by pre-treating HeLa cells with these agents that interfere with endosomal acidification. Similarly, pre-incubation of target cells with Bafilomycin A1 did not affect NK cell-mediated killing (Fig. 2c). Moreover, Bafilomycin A1 pre-treatment did not lead to more PFN-induced necrosis (Supplementary Fig. 4a). Therefore, PFN delivery of GzmB does not require endosomal acidification.
Figure 2. Endosome acidification is inhibited by PFN.
(a,b) Inhibiting endosome acidification does not alter PFN–GzmB-induced apoptosis assessed by M30 staining. HeLa cells, pre-incubated with bafilomycin A1 (a) or NH4Cl (b) for 1 h, were treated with GzmB ±sublytic rat PFN. Maintaining bafilomycin or NH4Cl in the medium during the assay did not affect apoptosis. Mean±s.d. from four independent experiments are shown. (c) Bafilomycin A1 also had no effect on NK cell-mediated killing. Data are representative of two independent experiments performed in triplicate. NS, not significant (d–g) PFN inhibits gigantosome acidification. EGFP-EEA-1-expressing HeLa cells were incubated with pHrodo dextran ±sublytic PFN. Live cells were imaged beginning 5 min later. pHrodo dextran fluorescence increases in normal endosomes, but decreases in gigantosomes formed after PFN treatment. Shown are representative images in (d) (pseudocolored in (e) to indicate pHrodo dextran flurorescence intensity) and mean±s.d. of six independent experiments in (f). Scale bars, 2 μm. pHrodo dextran fluorescence 5 min after adding PFN was defined as 0. AU, arbitrary units. (g) Confocal images 10 min after adding sublytic PFN and pHrodo dextran to EGFP-EEA-1-transfected cells. Scale bars, 10 μm. Dashed lines, plasma membrane. Data are representative of three independent experiments.
To assess whether PFN-containing gigantosomes acidify, which would interfere with PFN pore formation in the gigantosome membrane, we treated cells with sublytic PFN and pHrodo dextran, which emits a bright red-fluorescent signal in an acidic environment. In cells treated with pHrodo dextran without PFN, endosomal red fluorescence increased over a few minutes, as expected. However, in cells treated with sublytic PFN, red fluorescence within the gigantosomes progressively decreased with time (Fig. 2d–f). Within 10 min, most gigantosomes did not display any red fluorescence, unlike normal endosomes in the same cell or in cells not exposed to PFN (Fig. 2g). Thus gigantosomes do not acidify like normal endosomes. The lack of gigantosome acidification may be due to PFN pore formation in the gigantosome membrane, which would interfere with the maintenance of a pH gradient across the gigantosome membrane. To confirm these data, we also co-treated cells with PFN and Lysosensor Yellow/Blue, which fluoresces in the green channel only at low pH. At the earliest times, Lysosensor fluorescence was comparable in PFN-treated and control cells, suggesting that dye uptake is equivalent (not shown). However, compared to cells treated with medium, cells treated with sublytic PFN demonstrated a rapid progressive decrease in Lysosensor green fluorescence over 5 min. Moreover, reduced fluorescence was not due to dye leakage out of the cell, since plasma membrane integrity, assessed by lack of PI uptake, remained unimpaired (Supplementary Fig. 4b). Thus gigantosomes formed in PFN-treated cells do not acidify.
PFN forms pores in the endosomal membrane
The most likely explanation for PFN permeabilization of the gigantosome membrane is that PFN forms pores in the gigantosome membrane. Staining of target cells 7 min after treatment with PFN with the Pf80 antibody that we previously used to visualize PFN in target cell gigantosomes24 confirmed PFN localization within gigantosomes (Fig. 3a). High magnification images of gigantosomes stained with anti-human PFN (Pf80) mAb 7 min after adding PFN showed highly localized PFN staining in clumps on the endosomal membrane (Fig. 3b, Supplementary Fig. 5a,b), as might be expected to form as PFN multimerizes to form pores. However, Pf80 staining became virtually undetectable 15 min after PFN loading. This antibody recognizes an epitope that is exposed in monomeric PFN and in the first step of PFN binding to membranes, but disappears when PFN multimerizes to form transmembrane pores40. The disappearance of PFN staining could be due either to PFN pore formation or to PFN degradation within the target cell. However, PFN staining with the Pf344 mAb that recognizes an epitope that remains exposed throughout the various steps in PFN pore formation40 was still detectable even 15 min later (Fig. 3c,d and Supplementary Fig. 5c). Similarly by flow cytometry, Pf80 staining of PFN-treated cells was visible 5 min after adding PFN but was no longer detected at 10 or 15 min, while Pf344 staining persisted for as long as was measured (15 min) (Fig. 3e,f). Therefore PFN was not degraded, but PFN staining with Pf80 disappeared because PFN formed pores in the gigantosome membrane. To confirm this finding, we next tested whether we could detect PFN multimer formation within target cells using chemical crosslinking (Fig. 3g). Target cells were incubated with a sublytic concentration of native human PFN for 1–15 min before adding the membrane-permeable crosslinking agent, disuccinimidyl suberate (DSS). PFN immunoblots of the cross-linked cell lysates showed a gradual decrease of the 60 kDa PFN monomer, while two cross-linked bands appeared after 10 min and increased at 15 min. The lower band had an estimated size of ~420 kDa, consistent with the size of a PFN heptamer, and the top band migrated near the top of the gel, suggesting formation of a much larger multimer. At these time points PFN staining was almost exclusively in endosomes (Fig. 3a,c and 24), suggesting pore formation occurs within endosomal membranes. When PFN-treated cells were fractionated to isolate cytoplasmic vesicles before crosslinking with DSS, the same sized cross-linked bands were also seen (Supplementary Fig. 5d). Thus PFN pore formation increases over time in the gigantosome membrane.
