Abstract
Increasing evidence points to the functional importance of alternative splice variations in cancer pathophysiology with the alternative pre-mRNA processing of caspase 9 as one example. In this study, we delve into the underlying molecular mechanisms that regulate the alternative splicing of caspase 9. Specifically, the pre-mRNA sequence of caspase 9 was analyzed for RNA cis-elements known to interact with SRSF1, a required enhancer for caspase 9 RNA splicing. This analysis revealed thirteen possible RNA cis-elements for interaction with SRSF1 with mutagenesis of these RNA cis-elements identifying a strong intronic splicing enhancer located in intron 6 (C9-I6/ISE). SRSF1 specifically interacted with this sequence, which was required for SRSF1 to act as a splicing enhancer of the inclusion of the four exon cassette. To further determine the biological importance of this mechanism, we employed RNA oligonucleotides to redirect caspase 9 pre-mRNA splicing in favor of caspase 9b expression, which resulted in an increase in the IC50 of non-small cell lung cancer (NSCLC) cells to daunorubicin, cisplatinum, and paclitaxel. In contrast, downregulation of caspase 9b induced a decrease in the the IC50 of these chemotherapeutic drugs. Lastly, these studies demonstrated that caspase 9 RNA splicing was a major mechanism for the synergistic effects of combination therapy with daunorubicin and erlotinib. Overall, we have identified a novel intronic splicing enhancer that regulates caspase 9 RNA splicing and specifically interacts with SRSF1. Furthermore, we demonstrate that the alternative splicing of caspase 9 is an important molecular mechanism with therapeutic relevance to NSCLCs.
Keywords: ceramide, non-small cell lung cancer, RNA trans-factor, tumor repressor, oncogene, ASF/SF2, SRp30a, SRSF1, chemotherapy, erlotinib, daunorubicin, cisplatinum, paclitaxel
INTRODUCTION
Historically, the regulation of the alternative splicing of caspase 9 by signaling pathways began with the lipid second messenger, ceramide. This lipid second messenger is an important regulator of various stress responses and growth mechanisms including apoptosis, and the formation of ceramide from the hydrolysis of sphingomyelin or from de novo pathways has been observed in response to numerous apoptotic and stress agonists including chemotherapeutic agents (1–7). On mechanism by which ceramide imparts these biological actions is via a family of ceramide-regulated enzymes designated, ceramide-activated protein phosphatases, which include the serine/threonine-specific protein phosphatases PP1 and PP2A (8–11). Specific substrates of PP1 are the SR proteins (12), a family of arginine/serine-rich domain containing proteins that are major players in both constitutive and alternative pre-mRNA processing. Endogenous ceramide has been found to modulate the phosphorylation status of SR proteins in a PP1-dependent manner leading to dephosphorylation of the RS-domain (13). In regards to caspase 9 RNA splicing, a pathway linking the generation of de novo ceramide and the activation of PP1 to the regulation of the exclusion or inclusion of an exon 3, 4, 5, and 6 cassette of caspase-9 pre-mRNA was identified by our laboratory (14). Specifically, ceramide treatment resulted in an increase in the pro-apoptotic caspase 9a 1, mRNA and protein levels with concomitant decrease in caspase-9b (cassette exclusion) mRNA and protein levels in A549 cells (14). This effect required the generation of endogenous ceramide through the denovo pathway, and more importantly, inhibitors of protein phosphatase-1abolished the ability of ceramide to affect the alternative splicing of caspase 9 (14). Thus, both the phospho-state of SR proteins and the alternative splicing of caspase-9 are regulated by the generation of de novo ceramide and subsequent PP1 activation (14).
The involvement of PP1 and endogenous ceramide in the dephosphorylation of SR proteins and the effects on caspase 9 alternative splicing suggested that at least one SR protein isoform regulated the alternative splicing mechanism of caspase 9. Indeed, in further mechanistic studies, our laboratory identified one SR protein, SRSF1 (also known as SRp30a), as a critical splicing factor in the alternative splicing of caspase 9 pre-mRNA in A549 lung adenocarcinoma cells (15). Furthermore, we demonstrated that SRSF1 is a required RNA trans-acting factor for ceramide to affect the alternative splicing of caspase 9 pre-mRNA, thereby linking de novo ceramide generation, SRSF1, and the regulation of caspase 9 expression. More recently, our laboratory demonstrated that the phospho-status of SRSF1 on serine199, 210, 227, and 234 mediated the suppression of the exon 3,4,5,6 cassette, suggesting that ceramide activation of PP1 leads to dephosphorylation of these residues.
The linking of ceramide signaling to caspase 9 RNA splicing has significant relevance to cancer therapeutics as studies have shown that the ectopic expression of caspase 9b confers the opposite effect on apoptosis as full-length caspase 9 (caspase 9a); by inducing resistance to many apoptotic stimuli (e.g. FAS ligand and UV radiation) (16–18). Caspase 9b elicits this biological outcome at least in part by competing with the full length caspase 9 for binding to the apoptosome (e.g. APAF-1) (18, 19). Therefore, regulation of the inclusion of this four exon cassette is a critical determinant to decide whether a cell is susceptible or resistant to apoptosis (17, 18). Recently, our laboratory expanded upon both the mechanistic regulation of caspase 9 RNA splicing and the sensitivity of non-small cell lung cancer cells to the chemotherapeutic agent, erlotinib. Indeed, our laboratory found that the erlotinib induced an increase in the caspase 9a/9b ratio in NSCLC cells, which was important for the sensitivity of these cells to this chemotherapeutic agent (20). Therefore, the alternative splicing of caspase 9 has a role in the mechanism by which a clinically relevant therapy affects NSCLC cells.
In the presented study, we delve further into both the underlying molecular mechanisms that regulate the alternative splicing of caspase 9 as well as the biological relevance of this distal mechanism in the treatment of of non-small cell lung cancer. Specifically, we identify a novel and strong intronic splicing enhancer (C9-I6/ISE) for caspase 9 pre-mRNA and demonstrate that SRSF1 specifically interacts with this ISE. Furthermore, SRSF1 was demonstrated to regulate the alternative splicing of caspase 9 via this novel RNA cis-element. Lastly, we demonstrate that modulation of the alternative splicing of caspase 9 regulates the synergistic effects of daunorubicin and erlotinib, suggesting that this molecular splicing mechanism is a relevant therapeutic target for non-small cell lung cancer.
MATERIAL AND METHODS
Cell Culture
A549 cells were grown in 50% RPMI 1640 (BioWhittaker® Cambrex Bio Science Walkerville, Inc., Walkerville, MD) and 50% DMEM supplemented with L-glutamine (Invitrogen™, Carlsbad, CA), 10% (v/v) fetal bovine serum (Invitrogen™, Carlsbad, CA), 100 units/ml penicillin and 100 mg/ml streptomycin (BioWhittaker® Penicillin/Streptomycin, Cambrex Bio Science Walkerville, Inc., Walkerville, MD). H2030, H838 and HeLa cell lines were cultured with 100% RPMI 1640 supplemented with L-glutamine, 10% (v/v) fetal bovine serum, 100 units/ml penicillin and 100 mg/ml streptomycin. HBEC-3KT cells were cultured with keratinocyte serum-free medium containing bovine pituitary extract and recombinant epidermal growth factor (Life Technologies, Gaithersburg, MD). Cells were maintained at <80% confluence under standard incubator conditions (humidified atmosphere, 95% air, 5% CO2, 37°C). For transfection, cells were plated at 2.5 × 105 cells/35-mm plate in appropriate culture media. The chemotherapeutic drugs; Daunorubicin (DNR), Cisplatinum (Cp), and Paclitaxel (Pac) were purchased from Sigma-Aldrich, Inc. (Sigma, St. Louis, MD). Erlotinib was obtained from Genetech, Inc. (San Francisco, CA).
RNA transfections
siRNA
A549, H838, H2030, HeLa, and HBEC-3KT cell lines were transfected with the SRSF1 SMARTpool multiplex designed RNA (siRNA) (Dharmacon; Lafayette, CO) using Dharmafect 1 transfection reagent (Dharmacon; Lafayette, CO) following the manufacturer’s protocol. A549 and H2030 cells were also transfected with caspase 9b siRNA designed against following sequence; GATTTGGTGATGTCGAGCATT. The siRNA for caspase 9b (9b-si) and non targeting control siRNA using Dharmafect 1 transfection reagent (Dharmacon; Lafayette, CO) following the manufacturer’s protocol. Briefly, cell lines were plated in regular growth medium at 40–50% confluence in a six-well tissue culture dish 24 h before transfection. Cells were placed in Opti-Mem I medium without antibiotics/fetal bovine serum and transfected with 100 nM (dilution in 1X siRNA buffer) of the siRNA of choice (Dharmafect 1/Opti-MEM I reduced serum medium and incubated for 4 h at standard incubator conditions). After incubation, 0.5 ml Opti-MEM I reduced serum medium containing three times the normal concentration of antibiotics/fetal bovine serum was added to the transfected cells lines without removing the transfection mixture. After 48 h, total RNA isolation or total protein lysates were collected as previously described in our laboratory (15) for RT-PCR or western immunoblot analysis.
