Abstract
Dimorphic fungi collectively account for 5 to 10 million new infections annually worldwide. Ongoing efforts seek to clarify mechanisms of cellular resistance to these agents and develop vaccines. A major limitation in studying the development of protective T-cells in this group of organisms is the lack of tools to detect, enumerate and characterize fungus-specific T-cells during vaccination and infection. We generated a TCR transgenic (tg) mouse (Bd 1807) whose CD4+ T cells respond to a native epitope in Blastomyces dermatitidis and also in Histoplasma capsulatum. Here, we characterize the mouse, reveal its applications, and extend our analysis showing that 1807 cells also respond to the related dimorphic fungi Coccidioides posadasii and Paracoccidioides lutzii. On adoptive transfer into vaccinated wild-type mice, 1807 cells become activated, proliferate and expand in the draining lymph nodes, and differentiate into T1 effectors after trafficking to the lung upon lethal experimental challenge. 1807 cells confer vaccine-induced resistance against B. dermatitidis, H. capsulatum and C. posadasii. Transfer of naïve 1807 cells at serial intervals post-vaccination uncovered the prolonged duration of fungal Ag presentation. Using 1807 cells, we also found that the administration of vaccine only once induced a maximal pool of effector/memory CD4+ cells and protective immunity by four weeks after vaccination. The autologous adoptive transfer system described here reveals novel features of anti-fungal immunity and offers a powerful approach to study the differentiation of Ag-specific T cells responsive to multiple dimorphic fungi and the development of CD4+ T-cell memory needed to protect against fungal infection.
Keywords: TCR tg mouse, T-cell priming, Vaccine immunity, Fungi
Introduction
Although antibody-based therapies are undergoing a renaissance, the induction of durable T-cell memory is the cornerstone of vaccine immunity to fungi. The kinetics of induction and maintenance of Ag-specific CD4+ T-cell memory to fungi has not been amenable to study due to the lack of known protective antigens and clonally restricted T-cells with complementary TCRs. We recently created a TCR tg mouse (Bd 1807) to interrogate the Ag-specific CD4+ T-cell response to a natural and protective epitope of the fungal pathogen Blastomyces dermatitidis, a systemic fungal pathogen of humans and other mammals (1).
We report here that 1807 cells respond well to four systemic dimorphic fungi including B. dermatitidis, H. capsulatum, C. posadasii and P. lutzii, implying the cells recognize a shared epitope. Vaccine priming of 1807 cells protected recipient mice that lack endogenous CD4+ cells from infection with the three major systemic fungi of North America (B. dermatitidis, H. capsulatum and C. posadasii). Using this tg mouse, we established an adoptive transfer system to follow the activation of T cells that arise in response to protective antigen. TCR tg systems such as this are among the best immunological tools to analyze naïve T-cell activation in vivo, since the frequency of circulating Ag-specific T cells can be raised above the level of detection for flow cytometric and immuno-histological analysis (2-4). The development of this system allowed us to track key stages of activation, differentiation, and memory T-cell development during priming and recall phases of protective immunity. Naïve 1807 cells were activated, proliferated and expanded in the skin draining lymph nodes (sdLNs); however, they required trafficking to the site of vaccine delivery or the lung on experimental challenge to fully differentiate into effector cells. Bd 1807 cells also served as sentinels enabling us to define the duration of Ag persistence after vaccination and the conditions for inducing an optimal pool of effector/memory cells that mediate protective immunity to lethal experimental infection against dimorphic fungi.
Materials and Methods
Generation of the transgenic (tg) mouse
We described generation of the Blastomyces-specific TCR Tg 1807 mouse elsewhere (1). Briefly, we cloned the α and β chains of the TCR from CD4+ T-cell clone #5, which is specific for a protective epitope of B. dermatitidis (5). 1807 mice were bred to B6.PL (Thy1.1+) to obtain Thy1.1+1807 T cells to let us track transferred tg cells in Thy1.2+ recipient C57BL6 mice.
In vitro stimulated effector Bd 1807 cells
IFN-γ producing tg cells were generated by adding naïve, magnetic bead purified CD4 cells from 1807 mice on plate-bound anti-CD3 (5 μg/ml)(BD Bioscience, San Jose, CA) with soluble anti-CD28 (5 μg/ml) and human rIL-2 (50U/ml) in the presence or absence of mouse rIFN-γ (1000U/ml)(Sigma, St. Louis, MO) and anti-IL-4 mAb (10 μg/ml)(NCI Biological Research Branch, Rockville, MD) for 10-13 d at 37°C and 5% CO2. 2 × 107 effector tg cells were transferred i.v. into sublethally irradiated [5.5 gray (Gy)] C57BL/6 mice. Mice were rested for 10 wks before infection to allow lymphopenia-driven expansion of the transplant (6).
Fungi
Strains used were ATCC 26199 (7), a wild-type strain of B. dermatitidis, and the isogenic, attenuated mutant lacking BAD1, designated strain #55 (8), as well as H. capsulatum strain G217B, P. lutzii (Pb1) (9), P. brasiliensis (ATCC 60855) and Candida albicans strain #5314. B. dermatitidis was grown as yeast on Middlebrook 7H10 agar with oleic acid-albumin complex (Sigma) at 39°C. H. capsulatum was grown as yeast at 37°C and 5% CO2 on Histoplasma Macrophage Medium (HMM). P. lutzii was grown in liquid BHI (Difco, Detroit, MI) at 37°C and was rotated at 200 rpm. C. albicans was grown on YPD plates. The saprobic phase of Coccidioides posadasii (isolate C735) was grown on GYE medium (1% glucose, 0.5% yeast extract, 1.5% agar) at 30°C for 3 to 4 weeks to generate a confluent layer of arthroconidia (spores) on the agar surface. Formalin killed spherules (FKS) of C. posadasii were generated as described (10, 11).
Mouse strains
Inbred mice including C57BL/6, T lymphocyte-specific Thy 1.1 allele carrying congenic B6 strain B6.PL-Thy1a/Cy (stock # 000406) (12) and TCRα-/- deficient B6.129S2-Tcra tm1Mom (stock #002116) (13), OT1 C57BL/6-Tg(TcrαTcrβ)1100Mjb/J (stock#003831) (14) and OT2 B6.Cg-Tg (TcrαTcrβ) 425Cbn/J mice (stock#004194) (15) homozygous for a transgene that encodes a TCR specific for chicken ovalbumin 257-264 and 323-339 presented by MHC class I and II molecule, respectively, were obtained from Jackson laboratory, Bar Harbor, ME. TEa tg mice [C57BL/6J(B6) (I-Ab, I-E-) background] (16, 17) expressing the Thy 1.2 allele were provided by Dr. A.Y. Rudensky. TEa cells recognize residues 52-68 of the I-Eα chain (Eα peptide) bound to class II I-Ab. OT1 and TEa tg mice expressing the Thy 1.1 allele were made by backcrossing tg mice twice to wild-type B6 mice expressing the Thy 1.1 marker. Male mice were 7-8 weeks old at the time of these experiments. Mice were housed and cared for as per guidelines of the University of Wisconsin Animal Care Committee, who approved this work.