Figure 3. PFN multimerizes in gigantosome membranes.
(a–d) Single confocal sections of HeLa cells stained with Pf80 (a,b) or Pf344 (c,d) PFN mAbs after incubation for indicated times with sublytic human PFN. (b,d) show single high magnification confocal section of representative gigantosomes stained 7 min after HeLa cell treatment with sublytic human PFN. Pictures are representative of at least three independent experiments. Color bars and associated numbers indicate staining intensity. Scale bars: (a,c) 10 μm, (b,d) 5 μm. Dashed lines, plasma membrane. (e–f) PFN-treated HeLa cells from the same sample were stained with Pf80 or Pf344 at the indicated times. Representative flow cytometry histograms (e) indicating the percentage of PFN-positive cells and mean ± s.d. of three independent experiments (f) are shown. * P < 0.025, ** P < 0.002. (g) Detection of PFN aggregates by crosslinking with DSS. Target cells were incubated with native human PFN during the indicated time before adding the crosslinker DSS to the whole cells. PFN immunoblot shows PFN monomer (60 kDa) as well as formation with time of a PFN multimer of ~ 420 kDa and a large multimer near the top of the gel. Data are representative of three independent experiments.
GzmB and other cargo are released from gigantosomes
To test our hypothesis that PFN pore formation in the endosomal membrane is responsible for Gzm release, we investigated by co-staining for EEA-1 and GzmB the timing of GzmB uptake and cytosolic release following treatment with PFN and GzmB. In the absence of PFN, cells did not efficiently take up GzmB (Fig. 4a). After exposure to sublytic PFN and GzmB, GzmB-containing EEA-1+ gigantosomes formed within 5 min. After ~10–15 min, GzmB was released from gigantosomes to the cytosol as the bright vesicular staining of the endocytosed cargo dispersed into a faintly detected haze in the cytosol. Within 20 min, the majority of the GzmB signal concentrated in the nucleus, as expected41, and gigantosomes were no longer detected (Fig. 4a,b). Uptake of Alexa488-GzmB into gigantosomes was also seen within 2 min of adding PFN. Cytosolic fluorescence began to be visible within 5 min, but by 15 min gigantosome staining had disappeared and GzmB became cytosolic and nuclear (Fig. 4c). Therefore the release of GzmB from gigantosomes in PFN treated cells within ~15 min coincided temporally with PFN pore formation as judged by the disappearance of Pf80 staining and PFN cross-linking.
Figure 4. Endocytosed GzmB is released into the cytosol within ~10 min of PFN loading.
(a) Within 5–10 min of treatment with sublytic native rat PFN and native human GzmB, GzmB begins to be released from gigantosomes. HeLa cells were treated with GzmB ± sublytic PFN, fixed at the indicated time and stained for EEA-1 and GzmB. Representative single spinning disk confocal sections from three independent experiments are shown. Percentage of cells with GzmB in gigantosomes or in the cytosol (bottom row) is indicated (mean ± s.d.). (b) HeLa cells were treated with native human GzmB ±sublytic rat PFN, fixed at the indicated times and stained for GzmB and DAPI. Images were acquired by 3D-capture widefield microscopy followed by iterative deconvolution and projection. Pictures are representative of three independent experiments. (c) HeLa cells were treated with A488-labeled GzmB ± sublytic PFN and fixed at the indicated times. After release, GzmB accumulates in and around the nucleus. Pictures are representative of two independent experiments. Color bars and associated numbers indicate fluorescence intensity levels. Scale bars, 5 μm (a), 10 μm (b,c). Dashed lines, plasma membrane.