ASROs
2′-O-(2-methoxy) ethyl (MOE) 23-mer anti-sense RNA oligonucleotides (ASRO) were purchased from Dharmacon. This study focuses on E4 ASRO (2-O-methoxy derivative) (5′-GAGTGTACCTTGGCAGTCAGGTC-3′) that is targeted to the 5′splice site of Caspase 9 exon 4 (21). A549 cells were transfected with ASRO (400nM) using Dharmafect I reagent (Dharmacon, Lafayette, CO) and following the same protocol as the siRNA transfection.
Western Immunoblotting
Total protein was extracted by direct lysis with Laemmli buffer. Cells were lysed with 0.1 ml of 2x Laemmli buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.04% bromophenol blue, and 250 mM β-mercaptoethanol) after resuspension in 0.1 ml of ice-cold PBS. Samples were boiled for 10 min and either examined directly by SDS-PAGE or stored at −20°C. Protein samples (10 μg) were subjected to 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane (PDVF) (Bio-rad; Hercules, CA) and blocked in 5% milk/1 X PBS- 0.1% Tween (M-PBS-T) for 2 h. The membrane was incubated with one of the following primary antibodies, anti-SRSF1 (Zymed; San Francisco, CA) (1:1000 dilution), anti-T7 tag (1:10000, Novagen), anti-Caspase 9 (1:1000, Cell Signaling Technology), or anti-α-tubulin (1:5000, Sigma), or anti-β-actin (Sigma; St. Louis, MO) (1:5000 dilution) for 2 h in M-PBS-T followed by three washed with PBS-T. The membrane was then incubated with the appropriate secondary antibody (horseradish peroxidase-conjugated), either goat anti-mouse IgG antibody (Sigma; St. Louis, MO), goat anti-rabbit IgG antibody (1:10,000, Pierce), or goat anti-mouse IgM (1:10,000, Calbiochem) for 1 h in M-PBS-T followed by three washes with PBS-T. Immunoblots were developed using Pierce (Rockford, IL) enhanced chemiluminescence (ECL) reagents and Bio-max film.
Quantitative and Competitive Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA from A549 cells was isolated using the RNeasy® Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s protocol. Total RNA (1 μg) was reverse-transcribed using Superscript III reverse transcriptase (SuperScript™ First-Strand Synthesis System for RT-PCR, Invitrogen™, Carlsbad, CA) and oligo (dT) as the priming agent. After 50 min of incubation at 42°C, the reactions were stopped by heating at 70°C for 15 min. Template RNA was then removed using RNase H (Invitrogen, Carlsbad, CA). To evaluate the expression of endogenous caspase 9 splice variants, an upstream 5′ primer to caspase-9 (5′-GCT CTT CCT TTG TTC ATC TCC-3′) and a 3′ primer (5′-CAT CTG GCT CGG GGT TAC TGC-3′) (Integrated DNA Technologies, Inc., Coralville, IA) were used. Using these primers, 20% of the reverse transcriptase reaction was amplified for 25 cycles (94°C, 30 s melt; 58°C, 30 s anneal; 72°C, 1 min extension) using Platinum Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA). Gene products produced from endogenous caspase 9 PCR resulted in a 1248 bp caspase 9a splice variant and 798 bp caspase 9b splice variant. To specifically evaluate the expression of the splice variant products of the caspase 9 minigene (Figure 1), 5′ primer to caspase 9 minigene (5′-CAT GCT GGC TTC GTT TCT G-3′) and a 3′ primer (5′-AGG GGC AAA CAA CAG ATG G-3′) were used. Gene products produced from caspase 9 minigene PCR resulted in a 889 bp caspase 9a splice variant and a 443bp caspase 9b splice variant. Using these primers, 20% of the reverse transcriptase reaction was amplified for 20 cycles (94°C, 30 s denaturation; 57°C, 30 s anneal; 72°C, 1 min extension) using Platinum Taq DNA polymerase (Invitrogen™, Carlsbad, CA). The final PCR products were resolved on a 5% TBE acrylamide gel electrophoresis and visualized by staining with SYBR® Gold (Invitrogen™, Carlsbad, CA) and scanned using a Molecular Imager® FX (Bio-Rad) with a 488 nm EX (530 nm BYPASS) laser. These RT-PCR based assays are quantitative as to the ratio of caspase 9a/caspase 9b mRNA between analyzed samples as confirmed previously by our laboratory (14, 20, 21) and the initial reports by Larrick and co-workers (22).
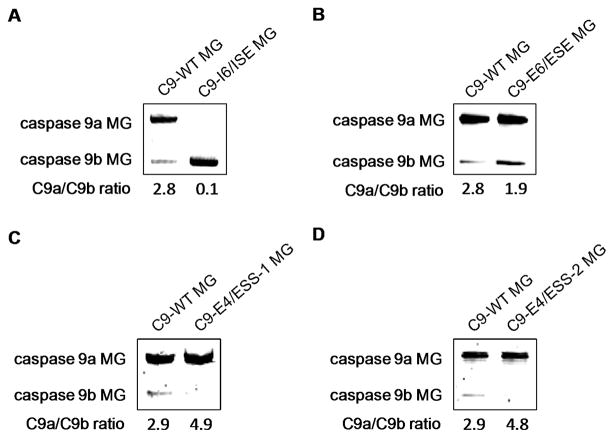
Figure 1. Identification of novel RNA cis-elements that regulate the inclusion of exon 3,4,5,6 cassette of caspase 9.
A–D) RT-PCR analysis of caspase 9 minigene-derived transcripts from A549s transfected with the WT caspase 9 minigene and A) C9-I6/ISE, B) C9-E6/ESE, C) C9-E4/ESS-1, or D) C9-E4/ESS-2. The caspase 9a/9b mRNA ratio was determined by densitometric analysis of RT-PCR fragments. Data represent 3 separate determinations on 3 separate occasions.
Mutation of the Caspase 9 Minigene
To mutate the 13 purine-rich sequences in the caspase 9 minigene (21) selected for replacement mutagenesis (following manufacturers protocol), the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used. All mutations were verified by DNA sequencing.
Plasmid Transfection
Cell lines were transfected with caspase 9 minigene constructs and additional plasmid vectors using Effectene® Transfection Reagent (Qiagen Inc., Valencia, CA) following the manufacturers protocol. Briefly, cell lines were plated at ~ 70% confluency in 6-well tissue culture dishes. Cells were transfected with plasmid constructs (2 μg total) followed by 24 h standard incubation. After 24 h standard incubation, cells were examined for minigene expression by quantitative/competitive reverse transcription RT-PCR. This procedure routinely gave >50% transfection efficiency for A549 cells. The generation of A549 cell lines stably expressing either caspase 9b cDNA or caspase 9b shRNA was previously described (20, 21).
Affinity Purification Assay
The following sequences were created for affinity purification assays: 5′-biotinylated-tagged RNA oligonucleotide for C9-I6/ISE: (5′Bio-CUC UGG GUG GGU CUG-3′); 5′-biotinylated-tagged RNA oligonucleotide for C9-I6/ISE MUT: (5′Bio-CUC UCG CUG CGU CUG-3′); 5′-biotinylated-tagged RNA oligonucleotide for non-specific competitor: Bio-NSC-2: (5′Bio-AGA GCU AGU CCU GU-3′); non-labeled specific competitor oligonucleotide: SC: (5′-CUG GGU GGG UC-3′); and non-labeled non-specific competitor oligonucleotide: NSC-1: (5′-GAA UUC GCA CGU UA-3′), (Dharmacon Inc., Lafayette, CO). To pre-clear, streptavidin-agarose beads (Invitrogen™, Carlsbad, CA) in the amount of 20 μl were placed in buffer containing 50 μg A549 nuclear extract, 40 U RNASIN, 11.3 μg tRNAs, 10 mM HEPES, 5 mM DTT, 120 mM KCl, 3 mM MgCl2, and 5% glycerol. Binding reactions were placed in 4°C for 2 hours with gentle agitation, and reaction mixtures were collected by centrifugation. Reaction mixtures were then incubated with 400 ng Bio-C9-I6/ISE, Bio-C9-I6/ISE MUT, or Bio-NSC-2 for 5 min following an addition of 100-fold molar excess of NSC or SC oligos and incubated on ice for 30 min. New streptavidin-agarose beads were then added to RNA-binding reactions and incubated at 4°C for 2 hours with gentle agitation. RNA:protein-binding reactions were washed with buffer containing 100 mM KCl, 20 mM Tris-HCl at pH 7.5, and 0.2 mM EDTA. After washing, the complex was pelleted by centrifugation. The pellet was resuspended in laemmli buffer, dry boiled for 10 min, and subjected to SDS-PAGE/western immunoblotting (anti-SRSF1 antibody) analysis.