Vaccination and infection
Mice were vaccinated as described (18) twice, two weeks apart, subcutaneously (s.c.) with 105 to 107 B. dermatitidis strain #55 yeast, 107 H. capsulatum G217B yeast, 107 P. lutzii yeast, 108 C. albicans yeast, 106 FKS of C. posadasii, or 200 μg whole cyst sonicate of Pneumocystis carinii (19) (gift of Dr. Jay Kolls) emulsified in complete Freund's adjuvant. Vaccine was injected at each of two sites. Mice were infected intratracheally (i.t.) with 2 × 103 isogenic wild-type yeast of B. dermatitidis strain 26199, 2 × 105 Hc G217B, 2 × 105 P. lutzii, 2 × 105 FKS or 60 spores of the virulent C. posadasii isolate C735 and 2 ×105 C. albicans as described (18, 20-24). To assess the infiltration of primed CD4 cells into the lungs, lung homogenates of challenged mice were analyzed at day 4 post-infection. To analyze the extent of lung infection, homogenized lungs were plated and yeast colony forming units enumerated (CFU) on BHI agar (Difco, Detroit, MI), sheep-blood containing Mycosel plates and on GYE plates containing 50 μg/ml of chloramphenicol (20).
Adoptive transfer of 1807 cells and experimental challenge
To see if 1807 cells mediate protection, we transferred 106 naïve 1807 or OT2 (control) tg cells into OT1 mice before vaccination. On the same day, recipients were vaccinated with 106 heat-killed B. dermatitidis and H. capsulatum yeast or 106 FKS of C. posadasii s.c.. Two weeks after vaccination, the mice were challenged.
Thy1.1 based enrichment and detection of lymph node cells and splenocytes
To detect transferred Thy1.1+ congenic tg cells, we used magnetic bead-based enrichment (25, 26). To reduce the background of contaminating recipient cells enriched cells were stained with the B-cell marker B220 and analyzed by FACS. By excluding cells that bound antibody to CD4 nonspecifically the background was reduced to virtually 0 as assessed in mice with no transfer (data not shown). This low background rate allowed detection of as few as 10 transferred cells (25, 26). Briefly, a single cell suspension of sdLN cells and splenocytes from one mouse was washed and resuspended in 1 ml of iMag® buffer containing PBS pH 7.4, 0.5% BSA and 2 mM EDTA, and 4 μl of biotinylated anti-Thy1.1 mAb (BD Biosciences). After 15 min of incubation on ice, cells were washed in iMag® buffer and the pellet suspended in 250 μl Streptavidin iMag® particles (BD Bioscience) and incubated at 4°C for 30 min. Bead-coated cells were enriched using direct magnetic bead separation as per the manufacturer. Enriched cells were resuspended in 250 μl iMag® buffer, counted, stained and analyzed by FACS.
Intracellular cytokine stain
Skin-tissue derived cells were collected serially after vaccination and cells from lung homogenates were harvested at day 4 post-infection. Cells (0.5 × 106 cells/ml) were stimulated for 4h with anti-CD3 (clone 145-2C11; 0.1μg/ml) and anti-CD28 (clone 37.51; 1μg/ml) in the presence of Golgi-Stop (BD Biosciences). Stimulation with fungal ligands yielded comparable cytokine production by transgenic T-cells compared to CD3/CD28 stimulation (data not shown). After cells were washed and stained for surface CD4 and CD8 using anti-CD4 PerCp, anti-CD8 PeCy7, anti-CD44-APC, anti-CD62L PE and B220 Alexa 700 (to gate out non-T-cell background) mAbs (Pharmingen), they were fixed and permeabilized in Cytofix/Cytoperm at 4° C overnight. Permeabilized cells were stained with anti-IFN-γ–Alexa 488 (clone XMG1.2) conjugated mAbs (Ebioscience) in FACS buffer for 30 min at 4° C, washed, and analyzed by FACS. Cells were gated on CD4 or CD8 and CD44hi, and cytokine expression in each gate analyzed. The number of cytokine positive CD4+ and CD8+ T cells per lung was calculated by multiplying the percent of cytokine-producing cells by the number of CD4+ and CD8+ cells in the lung.
Statistical Analysis
The number and percentage of activated, proliferating or cytokine producing T-cells and differences in number of CFU were compared among groups using ANOVA models (27). A two sided P value of < 0.05 was considered statistically significant.
Results
Bd 1807 cells recognize a shared antigen in systemic dimorphic fungi
B. dermatitidis, P. brasiliensis (and P. lutzii), H. capsulatum and C. immitis (and C. posadasii) are closely related at the level of 18S ribosomal DNA sequences (28, 29). We recently found that Bd 1807 cells respond to a shared immunodominant antigen in B. dermatitidis and H. capsulatum (1). Here, we extended this analysis and tested whether Bd 1807 cells may also respond to this shared antigen in C. posadasii and P. lutzii. After transferring 1807 cells into mice, we vaccinated them with C. posadasii, P. lutzii, H. capsulatum and B. dermatitidis. We also vaccinated mice with C. albicans and P. carinii as controls for agents that are not closely related to dimorphic fungi (http://timm.main.teikyo-u.ac.jp/pfdb/cover/taxonomic_list_eng.html). Bd 1807 cells were activated in response to the four systemic dimorphic fungi, but they were not activated in unvaccinated mice or mice vaccinated with C. albicans or P. carinii (Fig. 1A and data not shown). Over 98% of 1807 cells proliferated and became CFSElo in response to P. lutzii, C. posadasii, H. capsulatum and B. dermatitidis, and the pattern for CD44 and CD62L was also comparable for the groups. Vaccinated recipients of Bd 1807 cells were challenged i.t. with the homologous (vaccinating) fungus to see if the cells acquired memory, differentiated, and recruited back to the lung. 1807 Th1 effectors were recruited to lung in response to all four dimorphic fungi and 5 to 25% produced IFN-γ (Fig. 1B). 1807 responses were similar to endogenous, polyclonal CD4+ T-cell responses in each case, implying that the tg cells portray the endogenous response. Bd 1807 cells failed to recall in response to Candida, although endogenous CD4+ T-cells produced a robust IFN-γ response. These results imply that Bd 1807 cells recognize a shared antigen in four dimorphic fungi and can be used to study and compare T cell development in response to these organisms.
Fig. 1. Bd 1807 cells recognize a common antigen shared with P. lutzii, C. posadasi and H. capsulatum.
