Abstract
Recent studies in cell lines and genetically engineered mice have demonstrated that cytosolic double-stranded (ds) DNA could activate dendritic cells (DCs) to become effector antigen presenting cells. Recognition of DNA might be a major factor in antimicrobial immune responses against cytosolic pathogens and also in human autoimmune diseases such as systemic lupus erythematosus. However, the role of cytosolic dsDNA in human DC activation and its effects on effector T and B cells are still elusive. Here we demonstrate that intracellular dsDNA is a potent activator of human monocyte-derived DCs, as well as primary DCs. Activation by dsDNA depends on NF-κB activation, partially on the adaptor molecule IPS-1 and the novel cytosolic dsDNA receptor IFI16, but not on the previously recognized dsDNA sentinels AIM2, DAI, RNA polymerase III or HMGBs. More importantly, we report for the first time that human dsDNA-activated DCs, rather than LPS- or inflammatory cytokine cocktail-activated DCs, represent the most potent inducers of naïve CD4+ T cells to promote Th1-type cytokine production and to generate CD4+ and CD8+ cytotoxic T cells. dsDNA-, but not LPS- or cocktail-activated DCs induce B cells to produce complement fixing IgG1 and IgG3 antibodies. We propose that cytosolic dsDNA represents a novel, more effective approach to generate DCs to enhance vaccine effectiveness in reprogramming the adaptive immune system to eradicate infectious agents, autoimmunity, allergy and cancer.
Introduction
Dendritic cells (DCs) are central players in the initiation and regulation of effective immune responses against infectious agents. In addition, they are essential in the induction of tolerance and anti-tumor immunity, and prevention of autoimmunity. DCs are located in non-lymphoid and peripheral lymphoid tissues where they act as sentinels of environmental cues and orchestrate the interplay between the innate and the adaptive immune system to provoke a successful response. Non-activated, immature DCs (IDCs) specialize in antigen uptake, while activated mature DCs (MDCs) are professional antigen presenting cells (APCs) capable of activating T and B cells to become effector cells (1, 2). Antigen uptake and activation of DCs are mediated through the interaction of cell surface and intracellular receptors with antigens.
Several pattern-recognition molecules evolved to discriminate between foreign and self antigens (3, 4) and several receptors have been described to identify foreign nucleic acids. Endosomal Toll-like receptors (TLRs) such as TLR3 and 7/8 can detect microbe-derived double-stranded (ds) and single-stranded RNA, respectively (5). The RNA helicase domain-containing proteins retinoic acid-inducible gene-I (RIG-I) and the melanoma differentiation-associated gene 5 (Mda5) respond to negative-stranded viral RNA molecules present in the cytoplasm of infected cells (6). While unmethylated CpG-rich DNA sequences found in certain microbes can be easily recognized by TLR9 in the endosome of the host (7), the presence of naked DNA in the cytosol, a danger signal independent of its microbial or self origin, is of critical importance. The immunological detection of cytosolic naked dsDNA became evident during the past few years by demonstrating the presence of cytosolic DNA sensors that function independently of TLR9 and other TLRs (8–11) as well as from the RIG-I/Mda5 system (11). The cytoplasmic dsDNA induces many cell types to produce type I interferon (IFN) and this response requires signaling molecules such as the TANK-binding kinase-1 (TBK-1) and the inhibitor of nuclear factor κB (NF-κB) kinase (IKKi) (12). Several candidate receptors have been identified recently to recognize such cytosolic DNA. For example DNA-dependent activator of IFN-regulatory factor 3 (DAI) has been shown to associate with both TBK-1 and interferon regulatory transcription factor 3 (IRF3) (13). However, DAI-deficient cells and mice produce near wild-type amount of type I IFN in response to cytosolic DNA (12), suggesting that other receptors are involved. The pyrin and HIN200 domain–containing (PYHIN) protein absent in melanoma 2 (AIM2) has been identified as a cytoplasmic dsDNA receptor that induces the formation of a specific inflammasome and subsequent secretion of IL-1β and pyroptotic cell death (14–17). AIM2-deficient mice are defective in caspase-1 activation, IL-1β secretion and cell death in response to cytosolic DNA or Francisella tularensis infection (18) and has a partial role in the sensing of Listeria monocytogenes (19, 20). However, AIM2 does not appear to mediate type I IFN response to dsDNA. IFI16, another PYHIN protein was also identified as an intracellular DNA sensor that mediates the induction of IFN-β where STING, a critical mediator (21), was recruited to IFI16 after DNA stimulation (22). Recently, RNA polymerase III was shown to efficiently trigger the production of type I IFN by transcribing microbial DNA templates into dsRNA containing 5`-triphosphate, which is a potent activator of RIG-I (23, 24). Another cytosolic nucleic acid sensor leucine-rich repeat flightless-interacting protein 1 (LRRFIP1) could also mediate type I IFN production via a β-catenin-dependent pathway (25). Moreover, high-mobility group box (HMGB) proteins were reported also to function as universal sentinels for nucleic acids, modulating type I IFN as well as inflammatory cytokine production (26). Taken together, these observations suggest the existence of multiple cytosolic proteins with DNA sensor capability in various cell types; however it remains still elusive how human APCs can recognize dsDNA.
DNA binding to the cytosolic receptors activates not only the IRF but also the NF-κB transcriptional pathway to trigger the production of pro-inflammatory cytokines in addition to type I IFNs (4). The introduction of dsDNA, derived from either a pathogen or from the host, into the cytoplasm of macrophages and DCs induces phenotypic and functional maturation by activating a set of co-stimulatory genes, cytokines, chemokines and transcription factors (27–30), which are mediated by a TLR9-independent pathway (31–35). dsDNA activates Irf3 and the Ifn-β promoters via both TBK-1 and IKKi; whereas, NF-κB activation by dsDNA is independent of both TBK-1 and IKKi (11). Both pathways require the adaptor molecule IPS-1 but not TLRs or RIG-I (11, 36). The TNF receptor-associated factor 6 (TRAF6), which is essential for the activation of NF-κB and the production of type I IFNs, mediates antiviral responses triggered by cytosolic viral DNA (37). The recognition of dsDNA is sequence independent and requires the DNA to be present in the cytoplasm of the target cells (27, 28). These studies are seminal to the understanding the effects of cytosolic dsDNA on IFN production and response in cell lines and mouse gene knock-out systems. However, how cytosolic dsDNA activates human DCs and what types of effector T and B cells these DCs generate remains undetermined.
In this study we demonstrate that intracellular dsDNA is a potent activator of human monocyte-derived DCs, as well as of primary DCs. This activation is independent of the DNA sensors AIM2, DAI, RNA polymerase III or HMGBs; however, it depends in part on the cytosolic IFI16 dsDNA receptor. We report that cytosolic dsDNA-activated human DCs, unlike LPS- or inflammatory cytokine cocktail-activated DCs, can induce naïve CD4+ T cells to produce more IL-2, IFN-γ and granzyme but not IL-4, and that these effector CD4+ T cells efficiently kill tumor cells in in vitro cultures. In addition, we demonstrate that dsDNA-activated DCs generate cytotoxic CD8+ T cells that produce the effector proteins IFN-γ and granzyme. In the presence of T cells, dsDNA-activated DCs, but not LPS- or cocktail-activated DCs, induce B cells to produce complement fixing IgG1 and IgG3 but not the non-complement fixing IgG2 and IgG4 antibodies. Thus, synthetic cytosolic DNA represents a novel, more effective and safer means of generating DCs to use in vaccines to orchestrate and reprogram the adaptive immune system to eradicate infectious agents, autoimmunity, allergy and cancer.
Materials and methods
Generation of monocyte-derived dendritic cells and culturing
Human monocyte-derived DCs were generated from CD14+ blood monocytes isolated from peripheral blood mononuclear cells (PBMCs) separated from Buffy Coats by Ficoll-Paque (Fisher, Pittsburgh, PA) gradient centrifugation (38) followed by positive selection with anti-CD14-coated magnetic beads (Miltenyi Biotech, Auburn, CA). Purified CD14+ monocytes (≥95%) were plated at 2×106 cell/ml concentration and cultured in RPMI 1640 medium (Sigma, St. Louis, MO) supplemented with 2 mM Glutamine, 100 U/ml penicillin and 100 g/ml streptomycin in the presence of 100 ng/ml IL-4 and 75 ng/ml GM-CSF (Peprotech, Rocky Hill, NJ) given on days 0 and 2. Activation of IDCs were induced on day 5 by an inflammatory cocktail containing 10 ng/ml TNF, 5 ng/ml IL-1β, 20 ng/ml IL-6, 75 ng/ml GM-CSF (Peptrotech) and 1 µg/ml PGE2 (Sigma) or by lipopolysaccharide (LPS – 500 ng/ml) for 24 hours. For DC activation poly(dA:dT) (2.5 µg/ml) complexed with LyoVec transfection reagent (Invivogen, San Diego, CA) was used on 5-day IDCs for 24h. In some experiments the 5-day IDCs were pre-treated with 5 µg/ml chloroquine, 2 µg/ml ML-60218 or 1 µg/ml MG132 for 1h before the poly(dA:dT) treatment to inhibit the endosome acidification, RNA polymerase III or proteosome activity, respectively. The optimal inhibitor concentrations were titrated in preliminary experiments and the viability of the treated cells were analyzed by flow cytometry.