Immunodepletion Assay
For immunodepletion, Lamin-B2 (Abcam, Cambridge, MA), SRSF1 (Invitrogen™, Carlsbad, CA), or mouse IgG (Sigma, St. Louis, MO) was first coupled to protein-G-sepharose beads in lysis buffer (150 mM NaCl, 2 mM EDTA, 0.05% NP-40, 20 mM HEPES, and 5 mg/mL BSA) overnight at 4°C with little agitation. The protein-G coupled antibody beads were washed three times with lysis buffer and subsequently incubated with 100μg A549 nuclear extract in lysis buffer overnight at 4°C. After centrifugation, supernatants were either resolved by SDS-PAGE and analyzed by western blot analysis or utilized in EMSAs.
WST assay
A549 cells (1.0 × 105) were plated into each well of a 24-wellplate in a 1.0 ml media. After 24 h at standard incubator conditions (humidified atmosphere, 95% air, 5% CO2, 37°C), the cells were transfected with ASRO/siRNA as previously described (21). After 24 h of ASRO/siRNA treatment, cells were treated with DNR at its IC50 concentration (750 nM) for 48 h. After48 h incubation WST-1 reagent (Roche Applied Science, Indianapolis, IN) was added to these cells (final 1:10 dilution). The cells were then incubated along with the reagent for 30 min. The plates were then read against blank using a microplate (ELISA) reader at 420–480nm with the reference wavelength >600 nm.
Clonogenic assay
Clonogenic assays were performed with the indicated chemotherapeutic agents as previously described (20).
Apoptosis assay
A549 cells (2.0 × 105) were plated into each well of a 6-wellplate in 3.0 ml media. After 24 h at standard incubator conditions (humidified atmosphere, 95% air, 5% CO2, 37°C), the cells were transfected with ASRO following the manufacturer’s (Invitrogen Life Technologies, Carlsbad, CA) protocol (briefly described above). Cells were treated with the appropriate concentration of daunorubicin (150 nM) (Sigma, St. Louis, MD) (nine hours post-transfection) for 48 h. After the drug treatment cells were stained with Hoechst stain (10 μg/ml) (Sigma, St. Louis, MD) at room temperature for 10 min. The cells were later observed under fluorescent microscope (20X). Five fields were observed and in each field total numbers of cells and apoptotic cells were counted. Percentage apoptotic cells were counted over total number of cells.
Data Analysis
Comparison of the effects of various treatments was done using a 2-tailed, unpaired student t-test when appropriate. Differences with a P < 0.05 were considered statistically significant. Experiments shown are the means of multiple individual points (±SEM). Median dose-effect isobologram analyses to determine the synergism of drug interaction were done according to the methods of T-C. Chou and P. Talalay using the CalcuSyn program for Windows (Biosoft)(23). Cells were treated with agents at a fixed concentration dose. A combination index value of <1.00 indicates synergy of interaction between the drugs, a value of 1.00 indicates additivity, and a value of >1.00 indicates antagonism of action between the agents.
RESULTS
Identification of enhancing RNA cis-elements for the inclusion of the exon 3,4,5,6 cassette into caspase 9 mRNA
Previously, our laboratory reported that SRSF1 was a required enhancer for the inclusion of the exon 3,4,5,6 cassette of caspase 9 pre-mRNA (15). Many studies over two decades have shown that SRSF1 interacts with specific RNA cis-elements to mediate enhanced inclusion of exons. Therefore, the sequence (exonic and intronic) of the exon 3,4,5,6 cassette of caspase 9 was examined for purine-rich sequences, repeating purine-rich sequences, and consensus SRSF1 binding sites from the literature and those identified by ESE Finder 2.0 (24) and Splicing Rainbow (25). Analysis of the sequence revealed thirteen possible interaction sites for SRSF1 (Table I). To determine whether these RNA cis-elements played a role in regulating the alternative splicing of caspase 9 pre-mRNA, a functional minigene for caspase 9 splicing was utilized as previously described (21). The possible RNA cis-elements were modified in the minigene construct by replacement mutagenesis (Table I). Importantly, these mutations did not affect the distances between the possible RNA cis-elements and exonic splice sites, thus reducing the possibility of artifactual results. Of the thirteen possible RNA cis-elements, five mutations affected the caspase 9a/9b ratio (Table I and Figure 1). We have previously reported that mutation of the RNA cis-element, C9-E3/ESS, resulted in a dramatic increase in the caspase 9a/9b mRNA ratio and was demonstrated to be an exonic splicing silencer in exon 3 of caspase 9 pre-mRNA (Table I) (21). Therefore, disregarding C9-E3/ESS, four additional RNA cis-elements were identified. Specifically, two enhancer elements, one located in intron 6 termed C9-I6/ISE (Table I and Figure 1A) and the other located in exon 6 termed C9-E6/ESE (Table I and Figure 1B), and two additional silencer elements, both located in exon 4 and termed C9-E4/ESS-1 (Table I and Figure 1C) and C9-E4/ESS-2 (Table I and Figure 1D), were identified. Importantly, mutation of C9-I6/ISE induced a complete inversion of the caspase 9a/9b ratio from 2.8 ± 0.1 to 0.1 ± 0.0 (n = 6; P < 0.01; Figure 1A) whereas mutation of C9-E6/ESE decreased the caspase 9a/9b ratio from 2.8 ± 0.1 to 1.9 ± 0.1 (n = 4; P < 0.01; Figure 1B). Furthermore, mutation of C9-E4/ESS-1 induced an increase in the caspase 9a/9b ratio from 2.9 ± 0.1 to 4.9 ± 0.2 (n = 4; P < 0.01; Figure 1C) in a similar manner as mutation of C9-E4/ESS-2, which increased the caspase 9a/9b ratio from 2.9 ± 0.1 to 4.8 ± 0.1 (n = 4; P < 0.01; Figure 1D). The caspase 9 minigene mutants demonstrating no effect on the caspase 9a/9b ratio are also represented (Supplemental Figure 1). Therefore, these data demonstrate novel RNA cis-elements located in caspase 9 pre-mRNA required for the inclusion/exclusion of the exon 3,4,5,6 cassette. As the goal of this study was to define enhancing RNA cis-elements, the remainder of this report focuses on these RNA cis-elements.
Table I. Schematic representation of potential regulatory cis-elements involved in regulating the alternative splicing of caspase 9.
Depicted are the location, wild-type sequences, and corresponding mutagenic sequences (Sites 1–13) in the caspase 9 minigene construct of each of the potential regulatory cis-elements. Sites boxed represent RNA cis-elements that, when mutated, affected the caspase 9a/9b ratio. The sequence denoted with an “*” and termed C9-E3/ESS also affected the caspase 9a/9b ratio and was previously demonstrated to be an exonic splicing silencer in exon 3 of caspase 9 pre-mRNA(21).