(A) Thy1.2+ wild-type mice received 106 naïve Thy 1.1+ 1807 cells and were vaccinated s.c. with 107 P. lutzii, 107 B. dermatitidis (strain #55), 107 H. capsulatum G217B and 108 C. albicans yeast and 106 FKS of C. posadasii. 7 days after vaccination, skin-draining LNs were removed and Thy 1.1+ CD4+ cells assayed for proliferation (CFSE) and activation (CD44 and CD62L expression) by FACS. (B) Recall responses of 1807 cells to the lung. Four weeks after recipients of 106 1807 cells were vaccinated as in panel (A), they were challenged with the homologous fungus via the respiratory route: P. lutzii, wild-type B. dermatitidis, H. capsulatum, C. albicans and FKS (18). 4 days later, lung T-cells were harvested and analyzed for intra-cellular cytokine. The dot plots show IFN-γ producing T-cells from a representative mouse of 3 mice/group in one of three experiments performed.
Precursor frequency and vaccine dose dictate proliferation and activation of 1807 cells
To establish an adoptive transfer system, we titrated precursor frequency of naïve 1807 cells and vaccine dose of B. dermatitidis. The frequency of transferred T-cells can affect proliferative expansion of responding T cells (25, 30). We determined the minimum number of transferred 1807 cells enabling us to observe in vivo responses to the vaccine. Varied numbers of 1807 cells were transferred into Thy 1.2+ recipients on the day of vaccination and the expansion of tg cells was assessed in the sdLN 4 d post-vaccination (peak of expansion; data not shown) using a saturating dose of 107 vaccine yeast. Transfer of 103, 104, 105 and 106 naïve tg precursors yielded an expansion of 10-, 55-, 287- and 63-fold, respectively as measured by the total number of Thy1.1+ CD4+ T cells in vaccinated vs. unvaccinated recipient mice. The greatest in vivo expansion of 1807 cells in the sdLNs was seen when 105 precursors were transferred. The relative expansion of 1807 cells in vaccinated mice was 4, 25, 10 and 4, respectively, for the following comparisons: 106 vs. 105, 105 vs. 104, 104 vs. 103 and 103 vs. 102, again implying that the relative expansion was maximal using 105 precursors for adoptive transfer.
To see if the vaccine dose affects proliferation and activation of naïve tg precursors, we adoptively transferred 1807 cells and then vaccinated recipient mice with 105, 106 or 107 yeast. Immunization with increasing numbers of vaccine yeast increased expansion of 1807 cells in the sdLNs, according to the precursors transferred. This was evident in the number of calculated 1807 cells, the fold expansion of transferred 1807 cells, the percentage of cells that became CFSElo and the n-fold expansion of CFSElo cells (Fig. 2A+B). For example, the highest vaccine dose (107 yeast) yielded proliferation in >90% of the TCR tg cells in all the groups, whereas the lowest dose (105 yeast) limited T cell proliferation. A vaccine dose of 106 yeast induced proliferation of > 90% 1807 cells transferred at a frequency of 104 and 105 tg cells, and all CFSElo tg cells were activated as indicated by increased CD44 expression (data not shown). However, this vaccine dose yielded limited proliferation and activation in mice that received the highest precursor frequency of 106 cells. 107 yeast were needed to promote maximal expansion at this precursor frequency. To approximate the likely frequency of endogenous T cells and to use frequencies that yielded maximal proliferation, in experiments below we transferred 105 1807 cells unless otherwise stated. We vaccinated mice with 106 yeast since that dose induced maximal proliferation and activation at this frequency.
Fig. 2. Impact of precursor frequency and vaccine dose on T cell expansion and proliferation.
Mice received 104 to 106 1807 cells and were vaccinated with 105 to 107 B. dermatitidis vaccine yeast. Four days later, the draining lymph nodes were harvested and analyzed for the presence of Thy 1.1+ CD4+ T cells. The averaged number of total (A) and percentage of CFSElo (B) 1807 cells is depicted. The fold change in the number of total and CFSElo 1807 cells in vaccinated vs. unvaccinated control was calculated and depicted in white in the histogram. *, p < 0.05 vs. non-vaccinated mice receiving the same precursor frequency as vaccinated mice, **, p < 0.05 vs. mice vaccinated with ten-fold less yeast.
Bd 1807 cells develop memory, migrate to the lung and produce IFN-γ after challenge
The induction of effector function and the generation of memory requires a higher Ag level threshold per naïve T-cell precursor than proliferation (31, 32). For example, survival and memory generation of adoptively transferred TCR tg T cells is dependent on the number of transferred cells and the amount of Ag available per naïve precursor (25, 31, 32). Decreasing the input number of naïve CD4+ T cells can promote memory development; conversely, limiting the availability of Ag can impair memory development.
To see if Bd 1807 tg cells develop memory, we tested a precursor frequency of 105 transferred cells and a vaccine dose ranging from 105 to 107 yeast. At 31 days after vaccination, the number of transferred 1807 cells in the sdLNs was significantly higher in vaccinated mice vs. controls (Fig. 3A). The pool of primed CD44hi tg cells in all groups of vaccinated mice was 115-349 fold larger than in the unvaccinated controls. In contrast, the increase in the number of endogenous, polyclonal CD4+ CD44hi T cells was <2.4 fold higher in vaccinated vs. unvaccinated mice, and the percentage of CD44hi T cells was <15%. In vaccinated mice, 80-92% of the transferred 1807 cells expressed high levels of CD44 vs. 11% in unvaccinated control mice. Thus, the analysis of polyclonal cells did not permit detection or tracking of a memory pool, whereas transfer of 105 1807 precursors and ensuing vaccination induced a significant population of memory cells. This discrepancy between polyclonal and 1807 cells underscores the utility of this mouse and adoptive transfer system for interrogating Ag-specific T-cell memory immunity to fungi.
Fig. 3. Bd 1807 cells become memory T cells in the sdLNs and differentiate into IFN-γ producing cells upon recall to the lung.
(A) Memory development of 1807 tg and endogenous, polyclonal CD4+ cells. Mice received 105 1807 cells and were vaccinated with B. dermatitidis vaccine yeast on the same day. 31 days later, the skin draining lymph nodes (dLN) were harvested, surface stained and analyzed by FACS. The data represent an average of 3 mice/group from a representative experiment of three performed. The numbers above in the histogram bars indicate the fold-increase of the number of cells in vaccinated vs. unvaccinated mice. *, p < 0.05 vs. unvaccinated control mice. (B) Influx of IFN-γ producing TCR tg and polyclonal CD4 cells during the first 4 days of the recall response in the lung. Mice received 105 naïve TCR tg cells, and were vaccinated once and challenged with 2 × 103 virulent wild-type B. dermatitidis yeast. The number of 1807 tg and polyclonal CD4 cells that produce IFN-γ were enumerated and illustrated at serial time points post-infection. The data represent an average of 3 mice/group from one representative experiment of three performed. *, p < 0.05 vs. unvaccinated control mice and **, p < 0.05 vs. polyclonal T cells from mice vaccinated with 105, 106 and 107 yeast.