Gigantosomes leak cargo and then rupture
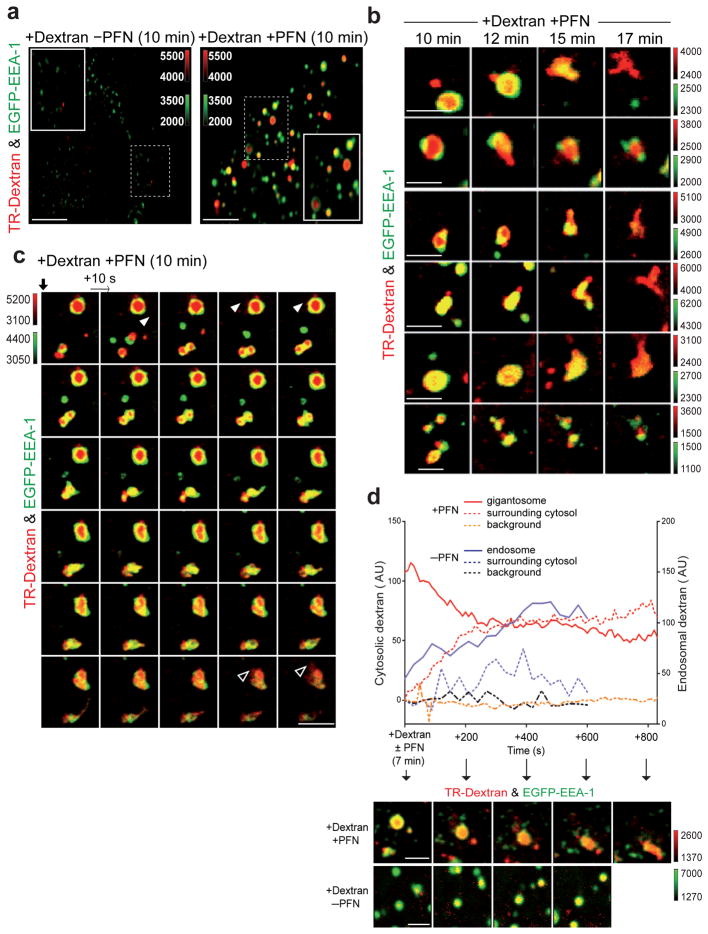
We next used live cell imaging to visualize the release of gigantosome cargo from PFN-treated cells. Time-lapse spinning disk confocal microscopy was used to image the trafficking of TR-Dextran in PFN-treated HeLa cells transfected to express EGFP-EEA-1. As previously described24, PFN enhanced 10 kDa TR-Dextran endocytosis, and TR-Dextran remained localized to gigantosomes after 10 min (Fig. 5a). Similar results were obtained when mRFP-EEA-1-transfected cells were treated with 10 kDa cationic rhodamine green-dextran and PFN (data not shown). After 10 min, we began to observe discrete and localized release of TR-Dextran from gigantosomes into the cytosol, while the gigantosome membrane appeared to remain intact (Fig. 5b and Supplementary Fig. 6a). A little later (~15–17 min after PFN–TR-Dextran loading), the gigantosome membrane became unstable. EEA-1 staining of gigantosomes disappeared and endosomal tubulations formed, which was followed by rupture of the gigantosome membrane, leading to complete release and diffusion of dextran into the cytosol (Fig. 5b,c, Supplementary Fig. 6b, Movies S1–S3). As dextran diffuses, it becomes difficult to detect. To confirm our impression that TR-Dextran was released from gigantosomes to the cytosol before they ruptured, we imaged PFN and dextran-treated cells by live cell 4D spinning disk confocal imaging beginning 7 min after adding PFN and dextran. TR-Dextran staining intensity was measured in the gigantosome or endosomes and in the surrounding cytoplasm (Fig. 5d). In the absence of PFN, the TR-Dextran signal in endosomes gradually increased as more dextran was incorporated, but the signal in the surrounding cytosol remained low and was stable with some fluctuation. However, in cells treated with PFN, TR-dextran signal intensity in the gigantosome gradually decreased as TR staining in the surrounding cytoplasm increased. As a control, we measured TR-Dextran background intensity in a region of the cytosol that did not contain gigantosomes or endosomes, and found that it did not change. Taken together, these data suggest that PFN pores in the gigantosome membrane allow slow release of endosomal cargo before completely destabilizing the endosomal membrane, which leads to endosomolysis and rapid release of the remaining cargo to the cytosol. A model for PFN delivery of Gzms is shown in Supplementary Fig. 7.
Figure 5. Endocytosed cargo is released from gigantosomes into the cytosol.
(a) Sublytic rat PFN induces rapid enhanced uptake of Texas Red (TR)-dextran in EGFP-EEA-1-transfected HeLa cells. Data are representative of six independent experiments. (b) Representative gigantosomes 10–17 min after EGFP-EEA-1-transfected HeLa cells were incubated with TR-dextran and sublytic PFN. Images obtained at 10–12 min suggest focal release of dextran, while at later times (15–17 min) dextran is released as gigantosomes rupture. (c) Time lapse confocal microscopy images acquired every 10 sec of EGFP-EEA-1+ HeLa cells beginning 10 min after treatment with sublytic PFN and TR-dextran. Data are representative of three different experiments. Supplementary Movie 1 shows the movie from which these images were extracted. Discrete TR-dextran release is observed initially (white arrowhead), but after ~15 min of PFN treatment, gigantosomes lose EEA-1 staining, form tubulations and rupture, leading to dextran dispersal (empty arrowhead). (d) Dextran intensity within a PFN-induced gigantosome or normal endosome (-PFN) and in the local surrounding area. Background dextran intensity was also measured in a region devoid of gigantosomes/endosomes. Corresponding images are shown below. Color bars indicate fluorescence intensity. Scale bars, 5 μm (a), 2μm (b–d).
GzmB is released from gigantosomes during NK attack
The most physiologically relevant system to study PFN’s actions is CTL- or NK-mediated lysis of target cells. However, no published studies have visualized PFN or Gzm trafficking in cells subjected to killer cell-mediated destruction, presumably because the amount of native enzyme that enters a target cell is limited. The data presented above were obtained by incubating target cells with sublytic amounts of PFN (with and without GzmB), which is considered a good surrogate for killer cell-mediated cell death, since it reproduces the apoptotic features of the target cell. We previously showed that the two components of the cellular membrane repair response (fusion of internal vesicles with the plasma membrane and rapid endocytosis of the damaged membrane) occur in cells targeted by CD8 T cells and NK cells23, 24. Moreover, EEA-1+ gigantosomes form in target cells during killer cell attack23, 24. To test further whether the two-step model of Gzm delivery by PFN via endosomes applies to the physiologically most relevant model of killer cell attack, we incubated YT-Indy NK cells with 721.221 target cells and examined NK:target cell conjugates at various times over 20 min on slides stained for GzmB or PFN. Within NK cells, GzmB and PFN stained in granules that concentrated at the interface with the target cell, as expected (Fig. 6a,b). Although GzmB or PFN staining was not apparent in most target cells, in a few cells we were able to visualize GzmB and PFN within enlarged cytosolic vesicles sized like gigantosomes. Target cells that stained with GzmB or PFN typically had one or a few gigantosomes visible near the killer cell:target interface. After 20 min incubation, in a few cells GzmB was detected dispersed in the target cell cytosol. At the same time we could not detect PFN staining using the conformation-sensitive Pf80 anti-PFN Ab in any target cell (Fig. 6b). To follow GzmB trafficking in target cells, we also imaged YT-Indy (F6) cells, expressing EGFP-GzmB, as they targeted 721.221 cells (Fig. 6c). EGFP-GzmB first concentrated in a gigantosome-like structure before dispersing in the cytosol about 10–17 min later. Therefore, GzmB and PFN endocytosis into gigantosomes and PFN-induced release of GzmB from gigantosomes to the cytosol of target cell also occurs during killer cell attack.