| Site | Location | Designation | WT Sequence | Mutated Sequence |
|---|---|---|---|---|
| 1 | Exon 3 | C9-E3/ESS | GAGAGTTTGAGGGGAAAT* | GAGAGTTTGCTACTAAAT |
| 2 | Exon 4 | TGGTGGAGGT | TGCTCGACGT | |
| 3 | Intron 4 | TGGAGGGAGAC | TCGACGCACAC | |
| 4 | Intron 4 | AGGGTGGGGGG | ACGCTCGCGCG | |
| 5 | Intron 4 | CAGTGGGTGGGGAAG | CAGTCGCTGCGCAAC | |
| 6 | Intron 4 | CATGGGAGGTAGGAC | CATCCGCTTTAGGAC | |
| 7 | Intron 5 | TGGGAGAGGGAGGGCAG | TGGGAGACGCTTTGCAG | |
| 8 | Exon 6 | CAGCCTGGGAGGG | CACCCTCGCAGCG | |
|
| ||||
| 9 | Intron 6 | C9-I6/ISE | TGGGTGGGT | TCGCTGCGT |
|
| ||||
| 10 | Intron 6 | CTGGTGGGGAGGGA | CTCGTCGCCAGCGA | |
|
| ||||
| 11 | Exon 4 | C9-E4/ESS-1 | CCTGAGCATGGAGC | CCTCACCCTCTCGC |
| 12 | Exon 4 | C9-E4/ESS-2 | TGAAGGGCG | TCCACCTCG |
| 13 | Exon 6 | C9-E6/ESE | AAGCCCAAGC | CCTCCCATCC |
SRSF1 specifically interacts with C9-I6/ISE
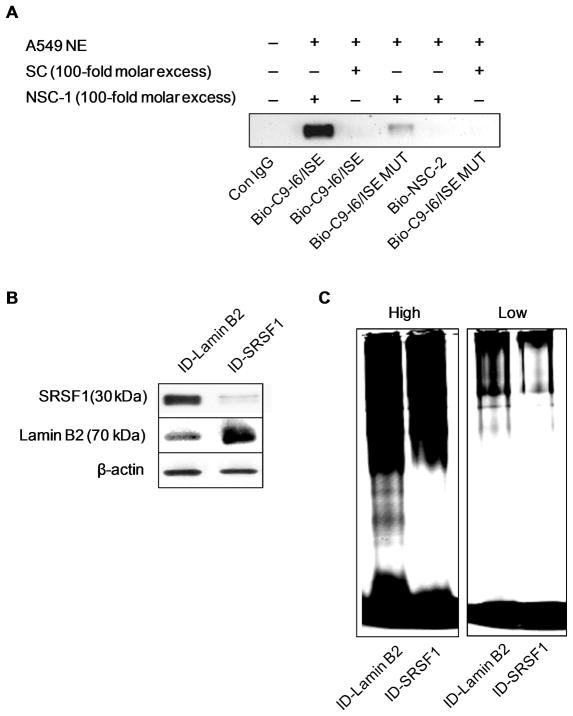
Previously, our laboratory reported that downregulation of SRSF1 by siRNA induced a dramatic reduction in the endogenous caspase 9a/9b mRNA ratio (1.67 ± 0.11 to 0.56 ± 0.08 (* P value < 0.005)) (15). Since mutation of the C9-I6/ISE enhancer element resulted in the more dramatic decrease in the caspase 9a/9b ratio and produced a comparable effect as siRNA downregulation of SRSF1 in A549 cells, we hypothesized that this C9-I6/ISE specifically interacted with SRSF1. To explore this hypothesis, C9-I6/ISE was subjected to streptavidin-agarose affinity purification with A549 nuclear extract. Biotinylated RNA oligonucleotide corresponding to C9-I6/ISE (Bio-I6/ISE), biotinylated RNA oligonucleotide corresponding to a mutant C9-I6/ISE (Bio-I6/ISE MUT), biotinylated RNA oligonucleotide corresponding to a non-specific competitor sequence (Bio-NSC-2), non-labeled specific competitor (SC) and non-labeled non-specific competitor (NSC-1) were utilized for this affinity assay. Of note, the biotinylated RNA oligonucleotide corresponding to mutant C9-I6/ISE possessed the same mutations as the mutant C9-I6/ISE minigene utilized in this study. As predicted, SRSF1 specifically interacted with the Bio-ISE-1 cis-element whereas no interaction of SRSF1 was observed with Bio-NSC-2. Importantly, the mutant C9-I6/ISE (Bio-I6/ISE) RNA oligonucleotide (Figure 2A) demonstrated a 89% reduction in the interaction with SRSF1. Furthermore, the addition of unlabeled SC completely abolished the observed SRSF1/ISE-1 interaction. Thus, SRSF1 specifically interacts with C9-I6/ISE in vitro.
Figure 2. SRSF1 specifically interacts with the C9-I6/ISE element.
A) A 5′ biotinylated C9-I6/ISE oligonucleotide (Bio-C9-I6/ISE), a 5′ biotinylated C9-I6/ISE mutant oligonucleotide (Bio-C9-I6/ISE MUT) or 5′ biotinylated non-specific competitor oligonucleotide (Bio-NSC-2) (negative control) were incubated in the presence of nuclear extract from A549 cells or IgG (control) and subjected to western immunoblot (anti-SRSF1 monoclonal antibody) analysis. Unlabeled competitor RNA oligonucleotides (e.g. NSC-1, SC) were also added to the reactions as indicated. The data are representative of 4 separate determinations on 2 separate occasions. B) Western blot analysis of immunodepleted A549 nuclear extracts (supernatants) to confirm Lamin B2 and SRSF1-immunodepletions. C) Antibodies to SRSF1 and Lamin B2 were coupled to protein-G-sepharose beads (see Materials and Methods). Protein-G coupled antibody beads were added to A549 nuclear extract to allow immunodepletion. Fluorescein-tagged C9-I6/ISE oligonucleotide was incubated in the presence of SRSF1-immunodepleted nuclear extracts or Lamin B2-immunodepleted nuclear extracts followed by electromoblilty shift assays. High = High intensity laser scan and Low = low intensity laser scan. The data are representative of 7 separate determinations on 3 separate occasions.
The specificity of this interaction was further demonstrated by immunodepletion (ID) of SRSF1 from A549 nuclear extracts (Figure 2B). Immunodepletion analysis of SRSF1 demonstrated a loss of a specific C9-I6/ISE protein complex in contrast to ID-nuclear lamin B2 extract (Figure 2C). The culmination of these data (e.g. competition/affinity purification and immunodepletion/affinity purification) demonstrates that SRSF1 specifically binds to C9-I6/ISE.
The regulation of the alternative splicing of caspase 9 pre-mRNA by SRSF1 translates to multiple cell lines and is via C9-I6/ISE
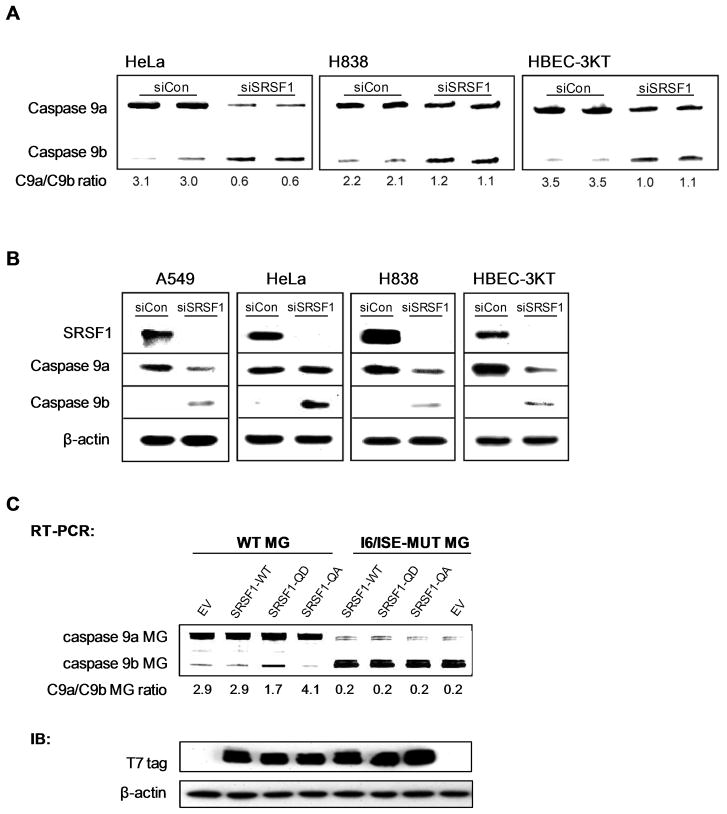
Based on previous findings from our laboratory, downregulation of SRSF1 in A549 cells induced an increase in the caspase-9b splice variant at the expense of caspase-9a, thereby inducing a decrease in the caspase 9a/9b mRNA ratio (15). We now demonstrate that the change in the caspase 9a/9b ratio translates to the protein level (Figure 3B). Additionally we demonstrate translatability of this mechanism into multiple cell lines (Figure 3A,B). Specifically, HeLa cells, H838 cells, and the non-transformed cell line, HBEC-3KT cells, were subjected to downregulation of SRSF1 utilizing siRNA. As with the A549 cells, downregulation of SRSF1 resulted in a dramatic decrease in the caspase 9a/b ratio in all cell lines examined (HeLa cells from 3.1 ± 0.12 to 0.6 ± 0.08 (n = 4; P < 0.01); H838 cells from 2.2 ± 0.11 to 1.2 ± 0.09 (n = 4; P < 0.01); and HBECK-3KT cells from 3.5 ± 0.07 to 1.1 ± 0.09 (n = 4; P < 0.01)) (Figure 3A). As with our previous data, this effect on the caspase 9a/9b mRNA ratio translated to the protein level (Figure 3B). Therefore, the regulation of the alternative splicing of caspase 9 by SRSF1 is a specific mechanism that translates to multiple cell lines. Of note, we have previously shown that the effect of SRSF1 on this mechanism was not due to a generalized effect on cellular RNA splicing (15).