Flow cytometry
The identification of DC and T cell activation were monitored by flow cytometric analysis using fluorochrome-conjugated anti-CD80, anti-CD83 and anti-CD86 (DC, B cell), anti-CD19 (B cell) and anti-CD3, anti-CD25 and anti-CD69 (T cell) antibodies as compared to isotype-matched control antibodies (Beckton Dickinson Pharmingen, San Diego, CA). T cell proliferation was measured by CFSE staining (5 µM, 5 min incubation on 37 °C, washing 2 times with medium containing 10% FBS). Intracellular granzyme production of the T cell subsets was detected by co-staining of fluorochrome-conjugated anti-granzyme with anti-CD4 or anti-CD8 antibodies in the CD3+ population. Fluorescence intensities were measured and analyzed by FACSCalibur flow cytometer (BD Biosciences Immunocytometry Systems, Franklin Lakes, NJ). Data analysis was performed using the FlowJo Flow Cytometry Analysis software (Ashland, OR).
Real-time quantitative reverse transcriptase-polymerase chain reaction (Q-RT-PCR)
Real-time PCR was performed as described previously (39). Total RNA was isolated from DCs by RNeasy Mini Kit (Qiagen, Valencia, CA). Reverse transcription was performed at 37 °C for 120 minutes from 100 ng total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR for AIM2, DAI, IRF3, IRF7, IFN-β, HMGB1, HMGB2, HMGB3, IFI16 and IPS-1 genes were performed (Light Cycler 480, Roche, Indianapolis, IN) with 40 cycles at 94 °C for 12 seconds and 60 °C for 60 seconds using Taqman assays (Applied Biosystems). All PCR reactions were run in triplicates with a control reaction containing no RT enzyme. The comparative Ct method was used to quantify transcripts relative to the endogenous control gene 36B4.
PCR arrays were performed using the Dendritic Cell - Antigen Presenting Cell and the Toll-like Receptor Pathway PCR Arrays (SA Biosciences, Frederick, MD) following the Manufacturer’s instructions. Reverse transcription was performed from 500 ng total RNA using the RT2 First Strand Kit (SA Biosciences). Quantitative real-time PCR were performed (Light Cycler 480, Roche) with 45 cycles at 94 °C for 15 seconds and 60 °C for 60 seconds. Fold changes were calculated for each gene using the Manufacturer’s web-based PCR Array Data Analysis. The data were deposited at the GEO database (GSE29131 - http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29131).
Enzyme-linked immunosorbent assay
IL-6, TNF, IFN-γ, IL-2, IL-4 and IL-1β proteins were measured from cell supernatants using DuoSet human immunoassay kits (R&D Systems, Minneapolis, MN); IFN-β cytokine was measured by VeriKine Human IFN Beta ELISA Kit (PBL, Piscataway, NJ) following the manufacturer’s instructions. Total IgG levels from DC-T-B cell co-cultures were measured by the Human IgG ELISA Kit (Immunology Consultants Lab, Inc., Newberg, OR) and the IgG subtypes were identified by the Human IgG Subclass Profile ELISA Kit (Invitrogen, Camarillo, CA). The optical density of the wells was determined using a microplate reader set at 450 nm.
Migration
DCs were suspended in migration medium (0.5 % BSA in RPMI 1640) at 106 cells/ml. Transmigration inserts (diameter 6.5 mm; pore size 5 µm) were obtained from Sigma. MIP3-β chemokine (Peprotech) were diluted at 200 ng/ml in migration medium and added to the lower chambers in a final volume of 600 µl. DCs were added to the upper chamber in a final volume of 250 µl, and chemotaxis assays were conducted for 4h in 5% CO2 at 37 °C. At the end of the assay, the inserts were discarded and cells migrated to the lower chamber were collected. Migrated cell numbers were counted by using polystyrene standard beads (Sigma) by flow cytometry.
Transfection of small interfering RNA
A mix of three different constructs of AIM2, DAI, IRF3, IRF7, HMGB1, HMGB2, IFI16 and IPS-1 siRNAs and control siRNAs (Applied Biosystems) were transfected into 2-day IDC to a final concentration of 2.5 nM using the GenePulser X Cell electroporator and 0.4 cm cuvettes (Bio-Rad Laboratories, Hercules, CA). After 4 additional days the knockdown of the genes were tested by Q-RT-PCR.
Mixed leukocyte reaction
The monocyte-derived DCs were activated as mentioned above and the MDCs were co-cultured with allogeneic naïve CD4+, CD8+ and total T cells or with B cells purified from peripheral blood using the Human Naïve CD4+ T cell Isolation Kit II (purity ≥ 98%), the Human Naïve CD8+ T cell Isolation Kit (purity ≥ 95%) (Miltenyi Biotech), the RosetteSep Human T cell Enrichment Cocktail (Stemcell Technologies, Vancouver, Canada) or anti-CD19-coated magnetic beads (purity ≥ 98%) (Miltenyi Biotech), respectively; in 1:5 DC-T/B cell ratio for different time points. In some of the experiments total T cells were replaced by soluble CD40 ligand (25 µg/ml, Peprotech) to activate B cells. T and B cells were labeled with CFSE. T and B cell activation was monitored by cytokine production (IFNγ, IL-2 and IL-4), proliferation (CFSE staining) and cell surface expression of activation molecules (CD25, CD69, CD86).
In vitro tumor killing assay
Monocyte-derived DCs were differentiated and matured as mentioned before. Naïve CD4+ and CD8+ T cells were purified using the above mentioned magnetic cell separation method and co-cultured with IDC or the DNA-, LPS- or inflammatory cytokine cocktail-activated DCs and the EBV-specific Burkitt lymphoma cell line Oku-1 (latency type I) (40) in different target:killer ratios (from 1:10 to 1:50). After 4 days of co-culture, fresh CFSE-labeled Oku-1 cells were added to the cultures. After additional 4h incubation, the cells were labeled with active caspase-3-specific antibodies (BioLegend) and the percentages of active caspase-3-CFSE double positive cells were measured by flow cytometry.
Statistical analysis
Statistical comparisons were made using unpaired t-test and level of significance was set to 0.05 (labeled by * in figures). The statistical calculations compared experimental samples to the IDC or non-treated (N.T.) samples unless otherwise noted. For all experiments, the mean and the standard deviation (SD) are reported for at least n=3 except in case of the gene expression experiments where one representative data set out of at least 3 independent experiments is shown.
Results
Characteristics of cytosolic DNA-activated human monocyte-derived and primary DCs
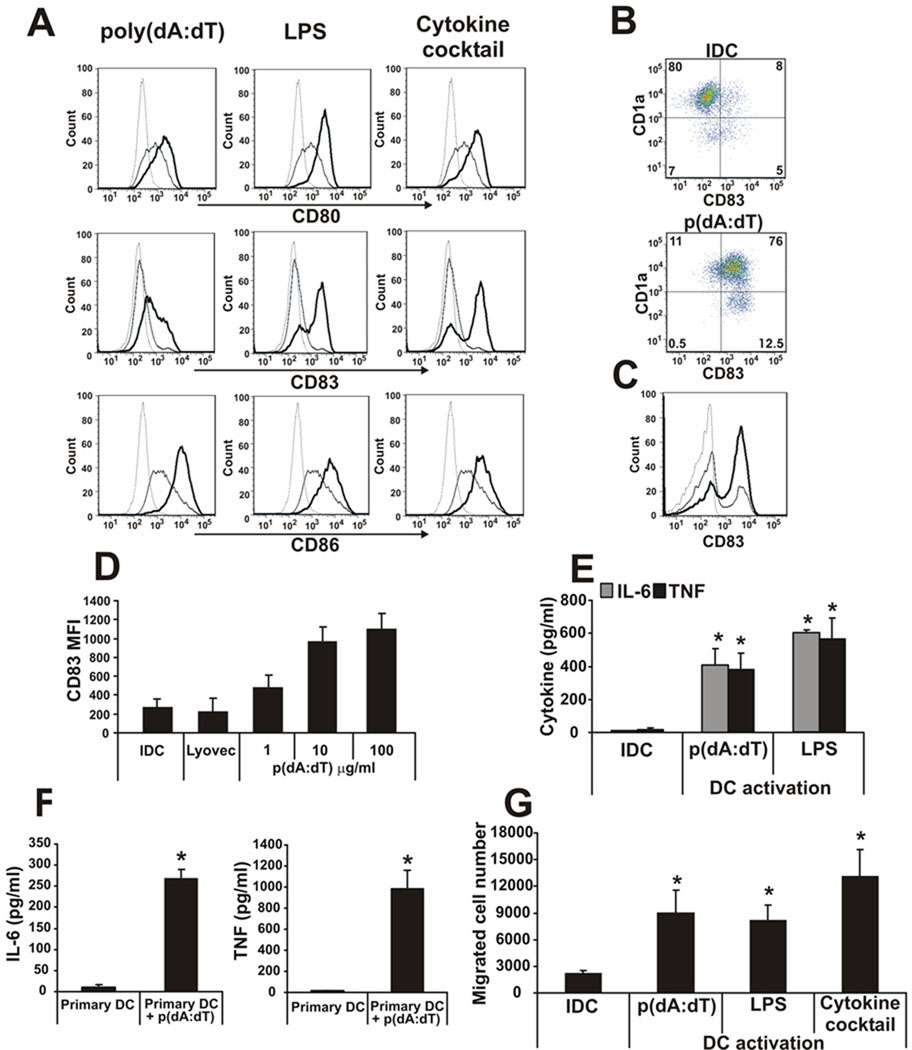
To determine the capacity of cytosolic dsDNA to activate and mature human DCs, poly(dA:dT) was transfected into monocyte-derived DCs (henceforth DNA-DCs) by the Lyovec transfection reagent and the level of activation was compared to two `classical` DC stimuli: LPS or an inflammatory cytokine (TNF, IL-1β, IL-6, GM-CSF and PGE2) cocktail. Poly(dA:dT) was as effective as LPS or the cytokine cocktail in inducing DC activation as shown by CD80, CD83 and CD86 expression (Fig. 1A). Cytosolic DNA activated both CD1a− and CD1a+ subpopulations of monocyte-derived DCs (Fig. 1B), which have been shown to exhibit different functional properties (41), as well as primary CD11c+ DCs, purified from human peripheral blood (Fig. 1C). The effect of poly(dA:dT) was dose-dependent (Fig. 1D). Monocyte-derived DCs (Fig. 1E), as well as primary DCs (Fig. 1F), produced significant amounts of the pro-inflammatory cytokines IL-6 and TNF after poly(dA:dT) treatment. The migratory capacity of DNA-DCs toward the MDC chemokine MIP3-β was comparable to that induced by other stimuli (Fig. 1G).