Figure 6. GzmB and PFN localizes in gigantosomes in target cells during NK cell lysis.
YT-Indy NK cells incubated with 721.221 target cells were stained at indicated times for GzmB (a) or PFN (b). Arrows indicate GzmB or PFN signal (pseudocolor) in target cells. After 10 min, GzmB-containing gigantosomes are visible, but at 20 min, GzmB staining is more dispersed. After 10 min, PFN staining (Pf80) in gigantosomes is visible, but disappears at 20 min. Images were acquired by spinning disk confocal microscopy. Representative z stack series projections from two independent experiments are shown. Color bars and associated numbers indicate fluorescence intensity. Scale bars, 10 μm. Dashed lines, plasma membrane. (c) YT-Indy NK cells expressing EGFP-GzmB were incubated with 721.221 target cells and imaged by widefield live imaging every minute. GzmB-containing gigantosomes are visible 2 min after conjugate formation, but after 15 min, GzmB staining is more dispersed. A representative time-lapse series from two independent experiments is shown. Numbers represent min after conjugate formation. Phase contrast is displayed in red. To visualize the low GzmB signal in the target cell, the EGFP channel was over-exposed. A control YT-Indy cell (bottom row) imaged with regular exposure time confirms the granular expression of EGFP-GzmB. Scale bars, 10 μm.
Discussion
We previously provided evidence that at physiologically relevant PFN concentrations and during CTL attack, PFN creates short-lived pores in the target cell plasma membrane. These pores cause a transient Ca2+ influx into the target cell that lasts a few hundred seconds and mobilizes the stereotypic cellular response to plasma membrane damage23. The membrane repair response, which is triggered by an increase in intracellular Ca2+ concentration, mends the damaged plasma membrane by the fusion of lysosomal and endosomal membranes23, 30 and by endocytosis to remove the damaged membrane24. The stimulation of endocytosis leads to internalization of PFN and Gzms into early endosomes24. In cells treated with physiologically relevant, sublytic concentrations of PFN, we observed formation of giant endosomes, we termed gigantosomes, that contain PFN and Gzms. Enlarged EEA-1+ vesicles were also seen in target cells during killer cell attack. A hallmark of the membrane repair response is exuberant heterotypic and homotypic fusion caused by Ca2+-dependent activation of vesicular trafficking molecules, such as the synaptotagmins and SNARE proteins27–29. We find that gigantosomes form by Rab5-dependent fusion of early endosomes. However, gigantosome formation is an extraneous phenomenon that does not contribute to target cell killing. Since target cells that do not form gigantosomes are equally susceptible to GzmB and PFN, these results suggest that Gzms also escape from smaller Gzm- and PFN-containing endosomes, via the action of PFN in endosomes. Although we could see Gzms in endosomes within a few minutes of exposure to PFN, Gzms were only found in the cytosol much later (about 15 min after treatment). This result suggested that the pores formed by PFN in the plasma membrane were either too small or too rapidly removed to deliver Gzms directly to the cytosol.
We next looked at how gigantosome cargo is delivered to the cytosol. We provide evidence that Gzms and other cargo are released into the cytosol through PFN acting in the gigantosome membrane. Imaging of cells treated with PFN and Gzms showed PFN concentration in discrete foci that form on the gigantosome membrane suggesting that PFN forms pores in the endosomal membrane. However, PFN pore formation does not occur at acidic pH36. Here we show that unlike normal early endosomes35, gigantosomes do not acidify, thereby facilitating PFN pore formation in the gigantosome. The lack of acidification is likely secondary to PFN pore formation, which would interfere with maintaining a pH gradient across the gigantosome membrane and lead to equilibration of gigantosome pH with the neutral cytosolic pH. In further support of PFN pore formation, gigantosome staining with the PFN antibody Pf80 that recognizes an epitope obscured during pore formation40 disappears. This lack of staining is not because PFN is degraded, since staining with an antibody that recognizes both monomeric and multimerized PFN (Pf344) persists. Moreover, crosslinking studies showed that PFN assembles into larger complexes within gigantosomes over 15 min, coincident with gigantosome cargo release. Cross-linking suggested that there might be two types of PFN pores – a smaller multimer made up of about 7 PFN monomers and a much larger multimer of indeterminate size. However, this finding needs to be confirmed by other methods. Fluorescent cargo (fluorescently labeled and unlabeled GzmB and fluorescently labeled dextrans) was imaged as it was released from gigantosomes. Cargo was first released at a slow, but steady, rate beginning about 10 min after cells were exposed to PFN. After about 15 min, the endosomal membrane developed tubulations and eventually ruptured leading to complete release of cargo. It is worth noting that treatment of cells with the vacuolar ATPase inhibitor bafilomycin A, which like PFN prevents endosomal acidification, also causes endosomal tubulations like we visualized with PFN42. It is not clear what triggers the final rupture of the gigantosome or what proportion of GzmB might be released via pores versus at the time of “gigantosomolysis”. The release of 10 kDa dextran, which we measured before rupture, may not exactly mimic what happens with 32 kDa GzmB or the twice as large GzmA dimer. From live cell imaging of PFN-treated cells it is likely that most Gzm release occurs at the time of gigantosome rupture. Our images of GzmB in NK-targeted cells also suggest that some GzmB is released prior to gigantosome rupture, but the resolution is not good enough to be certain.