To see if memory 1807 cells migrate to the lung during pulmonary recall, we analyzed lung T cells during the first 4 days post-infection when memory T-cells arrive and peak in the lung (21, 33). In vaccinated mice, 1807 cells that produced IFN-γ migrated into the lungs by day 2 and increased sharply by day 3 post-infection (Fig. 3B). Unvaccinated mice showed no influx of IFN-γ producing tg cells during this interval. Polyclonal IFN-γ producing CD4+ cells from vaccinated mice showed a similar kinetics of influx, indicating that 1807 cells accurately report the behavior of endogenous Ag-specific CD4+ T cells. In unvaccinated mice infected i.t. with wild-type yeast, polyclonal CD4+ cells that produced IFN-γ were detected by day 1 of infection, whereas 1807 cells were not detected. It is unclear if these polyclonal CD4+ cells are fungus-specific. Thus, Bd 1807 cells acquired memory, were recalled to the lung upon experimental challenge, and showed fidelity with endogenous CD4+ T cells.
Persistence of fungus-derived Ag presentation
The kinetics of presentation of fungal Ag and its impact on the differentiation of CD4+ T-cell effectors and generation of memory remains undefined. During an experimental influenza infection, live virus is cleared within the first 10 days of infection, but adoptive transfer of naïve influenza-specific CD4 T cells indicated Ag persistence for ≥ 3 wks after virus clearance (34). To assess the duration and relative amount of persistent vaccine Ag here, we used 1807 cells. We transferred the tg cells into mice at various times after Blastomyces vaccination and analyzed the donor cells for cell division and activation 7 days after transfer (34). This approach also enabled us to compare persistence of Ag using heat-killed vs. live vaccine. To confirm that 1807 cells report proliferation and activation in an Ag-specific manner, we analyzed 1807 phenotypes in vaccinated and unvaccinated recipients. Naïve 1807 cells transferred into unvaccinated mice remained largely CFSEhi, CD44lo and CD62Lhi for most time points, but there were minor fluctuations of the phenotypic markers for individual batches of donor cells. Consequently, this unvaccinated control group was used as a baseline. To establish that the division of donor 1807 cells was Ag-specific, Eα-specific TEa cells were transferred into vaccinated controls. TEa cells did not divide or become activated, establishing that 1807 cells behaved in an Ag-specific manner (data not shown).
In response to live vaccine, 105 1807 cells became activated within the first 3 weeks after vaccination, as noted by loss of CFSE (>90%) and expression of CD44 (>80%) (Fig. 4A and data not shown). Thereafter, 1807 T-cell activation dwindled, but was still evident by day 89 post-vaccination (Fig. 4B). At day 28 post-vaccination and thereafter, T-cell activation was hardly detectable with 105 precursors, and the lower frequency of 104 cells plus Thy1.1-based enrichment (25, 26) was required to detect T-cell activation. Thus, the amount of vaccine Ag became limiting by four weeks after vaccination. In mice vaccinated with heat killed yeast, T-cell priming during the first three weeks after vaccination was comparable to that in mice with live vaccine; it was reduced thereafter and not detected more than 35 days after vaccination. Thus, fungal Ag persisted longer with live vs. dead vaccine, and 1807 cells reported extended persistence of vaccine Ag.
Fig. 4. Bd 1807 cells are a sentinel of persistent fungal Ag.
(A) C57BL6 wild-type mice were vaccinated with 106 live or heat-killed attenuated B. dermatitidis and proliferation of sentinel 1807 cells measured. Beginning at day 0 and bi-weekly thereafter, naïve, CFSE- labeled 1807 cells were transferred into vaccinated mice or unvaccinated controls. 105 cells per mouse were transferred at day 0 and 14 post-vaccination and 104 cells per mouse at the later time points. 7 days after transfer, Thy1.1+ CD4+ T cells from the skin draining lymph nodes were analyzed for proliferation. The numbers depict the percentages of CFSElo donor CD4+ T cells in vaccinated mice (bold number and histogram) and unvaccinated controls (grey number and histogram). (B) The average numbers of CFSElow 1807 cells were calculated for each group and time point. The adoptive transfer, harvest and analysis of 1807 cells were identical as described in panel A. The data represent averages ± SEM of 4-5 mice per group. *, p < 0.05 vs. 1807 cells from all three groups of vaccinated mice, **, p < 0.05 vs. 1807 cells from unvaccinated control mice. (C) Skin CFU correlate with T-cell proliferation and activation. Skin tissue from the site of vaccination was harvested, homogenated and plated for CFU.
We measured Ag persistence using another approach. We vaccinated mice with live yeast measured CFU at the vaccine site (skin). Fungal CFU persisted during the first three weeks post-vaccination, they fell consistently thereafter and were cleared completely by day 104 post-vaccination (Fig. 4C). Thus, the findings revealed with 1807 cells are consistent with these results.
Prolonged Ag exposure drives optimal T cell development
Since fungal Ag persists up to three months after vaccination, we wondered what parameters are needed to drive optimal T-cell priming and protection. We have used an empirical schedule of two vaccinations over four weeks, which induces sterilizing immunity in most vaccinated mice (18, 33). The generation of the 1807 tg mouse and an adoptive transfer system enables us to study the parameters for optimal priming of protective T cells. We studied the time interval after vaccination needed to induce maximal protection and T-cell memory. In view of the persistence of vaccine Ag, we postulated that vaccine boosting might not be required. We vaccinated mice over the course of two, four or eight weeks and boosted them or not to measure the size of the effector/memory T-cell pool in the sdLNs. The number of primed (CD44hi) 1807 cells was significantly greater in all groups of vaccinated mice vs. unvaccinated controls (Fig. 5A). Mice vaccinated over four weeks, independent of a boosting dose, harbored a significantly larger pool of effector/memory T cells than the groups vaccinated over two or eight weeks. Remarkably, the number of polyclonal CD4+ CD44hi T cells evinced only subtle increases in vaccinated mice vs. unvaccinated controls. Thus, Bd 1807 cells let us monitor and quantify the emergence of Ag-specific effector/memory T-cells in the sdLNs in a setting where the analysis of polyclonal T cells was uninformative.