Figure 1. Phenotypic characterization of dsDNA-activated DCs.
A, Poly(dA:dT) was transfected into monocyte-derived DCs using Lyovec transfection reagent and the level of activations were compared to LPS- and inflammatory cytokine cocktail-activated DCs by CD80, CD83 and CD86 surface expression. Dashed line: isotype control, thin line: immature DC, thick line: activated DC. B, Cytosolic dsDNA activation of CD1a− and CD1a+ subpopulations of monocyte-derived DCs and C, primary CD11c+ blood DCs monitored by CD83 upregulation. D, Dose-dependent activation of DCs by poly(dA:dT) monitored by CD83 upregulation. E, IL-6 and TNF pro-inflammatory cytokine production of monocyte-derived DCs and F, primary DCs after poly(dA:dT) transfection. G, Migratory capacity of poly(dA:dT)-, LPS- or inflammatory cytokine cocktail-DCs towards the MDC chemokine MIP3-β.
n=5 independent experiments.
The length of poly(dA:dT) did not dictate the magnitude of maturation of the DCs (Supplemental Fig. 1A and C). Besides synthetic DNA poly(dA:dT), natural DNA from different sources (human or foreign) was also able to activate DCs after transfection (Supplemental Fig. 1B and D). DNA treatment without transfection (data not shown) or with the Lyovec transfection reagent alone (Fig. 1D) did not activate DCs.
To identify and compare the spectrum of genes induced by cytosolic dsDNA, gene expression arrays focusing on DC-related genes and those implicated in APC functions or in TLR pathways were performed. The heat maps showed similar expression profiles in DCs treated with poly(dA:dT), LPS or the inflammatory cytokine cocktail (Supplemental Fig. 2A and B). However, DNA- and LPS-DCs appeared to share more commonly regulated genes (Supplemental Fig. 2C and D) suggesting that these stimuli employ similar activation mechanisms and/or signaling pathways. Although most genes were induced similarly by all three stimuli, IFN-β expression was markedly upregulated in poly(dA:dT)-activated DCs (fold induction 189x) compared to LPS- (18x) or cytokine cocktail-activated DCs (1.7x).
Taken together, the data demonstrate that synthetic or natural cytosolic dsDNA can activate/mature human DCs and suggest that during inflammatory processes such as viral infection or tissue injury, virus-derived or damaged cell-derived dsDNA once inside DCs may trigger their maturation to become full-fledged professional APCs.
DNA-DCs confer killing ability on CD4+ and CD8+ T cells
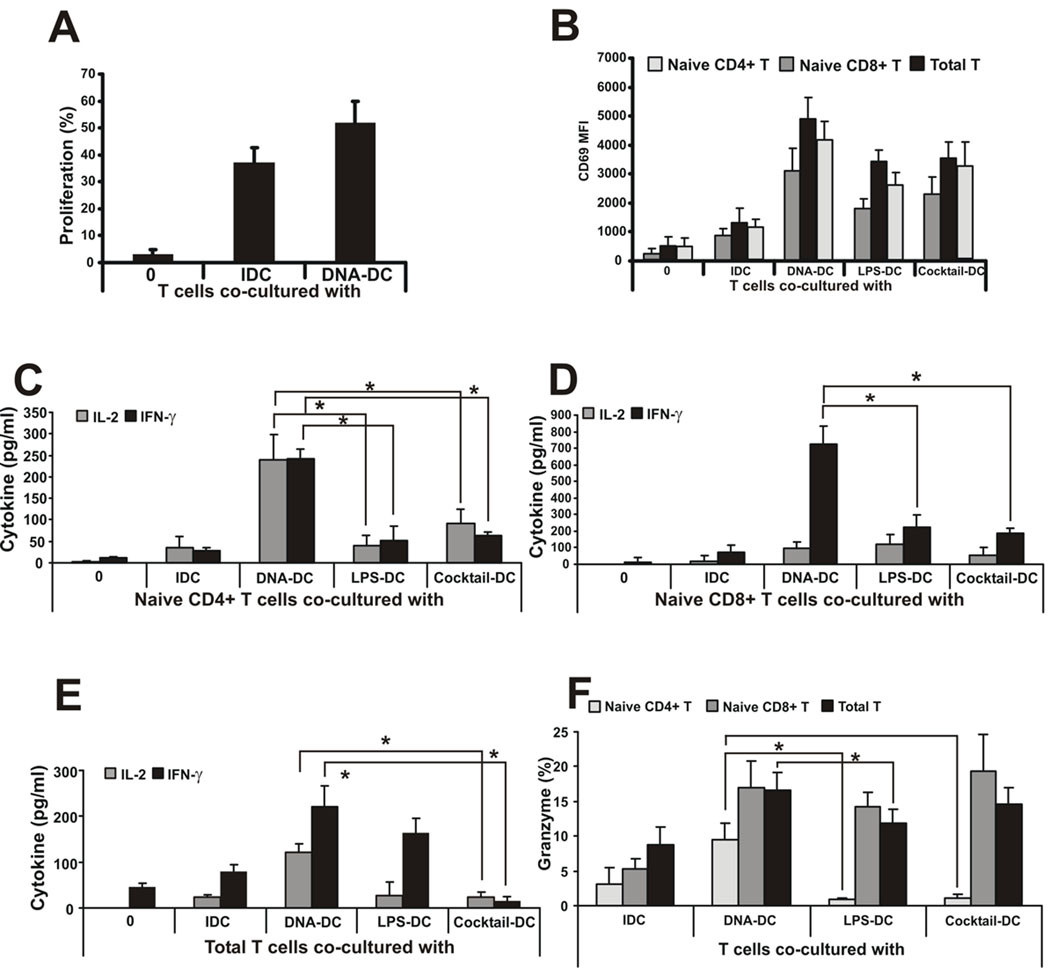
The above data clearly demonstrated the effectiveness of cytosolic DNA in producing activated, mature human DCs. Whether these DNA-DCs could function as professional APCs to invoke a fully functional cellular immunity has not been explored. To determine the type of effector functions that DNA-DCs would impart on CD4+ and CD8+ T cells, mixed leukocyte cultures using allogeneic total, naïve CD4+ or CD8+ T cells from peripheral blood were performed. As shown in Fig. 2A, total allogeneic T cells proliferated slightly better when co-cultured with DNA-DCs compared to IDCs. To dissect which subpopulation of T cells were most responsive to DNA-DC signals, total T cells were fractionated into naïve CD4+ or CD8+ T cells using magnetic cell sorting. As shown in Fig. 2B, naïve CD4+ or CD8+ T cells, as well as total T cells, co-cultured with DNA-DCs, displayed higher expression levels of the surface activation marker CD69 compared to LPS-DCs, inflammatory cytokine cocktail-DCs or IDCs. Naïve CD4+ or CD8+ T cells as well as total T cells co-cultured with DNA-DCs produced higher amount of IL-2 and IFN-γ compared to LPS-DCs, cocktail-DCs or IDCs (Fig. 2C–E). Furthermore, DNA-DC-conditioned CD8+ cells produced similar levels of granzyme compared to LPS-DCs, cocktail-DCs or IDCs, and total T cells produced the highest levels of granzyme when activated by DNA-DC (Fig. 2F). Surprisingly a population of DNA-DC-conditioned CD4+ T cells was also granzyme positive, suggesting that these T cells may be primed to perform killing functions (Fig. 2F).
Figure 2. dsDNA-activated DCs function as professional APCs.
A, Total allogeneic T cells were co-cultured with IDCs or poly(dA:dT)-activated DCs and cell proliferation was measured by CFSE labeling. Proliferation is shown as percentage of cells with low CFSE staining. B, Naïve CD4+, naïve CD8+ and total allogeneic T cells were co-cultured with IDCs or poly(dA:dT)-, LPS- or inflammatory cytokine cocktail-activated DCs and the surface expression of the activation molecule CD69 was measured by flow cytometry. C, Naïve CD4+,D, naïve CD8+ and E, total allogeneic T cells were co-cultured with IDCs or poly(dA:dT)-, LPS- or inflammatory cytokine cocktail-activated DCs and IL-2 and IFN-γ cytokine production by T cells was measured by ELISA. F, Granzyme positive CD4+, CD8+ and total allogeneic T cells co-cultured with differentially activated DCs measured by flow cytometry.
N.T.: non-treated. n=4 independent experiments.