In our studies of PFN loading of Gzms and killer cell lysis by staining fixed cells or by videomicroscopy we never saw evidence of Gzm entry directly into the cytosol via the plasma membrane. We therefore think it unlikely. However, we cannot rule out the possibility that small amounts of Gzms enter through transient plasma membrane pores, but are not detected because they diffuse rapidly and the fluorescent signal is not concentrated enough to be detected above background. The damaged plasma membrane response to bacterial pore-forming toxins and complement has been shown to include blebbing or exocytosis of damaged membranes43, 44 in addition to the two phenomena we have found in cells exposed to sublytic PFN (patching by fusion of vesicular membranes and endocytosis of damaged membrane23, 24). In some cells plasma membrane blebbing is a prominent feature after treatment with sublytic PFN and Gzms. Examination of culture supernatants of targeted cells for Gzm- and PFN-containing exosomes may indicate whether exocytosis of PFN-damaged membranes is prominent in some cells and promotes apoptotic cell death.
Our results suggest a two-step model for PFN delivery of Gzms in which PFN first forms transient pores in the cell membrane that trigger the target cell membrane repair response leading to coendocytosis of Gzms and PFN. PFN then forms larger, more stable, pores in the endosomal membrane to trigger release of Gzms. Although the experiments presented here used GzmB, we obtained similar results when the other major Gzm (GzmA) was substituted (data not shown). Therefore we expect this model applies to all the Gzms. This model, which suggests that PFN can form at least two types of pores of different size and stability, is supported by a recent study45 that measured conductance through various sorts of membranes (planar lipid bilayers and unilamellar vesicles of different lipid composition and size) treated with PFN. There was a good deal of heterogeneity in PFN pores; in particular formation of small, highly unstable pores preceded the development of more stable and larger pores with a distribution in size. Heterogeneous pore formation was confirmed by cryoelectron microscopy. We therefore hypothesize that the rapid membrane repair response interferes with the formation of larger pores on the plasma membrane, but that PFN multimerizes into larger stable pores on the gigantosome membrane that increase in size within 5–15 min after adding PFN. Based on the kinetics of early endosome acidification, our data suggest that the smaller pores form within the gigantosome membrane almost immediately to interfere with acidification and allow PFN to remain active.
Our evidence for replacing the old model of Gzm delivery through plasma membrane pores with a more complicated two-step model of PFN delivery is based mostly on experiments that used PFN and Gzm treatment of target cells at sublytic concentrations of PFN, which is considered a good model for cell-mediated cytotoxicity. It could be argued, however, that what happens during killer cell attack might be different. During PFN and Gzm loading experiments the killer molecules are delivered across the plasma membrane, while in cell-mediated lysis PFN and Gzms are delivered to a localized area of the target cell membrane within the immune synapse. In the former case, repair of diffuse membrane damage might become more important than in the latter, when membrane damage is localized. Until now it has been impossible to visualize PFN and Gzms within target cells undergoing killer cell attack, making it difficult to assess whether it is really necessary to revise the old plasma membrane pore model. We previously showed that EEA-1+ gigantosomes do in fact form in the target cell during CTL attack23, 24. Because of improved imaging resolution using a highly sensitive, spinning disk confocal microscope, we were now able to detect GzmB and PFN in target cells as they are being killed. The killer molecules do in fact localize rapidly to giant endosomes that form near the immune synapse, before they are detected throughout the cytosol. Rather than multiple gigantosomes being formed, as was seen in cells treated with PFN and GzmB in solution, presumably in response to diffuse membrane damage, it is likely that only one or a few gigantosomes form when the damage is localized to the immune synapse. PFN and Gzm concentration in gigantosomes in target cells followed by Gzm release during killer cell attack strongly suggests that the two-step model for PFN delivery of Gzms accurately reflects what happens in vivo.
Methods
PFN and GzmB purification
Native human PFN and GzmB were purified from YT-Indy NK cells and native rat PFN was purified from RNK16 cells as described46. Animal use was approved by the Animal Care and Use Committees of the Immune Disease Institute and Harvard Medical School. Recombinant GzmB was produced using the mammalian expression vector pHLseq47. GzmB cDNA was cloned into pHLseq at Age-1 and Kpn-1 sites using the forward primer 5′-GAAACCGGTGACGACGACGACAAGATCATCGGGGGACATGAG-3′ (which introduces an enterokinase site before the N-terminus of the active protease) and the reverse primer 5′-GTGCTTGGTACCGTAGCGTTTCATGGTTTTCTT-3′. The cell supernatant from transfected 293T cells, grown in ExCell 293 medium (Sigma) for 4 days, was purified by immobilized metal affinity chromatography using Nickel-NTA (QIAGEN). Eluted GzmB was treated with enterokinase (Sigma, 0.05 IU/ml cell supernatant) for 16 h at 20°C. Active GzmB was purified on an S column, concentrated and quality tested as previously described46.