Figure 3. Downregulation of SRSF1 decreases the caspase 9a/9b splice ratio in multiple cell lines.
A) The indicated cell lines were transfected with 100 nM control siRNA or 100 nM SRSF1 SMARTpool siRNA for 48 h. Total RNA was isolated and analyzed by competitive/quantitative RT-PCR for caspase 9 splice variants(15). The C9a/C9b mRNA ratio was determined by densitometric analysis of RT-PCR fragments. B) Simultaneously, total protein lysates were also produced, subjected to SDS-PAGE analysis, and immunoblotted for SRSF1, caspase 9a, caspase 9b, and β-actin. C) RT-PCR analysis of Casp9 minigene-derived transcripts from A549s transfected with the wild-type caspase 9 minigene (WT MG) or mutant C9-I6/ISE caspase 9 minigene (I6/ISE MUT MG) (1 μg) in conjunction with either wild-type SRSF1 (SRSF1-WT) (1 μg), mutant SRSF1-QD (SRSF1-QD) (1 μg), or mutant SRSF1-QA (SRSF1-QA) (1 μg). All ratios of caspase 9a/9b were determined by densitometric analysis of RT-PCR fragments. Data are expressed as means ± S.E. The protein expression levels of SRSF1-WT, SRSF1-QD, and SRSF1-QA were also assayed by western immuoblot analysis using an anti-T7 tag antibody.
Previous findings from our laboratory have found that ceramide treatment led to the dephosphorylation of SR proteins, and SRSF1 was required for ceramide to affect the alternative splicing of caspase 9 pre-mRNA (15). Furthermore, our laboratory has recently demonstrated that the phosphorylation status of SRSF1 on serine199, 201, 227, and 234 regulates the alternative splicing of caspase 9 pre-mRNA in A549 cells downstream of Akt (20). Specifically, ectopic expression of the SRSF1 quadruple (SRSF1-QD) mutant, which harbors serine to aspartic acid mutations at serine residues 199, 201, 227 and 234, induced a significant decrease in the caspase 9a/9b ratio (20). Conversely, ectopic expression of the SRSF1 quadruple (SRSF1-QA) mutant, which harbors serine to alanine mutations at serine residues 199, 201, 227 and 234, induced a significant increase in the caspase 9a/9b ratio (20). Based on these results, we hypothesized that mutated C9-I6/ISE co-transfected with either wild-type SRSF1 or SRSF1-QD would abolish any observed effect on the caspase 9a/9b mRNA ratio. To test this hypothesis, A549 cells were transfected with the mutated C9-I6/ISE minigene in conjunction with SRSF1-WT, SRSF1-QD, or SRSF1-QA. Neither the wild-type nor mutated SRSF1 were able to affect the minigene-derived caspase 9a/9b mRNA ratio when co-transfected with the mutated C9-I6/ISE, in contrast to co-transfection with wild-type caspase 9 miniene (Figure 3C). These data support the hypothesis that SRSF1 regulates the alternative splicing of caspase 9 via the strong, intronic splicing enhancer, C9/I6-ISE.
Clonogenic survival and chemotherapy sensitivity is altered in cells by direct manipulation of the alternative splicing of caspase 9
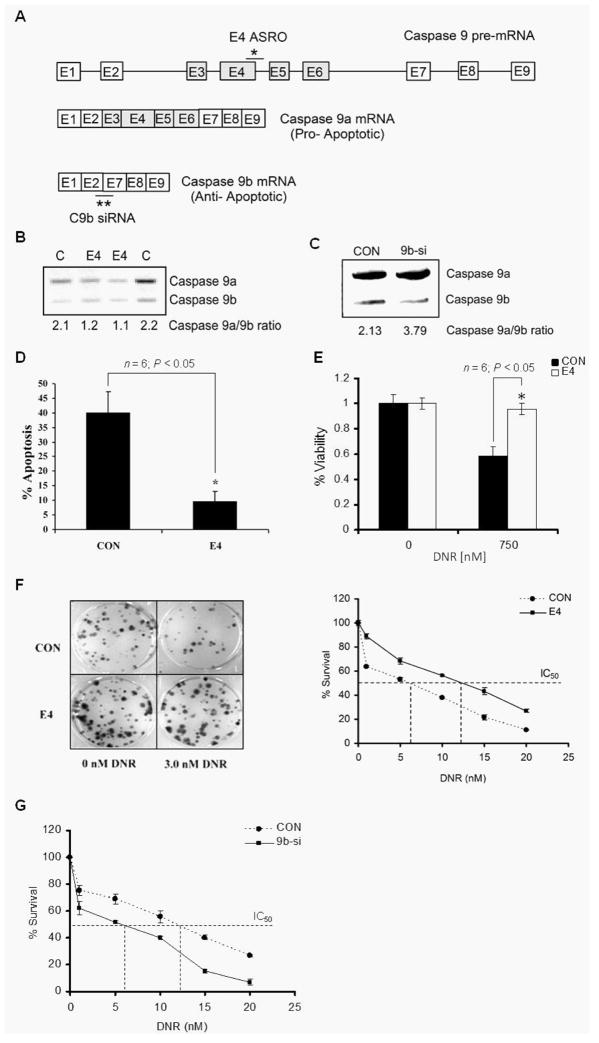
NSCLC cells in general demonstrate high resistance to chemotherapy (26) due to a lack of caspase activation (27, 28), and we have reported that the ratio of caspase 9 splice variants (C9a/C9b) is dysregulated in NSCLC tumors and lung cancer cell lines (also see Supplemental Figure 2) (20, 21). Furthermore, our laboratory reported that the clinically relevant therapeutic, erlotinib, and the lipid second messenger, ceramide, induced an increase in the caspase 9a/9b mRNA ratio at concentrations below the IC50 of either agent via a pathway involving the phospho-state of SRSF1 and likely via the RNA splicing enhancers reported here (15, 20). As erlotinib and de novo ceramide will synergize with various chemotherapies such as adriamycin-derivatives in vitro (14, 29), we hypothesized that the caspase 9a/9b ratio may be significant in chemotherapeutic sensitivity induced by erlotinib towards chemotherapies that do not induce de novo ceramide (e.g. daunorubicin). In order to study the role of alternative splicing of caspase 9 in chemotherapy sensitivity of lung carcinoma cells, anti-sense RNA oligonucleotides (ASROs) and small interfering RNA (siRNA) were utilized to directly manipulate the alternative splicing of caspase 9 as we have previously described (21). Specifically, an ASRO designed to hybridize to 5′SS of exon 4 (E4 ASRO) was effective in reducing the caspase 9a/9b splice ratio (Figure 4A,B). Specifically, E4 ASRO shifts the caspase 9 splice ratio to favor caspase 9b with a 57% reduction in the caspase 9a/9b mRNA ratio (Figure 4B). To achieve an increase in the ratio of C9a/C9b mRNA, siRNA to specifically downregulate caspase 9b (9b-si) was developed (Figure 4C). RT-PCR analysis revealed that transfection of A549s with 9b-siRNA resulted in an increase in the caspase 9a/9b ratio (Figure 4C).
Figure 4. The E4-ASRO treatment increases the survival and chemotherapy resistance of A549 cells.