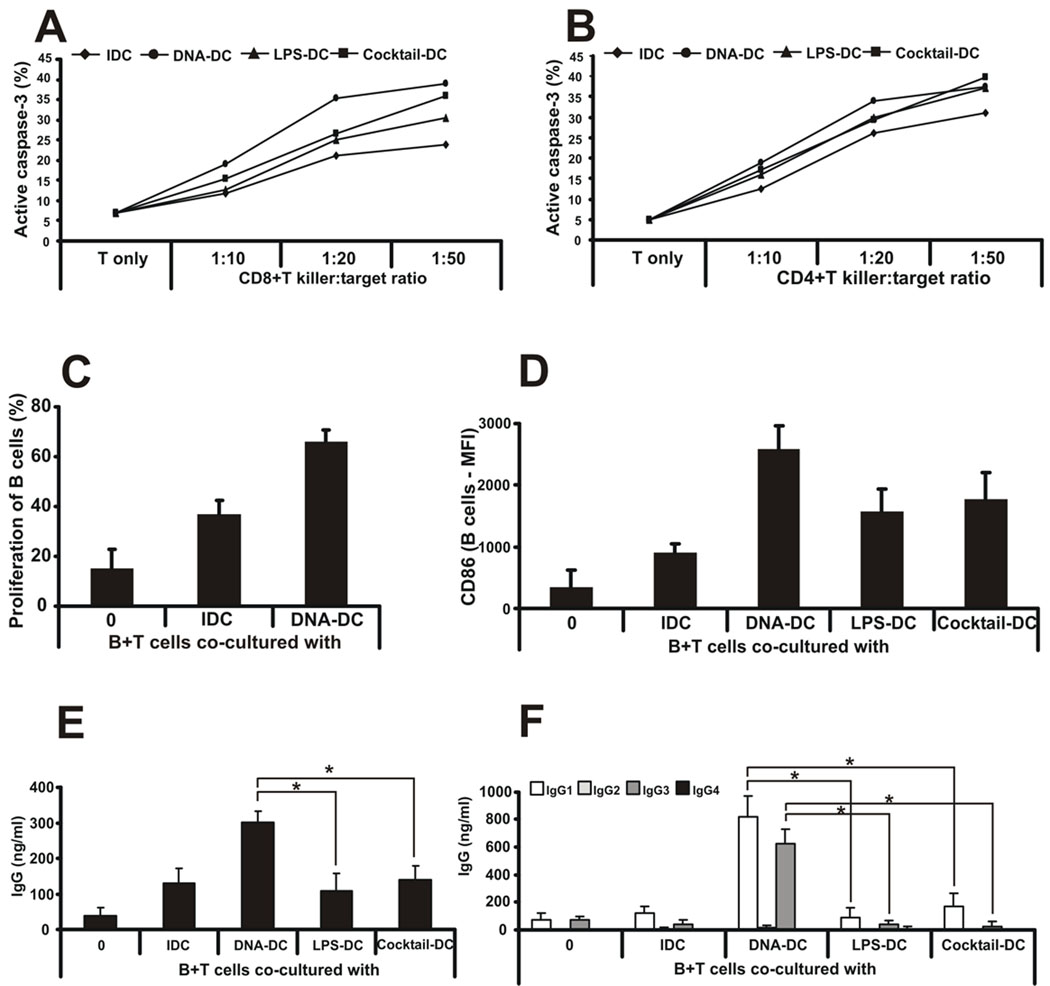
To test the ability of DNA-DC-primed CD4+ and CD8+ T cells to kill target cells, the EBV+ Burkitt’s lymphoma cell line Oku-1 (latency type I) was chosen as the target. Oku-1 cells were treated with mitomycin C to prevent overgrowth and then co-cultured with T cells and DCs for 4 days at different target:killer ratios (from 1:10 to 1:50). To measure the killing capacity of DC-activated T cells, CFSE-labeled Oku-1 cells were added to the co-cultures for an additional 4h after the 4-day pre-sensitization. To identify apoptotic target cells, flow cytometry was used to detect intracellular active caspase-3, which was processed from an inactive pro-enzyme form. Indeed, DNA-DC-primed CD8+ T cells were more proficient in killing target cells compared to other MDCs or IDCs (Fig. 3A). Surprisingly, DNA-DC-primed CD4+ T cells showed similar killing capacity compared to primed CD8+ T cells. DNA-DCs were slightly better at generating killer CD4+ T cells than LPS-DCs, cocktail-DCs or IDCs, especially when 1:20 killer:target cell ratio was used (Fig. 3B). These findings support the notion that the quality and type of CD4+ T cell immunity generated were directly linked to the type of DCs they encountered during an immune response.
Figure 3. dsDNA-activated DCs confer killing ability on CD4+ and CD8+ T cells and collaborate with T cells to generate a specific humoral response.
A, Oku-1 cells were co-cultured with naïve CD8+ or B, CD4+ T cells and differentially activated DCs for 4 days at different killer:target ratios (from 1:10 to 1:50). To measure the killing capacity of DC-activated T cells, fresh, CFSE-labeled Oku-1 cells were added to the co-cultures for an additional 4h. To identify apoptotic target cells, flow cytometry was used to detect intracellular active caspase-3 in CFSE-positive cells. n=5 independent experiments. One representative data set is shown. C, Allogeneic B cells were co-cultured with IDCs or poly(dA:dT)-activated DCs and cell proliferation was measured by CFSE labeling. Proliferation is shown as percentage of cells with low CFSE staining. D, Allogeneic B cells were co-cultured with IDCs or poly(dA:dT)-, LPS- or inflammatory cytokine cocktail-activated DCs and the surface expression of CD86 molecule was measured by flow cytometry. E, Allogeneic B and T cells were co-cultured with IDCs or poly(dA:dT)-, LPS- or inflammatory cytokine cocktail-activated DCs and total immunoglobulin G production was measured from the supernatants. F, Immunoglobulin G subclass profiling was done from co-culture supernatants by ELISA. n=3 independent experiments.
DNA-DCs enable T cells to help B cells to generate a specific humoral response
Another function of APCs is to collaborate with T cells to induce an effective B cell humoral immunity. To test the efficiency of DNA-DCs to induce B cell responses, mixed lymphocyte reactions using allogeneic DCs co-cultured with B and T cells were performed. Both B and T cells were obtained from the same donor. B cells proliferated well in the presence of DNA-DCs compared to IDCs (Fig. 3C) and expressed more CD86 on their surface (Fig. 3D). DNA-DCs, in collaboration with T cells, were capable of triggering B cells to produce higher levels of immunoglobulin G (IgG) compared to co-cultures with IDCs or with any other MDCs (Fig. 3E). Interestingly, soluble CD40 ligand with DCs or DCs alone could not induce B cells to produce IgG (data not shown) suggesting that the effects of DNA-DCs were manifested mainly through the direct interaction between T cells, DNA-DCs and B cells. Cytokines reportedly responsible for the antibody production did not appear to participate in the induction of IgG since neither IL-4 nor IL-21 cytokines were detected in the supernatants of these co-cultures (data not shown). More importantly, IgG subclass profiling revealed that the main IgG subclasses produced by DNA-DC-activated B cells were the effector IgG1 and IgG3 isotypes, which are known potent activators of the complement system, compared to other DCs (Fig. 3F). The non-complement fixing IgG2 and IgG4 were not induced (Fig. 3F). Thus, the type and quality of effector B cells generated during a humoral response depended on the quality and type of DCs they encountered. Again, they underscored the plasticity of DCs in shaping the adaptive immune system.
DNA-DC-produced interferon-β is dispensable for dsDNA-induced DC activation
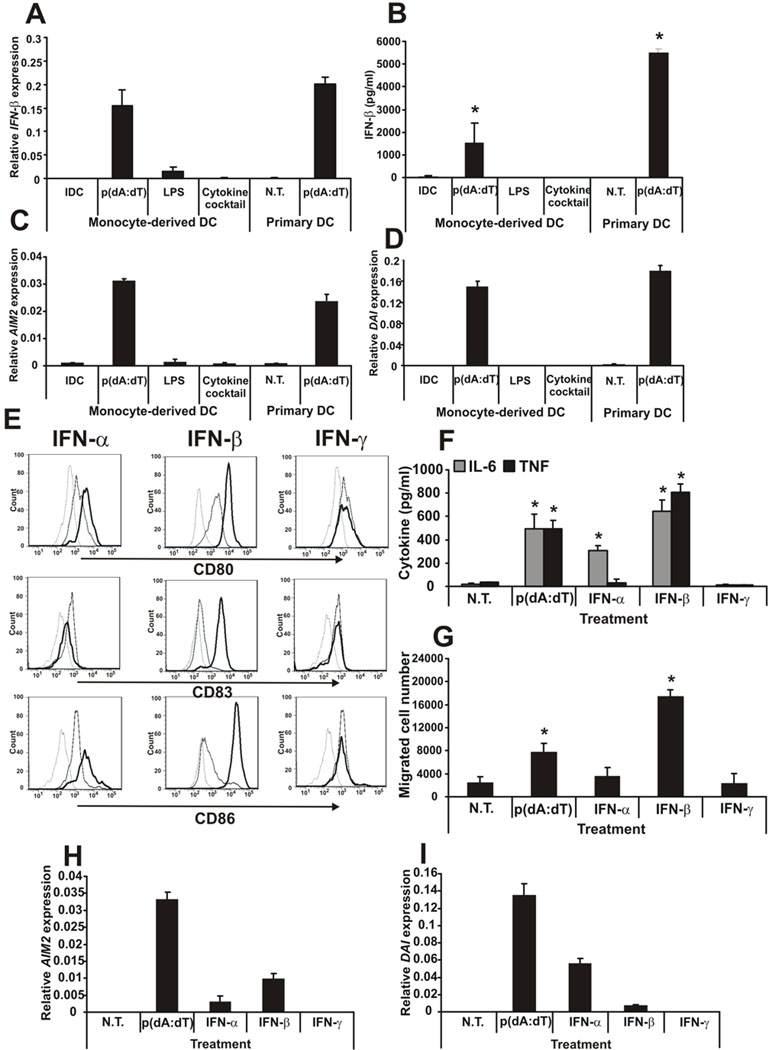
The experiments above clearly showed that dsDNA can effectively activate human DCs and trigger potent adaptive immune system activation. Therefore, next we investigated the mechanism of this activation. Because the gene array experiments showed a significantly increased level of IFN-β expression in poly(dA:dT)-activated DCs compared to LPS- or cytokine cocktail-activated DCs (Supplemental Fig. 2B) next we investigated the role of IFN-β in the maturation and function of DCs. First, the IFN-β production was confirmed at the gene (Fig. 4A) and protein (Fig. 4B) levels. The upregulation of IFN-β resulted in the induction of two IFN-regulated genes, AIM2 (Fig. 4C) and DAI (Fig. 4D).