Treatment with PFN and GzmB
Cells were washed and equilibrated for 5 min in cell loading buffer (HBSS, 10 mM HEPES pH 7.5, 4 mM CaCl2, 0.4% BSA) before adding a sublytic concentration of rat or human PFN (as indicated) and/or native or recombinant GzmB, diluted in PFN buffer (HBSS, 10 mM HEPES pH 7.5). The sublytic PFN concentration was determined for each experiment as the concentration that induced 5–15% PI uptake (2 μg/mL) (Sigma) measured 20 min later by flow cytometry (FACScalibur; Becton Dickinson)46, 48.
Uptake of fluorescent native human GzmB and dextran
The AlexaFluor 488 microscale protein labeling kit (Molecular Probes) was used to label native human GzmB. A488-GzmB (10 μg/mL) was added with sublytic native rat PFN. Cells were then washed with PBS and fixed before analysis by microscopy. Internalization of 10 kDa cationic Texas Red-dextran (1.25 mg/mL, Molecular Probes) was analyzed by live cell imaging. Dextran was added with sublytic native rat PFN for the indicated times. Cells were then washed with cell loading buffer before image acquisition with a spinning disk confocal microscope (see supplementary methods).
Time lapse videomicroscopy and live cell imaging
HeLa cells were grown on 25 mm collagen-coated coverslips and transfected overnight with EGFP-EEA-1 or mRFP-EEA-1 alone or in combination with EGFP-Rab5(WT), EGFP-Rab5(S34N), EGFP-Rab5(Q79L)49 using FuGENE® 6. Coverslips were transferred to a sample holder (20/20 Technology, Inc.) inside an environmental chamber, maintained at 37°C, 5% CO2 and 100% humidity, containing the objective lenses. PFN treatment was performed under the microscope after starting acquisition with a spinning disk confocal microscope (see supplementary methods).
Detection of PFN aggregation by crosslinking
K562 cells (2 × 105) were washed and equilibrated for 5 min in cell loading buffer before adding sublytic native human PFN diluted in PFN buffer (HBSS, 10 mM HEPES pH 7.5). After incubation at 37°C, cells were transferred immediately to 4°C and 2 mM of freshly prepared membrane-permeable crosslinker disuccinimidyl suberate (DSS, Pierce) was added. For cell fractionation, K562 cells (4 × 105) were washed and treated with native human PFN as above. At indicated times, cells were transferred to 4°C and spun at 300g for 5 min. The cell pellet was incubated 5 min in ice-cold cell fractionation buffer (Ambion) before centrifugation at 500g for 5 min to remove plasma membrane and nuclei. The remaining cytoplasmic fraction (supernatant) was centrifuged at 15000g for 10 min to pellet cytosolic vesicles. Vesicles were resuspended in PBS before adding 2 mM of freshly prepared DSS. After 30 min incubation at 4°C, 5X SDS-PAGE loading buffer (200 mM Tris-HCl pH 6.8, 5% SDS, 30% glycerol,0.05% bromophenol blue, 10 mM β-mercaptoethanol) was added and samples were boiled for 5 min. PFN was detected by immunoblot after electrophoresis through a 4–20% denaturing gradient gel, probed with mouse anti-human PFN, clone 2d4.
Imaging of NK/target cell conjugates
YT-Indy NK cells were added to 721.221 target cells (effector:target ratio 2:1) in RPMI 1640 containing 10% FCS in 96-well V bottom plates, which were spun briefly and incubated for 20 min at 37°C to allow conjugate formation (T=0). Conjugates were then spun on poly-L-lysine-coated coverslips and fixed with PBS in 4% PFA and stained with PFN (Pf80) or GzmB (GB11) mAbs as above, before spinning disk confocal imaging. For live cell imaging, YT-Indy (F6) expressing EGFP-GzmB were added to 721.221 target cells, adhered to poly-L-lysine-coated coverslips, before widefield imaging.
Quantification of dextran release from endosomes/gigantosomes by fluorescent live cell confocal microscopy
HeLa cells, seeded on collagen-coated 25 mm glass coverslips, were transfected overnight with a plasmid encoding EGFP-EEA1. The next day TR-dextran was added as above with or without sublytic native rat PFN. The coverslips were placed in an environmental chamber and imaged using the spinning disk confocal microscope as above. Three-dimensional movies were obtained from a cross section of the cells that correspond to a Z-stack series of 5 consecutive optical planes spaced by 0.7 μm acquired at a frequency of 0.05 Hz per stack series with a 100 ms exposure time for GFP and a 200 ms exposure time for dextran. 2D movies were then obtained by generating for each time point a maximum intensity z-projection. Gigantosomes containing fluorescent dextran were identified, tracked and analyzed using Slidebook software (Intelligent Imaging) and a lab-developed MATLAB routine (Boulant et al., submitted). Briefly, three sequential steps (2D Gaussian and Laplacian filtering followed by a local maximum-finding algorithm) were used to detect dextran-containing endosomes. Masks corresponding to the local cytosolic area surrounding the selected dextran-containing endosome/gigantosome were created by expanding the endosome mask by 10 pixels in diameter and subtracting the original endosome mask. The resulting mask had a donut shape. An identical mask was created far from any dextran-staining objects to capture far background intensity. The intensity of the local peri-endosome and far background regions was arbitrarily set to 0 at the beginning of the time lapse movie. The intensity profile as a function of time was plotted.