A) Schematic representation of the alternative splicing mechanism of caspase 9 and binding sites for the E4 ASRO and caspase 9b siRNA utilized in these studies. * = binding site for E4 ASRO and ** = binding site for caspase 9b siRNA. B) RT-PCR analysis of caspase 9 splice variants from A549s transfected with control ASRO (CON) or E4 ASRO (E4) (400nM) and C) control siRNA or caspase 9b siRNA. All ratios of caspase 9a/9b were determined by densitometric analysis of RT-PCR fragments. Data are expressed as means ± S.E. D) A549 cells were transfected with the indicated ASROs (400 nM). Twenty-four hours following ASRO transfection, cells were treated with or without DNR (150 nM) for 48h. After drug treatment, apoptosis assays were performed as described in Materials and Methods section. The graph depicts percentage apoptosis for cells treated ± DNR. Data are expressed as mean ± SE, * = P<0.05. E) The viability of A549 cells transfected with the indicated ASROs was monitored as mitochondrial function using WST assays as described in materials and methods. A549 cells were treated with DNR (150 nM) or sham control for 48h. Data are expressed as mean ± SE and are representative of four separate determinations on two separate occasions. F & G) A549 cells were transfected with the indicated ASROs or siRNAs. Twenty-four hours post-transfection, cells were replated (500 cells/well of a 6-well dish) and treated with DNR (0.0 nM to 20.0 nM) for 48h. Cells were then allowed to form colonies for 10 days. The colonies were scored and percentage survival was calculated with respect to the control treated cells. Panel F also includes representative photographs of stained tissue culture plates following treatment with 3 nM DNR to demonstrate the increased survival imparted by the E4 ASRO in the presence of DNR. Data are expressed as mean ±SE and are representative of three separate determinations on four separate occasions.
In order to examine the role that direct manipulation of the alternative splicing of caspase 9 on the sensitivity of chemothereutic agents for NSCLC cells that do not generate appreciable amounts of de novo ceramide, three clinically relevant agents used in clinical combinations therapies for cancer were chosen, Daunorubicin (DNR), Cisplatinum (Cp) and Paclitaxel (Pac). These chemotherapies were also chosen because none of these agents had an appreciable effect on the caspase 9a/9b mRNA ratio individually (Supplemental Figure 3). Both, DNR and Cp, are similar in that they are DNA damaging agents with DNR being an intercalating agent versus Cp being an alkylating agent. Pac is the traditional cytotoxic agent targeting spindle fibers, and thus blocking cell division. Lowering the caspase 9a/9b ratio using the E4 ASRO induced significant resistance to daunorubicin (DNR)-induced apoptosis (n = 6; P < 0.05; Figure 4D) as well as suppression of cell viability (n = 6; P < 0.05; Figure 4E) and cell survival (increased the IC50 from 6.0 nM to 13.0 nM) (Figure 4F). This effect was also observed for Cp (Supplemental Figure 4A) and to a lesser, but significant extent, for Pac treatment (Supplemental Figure 4B).
Based on the above findings, we also hypothesized that an increase in the caspase 9a/9b ratio would produce the opposite effect and sensitize NSCLC cells to the same chemotherapeutic agents. More importantly, 9b-siRNA treatment of A549 cells demonstrated a reduced DNR IC50 of 6.0 nM ± 0.7 compared to cells transfected with control siRNA (12.5 nM) as measured by the clonogenic survival assays (Figure 4G). In addition, 9b-siRNA treatment also reduced the IC50 for Cp (from 0.25 μg/ml to 0.08 μg/ml) (Supplemental Figure 5A). A similar reduction in the IC50 of Pac was also observed in A549 cells, although to a slightly lesser extent as the DNA-damaging agents (Supplemental Figure 5B). Importantly, these effects translated to more than one NSCLC cell line as similar increases in the drug de-sensitization were observed in H2030 cells when transfected with E4 ASRO and treated with DNR and Pac (Supplemental Figure 6). Similarly, H2030 cells transfected with 9b-siRNA also resulted in a reduction of the IC50 as compared to Con-siRNA-treated cells (Supplemental Figure 7). Overall, these data demonstrate that the ratio of caspase 9a/9b mRNA is an indicator of the resistance/sensitivity of NSCLC cells to broad-types of chemotherapeutic agents.
Modulation of the alternative splicing of caspase 9 regulates the synergistic effects of Daunorubicin and Erlotinib
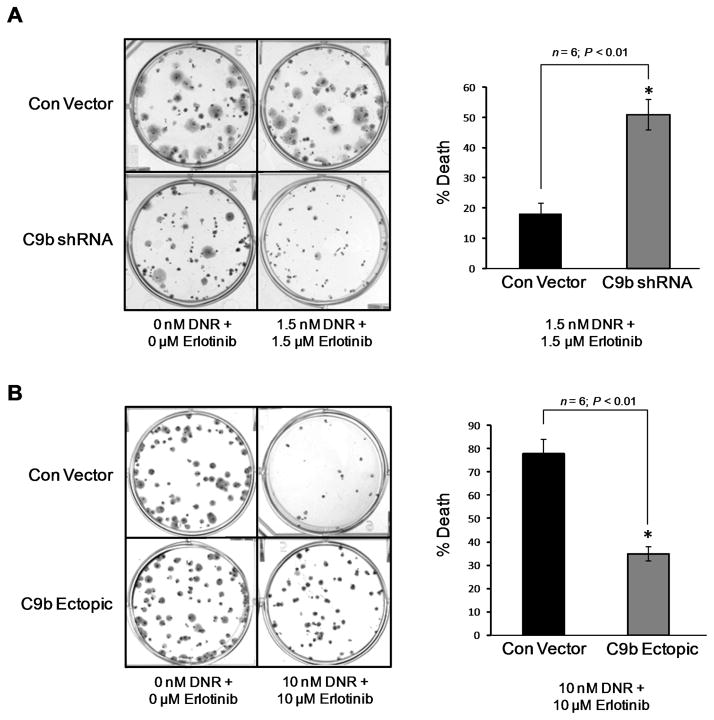
Since modulation of the caspase 9a/9b mRNA ratio sensitized NSCLC cells to DNA damaging agents currently utilized in combination therapies for NSCLC patients, we next tested our overall hypothesis that erlotinib sensitized NSCLC cells to adriamycin-derivatives via manipulation of the caspase 9a/9b ratio. Specifically, we examined if genetic manipulation of the alternative splicing of caspase 9 would modulate the synergistic effects of erlotinib and DNR in low and high dose combinations. As demonstrated by colony formation assays, erlotinib and DNR interacted in a synergistic manner to inhibit the colony formation of A549 cell as indicated by CI values of less than 1.00 (Table II) and a further reduction of cell survival was observed when the NSCLC cells were treated with sub-IC50 concentrations of these drugs (Figure 5A,B). Importantly and validating our hypothesis, the synergism of erlotinib and DNR in combination at low concentrations (sub-IC50 values) was dramatically diminished in NSCLC cells expressing C9b-shRNA and presenting with a low caspase 9a/9b ratio (Table II and Figure 5A). Low ectopic expression of caspase 9b (a 2-fold decrease in the caspase 9a/9b ratio) also dramatically inhibited the synergism of DNR and erlotinib irrespective of dose (Table II and Figure 5B). Therefore, these data support the hypothesis that erlotinib acts to synergize with DNA-damaging agents, such as DNR, through its ability to modulate the alternative splicing of caspase 9.
Table II. Synergistic effects of Daunorubicin and Erlotinib in A549 cells.
A549 cells expressing vector control, ectopic Casp9b cDNA or Casp9b shRNA were treated 2 hours after plating as single cells (150–400 cells/well) with vehicle (DMSO), daunorubicin (1.5–20 nM), erlotinib (1.5–20 μM), or with both drugs combined, as indicated at a fixed concentration ratio, to perform median dose-effect analyses for the determination of synergy. After drug exposure the medium was changed, and cells were cultured in drug-free media for an additional 10 to 14 days. Cells were fixed, stained with crystal violet, and counted. Colony formation data were entered into the Calcusyn program, and CI values were determined. A CI value of less than 1.00 indicates synergy; a CI value of greater than 1.00 indicates antagonism.
| Cell Line | Daunorubicin | Erlotinib | CI |
|---|---|---|---|
| nM | μM | ||
| A549 Con Vector | 1.5 | 1.5 | 0.42 |
| 5 | 5 | 0.28 | |
| 10 | 10 | 0.27 | |
| 20 | 20 | - | |
| A549 C9b shRNA | 1.5 | 1.5 | 0.91 |
| 5 | 5 | 0.29 | |
| A549 C9b Ectopic | 10 | 10 | 0.57 |
| 20 | 20 | 0.50 |
Figure 5. The alternative splicing of caspase 9 regulates the synergistic effects of erlotinib and DNR in combination.