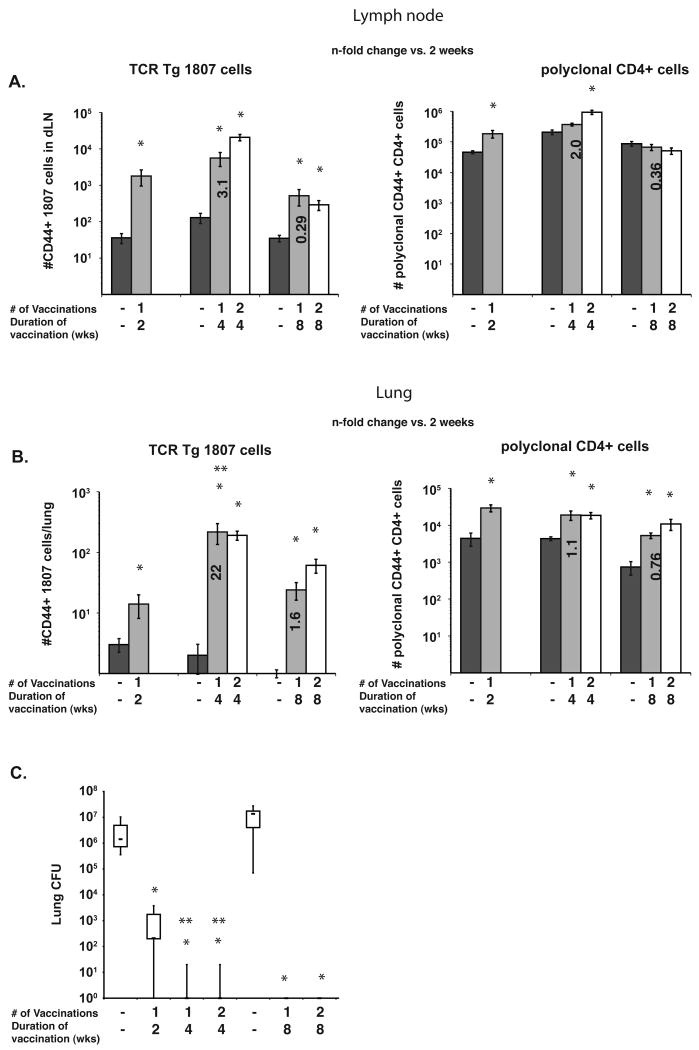
Fig. 5. Bd 1807 cells enable rational vaccine design, showing that 4 weeks of vaccination independent of boosting is needed to drive optimal T-cell priming and protective immunity.
(A) T-cell priming in the skin draining LN was analyzed for 1807 and polyclonal CD4+ T cells. The number of CD44hi 1807 and polyclonal T cells in the LN was determined by FACS after 2, 4 and 8 weeks of vaccination with B. dermatitidis. The data are averages ± SEM of 8 mice/group. *, p < 0.05 vs. 1807 or polyclonal CD4 T cells from unvaccinated mice. Bold numbers represent the n-fold increase in CD44+ T-cells vs. mice vaccinated over the course of two weeks. (B) T-cell influx into the lung on challenge. Mice received 105 1807 cells before vaccination and were challenged i.t. at the time indicated with wild-type B. dermatitidis. At day 4 post-infection, lung influx of 1807 and polyclonal T cells was analyzed. The data represent averages ± SEM of 4-8 mice per group. *, p < 0.05 vs. 1807 or polyclonal CD4+ T cells from unvaccinated control mice. **, p < 0.05 vs. the number of T cells from mice that were vaccinated over the course of two weeks. Bold numbers represent the n-fold increase in CD44+ T-cells vs. mice vaccinated over the course of two weeks. (C) Lung CFU was analyzed at day 16 post-infection. Mice were challenged as in (B). *, p < 0.001 vs. unvaccinated control mice. **, p < 0.001 vs. mice that were vaccinated over the course of two weeks.
After we challenged vaccinated mice i.t., we enumerated 1807 cells that migrated to the lung, and measured lung CFU in unvaccinated and vaccinated mice. Mice vaccinated over four weeks, whether they were boosted or not, harbored 22-fold more primed 1807 cells in their lungs than mice vaccinated over two weeks (Fig. 5B). Whereas the number of primed polyclonal CD4+ T cells in the lung was increased in all groups of vaccinated and unvaccinated mice, the analysis of the polyclonal pool again revealed only small differences between groups. Thus, polyclonal and 1807 tg cells migrated to the lung in an Ag-specific manner, but only 1807 tg cells offered the resolving power needed to determine the vaccine conditions that optimally induce and recall memory T cells to the lung.
The number of primed 1807 cells recruited to the lung correlated with the lung burden. Mice challenged four weeks post-vaccination cleared yeast from their lungs, whereas mice that had a shorter two-week interval of vaccination yielded 2-3 logs higher lung CFU (Fig. 5C). When vaccinated mice were challenged eight weeks after vaccination, they still cleared the yeast from their lungs, despite the fact that the influx of primed 1807 cells was smaller than in mice challenged four weeks after vaccination. By eight weeks the pool of primed 1807 cell had likely contracted but retained the functional capacity of a long-term memory pool. Thus, a four week interval after vaccination, independent of boosting, is necessary and sufficient to maximally induce a pool of effector/memory T cells in the sdLNs that can efficiently migrate to the lung upon challenge to mediate sterilizing immunity against a lethal pulmonary infection.
Bd 1807 cells confer protection against multiple dimorphic fungi
Since the 1807 mouse was made from a TCR that mediates resistance to infection with B. dermatitidis (5), we sought to determine whether 1807 cells confer protection against infection. We used two approaches: First, we generated T1-polarized effector cells in vitro and transfered them into naïve wild-type mice before challenge. In vitro primed TCR tg CD4+ effector cells confer protection against lethal influenza infection (35). Naïve 1807 cells were stimulated in vitro with anti-CD3 and anti-CD28 mAb in the presence of recombinant IFN-γ and anti-IL-4 mAb to polarize them into a T1 phenotype (36). After 10 days of culture, 1807 cells were polarized and produced chiefly IFN-γ in cell culture supernatants, whereas non-polarized 1807 cells produced both Th1 and T2 cytokines. Upon adoptive transfer and challenge, all three effector T cell populations reduced lung CFU significantly compared to mice that did not receive any T cells (Fig. 6A). Thus, 1807 effector cells confer protection.
Fig. 6. Bd 1807 cells mediate protection against experimental infection with B. dermatitidis, C. posadasi and H. capsulatum.
(A) In vitro activated, adoptively transferred effector T cells mediate protection. 10 weeks after adoptive transfer, mice were challenged with B. dermatitidis and 21 days later analyzed for lung CFU. Data represent the geometric mean CFU ± SEM; n= 9-13 mice/group. *, p < 0.001 vs. no transfer control group. Bold numbers indicate the n-fold reduction in lung CFU vs. control mice that received no T cells. (C) In vivo primed 1807 cells engender resistance to B. dermatitidis infection. (B) TCR-α deficient mice received naïve 1807 or TEa cells as a control and were vaccinated with 106 heat-killed B. dermatitidis yeast twice two weeks apart. 4 weeks after the boost, mice were challenged with 103 26199 wild-type yeast and 25 days later, when unvaccinated mice were moribund, analyzed for lung CFU. Wild-type mice were vaccinated or not and challenged in parallel. Data represent the geometric mean CFU ± SEM; n= 8 mice/group. *, p < 0.001 vs. unvaccinated OT-1 and wild-type mice that received no TCR tg cells. **, p < 0.005 vs. unvaccinated mice that received 1807 or TEa cells. Bold numbers indicate the n-fold reduction in lung CFU vs. control mice that received no T cells or vaccine. (C) OT-1 mice received TCR tg 1807 or OT-2 cells as a control and were vaccinated (or not) with 106 heat-killed yeast on the day of transfer and boosted two weeks later. Two weeks after the boost, mice were challenged with 2 × 103 26199 yeast. 19 and 25 days post-infection when the unvaccinated wild-type and OT-1 mice, respectively, were moribund the lungs were harvested and plated for CFU. *, p < 0.05 vs. unvaccinated wild-type and OT-1 mice that received no TCR tg cells. **, p < 0.05 vs. vaccinated OT-1 mice that received OT-2 cells prior to vaccination. (D+E) OT-1 mice received TCR tg 1807 or OT-2 cells as a control and were vaccinated (or not) with 106 heat-killed H. capsulatum yeast or 106 FKS from C. posadasi on the day of transfer and boosted two weeks later. Two weeks after the boost, mice were challenged with 2 × 105 H. capsulatum yeast (strain G217B) or 60 viable spores of C. posadasii (isolate C735). 14 days post-infection when the unvaccinated wild-type and OT-1 mice, respectively, were moribund the lungs were harvested and plated for CFU. *, p < 0.05 vs. unvaccinated wild-type and OT-1 mice that received no TCR tg cells. **, p < 0.05 vs. vaccinated OT-1 mice that received OT-2 cells prior to vaccination.