Supplementary Material
Acknowledgments
We thank E. Marino for assistance with microscopy and image analysis. This work was supported by NIH AI063430 (JL), NERCE grant (TK), Canadian Institutes of Health Research and Canadian Cancer Society (RCB), fellowships from the Stiefel-Zangger Foundation (MW) and the Human Frontier Science Program Organization (EB). We thank Y. Jones (University of Oxford, UK) for kindly providing the mammalian expression vector pHLseq.
Footnotes
Authorship
Contribution: J.T. designed and performed experiments, analyzed data and wrote the manuscript. S.B., D.K. and E.B. also performed and helped analyze some experiments. M.W. and D.M. purified GzmB and helped with PFN purification. I.S.G. and R.C.B. developed the EGFP-GzmB expressing NK cell line. T.K. and J.L. conceived and supervised the project, helped design experiments and coordinated the writing of the manuscript.
Conflict of interest disclosure
The authors declare no competing financial interests.
References
- 1.de Saint Basile G, Menasche G, Fischer A. Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat Rev Immunol. 2010;10:568–579. doi: 10.1038/nri2803. [DOI] [PubMed] [Google Scholar]
- 2.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 3.Stinchcombe JC, Bossi G, Booth S, Griffiths GM. The immunological synapse of CTL contains a secretory domain and membrane bridges. Immunity. 2001;15:751–761. doi: 10.1016/s1074-7613(01)00234-5. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 5.Dustin ML, Long EO. Cytotoxic immunological synapses. Immunol Rev. 2010;235:24–34. doi: 10.1111/j.0105-2896.2010.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthony DA, Andrews DM, Watt SV, Trapani JA, Smyth MJ. Functional dissection of the granzyme family: cell death and inflammation. Immunol Rev. 2010;235:73–92. doi: 10.1111/j.0105-2896.2010.00907.x. [DOI] [PubMed] [Google Scholar]
- 7.Bovenschen N, Kummer JA. Orphan granzymes find a home. Immunol Rev. 2010;235:117–127. doi: 10.1111/j.0105-2896.2010.00889.x. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman J. Granzyme A activates another way to die. Immunol Rev. 2010;235:93–104. doi: 10.1111/j.0105-2896.2010.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasternack MS, Eisen HN. A novel serine esterase expressed by cytotoxic T lymphocytes. Nature. 1985;314:743–745. doi: 10.1038/314743a0. [DOI] [PubMed] [Google Scholar]
- 10.Podack ER, Young JD, Cohn ZA. Isolation and biochemical and functional characterization of perforin 1 from cytolytic T-cell granules. Proc Natl Acad Sci U S A. 1985;82:8629–8633. doi: 10.1073/pnas.82.24.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gershenfeld HK, Weissman IL. Cloning of a cDNA for a T cell-specific serine protease from a cytotoxic T lymphocyte. Science. 1986;232:854–858. doi: 10.1126/science.2422755. [DOI] [PubMed] [Google Scholar]
- 12.Bleackley RC, et al. The isolation and characterization of a family of serine protease genes expressed in activated cytotoxic T lymphocytes. Immunol Rev. 1988;103:5–19. doi: 10.1111/j.1600-065x.1988.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 13.van den Broek ME, et al. Decreased tumor surveillance in perforin-deficient mice. J Exp Med. 1996;184:1781–1790. doi: 10.1084/jem.184.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stepp SE, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286:1957–1959. doi: 10.1126/science.286.5446.1957. [DOI] [PubMed] [Google Scholar]
- 15.Katano H, Cohen JI. Perforin and lymphohistiocytic proliferative disorders. Br J Haematol. 2005;128:739–750. doi: 10.1111/j.1365-2141.2004.05305.x. [DOI] [PubMed] [Google Scholar]
- 16.Pipkin ME, Lieberman J. Delivering the kiss of death: progress on understanding how perforin works. Curr Opin Immunol. 2007;19:301–308. doi: 10.1016/j.coi.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voskoboinik I, Smyth MJ, Trapani JA. Perforin-mediated target-cell death and immune homeostasis. Nat Rev Immunol. 2006;6:940–952. doi: 10.1038/nri1983. [DOI] [PubMed] [Google Scholar]
- 18.Baran K, et al. The molecular basis for perforin oligomerization and transmembrane pore assembly. Immunity. 2009;30:684–695. doi: 10.1016/j.immuni.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Podack ER, Dennert G. Assembly of two types of tubules with putative cytolytic function by cloned natural killer cells. Nature. 1983;302:442–445. doi: 10.1038/302442a0. [DOI] [PubMed] [Google Scholar]
- 20.Podack ER, Konigsberg PJ, Acha-Orbea H, Pircher H, Hengartner H. Cytolytic T-cell granules: biochemical properties and functional specificity. Adv Exp Med Biol. 1985;184:99–119. doi: 10.1007/978-1-4684-8326-0_8. [DOI] [PubMed] [Google Scholar]
- 21.Trapani JA, et al. Genomic organization of the mouse pore-forming protein (perforin) gene and localization to chromosome 10. Similarities to and differences from C9. J Exp Med. 1990;171:545–557. doi: 10.1084/jem.171.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tschopp J, Masson D, Stanley KK. Structural/functional similarity between proteins involved in complement- and cytotoxic T-lymphocyte-mediated cytolysis. Nature. 1986;322:831–834. doi: 10.1038/322831a0. [DOI] [PubMed] [Google Scholar]