Figure 4. dsDNA-activated DCs produce IFN-β and mature upon type I IFN treatment.
A, Gene expression and B, protein secretion of IFN-β upon different activation of monocyte-derived and primary DCs. C, AIM2 and D, DAI gene expression upon different activation of monocyte-derived and primary DCs. E, DCs were treated with type I (α and β) and type II (γ) IFNs and the levels of activation were compared by CD80, CD83 and CD86 surface expression. Dashed line: isotype control, thin line: immature DC, thick line: activated DC. F, IL-6 and TNF production and G, migratory capacity of the IFN-activated DCs compared to poly(dA:dT) activation. H, AIM2 and I DAI gene expression in IFN-activated DCs compared to poly(dA:dT) activation.
N.T.: non-treated. n=3 independent experiments.
Next, we asked how the autocrine-produced IFN-β affects DNA-DC functions as type I INFs are known to induce DC activation (42). To answer this question, cells were treated with different concentrations of type I IFN-α and -β or the type II IFN-γ (at 1 ng/ml to 100 ng/ml – data not shown). The low concentration of exogenous recombinant IFN-β, comparable to the levels of IFN-β produced by DCs upon poly(dA:dT) treatment (Fig. 4B), was able to fully activate DCs, monitored by cell surface molecule expression (Fig. 4E), pro-inflammatory cytokine production (Fig. 4F) and cell migration (Fig. 4G). In contrast, IFN-γ could not activate monocyte-derived DCs and IFN-α activated the DCs moderately compared to IFN-β (Fig. 4E–G), even at the highest used concentration (data not shown). Both IFN-α and IFN-β upregulated mRNA expression of AIM2 (Fig. 4H) and DAI (Fig. 4I) but the efficiency of the upregulation was remarkably lower than upon poly(dA:dT) treatment, even at high (100 ng/ml) IFN concentration. The results indicated that cytosolic DNA skewed DCs toward IFN-β (but no IFN-α – Supplemental Fig. 2B) production, thus the expression of the appropriate effector genes.
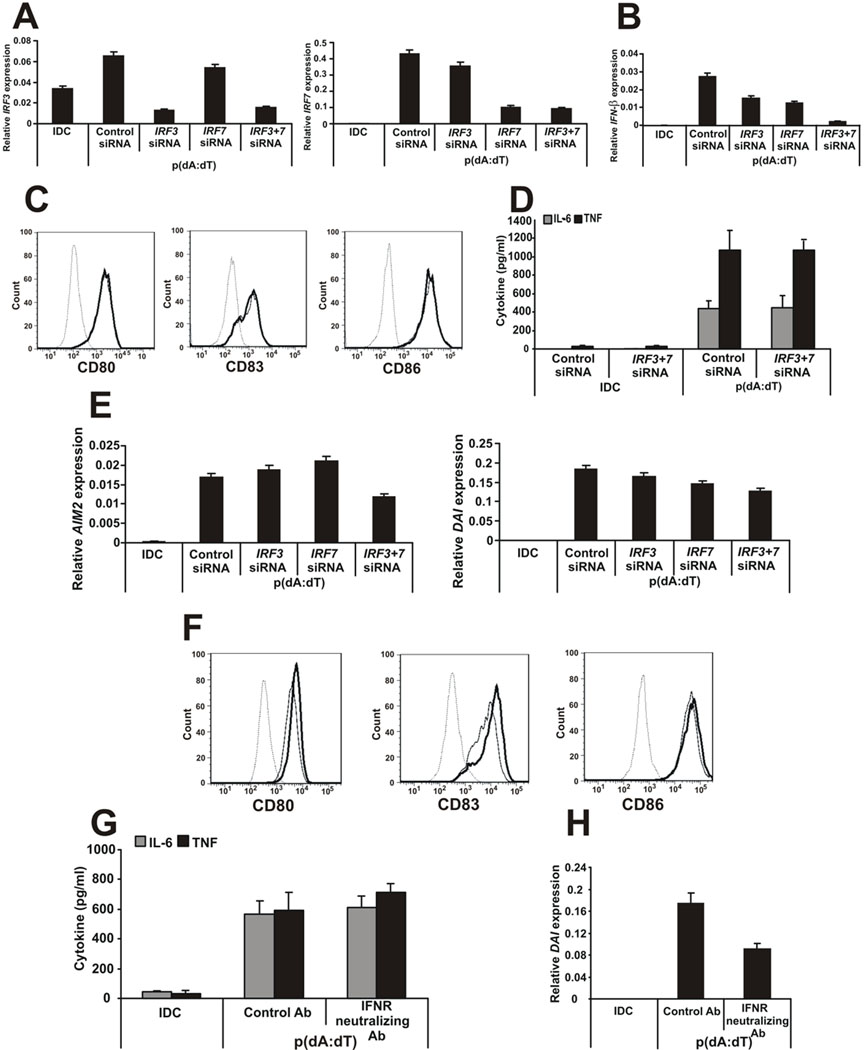
To evaluate whether IFN-β was the main trigger factor for DC activation by cytosolic DNA, the two molecules responsible for IFN signaling and the production of type I IFNs, the interferon regulatory factor (IRF) 3 and IRF7 were silenced using small interfering RNAs (siRNAs). The siRNAs were effective in specifically silencing both genes (Fig. 5A) and the expression of IFN-β was significantly downregulated by the specific siRNA treatment especially in the case of the IRF3 and IRF7 double knock-down (Fig. 5B) showing that the IRF3/7 pathway is needed for the IFN-β production in DNA-activated DCs. In contrast, the activation of DCs by poly(dA:dT) was unaffected even when both IRF3 and IRF7 were knocked down simultaneously (Fig. 5C and D) and had no effect on the ability of DCs to activate T cells (data not shown). Interestingly, the expression of AIM2 and DAI were only slightly lower than in case of the control siRNA treatment (Fig. 5E) suggesting that other mechanisms may also be responsible for the expression of these genes. Furthermore, blocking type I INF receptor A with neutralizing antibodies did not prevent the activation of DNA-DCs by poly(dA:dT) (Fig. 5F and G). Poly(dA:dT) could still induce the induction of IFN-β response genes DAI and AIM2 even in the presence of IFNRA neutralizing antibodies; however, we could detect a slight decrease in the induction of these genes (Fig. 5H and data not shown).
Figure 5. The type I IFN pathway is not required for DC activation by cytosolic dsDNA.
A, Effectiveness of IRF3- and IRF7-specific knock-downs. B, IFN-β gene expression in siRNA-treated DNA-DCs. C, CD80, CD83 and CD86 surface expression of the siRNA-treated, DNA-activated DCs. Dashed line: isotype control, thin line: DNA-DC treated with control siRNA, thick line: DNA-DC treated with IRF3- and IRF7-specific siRNAs. D, IL-6 and TNF production by siRNA-treated DNA-DCs. E, AIM2 and DAI gene expression in siRNA-treated DNA-DCs. F, CD80, CD83 and CD86 surface expression of the interferon receptor neutralizing antibody-treated, DNA-activated DCs. Dashed line: isotype control, thin line: DNA-DC treated with control antibody, thick line: DNA-DC treated with interferon receptor-specific neutralizing antibody. G, IL-6 and TNF production of control or neutralizing antibody-treated DNA-DCs. H, DAI gene expression in control or neutralizing antibody-treated DNA-DCs.
n=3 independent experiments.
Thus, dsDNA can activate DCs even if the autocrine production of IFN-β is inhibited suggesting that IFN-β is dispensable for the activation of DCs by cytosolic dsDNA. However, we could not exclude the possibility that it may contribute to the maturation of DCs at least by inducing some of its target genes.
Cytosolic DNA activation of DCs is independent of AIM2, DAI, the endosome or the RNA polymerase III pathways but depends in part on IFI16
The induction of AIM2 and DAI in MDCs after cytosolic DNA treatment (Fig. 4C and D) appears to be, at least partially, indirect and due to the autocrine effect of the secreted IFN-β by the MDCs. Yet, it is not clear whether the induced AIM2 and DAI proteins function as cytosolic DNA sensors in DNA-DCs. To answer this question, specific siRNAs were used to downregulate the expression of these two molecules. Surprisingly, silencing of AIM2 or DAI did not abrogate the activation of DCs by cytosolic DNA, even when both genes were knocked-down simultaneously (Supplemental Fig. 3A–C) and had no effect on the ability of DCs to activate T cells (data not shown). However both AIM2 and DAI siRNAs were effective in knocking down these genes (Supplemental Fig. 4D). To further show that the siRNA knock down was specific, the production of IL-1β was monitored in activated DCs. As shown in Supplemental Fig. 3E, the production of IL-1β was reduced when AIM2 was downregulated showing its role in forming an inflammasome responsible for processing IL-1β (14–17). The data imply that AIM2 and DAI do not function as intracellular sensors responsible for the activation of human DCs by cytosolic DNA in this case.