A,B) Clonogenic survival of A549 cells expressing vector control, ectopic caspase 9b cDNA or Casp9b shRNA. Depicted are representative photographs of stained tissue culture plates of the indicated cells treated with DMS vehicle control or DNR and Erlotinib in combination. Bar graphs represent the percent death of the indicated cell line following treatment with DNR and Erlotinib in combination as determined by the number of colonies counted in comparison to DMSO control treated (n = 6; P < 0.01). Data are mean ± S.E. represented as percent control.
DISCUSSION
The importance of the presented study lies in several key findings. First, our mechanistic knowledge of how the alternative splicing of caspase 9 is regulated has been expanded by our identification of a novel RNA cis-element via which SRSF1 (ASF/SF2) enhances the inclusion of the exon 3,4,5,6 cassette. Furthermore, SRSF1 was shown to specifically interact with this RNA cis-element, and regulate the alternative splicing of caspase 9 via this novel RNA cis-element. Second, we demonstrate that the alternative splicing of caspase 9 is a relevant therapeutic target as demonstrated by direct manipulation of this splicing cascade having significant effect on the sensitivity NSCLC cells to clinically relevant chemotherapeutics. Lastly, one of the major findings of the report is our data showing that the synergism of erlotinib combination therapy is in part via modulation of the alternative splicing of caspase 9.
In regards to the RNA cis-element that specifically interacts with SRSF1, a purine-rich RNA cis-element was identified in intron 6, 24 bp downstream of the 5′ splice site of exon 6. The position of this RNA cis-element makes logical sense as it is localized near a juxta-exon for the large exon 3,4,5,6 cassette. We had initially anticipated that regulatory elements for the exon 3,4,5,6 cassette would be positioned in or near exon 3 and 6, and indeed, this study showed that an intronic splicing enhancer (now termed C9-I6/ISE) was immediately downstream of exon 6. There is still a possibility that C9-I6/ISE is only one of several required splicing enhancers for SRSF1 within the exon 3,4,5,6 cassette, and indeed, mutation of one other possible RNA cis-element for SRSF1 interaction in exon 6 had a significant, albeit smaller effect on the caspase 9a/9b mRNA ratio. Still, C9-I6/ISE is the strongest enhancer element identified to date, and mutation of this RNA cis-element produced the same results as downregulation of SRSF1 by siRNA (15). This is another key indicator that C9-I6/ISE is the major enhancer element for inclusion of the 3,4,5,6 cassette. Furthermore, our studies demonstrate that SRSF1 specifically binds C9-I6/ISE, and wild-type SRSF1 as well as the phospho-mutants of SRSF1 could not affect the ratio of minigene caspase 9a/9b mRNA when C9-I6/ISE was mutated. Therefore, the culmination of these date demonstrate that C9-I6/ISE is the major splicing enhancer for the inclusion of the exon 3,4,5,6 cassette via specific interaction with SRSF1.
As more data is accumulated on the regulation of the exon 3,4,5,6 cassette of caspase 9, a complex and novel mechanism is beginning to emerge. For example, we recently demonstrated that an exonic splicing silencer (ESS) was located in exon 3 termed C9/E3-ESS. Indeed, we further showed that the RNA trans-factor, hnRNP L, bound this sequence in non-small cell lung cancer (NSCLC) cells to repress the inclusion of the exon 3,4,5,6 cassette into the mature transcript. In this same report, we addressed the possibility that this was an oversimplification, and indeed, we identified two additional ESS sequences in the exonic cassette, both in exon 4. These sequence share homology to the C9/E3-ESS sequence suggesting that hnRNP L may also associate with these sequences to repress the entirety of the cassette. This repression by hnRNP L activated by phosphorylation along with the inactivation of SRp30a by phosphorylation on ser199, 201, 227, 234 bound to exon 6 is a logical mechanism to rapidly regulate the alternative splicing of caspase 9. Overall, the additional regulatory RNA cis-elements identified in these studies suggest a complex mechanism involving a number of RNA cis-elements along the exon 3,4,5,6 cassette required for both inclusion and exclusion (repression). For example, repression of the exonic cassette requires not one, but a number of exonic splicing silencers bound to inhibitory RNA trans-factors (e.g. phosphorylated hnRNP L). Hence, the default splicing paradigm is inclusion of the exonic cassette as long as SRSF1 is expressed and not phosphorylated on ser199, 201, 227, 234, peripheral phosphorylation sites to the standard RS domain of the RNA trans-factor. Taken together with our recent reports on SRSF1 and hnRNP L in this alternative splicing mechanism, these new results suggest that RNA trans-factors not only play constitutive roles in RNA splicing, but also activated roles in modulating alternative splicing induced by post-translational modifications in contrast to simple expression gradients.
In regards to biology, the role of SRSF1 in this mechanism is translatable to more than one NSCLC cell line as well as other cell types. Our laboratory previously published that downregulation of SRSF1 by siRNA induced a dramatic decrease in the caspase 9a/9b mRNA ratio of A549 cells (15). Essentially, the inclusion of the exon 3,4,5,6 cassette was abolished into the mature transcript. Here, we expanded this early study and demonstrated that downregulation of SRSF1 in the NSCLC cell lines, H2030 and H838 cells, the cervical cancer cell line, HeLa cells, and in the non-transformed immortalized human bronchial epithelial cell line, HBEC-3 cells, also lowered the caspase 9a/9b mRNA ratio. Whereas the effect of SRSF1 in the NSCLC cell lines and HBEC-3 cells was not unexpected, the result in HeLa cells was surprising due to a previous report by Krainer and co-workers (30). Specifically, downregulation of SRSF1 was reported to have no effect on the alternative splicing of caspase 9 (30). Unfortunately, our findings are not in agreement. This may be due to the extent of downregulating SRSF1 in HeLa cells or other technical differences. During the preparation of this report, an additional study published by Silver and co-workers, which also reported that the downregulation of SRSF1 affected the alternative splicing of caspase 9 in HeLa cells, but in this case, the conclusion was an increase in the caspase 9a/9b mRNA ratio (31). This difference is likely due to this report examining only the levels of caspase 9a for their conclusions (31). Regardless of these contrasting findings between research groups, all of the data reported in this manuscript demonstrate that SRSF1 is a required RNA trans-factor for the inclusion of the 3,4,5,6 exon cassette of caspase 9 pre-mRNA in all cell lines examined to date, which is further supported by the corroborating effect observed by mutation of the SRSF1 binding site in the caspase 9 minigene (e.g. also lowered the caspase 9a/9b ratio).
Also in this study, we continued to address the biological consequences of a dysregulated caspase 9a/caspase9b ratio, and demonstrate that this distal splicing mechanism was related to the sensitivity of NSCLC cells to additional types of chemotherapies beyond EGFR inhibitors like erlotinib. For example, C9b siRNA induced a 2.7-fold increase in the caspase 9a/9b mRNA ratio, which translated to a significant decrease in the IC50 of DNA-damaging agents. Thus, the ratio of caspase 9a/9b mRNA may be a prognostic/predictive indicator of chemotherapy response to DNA-damaging agents. On the other hand, the ratio of caspase 9a/9b mRNA may not function as an indicator of response to classical cytotoxic agents such as paclitaxel. As this study shows, the shift in IC50 after either E4 treatment or 9b-si treatment in the case of Paclitaxel was significant, but not as nearly as dramatic when compared to the effects on the IC50 of DNR and Cp. This could be attributed to the mode of action for Pac versus the DNA damaging agents. For example, DNR sensitivity is highly linked to apoptosis capacity, thus manipulation of caspase 9 activity dramatically affecting the IC50 of this agent is logical. Regardless of the limited effect of the manipulating the caspase 9a/9b ratio on Pac, the effect on the IC50 of these chemotherapeutic agents translated to more than one NSCLC cell line suggesting a broad-based mechanism. Therefore, this study demonstrates that a single mechanism in these cells (e.g. the alternative splicing of caspase 9) has a large impact on the sensitivity of non-small cell lung cancer cells to several chemotherapeutic agents.
These findings have direct implications to the resistance observed in NSCLC patients to these compounds in the clinic. Specifically, our results suggest that direct manipulation of the caspase 9a/9b ratio would sensitize NSCLC tumors to already available chemotherapies. Furthermore, direct manipulation of this splicing event may allow for the use of one anti-cancer agent limiting side-effects and possibly toxic deaths as synergism between erlotinib and the DNA-damaging agent, daunorubicin, was lost by direct manipulation of the caspase 9a/b ratio. This is further highlighted as caspase 9b is only appreciably expressed in transformed cells, and hence, should have essentially no unwanted side-effects in patients. Currently, pre-clinical animal studies are being undertaken to assess the efficacy of this treatment regime.