- 23.Keefe D, et al. Perforin triggers a plasma membrane-repair response that facilitates CTL induction of apoptosis. Immunity. 2005;23:249–262. doi: 10.1016/j.immuni.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Thiery J, et al. Perforin activates clathrin- and dynamin-dependent endocytosis, which is required for plasma membrane repair and delivery of granzyme B for granzyme-mediated apoptosis. Blood. 2010;115:1582–1593. doi: 10.1182/blood-2009-10-246116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Froelich CJ, et al. New paradigm for lymphocyte granule-mediated cytotoxicity. Target cells bind and internalize granzyme B, but an endosomolytic agent is necessary for cytosolic delivery and subsequent apoptosis. J Biol Chem. 1996;271:29073–29079. doi: 10.1074/jbc.271.46.29073. [DOI] [PubMed] [Google Scholar]
- 26.Browne KA, et al. Cytosolic delivery of granzyme B by bacterial toxins: evidence that endosomal disruption, in addition to transmembrane pore formation, is an important function of perforin. Mol Cell Biol. 1999;19:8604–8615. doi: 10.1128/mcb.19.12.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 28.McNeil PL, Steinhardt RA. Plasma membrane disruption: repair, prevention, adaptation. Annu Rev Cell Dev Biol. 2003;19:697–731. doi: 10.1146/annurev.cellbio.19.111301.140101. [DOI] [PubMed] [Google Scholar]
- 29.McNeil PL, Kirchhausen T. An emergency response team for membrane repair. Nat Rev Mol Cell Biol. 2005;6:499–505. doi: 10.1038/nrm1665. [DOI] [PubMed] [Google Scholar]
- 30.Idone V, et al. Repair of injured plasma membrane by rapid Ca2+-dependent endocytosis. J Cell Biol. 2008;180:905–914. doi: 10.1083/jcb.200708010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bucci C, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 32.Olkkonen VM, Stenmark H. Role of Rab GTPases in membrane traffic. Int Rev Cytol. 1997;176:1–85. doi: 10.1016/s0074-7696(08)61608-3. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong J. How do Rab proteins function in membrane traffic? Int J Biochem Cell Biol. 2000;32:303–307. doi: 10.1016/s1357-2725(99)00112-0. [DOI] [PubMed] [Google Scholar]
- 34.Li G, Barbieri MA, Colombo MI, Stahl PD. Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J Biol Chem. 1994;269:14631–14635. [PubMed] [Google Scholar]
- 35.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 36.Tschopp J, Masson D. Inhibition of the lytic activity of perforin (cytolysin) and of late complement components by proteoglycans. Mol Immunol. 1987;24:907–913. doi: 10.1016/0161-5890(87)90002-2. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 38.Recchi C, Chavrier P. V-ATPase: a potential pH sensor. Nat Cell Biol. 2006;8:107–109. doi: 10.1038/ncb0206-107. [DOI] [PubMed] [Google Scholar]
- 39.Galloway CJ, Dean GE, Marsh M, Rudnick G, Mellman I. Acidification of macrophage and fibroblast endocytic vesicles in vitro. Proc Natl Acad Sci U S A. 1983;80:3334–3338. doi: 10.1073/pnas.80.11.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praper T, et al. Human perforin permeabilizing activity, but not binding to lipid membranes, is affected by pH. Mol Immunol. 2010;47:2492–2504. doi: 10.1016/j.molimm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Jans DA, Jans P, Briggs LJ, Sutton V, Trapani JA. Nuclear transport of granzyme B (fragmentin-2). Dependence of perforin in vivo and cytosolic factors in vitro. J Biol Chem. 1996;271:30781–30789. doi: 10.1074/jbc.271.48.30781. [DOI] [PubMed] [Google Scholar]
- 42.D’Arrigo A, Bucci C, Toh BH, Stenmark H. Microtubules are involved in bafilomycin A1-induced tubulation and Rab5-dependent vacuolation of early endosomes. Eur J Cell Biol. 1997;72:95–103. [PubMed] [Google Scholar]
- 43.Husmann M, et al. Elimination of a bacterial pore-forming toxin by sequential endocytosis and exocytosis. FEBS Lett. 2009;583:337–344. doi: 10.1016/j.febslet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 44.Pilzer D, Gasser O, Moskovich O, Schifferli JA, Fishelson Z. Emission of membrane vesicles: roles in complement resistance, immunity and cancer. Springer Semin Immunopathol. 2005;27:375–387. doi: 10.1007/s00281-005-0004-1. [DOI] [PubMed] [Google Scholar]
- 45.Praper T, et al. Human perforin employs different avenues to damage membranes. J Biol Chem. 2010 doi: 10.1074/jbc.M110.169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiery J, Walch M, Jensen DK, Martinvalet D, Lieberman J. Isolation of cytotoxic T cell and NK granules and purification of their effector proteins. Curr Protoc Cell Biol. 2010;Chapter 3(Unit3):37. doi: 10.1002/0471143030.cb0337s47. [DOI] [PubMed] [Google Scholar]
- 47.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 48.Martinvalet D, Thiery J, Chowdhury D. Granzymes and cell death. Methods Enzymol. 2008;442:213–230. doi: 10.1016/S0076-6879(08)01411-0. [DOI] [PubMed] [Google Scholar]
- 49.Barbieri MA, Li G, Mayorga LS, Stahl PD. Characterization of Rab5:Q79L-stimulated endosome fusion. Arch Biochem Biophys. 1996;326:64–72. doi: 10.1006/abbi.1996.0047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.