A recent study suggested that the high-mobility group box (HMGB) proteins function as universal sentinels for nucleic acids, modulate type I IFN as well as inflammatory cytokine production by DNA or RNA (26). However, it is not known whether activated human monocyte-derived DCs express HMGB proteins and whether they serve as DNA sensors. Here we show that DCs express HMGB1 and HMGB2 but no detectable amount of HMGB3. Moreover, the expression levels of HMGB2 were significantly upregulated after treatment with poly(dA:dT) or IFN-β (Supplemental Fig. 4A). However, in human DCs, HMGB1 and HMGB2 did not appear to serve as cytosolic DNA sensors and regulate MDC functions including activation, cytokine production, migration (Supplemental Fig. 4C–E) as well as T cell activation (data not shown) using specific siRNA treatments (Supplemental Fig. 4B). Similarly, the inhibition of the endosome formation by chloroquine, thus endosomal TLR activation, and of the RNA polymerase III nucleic acid sensing pathway with the specific inhibitor ML-60218 did not appear to affect human DC activation by cytosolic DNA and DNA-DC functions (Supplemental Fig. 4C–E) or the capacity of DCs to activate T cells (data not shown).
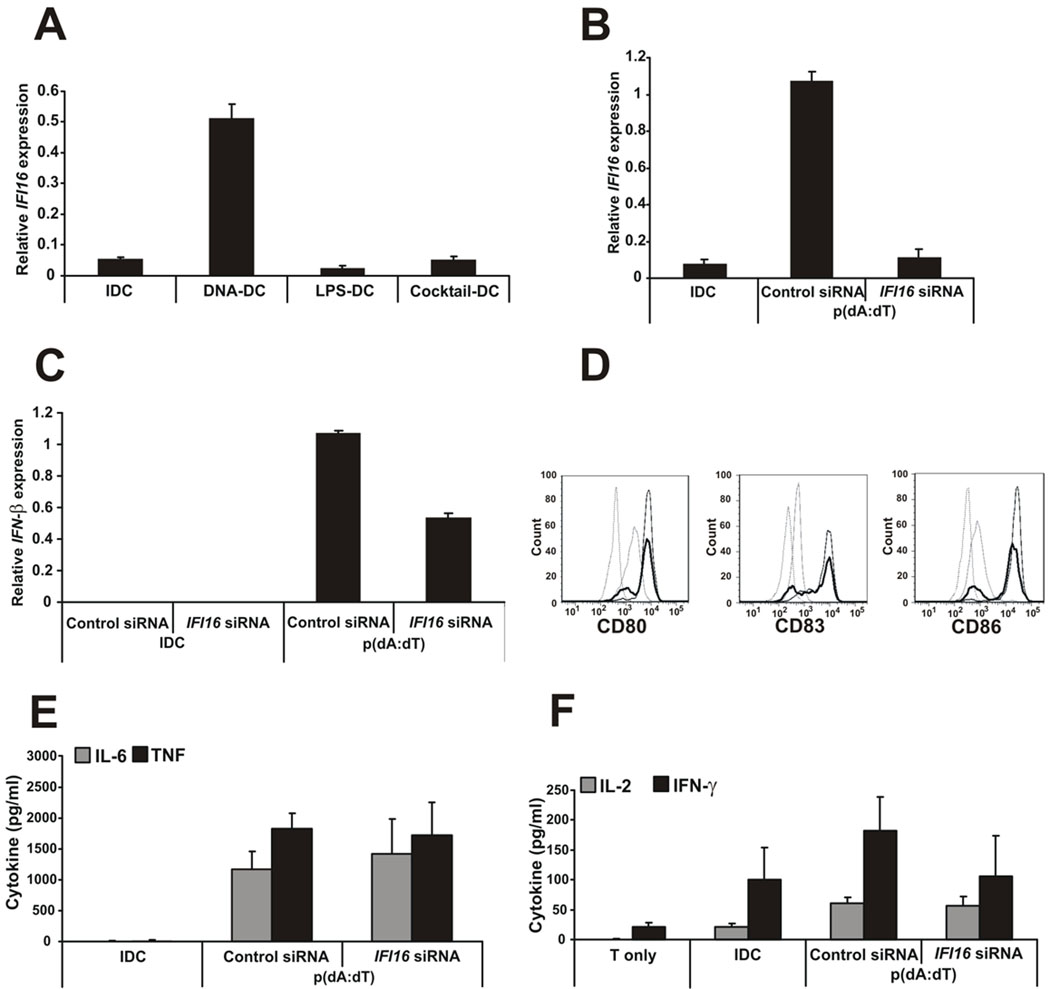
IFI16, the newly characterized IFN-inducible gene and cytosolic DNA sensor (22) was upregulated in DCs treated with poly(dA:dT) but not in LPS- or inflammatory cytokine cocktail-activated DCs (Fig. 6A). Silencing IFI16 in DNA-DCs by specific siRNA was efficient (Fig. 6B) and the production of IFN-β by IFI16-silenced DCs was downregulated (Fig. 6C). Inhibition of IFI16 associated with slight inhibition of the upregulation of cell surface activation molecules CD80, CD83 and CD86 (Fig. 6D); however, the production of pro-inflammatory cytokines by these DCs was not inhibited (Fig. 6E) and the T cell activation provided by the IFI16-deficient DCs was only slightly downregulated (Fig. 6F).
Figure 6. Cytosolic DNA activation of DCs partly depends on the IFI16 sensor.
A, IFI16 gene expression upon DC maturation with different activation stimuli. B, Effectiveness of IFI16-specific knock-down. C, IFN-β gene expression by siNA-treated DCs measured after 4h of p(dA:dT) activation by Q-PCR. D, CD80, CD83 and CD86 surface expression of the siRNA-treated, DNA-activated DCs. Dashed line: isotype control, grey thin line: IDC, black thin line: DNA-DC treated with control siRNA, thick line: DNA-DC treated with IFI16-specific siRNA. E, IL-6 and TNF production by siRNA-treated DNA-DCs. F, Total allogeneic T cells were co-cultured with IDCs or siRNA-treated, poly(dA:dT)-activated DCs and IL-2 and IFN-γ cytokine production by T cells was measured by ELISA.
n=3 independent experiments.
Taken together, the data indicate that, in humans, activation of DCs by dsDNA is independent of the previously recognized dsDNA sentinels AIM2, DAI, RNA polymerase III or HMGBs. On the other hand, IFI16, the novel cytosolic dsDNA receptor may play a role in DC activation; however, knocking down this molecule did not significantly affect DC capability to activate T cells. Because the exact role of the IFI16 molecule as an exclusive dsDNA sensor in DCs has not been definitely proven, we cannot exclude the existence of a yet to be identified cytosolic DNA sensor(s) in human DCs.
Cytosolic DNA activation of monocyte-derived DCs depends on the proteosome and IPS-1 pathways
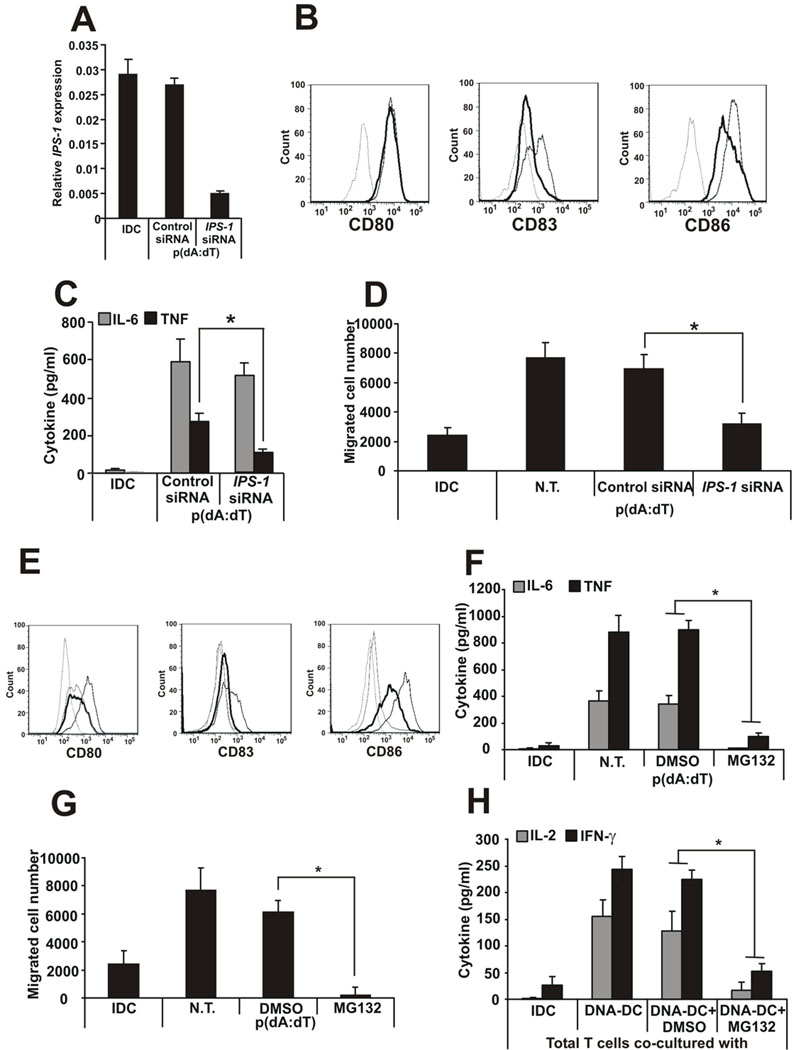
Previously published data have shown that signaling and transcription factors such as STING, TRAF6, IPS-1, TBK-1 and NF-κB mediated the genetic reprogramming of DCs activated by dsDNA. To identify which signaling and transcriptional pathways participated in human DC activation by cytosolic DNA, specific siRNAs against STING, TRAF6, TBK-1 and IPS-1 were used to disrupt the respective pathways. Effective silencing of IPS-1 (Fig. 7A) resulted in the downregulation of the cell surface molecules CD83 and CD86 (Fig. 7B), a slight downregulation of IL-6 and a significant inhibition of TNF production (Fig. 7C) as well as the migratory capacity of the DCs (Fig. 7D) compared to the control siRNA-treated cells. The T cell activation capacity of IPS-1-silenced DCs was not affected (data not shown). It was not clear whether STING, TRAF6 and TBK-1 regulated cytosolic DNA signaling in human monocyte-derived DCs, since the knock-down siRNAs were not effective (data not shown).