In conclusion, this study links our previous findings on the phospho-status of SR proteins and the alternative splicing of caspase 9 during apoptotic signaling (15) by establishing a mechanistic link between the SR proteins, SRSF1, and the alternative splicing of caspase 9 via a novel ISE in intron 6 that regulates the inclusion of the exon 3,4,5,6 cassette. These current studies also strongly suggest that the alternative splicing of caspase 9 is an important regulatory mechanism that influences the chemotherapy sensitivity of NSCLC cells. The direct manipulation of this mechanism with the help of ASROs, siRNA, and RNA scavengers gives us the promise of a new generation of molecular-therapeutic agents for NSCLC that sensitizes NSCLCs to a variety of chemotherapeutic agents with little or no unwanted side-effects.
Supplementary Material
Acknowledgments
This work was supported by grants from the Veteran’s Administration (VA Merit Review I to C.E.C. and a Research Career Scientist Award to C.E.C.), from the National Institutes of Health via HL072925 (C.E.C), CA117950 (C.E.C), NH1C06-RR17393 (to Virginia Commonwealth University for renovation), and a National Research Service Award-T32 Post Doctoral Fellowship in Wound Healing (DSW). We would like to thank Drs. John D. Minna and Jerry Shay of the University of Texas Southwestern Medical Center, Dallas, Texas for their gracious gift of non-transformed immortalized HBEC-3KT cells, which were created using funds from the National Institutes of Health via an NCI Lung Cancer SPORE P50CA70907 (J.D.M.) and from the National Aeronautics and Space Agency (NASA) NNJO5HD36G (J.D.M). We also thank Dr. Paul Dent and Clint Mitchell of Department of Neurosurgery, Virginia Commonwealth University for their help with clonogenic survival assays. Lastly, we would like to thank Dr. Javier Cacares for the gracious gift of the SRSF1 wild-type and phospho-mutant constructs utilized in this study.
References
- 1.Quintans J, Kilkus J, McShan CL, Gottschalk AR, Dawson G. Ceramide mediates the apoptotic response of WEHI 231 cells to anti-immunoglobulin, corticosteroids and irradiation. Biochem Biophys Res Commun. 1994 Jul 29;202(2):710–4. doi: 10.1006/bbrc.1994.1988. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki T, Bielawska A, Bell RM, Hannun YA. Role of ceramide as a lipid mediator of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J Biol Chem. 1990 Sep 15;265(26):15823–31. [PubMed] [Google Scholar]
- 3.Liscovitch M. Crosstalk among multiple signal-activated phospholipases. Trends Biochem Sci. 1992 Oct;17(10):393–9. doi: 10.1016/0968-0004(92)90007-v. [DOI] [PubMed] [Google Scholar]
- 4.Kim MY, Linardic C, Obeid L, Hannun Y. Identification of sphingomyelin turnover as an effector mechanism for the action of tumor necrosis factor alpha and gamma-interferon. Specific role in cell differentiation. J Biol Chem. 1991 Jan 5;266(1):484–9. [PubMed] [Google Scholar]
- 5.Hannun YA. The sphingomyelin cycle and the second messenger function of ceramide. J Biol Chem. 1994 Feb 4;269(5):3125–8. [PubMed] [Google Scholar]
- 6.Cifone MG, De Maria R, Roncaioli P, et al. Apoptotic signaling through CD95 (Fas/Apo-1) activates an acidic sphingomyelinase. J Exp Med. 1994 Oct 1;180(4):1547–52. doi: 10.1084/jem.180.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballou LR, Chao CP, Holness MA, Barker SC, Raghow R. Interleukin-1-mediated PGE2 production and sphingomyelin metabolism. Evidence for the regulation of cyclooxygenase gene expression by sphingosine and ceramide. J Biol Chem. 1992 Oct 5;267(28):20044–50. [PubMed] [Google Scholar]
- 8.Chalfant CE, Kishikawa K, Mumby MC, Kamibayashi C, Bielawska A, Hannun YA. Long chain ceramides activate protein phosphatase-1 and protein phosphatase-2A. Activation is stereospecific and regulated by phosphatidic acid. J Biol Chem. 1999 Jul 16;274(29):20313–7. doi: 10.1074/jbc.274.29.20313. [DOI] [PubMed] [Google Scholar]
- 9.Dobrowsky RT, Kamibayashi C, Mumby MC, Hannun YA. Ceramide activates heterotrimeric protein phosphatase 2A. J Biol Chem. 1993 Jul 25;268(21):15523–30. [PubMed] [Google Scholar]
- 10.Law B, Rossie S. The dimeric and catalytic subunit forms of protein phosphatase 2A from rat brain are stimulated by C2-ceramide. J Biol Chem. 1995 May 26;270(21):12808–13. doi: 10.1074/jbc.270.21.12808. [DOI] [PubMed] [Google Scholar]
- 11.Wolff RA, Dobrowsky RT, Bielawska A, Obeid LM, Hannun YA. Role of ceramide-activated protein phosphatase in ceramide-mediated signal transduction. J Biol Chem. 1994 Jul 29;269(30):19605–9. [PubMed] [Google Scholar]
- 12.Mermoud JE, Cohen PTW, Lamond AI. Regulation of Mammalian Spliceosome Assembly by a Protein-Phosphorylation Mechanism. Embo J. 1994 Dec;13(23):5679–88. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalfant CE, Ogretmen B, Galadari S, Kroesen BJ, Pettus BJ, Hannun YA. FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phosphatase 1. J Biol Chem. 2001 Nov 30;276(48):44848–55. doi: 10.1074/jbc.M106291200. [DOI] [PubMed] [Google Scholar]
- 14.Chalfant CE, Rathman K, Pinkerman RL, et al. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem. 2002 Apr 12;277(15):12587–95. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 15.Massiello A, Chalfant CE. SRp30a (ASF/SF2) regulates the alternative splicing of caspase-9 pre-mRNA and is required for ceramide-responsiveness. J Lipid Res. 2006 May;47(5):892–7. doi: 10.1194/jlr.C600003-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005 Jul 1;19(1):1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Seol DW, Billiar TR. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. J Biol Chem. 1999;274(4):2072–6. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 18.Srinivasula SM, Ahmad M, Guo Y, et al. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer research. 1999 Mar 1;59(5):999–1002. [PubMed] [Google Scholar]
- 19.Seol DW, Billiar TR. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. The Journal of biological chemistry. 1999 Jan 22;274(4):2072–6. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 20.Shultz JC, Goehe RW, Wijesinghe DS, et al. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer research. Nov 15;70(22):9185–96. doi: 10.1158/0008-5472.CAN-10-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goehe RW, Shultz JC, Murudkar C, et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. The Journal of clinical investigation. Nov 1;120(11):3923–39. doi: 10.1172/JCI43552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siebert PD, Larrick JW. Competitive PCR. Nature. 1992 Oct 8;359(6395):557–8. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]
- 23.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 24.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003 Jul 1;31(13):3568–71. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koscielny G, Le Texier V, Gopalakrishnan C, et al. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009 Mar;93(3):213–20. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Joseph B, Lewensohn R, Zhivotovsky B. Role of apoptosis in the response of lung carcinomas to anti-cancer treatment. Ann N Y Acad Sci. 2000;926:204–16. doi: 10.1111/j.1749-6632.2000.tb05613.x. [DOI] [PubMed] [Google Scholar]
- 27.Joseph B, Ekedahl J, Lewensohn R, Marchetti P, Formstecher P, Zhivotovsky B. Defective caspase-3 relocalization in non-small cell lung carcinoma. Oncogene. 2001 May 24;20(23):2877–88. doi: 10.1038/sj.onc.1204402. [DOI] [PubMed] [Google Scholar]
- 28.Gallego MA, Ballot C, Kluza J, et al. Overcoming chemoresistance of non-small cell lung carcinoma through restoration of an AIF-dependent apoptotic pathway. Oncogene. 2008 Mar 27;27(14):1981–92. doi: 10.1038/sj.onc.1210833. [DOI] [PubMed] [Google Scholar]
- 29.Landriscina M, Maddalena F, Fabiano A, Piscazzi A, La Macchia O, Cignarelli M. Erlotinib enhances the proapoptotic activity of cytotoxic agents and synergizes with paclitaxel in poorly-differentiated thyroid carcinoma cells. Anticancer research. Feb;30(2):473–80. [PubMed] [Google Scholar]
- 30.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007 Mar;14(3):185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. Aug 20;142(4):625–36. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.