Second, we tested whether in vivo priming of 1807 cells could promote vaccine immunity. We transferred naïve 1807 and TEa (irrelevant control) cells into TCR-α-/- mice and vaccinated them with heat-killed yeast. Heat-killed yeast were used to avoid dissemination in immune-deficient mice (37). Vaccination engendered robust immunity (Fig. 6B). Transfer of naïve 1807 cells into vaccinated or unvaccinated TCR-α-/- mice reduced lung CFU by 6 and 3 logs, respectively. Since transfer of naïve T cells into lymphopenic hosts can activate T cells and promote memory (38), 1807 cells transferred into unvaccinated TCR-α-/- mice could have promoted immunity due to non-specific activation. To demonstrate that vaccine priming and resistance of 1807 cells was Ag-specific, we transferred naïve TEa cells specific for Eα Ag (17), which was not present in the vaccine strain. Vaccinated mice that received TEa cells before vaccination had 3 logs less lung CFU than unvaccinated controls that did not receive TEa cells. Thus, vaccine-primed 1807 cells reduced lung CFU by 3 logs more than TEa cells, indicating that primed Ag-specific 1807 cells contributed significantly to vaccine resistance.
Why did transferred CD4+ T cells from TEa mice reduced lung CFU? The frequency of tg cells in TEa mice (and 1807 mice) is 90-95% of the total CD4+ T-cell pool, indicating that the mice harbor endogenous non-TEa CD4+ cells. Those cells may have become activated during vaccination or due to lymphopenia-induced expansion and contributed to vaccine immunity.
To circumvent the limitation above, we used another design. To avoid lymphopenia-induced activation of transferred T cells, we transferred tg cells into OT-1 mice that harbor chiefly SIINFEKL-specific CD8 T cells and few endogenous CD4+ T cells (14). Because OT-1 mice are not lymphopenic, transferred tg cells should not expand and become non-specifically activated. To see if OT-1 mice that received non-specific OT-2 cells acquire vaccine immunity, we vaccinated these mice with B. dermatitidis yeast and analyzed lung CFU after challenge. Vaccinated and non-vaccinated mice showed similar lung CFU, indicating that any endogenous cells in OT-1 mice or OT-2 donor cells do not confer immunity (Fig. 6C). This transfer system was thus deemed suitable to assess in vivo protection by 1807 cells. OT-1 mice that received naïve 1807 cells before vaccination and challenge reduced lung CFU by ≈3 logs vs. control mice that received OT-2 cells. Transfer of naïve 1807 cells without vaccination reduced lung CFU 33-fold, while OT-2 cells had no effect. Thus, in vivo primed 1807 cells confer vaccine protection to B. dermatitidis and naïve 1807 cells confer resistance to primary infection, but much less so. Since 1807 cells recognize a protective antigen in Blastomyces yeast that is shared in other dimorphic fungi, we tested whether 1807 cells protect against Histoplasma or Coccidioides infection. Transfer of 1807 cells into OT1 mice vaccinated with H. capsulatum or C. posadasii reduced lung CFU significantly by 8- and 31–fold respectively vs. vaccinated OT1 controls (Fig. 6D&E). Thus, vaccine induced 1807 cells are sufficient to protect against experimental coccidioidomycosis and histoplasmosis.
Discussion
We describe a TCR tg mouse and its use in studying the developmental progression of fungal Ag-specific T cells. This mouse is unique and will benefit the field of medical mycologists studying cellular mechanisms of CD4 T-cell immunity and memory to dimorphic fungi. First, Bd 1807 cells recognize a shared antigen in B. dermatitidis, H. capsulatum, P. brasiliensis, and C. posadasii. Since T-cell responses are requisite for vaccine immunity against dimorphic fungi, investigators studying them will now have a tool to track and precisely enumerate Ag-specific T cells during development. Second, Bd 1807 cells report a biologically vital endpoint, as they harbor a TCR from cells that protect against experimental infection. Bd 1807 cells themselves protect vaccinated mice against infection. Thus, Bd 1807 cells and the adoptive transfer system we developed produce functional T cells that differentiate, develop memory and protect against experimental pulmonary blastomycosis, histoplasmosis and coccidiomycosis. To our knowledge, this is the first anti-fungal TCR tg mouse that harbors protective T cells. Although Af3.16 TCR tg T cells are specific for A. fumigatus and migrate, proliferate and differentiate in an Ag-specific manner, their protective role was not investigated (4). The ability of Bd 1807 cells to confer resistance to infection with the three major endemic, dimorphic fungi of North America should allow us and others to study the role of CD4+ T-cell differentiation down varied T helper pathways in generation of long-term memory and immunity to fungal disease.
The TCR tg mouse and adoptive transfer system here enabled us to analyze T cells at a level that has not been readily attainable in fungal immunology (4). We used 1807 tg cells to address two simple questions about Ag-persistence and optimal conditions for vaccine immunity. First, we studied how long Ag persists after immunization with live or heat-killed vaccine. We used naïve 1807 cells as a sentinel transferring them into vaccinated mice at serial time points post-vaccination to assess T-cell activation and proliferation one week later. T-cells proliferated and became activated for up to two or three months post-vaccination using heat-killed or live vaccine, respectively. In the case of the live vaccine, CFU plating of skin tissue harvested from the vaccine site confirmed the prolonged presence of Ag reported by sentinel 1807 cells. The persistence of fungal Ag contrasts with influenza where viral Ag persisted for several weeks after viral clearance as detected by HNT-specific TCR tg T cells (34). Bd 1807 cells did not report persistent fungal Ag after CFU clearance. The Ag load might be higher after systemic viral infection as opposed to a local fungal vaccine administration. If so, the amount of Ag available per naïve TCR tg precursor would be lower in our fungal model. This hypothesis was supported by the fact that T-cell proliferation and activation after 21 days post-vaccination was only detectable when we transferred fewer (104) naïve 1807 cells and enriched them with beads (25).