Figure 7. Cytosolic DNA activation of DCs depends on the IPS-1 and proteosome pathways.
A, Effectiveness of IPS-1-specific knock-down. B, CD80, CD83 and CD86 surface expression of the siRNA-treated, DNA-activated DCs. Dashed line: isotype control, thin line: DNA-DC treated with control siRNA, thick line: DNA-DC treated with IPS-1-specific siRNA. C, IL-6 and TNF production by siRNA-treated DNA-DCs. D, Migratory capacity of siRNA-treated DNA-DCs. E, CD80, CD83 and CD86 surface expression of the proteosome inhibitor MG132-treated DCs. Dashed line: isotype control, grey thin line: IDC, black thin line: DNA-DC treated with DMSO, thick line: DNA-DC treated with MG132. F, IL-6 and TNF production and G, migratory capacity of MG132-treated DNA-DCs. H, cytokine production by total T cells co-cultured with MG132-treated DNA-DCs.
N.T.: non-treated. n=3 independent experiments.
To inhibit the NF-κB pathway, a cell permeable proteosome inhibitor MG132 was used at a concentration that did not affect cell viability. Treatment of DNA-DCs with the inhibitor resulted in a complete inhibition of DC activation and function monitored by cell surface molecule expression (Fig. 7E), pro-inflammatory cytokine production (Fig. 7F), migration (Fig. 7G) and total T cell activation (Fig. 7H). The data suggest that the activation of human monocyte-derived DCs by cytosolic dsDNA required the IPS-1 pathway and a functional NF-κB pathway.
Discussion
At present there is an emerging interest in understanding the mechanisms by which the innate immune system is able to detect nucleic acids as danger signals. Much of the efforts focus on identifying DNA receptors and pathways leading to cell activation, especially in cells with antigen presenting capacity. Recent studies, in cell lines and mouse knock out systems, have elegantly demonstrated that cytosolic dsDNA activates APCs by interacting with unknown intracellular nucleic acid sensor(s) (11, 27, 34, 36, 43). In humans, myeloid DC activation by cytosolic DNA has been observed but only partly characterized (35, 44).
Our results clearly indicate that naturally-derived or synthetic cytosolic DNAs induce activation/maturation of human monocyte-derived DCs as well as of primary CD11c+ blood DCs. Activation of DCs by dsDNA resulted in the induction of DC-related genes similar to those present in LPS-DCs and inflammatory cocktail-DCs. DNA-DCs and LPS-DCs appeared to share commonly regulated genes suggesting that these stimuli employed similar activation mechanisms and/or signaling pathways. Although most genes were induced at comparable levels by all three stimuli, IFN-β expression was markedly upregulated in DNA-DCs. In turn, the induced IFN-β but not IFN-α or IFN-γ, cooperated with cytosolic DNA to mediate the full spectrum of DC activation and function including CD86 upregulation, pro-inflammatory cytokine production and the acquisition of migratory capacity. Interestingly, IRF3/IRF7-deficient DCs could be fully activated by cytosolic DNA. Thus, the signaling/transcription pathway(s) by which cytosolic DNA mediated the production of IFN-β was distinct from those that cooperated with cytosolic DNA to induce the expression of IFN-β response genes and function. Furthermore, the data suggested that IFN-β is important but not required for DC activation/maturation by cytosolic DNA.
In contrast, the activation of the NF-κB pathway was absolutely required for the acquisition of the full spectrum of DNA-DC function such as CD86 upregulation, pro-inflammatory cytokine production, migration and T cell activation. On the other hand, the adaptor protein IPS-1 was necessary for the induction of CD83 and CD86, TNF production and migration of DC by cytosolic DNA but it was not required for the activation of T cells by DNA-DCs. Thus, in human DCs, the interaction of cytosolic DNA with the receptor activated the NF-κB pathway, in part via the IPS-1 adaptor molecule, which then mediated the induction of all the functional characteristics of mature APCs.
Systemic elimination of the known nucleic acid sensors by siRNA knock down or by specific inhibition showed that AIM2 and DAI (13–17) did not abrogate the activation of DCs by cytosolic DNA. Similarly, the inhibition of the endosome formation by chloroquine, thus endosomal TLR activation (5), and of the RNA polymerase III nucleic acid sensing pathway with ML-60218 (23, 24) did not appear to affect human DC activation by cytosolic DNA and DNA-DC functions. A recent study had suggested that the high-mobility group box (HMGB) proteins function as universal sentinels for nucleic acids and modulate type I IFN as well as inflammatory cytokine production by DNA or RNA (26). However, it was not known if human monocyte-derived DCs expressed HMGB proteins and if they served as DNA sensors. Indeed, DCs were found to express HMGB1 and HMGB2 but not HMGB3. Moreover, the expression levels of HMGB2 were significantly upregulated after treatment with poly(dA:dT) or IFN-β. However, in humans, HMGB1 and HMGB2 did not appear to serve as cytosolic DNA sensors and to regulate DC functions including activation, cytokine production, migration and T cell activation.
The newly characterized cytoplasmic DNA sensor, IFI16, (22) was exclusively upregulated in DCs treated with poly(dA:dT) but not in LPS- or inflammatory cytokine cocktail-activated DCs. IFI16-deficient DCs produced less IFN-β and cell surface activation molecules showing the functional effectiveness of the gene knock down. However, the production of pro-inflammatory cytokines (IL-6 and TNF) by these DCs was not inhibited and the T cell activation provided by the IFI16-deficient DCs was only slightly downregulated. In contrast, IFI16 deficiency resulted in a significant downregulation of IL-6 and TNF gene expression in mouse RAW264.7 cells and embryonic fibroblasts (22). The difference between our results and those of Unterholzner et al. may represent the difference between humans and mice and/or the type of DNA used (a 70 base-pair long vaccinia virus DNA transfection and infection with HSV-1 vs. poly(dA:dT)).
Our data showed that IFI16 played a minor role in human DC activation by dsDNA and in the subsequent activation of the adaptive immune system thus we could not exclude the existence of a yet to be identified cytosolic DNA sensor(s).
In humans the increasing number of DC subsets residing in unique niches, exhibiting distinct activation and maturation states suggested that, depending on the nature of the extrinsic or intrinsic signals, each subset of DCs should induce a specific adaptive immune response. This hypothesis was further supported by our results showing that in mixed lymphocyte cultures, dsDNA-activated DCs, but not LPS- or inflammatory cytokine cocktail-activated DCs, induce naïve CD4+ T cells to produce more IL-2, granzyme and IFN-γ but not IL-4, and these effector CD4+ T cells efficiently kill tumor cells in in vitro cultures. The existence of CD4+ cytotoxic T cells, capable of killing tumor and virally infected cells, had been documented in the mouse and cell lines (45–47) as well as in humans (48–50). However, this was the first demonstration that human DCs could be conditioned by cytosolic DNA to endow in vitro primed naïve CD4+ T cells with killing capacity. In addition, in the presence of T cells, dsDNA-activated DCs, but not LPS- or cocktail-activated DCs, activated B cells to produce complement fixing IgG1 and IgG3 but not non-complement fixing IgG2 and IgG4 antibodies. This finding extended the knowledge on how DCs were able to enhance plasma cell differentiation (51–53). DNA-DCs could also generate CD8+ cytotoxic T cells that produced the effector proteins IFN-γ and granzyme.
Interestingly, the activation/maturation of DCs by cytosolic DNA was dose-dependent suggesting that the amount of cytosolic nucleic acids released by intracellular viruses or parasites dictated the functional outcome of DNA-activated DCs. These data further emphasized the functional plasticity nature of human DCs and their monocyte precursors imparted by different stimuli.
In recent years, the recognition that DCs play a pivotal role in the initiation and regulation of the adaptive immunity and the realization that adjuvants acted primarily as DC activators lead to the development of preventive and therapeutic vaccines using DCs (54). Despite important breakthroughs that murine studies had contributed to our understanding of DC biology, subtle, but highly relevant, differences between the human and mouse immune systems have been identified (55, 56). Therefore, in order to successfully generate effective human DC vaccines, complete understanding of the diversity and biology of human DCs is needed. In this respect, our studies provided a simple, but highly relevant system to further our understanding of human DC function. The use of synthetic cytosolic DNA might represent a novel, more effective and safer means of generating DCs for use in human vaccine design to orchestrate and reprogram the adaptive immune system to eradicate infectious agents, autoimmunity, allergy or cancer.
Supplementary Material
Abbreviations
- IDC
immature dendritic cell
- MDC
mature dendritic cell
- TLR
Toll-like receptor
- AIM2
absent in melanoma 2
- DAI
DNA-dependent activator of IFN-regulatory factor 3
- HMGB
high-mobility group box
- IRF
interferon regulatory factor
Footnotes
This work was supported by the Mary Kirkland Center for Lupus at the Hospital for Special Surgery funded by Rheuminations, and NIH grants RO1 AI 49954 and RO1 AI 68787 to GCT.
REFERENCES
- 1.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Lanzavecchia A, Sallusto F. The instructive role of dendritic cells on T cell responses: lineages, plasticity and kinetics. Curr Opin Immunol. 2001;13:291. doi: 10.1016/s0952-7915(00)00218-1. [DOI] [PubMed] [Google Scholar]
- 3.Kaisho T, Akira S. Critical roles of Toll-like receptors in host defense. Crit Rev Immunol. 2000;20:393. [PubMed] [Google Scholar]
- 4.Jefferies CA, Fitzgerald KA. Interferon gene regulation: not all roads lead to Tolls. Trends Mol Med. 2005;11:403. doi: 10.1016/j.molmed.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 8.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Hochrein H, Schlatter B, O'Keeffe M, Wagner C, Schmitz F, Schiemann M, Bauer S, Suter M, Wagner H. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc Natl Acad Sci U S A. 2004;101:11416. doi: 10.1073/pnas.0403555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 12.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 13.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 14.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 15.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 16.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren SE, Armstrong A, Hamilton MK, Mao DP, Leaf IA, Miao EA, Aderem A. Cutting Edge: Cytosolic Bacterial DNA Activates the Inflammasome via Aim2. J Immunol. 2010 doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 26.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, Savitsky D, Ronfani L, Akira S, Bianchi ME, Honda K, Tamura T, Kodama T, Taniguchi T. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 27.Ishii KJ, Suzuki K, Coban C, Takeshita F, Itoh Y, Matoba H, Kohn LD, Klinman DM. Genomic DNA released by dying cells induces the maturation of APCs. J Immunol. 2001;167:2602. doi: 10.4049/jimmunol.167.5.2602. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, Mori A, Ishii KJ, Saito J, Singer DS, Klinman DM, Krause PR, Kohn LD. Activation of target-tissue immune-recognition molecules by double-stranded polynucleotides. Proc Natl Acad Sci U S A. 1999;96:2285. doi: 10.1073/pnas.96.5.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu FG, Reich CF, Pisetsky DS. Effect of cytofectins on the immune response of murine macrophages to mammalian DNA. Immunology. 2003;109:255. doi: 10.1046/j.1365-2567.2003.01653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang W, Reich IC, Pisetsky DS. Mechanisms of activation of the RAW264.7 macrophage cell line by transfected mammalian DNA. Cell Immunol. 2004;229:31. doi: 10.1016/j.cellimm.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Wilkinson M, Xia X, David M, Xu L, Purkel-Sutton A, Bhardwaj A. Induction of IFN-regulated factors and antitumoral surveillance by transfected placebo plasmid DNA. Mol Ther. 2005;11:112. doi: 10.1016/j.ymthe.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda K, Yu P, Kirschning CJ, Schlatter B, Schmitz F, Heit A, Bauer S, Hochrein H, Wagner H. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J Immunol. 2005;174:6129. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 33.Ishii KJ, Akira S. Innate immune recognition of nucleic acids: beyond toll-like receptors. Int J Cancer. 2005;117:517. doi: 10.1002/ijc.21402. [DOI] [PubMed] [Google Scholar]
- 34.Martin DA, Elkon KB. Intracellular mammalian DNA stimulates myeloid dendritic cells to produce type I interferons predominantly through a toll-like receptor 9-independent pathway. Arthritis Rheum. 2006;54:951. doi: 10.1002/art.21677. [DOI] [PubMed] [Google Scholar]
- 35.Katashiba Y, Miyamoto R, Hyo A, Shimamoto K, Murakami N, Ogata M, Amakawa R, Inaba M, Nomura S, Fukuhara S, Ito T. Interferon-alpha and interleukin-12 are induced, respectively, by double-stranded DNA and single-stranded RNA in human myeloid dendritic cells. Immunology. 2010 doi: 10.1111/j.1365-2567.2010.03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirota H, Ishii KJ, Takakuwa H, Klinman DM. Contribution of interferon-beta to the immune activation induced by double-stranded DNA. Immunology. 2006;118:302. doi: 10.1111/j.1365-2567.2006.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konno H, Yamamoto T, Yamazaki K, Gohda J, Akiyama T, Semba K, Goto H, Kato A, Yujiri T, Imai T, Kawaguchi Y, Su B, Takeuchi O, Akira S, Tsunetsugu-Yokota Y, Inoue J. TRAF6 establishes innate immune responses by activating NF-kappaB and IRF7 upon sensing cytosolic viral RNA and DNA. PLoS One. 2009;4:e5674. doi: 10.1371/journal.pone.0005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurner B, Roder C, Dieckmann D, Heuer M, Kruse M, Glaser A, Keikavoussi P, Kampgen E, Bender A, Schuler G. Generation of large numbers of fully mature and stable dendritic cells from leukapheresis products for clinical application. J Immunol Methods. 1999;223:1. doi: 10.1016/s0022-1759(98)00208-7. [DOI] [PubMed] [Google Scholar]
- 39.Szatmari I, Gogolak P, Im JS, Dezso B, Rajnavolgyi E, Nagy L. Activation of PPARgamma specifies a dendritic cell subtype capable of enhanced induction of iNKT cell expansion. Immunity. 2004;21:95. doi: 10.1016/j.immuni.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Nonkwelo C, Ruf IK, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gogolak P, Rethi B, Szatmari I, Lanyi A, Dezso B, Nagy L, Rajnavolgyi E. Differentiation of CD1a- and CD1a+ monocyte-derived dendritic cells is biased by lipid environment and PPARgamma. Blood. 2007;109:643. doi: 10.1182/blood-2006-04-016840. [DOI] [PubMed] [Google Scholar]
- 42.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 43.Karayel E, Burckstummer T, Bilban M, Durnberger G, Weitzer S, Martinez J, Superti-Furga G. The TLR-independent DNA recognition pathway in murine macrophages: Ligand features and molecular signature. Eur J Immunol. 2009;39:1929. doi: 10.1002/eji.200939344. [DOI] [PubMed] [Google Scholar]
- 44.Chen R, Zhang L, Zhong B, Tan B, Liu Y, Shu HB. The ubiquitin-specific protease 17 is involved in virus-triggered type I IFN signaling. Cell Res. 2010;20:802. doi: 10.1038/cr.2010.41. [DOI] [PubMed] [Google Scholar]
- 45.Martorelli D, Muraro E, Merlo A, Turrini R, Rosato A, Dolcetti R. Role of CD4+ cytotoxic T lymphocytes in the control of viral diseases and cancer. Int Rev Immunol. 2010;29:371. doi: 10.3109/08830185.2010.489658. [DOI] [PubMed] [Google Scholar]
- 46.Mumberg D, Monach PA, Wanderling S, Philip M, Toledano AY, Schreiber RD, Schreiber H. CD4(+) T cells eliminate MHC class II-negative cancer cells in vivo by indirect effects of IFN-gamma. Proc Natl Acad Sci U S A. 1999;96:8633. doi: 10.1073/pnas.96.15.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, Restifo NP, Allison JP. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010;207:637. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paludan C, Bickham K, Nikiforow S, Tsang ML, Goodman K, Hanekom WA, Fonteneau JF, Stevanovic S, Munz C. Epstein-Barr nuclear antigen 1-specific CD4(+) Th1 cells kill Burkitt's lymphoma cells. J Immunol. 2002;169:1593. doi: 10.4049/jimmunol.169.3.1593. [DOI] [PubMed] [Google Scholar]
- 49.Hegde NR, Dunn C, Lewinsohn DM, Jarvis MA, Nelson JA, Johnson DC. Endogenous human cytomegalovirus gB is presented efficiently by MHC class II molecules to CD4+ CTL. J Exp Med. 2005;202:1109. doi: 10.1084/jem.20050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spaapen RM, Lokhorst HM, van den, Oudenalder K, Otterud BE, Dolstra H, Leppert MF, Minnema MC, Bloem AC, Mutis T. Toward targeting B cell cancers with CD4+ CTLs: identification of a CD19-encoded minor histocompatibility antigen using a novel genome-wide analysis. J Exp Med. 2008;205:2863. doi: 10.1084/jem.20080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fayette J, Durand I, Bridon JM, Arpin C, Dubois B, Caux C, Liu YJ, Banchereau J, Briere F. Dendritic cells enhance the differentiation of naive B cells into plasma cells in vitro. Scand J Immunol. 1998;48:563. doi: 10.1046/j.1365-3083.1998.00471.x. [DOI] [PubMed] [Google Scholar]
- 52.Dubois B, Vanbervliet B, Fayette J, Massacrier C, Van Kooten C, Briere F, Banchereau J, Caux C. Dendritic cells enhance growth and differentiation of CD40-activated B lymphocytes. J Exp Med. 1997;185:941. doi: 10.1084/jem.185.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teichmann LL, Ols ML, Kashgarian M, Reizis B, Kaplan DH, Shlomchik MJ. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 2010;33:967. doi: 10.1016/j.immuni.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palucka K, Banchereau J, Mellman I. Designing vaccines based on biology of human dendritic cell subsets. Immunity. 2010;33:464. doi: 10.1016/j.immuni.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 56.Gordon CJ, Grafton G, Wood PM, Larche M, Armitage RJ. Modelling the human immune response: can mice be trusted? Commentary. Curr Opin Pharmacol. 2001;1:431. doi: 10.1016/s1471-4892(01)00074-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.