We asked what vaccine schedule drives the largest effector/memory pool of Ag-specific T cells that mediates protective immunity. The increase in number of primed Ag-specific T cells in vaccinated vs. unvaccinated mice was more than one magnitude larger for the 1807 cells than for polyclonal CD4+ cells. The number of primed 1807 cells detected in the sdLNs and lung after recall was the largest in mice vaccinated over a period of four weeks, demonstrating that this interval induces a maximal pool of effector/memory cells. A two-week interval was too short and by eight weeks effector T cells likely contracted. The analysis of polyclonal CD4+ T cells did not reveal a difference in the number of primed T cells in the sdLNs or the lung upon challenge over the course of four weeks of vaccination. Thus, only 1807 cells offered the resolution to track the development and recruitment of protective T cells upon vaccination, experimental challenge, and recall to the lung.
To evaluate the utility and fidelity of the mouse in monitoring Ag-specific T cells, we compared 1807 tg cells and polyclonal CD4+ T cells during sequential stages after vaccination: activation, differentiation, memory and protection. We established an adoptive transfer system and defined the conditions that induce optimal activation and expansion of the TCR tg T cells. 1807 tg cells expanded maximally using a precursor frequency of 105 tg cells and a vaccine dose of 106 to 107 yeast. Therein, >90% of 1807 cells were activated, and proliferated and expanded at maximum by 2,000- to 5,800-fold. At either higher or lower precursor frequencies, the maximal expansion of T cells and their activation and proliferation, as denoted by the number and percentage of CD44hi cells or CFSElo cells, were all reduced. Considering that about 10 to 15% of the transferred precursors (∼ 104 cells) survive transplantation (25, 30, 39), we could have created a non-physiological situation wherein transferred 1807 tg cells exceeded the pool size of endogenous naïve Ag-specific T cells. The largest pool of such cells known so far is around 1,200 cells per mouse (26, 40-42), however only a handful of the estimated 106 endogenous Ag-specific precursor populations have been measured (43). Thus, it is unknown whether larger populations exist in the normal repertoire. Since 105 precursors induced the most robust expansion and activation of 1807 cells in our model, we used this frequency in our studies to evaluate downstream T-cell functions of differentiation, memory and protection.
Memory CD4+ T cells developed in the sdLNs when we transferred 105 1807 cells prior to vaccination. Vaccinated mice had over 100-fold more primed (CD44hi) TCR tg cells than did unvaccinated mice, and the percentage of CD44hi cells in the respective groups was >80% vs. <15%. Importantly, analysis of polyclonal CD4+ T cells did not permit the detection of Ag-specific memory. The number of primed polyclonal CD4+ T cells increased by less than 2.4-fold by 30 days after vaccination, when less than 15% of the cells were CD44hi. Primed 1807 effector/memory T cells migrated to the lung and produced IFN-γ upon pulmonary challenge. Transferring 105 naïve precursors yielded 500 to 1,500 CD44hi 1807 cells in the lung of which 58 to 170 cells produced IFN-γ. A lower precursor frequency did not yield a detectable number of IFN-γ producing 1807 cells in the lung upon challenge. Thus, even though we transferred 1807 cells at a frequency that may have exceeded a natural precursor frequency, the tg cells differentiated, became memory cells and migrated to the lung upon recall, indicating that they behaved like functional memory T cells. That is, they produced IFN-γ and mediated resistance against a lethal pulmonary infection
The availability of this tg mouse will enable investigators to study the earliest stages of priming and differentiation of fungal Ag-specific T cells and examine the requirements for maintaining long-term memory of protective T cells. Our work demonstrates that insights from a fungal model differ from viral or bacterial models (34, 44), for example vis-à-vis duration of Ag persistence. Other tg models also emphasize using very low precursor frequencies to portray Ag-specific T-cell responses, but we found that a frequency of 105 precursors enabled optimal in vivo expansion. We also found that this frequency was needed to analyze recall responses in the lung and preserve key functions such as IFN-γ production in vivo and resistance to infection. We expect that the better resolving power of 1807 cells over polyclonal analysis will enable us to explore and answer critical questions in fungal immunology: 1) the occurrence and importance of Ag-specific T-cell differentiation down various T helper pathways in establishing protective immunity; 2) the contribution of distinct APC populations in vaccine priming of Ag-specific T-cell immunity to fungi; 3) the tissue specific influences on priming T-cell immunity, for example in lung vs. skin; and 4) the presence of shared antigens in the fungal kingdom that are recognized by protective T cells, such as those used to generate the Bd 1807 mouse described here.
Acknowledgments
We thank Dr. Mary Lindstrom from the Department of Biostatistics and Medical Informatics for assisting with statistical analysis.
This work was supported by NIH grants R21 AI076700 (MW), R01 AI40996-10 (BSK), and R01 AI 071118 (GTC).
References
- 1.Wüthrich M, Ersland K, Galles KJ, Sullivan TD, Klein BS. Fungi subvert vaccine T-cell priming at the respiratory mucosa by inducing MMP2 and retarding CCL7-mediated influx of inflammatory monocytes. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 3.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 4.Rivera A, Ro G, Van Epps HL, Simpson T, Leiner I, Sant'Angelo DB, Pamer EG. Innate immune activation and CD4+ T cell priming during respiratory fungal infection. Immunity. 2006;25:665–675. doi: 10.1016/j.immuni.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Wüthrich M, Filutowicz HI, Allen HL, Deepe GS, Klein BS. V{beta}1+ J{beta}1.1+/V{alpha}2+ J{alpha}49+ CD4+ T Cells Mediate Resistance against Infection with Blastomyces dermatitidis. Infect Immun. 2007;75:193–200. doi: 10.1128/IAI.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldrath AW, Bogatzki LY, Bevan MJ. Naive T cells transiently acquire a memory-like phenotype during homeostasis-driven proliferation. J Exp Med. 2000;192:557–564. doi: 10.1084/jem.192.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey RP, Schmid ES, Carrington CC, Stevens DA. Mouse model of pulmonary blastomycosis: utility, simplicity, and quantitative parameters. American Review of Respiratory Disease. 1978;117:695–703. doi: 10.1164/arrd.1978.117.4.695. [DOI] [PubMed] [Google Scholar]
- 8.Brandhorst TT, Wüthrich M, Warner T, Klein B. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med. 1999;189:1207–1216. doi: 10.1084/jem.189.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teixeira MM, Theodoro RC, de Carvalho MJ, Fernandes L, Paes HC, Hahn RC, Mendoza L, Bagagli E, San-Blas G, Felipe MS. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol. 2009;52:273–283. doi: 10.1016/j.ympev.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Levine HB, Cobb JM, Smith CE. Immunity to coccidioi-domycosis induced in mice by purified spherule, arthrospore, and mycelial vaccines. Trans N Y Acad Sci. 1960;22:436–449. doi: 10.1111/j.2164-0947.1960.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 11.Levine HB, Kong YC, Smith C. Immunization of Mice to Coccidioides Immitis: Dose, Regimen and Spherulation Stage of Killed Spherule Vaccines. J Immunol. 1965;94:132–142. [PubMed] [Google Scholar]
- 12.Fabien N, Bergerot I, Maguer-Satta V, Orgiazzi J, Thivolet C. Pancreatic lymph nodes are early targets of T cells during adoptive transfer of diabetes in NOD mice. Journal of Autoimmunity. 1995;8:323–334. doi: 10.1006/jaut.1994.0025. [DOI] [PubMed] [Google Scholar]
- 13.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper ML, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages [published erratum appears in Nature 1992 Dec 3;360(6403):491] Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 14.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 15.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 16.Firpo EJ, Kong RK, Zhou Q, Rudensky AY, Roberts JM, Franza BR. Antigen-specific dose-dependent system for the study of an inheritable and reversible phenotype in mouse CD4+ T cells. Immunology. 2002;107:480–488. doi: 10.1046/j.1365-2567.2002.01540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grubin CE, Kovats S, deRoos P, Rudensky AY. Deficient positive selection of CD4 T cells in mice displaying altered repertoires of MHC class II-bound self-peptides. Immunity. 1997;7:197–208. doi: 10.1016/s1074-7613(00)80523-3. [DOI] [PubMed] [Google Scholar]
- 18.Wüthrich M, Filutowicz HI, Klein BS. Mutation of the WI-1 gene yields an attenuated Blastomyces dermatitidis strain that induces host resistance. J Clin Invest. 2000;106:1381–1389. doi: 10.1172/JCI11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng M, Shellito JE, Marrero L, Zhong Q, Julian S, Ye P, Wallace V, Schwarzenberger P, Kolls JK. CD4+ T cell-independent vaccination against Pneumocystis carinii in mice. J Clin Invest. 2001;108:1469–1474. doi: 10.1172/JCI13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wüthrich M, Gern B, Hung CY, Ersland K, Rocco N, Pick-Jacobs J, Galles K, Filutowicz H, Warner T, Evans M, Cole G, Klein B. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J Clin Invest. 2011;121:554–568. doi: 10.1172/JCI43984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wüthrich M, Filutowicz HI, Warner T, Deepe GS, Jr, Klein BS. Vaccine Immunity to Pathogenic Fungi Overcomes the Requirement for CD4 Help in Exogenous Antigen Presentation to CD8+ T Cells: Implications for Vaccine Development in Immune-deficient Hosts. J Exp Med. 2003;197:1405–1416. doi: 10.1084/jem.20030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braga CJ, Rittner GM, Munoz Henao JE, Teixeira AF, Massis LM, Sbrogio-Almeida ME, Taborda CP, Travassos LR, Ferreira LC. Paracoccidioides brasiliensis vaccine formulations based on the gp43-derived P10 sequence and the Salmonella enterica FliC flagellin. Infect Immun. 2009;77:1700–1707. doi: 10.1128/IAI.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loures FV, Pina A, Felonato M, Calich VL. TLR2 is a negative regulator of Th17 cells and tissue pathology in a pulmonary model of fungal infection. J Immunol. 2009;183:1279–1290. doi: 10.4049/jimmunol.0801599. [DOI] [PubMed] [Google Scholar]
- 24.Aratani Y, Koyama H, Nyui S, Suzuki K, Kura F, Maeda N. Severe impairment in early host defense against Candida albicans in mice deficient in myeloperoxidase. Infect Immun. 1999;67:1828–1836. doi: 10.1128/iai.67.4.1828-1836.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 26.Moon JJ, Chu HH, Hataye J, Pagan AJ, Pepper M, McLachlan JB, Zell T, Jenkins MK. Tracking epitope-specific T cells. Nat Protoc. 2009;4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher LD, van Belle G. Biostatistics: A Methodology for the Health Sciences. John Wiley & Sons; New York: 1993. pp. 611–613. [Google Scholar]
- 28.Bialek R, Ibricevic A, Fothergill A, Begerow D. Small subunit ribosomal DNA sequence shows Paracoccidioides brasiliensis closely related to Blastomyces dermatitidis. J Clin Microbiol. 2000;38:3190–3193. doi: 10.1128/jcm.38.9.3190-3193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowman BH, Taylor JW, White TJ. Molecular evolution of the fungi: human pathogens. Mol Biol Evol. 1992;9:893–904. doi: 10.1093/oxfordjournals.molbev.a040766. [DOI] [PubMed] [Google Scholar]
- 30.Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blair DA, Lefrancois L. Increased competition for antigen during priming negatively impacts the generation of memory CD4 T cells. Proc Natl Acad Sci U S A. 2007;104:15045–15050. doi: 10.1073/pnas.0703767104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yarke CA, Dalheimer SL, Zhang N, Catron DM, Jenkins MK, Mueller DL. Proliferating CD4+ T cells undergo immediate growth arrest upon cessation of TCR signaling in vivo. J Immunol. 2008;180:156–162. doi: 10.4049/jimmunol.180.1.156. [DOI] [PubMed] [Google Scholar]
- 33.Wüthrich M, Filutowicz HI, Warner T, Klein BS. Requisite elements in vaccine immunity to Blastomyces dermatitidis: plasticity uncovers vaccine potential in immune-deficient hosts. J Immunol. 2002;169:6969–6976. doi: 10.4049/jimmunol.169.12.6969. [DOI] [PubMed] [Google Scholar]
- 34.Jelley-Gibbs DM, Brown DM, Dibble JP, Haynes L, Eaton SM, Swain SL. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J Exp Med. 2005;202:697–706. doi: 10.1084/jem.20050227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006;177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 36.Jelley-Gibbs DM, Dibble JP, Filipson S, Haynes L, Kemp RA, Swain SL. Repeated stimulation of CD4 effector T cells can limit their protective function. J Exp Med. 2005;201:1101–1112. doi: 10.1084/jem.20041852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wüthrich M, Warner T, Klein BS. IL-12 Is Required for Induction but Not Maintenance of Protective, Memory Responses to Blastomyces dermatitidis: Implications for Vaccine Development in Immune-Deficient Hosts. J Immunol. 2005;175:5288–5297. doi: 10.4049/jimmunol.175.8.5288. [DOI] [PubMed] [Google Scholar]
- 38.Jameson SC. Maintaining the norm: T-cell homeostasis. Nat Rev Immunol. 2002;2:547–556. doi: 10.1038/nri853. [DOI] [PubMed] [Google Scholar]
- 39.Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis MM. The alphabeta T cell repertoire comes into focus. Immunity. 2007;27:179–180. doi: 10.1016/j.immuni.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Winslow GM, Roberts AD, Blackman MA, Woodland DL. Persistence and turnover of antigen-specific CD4 T cells during chronic tuberculosis infection in the mouse. J Immunol. 2003;170:2046–2052. doi: 10.4049/jimmunol.170.4.2046. [DOI] [PubMed] [Google Scholar]