Abstract
Development of targeted therapy for hepatocellular carcinoma (HCC) remains a major challenge. We have recently identified an elevated expression of the fifth subunit of COP9 signalosome (CSN5) in early HCC as compared to dysplastic stage. In the present study, we explored the possibility of CSN5 being a potential therapeutic target for HCC. Our results show that CSN5 knockdown by small interfering (si) RNA caused a strong induction of apoptosis and inhibition of cell cycle progression in HCC cells in vitro. The downregulation of CSN5 was sufficient to interfere with CSN function as evidenced by the accumulation of neddylated Cullin1 and changes in the protein levels of CSN controlled substrates SKP2, p53, p27 and NF-kB, albeit to a different degree depending on the HCC cell line, which could account for the CSN5 knockdown phenotype. The transcriptomic analysis of CSN5 knockdown signature showed that the anti-proliferative effect was driven by a common subset of molecular alterations including downregulation of CDK6 and ITGB1, which were functionally interconnected with key oncogenic regulators MYC and TGFβ1 involved in the control of proliferation, apoptotic cell death and HCC progression. Consistent with microarray analysis, western blotting revealed that CSN5 depletion increased phosphorylation of Smad 2/3, key mediators of TGFβ1 signaling, decreased the protein levels of ITGB1, CDK6, and cyclin D1 and caused reduced expression of anti-apoptotic Bcl-2 while elevating the levels of pro-apoptotic Bak. A chemically modified variant of CSN5 siRNA was then selected for in vivo application based on the growth inhibitory effect and minimal induction of unwanted immune response. Systemic delivery of the CSN5 3/8 variant by stable-nucleic-acid-lipid-particles (SNALP) significantly suppressed the tumor growth in Huh7-luc+ orthotopic xenograft model. Taken together, these results indicate that CSN5 plays a pivotal role in HCC pathogenesis and maybe an attractive molecular target for systemic HCC therapy.
Keywords: hepatocellular carcinoma, CSN5, targeted siRNA therapeutics, SNALP
Introduction
HCC is the third most lethal neoplasm causing an estimated 600,000 deaths annually due to late diagnosis underlying liver disease and lack of effective treatment options (Parkin et al., 2005; Llovet and Bruix, 2008; Bruix and Sherman, 2005; Llovet et al., 2008). However, recent advances in the understanding the complex molecular pathways driving HCC indicate that the early stages of HCC development are characterized by selection of certain common traits (Thorgeirsson and Grisham, 2002). The progressive accumulation of genomic and epigenetic alterations during the course of chronic liver disease affects numerous regulatory pathways resulting in gradual loss of differentiation (Avila et al., 2000; Okabe et al., 2001; Berasain et al., 2003) and selection of clones characterized by unrestrained proliferation and resistance to cell death (Rust and Gores, 2000; Feitelson et al., 2002; Lee and Thorgeirsson, 2004; Arsura and Cavin, 2005). The comprehensive analysis of genomic changes during the process of hepatocarcinogenesis may reveal suitable targets for preventing and treating HCC through targeted therapy (Roberts and Gores, 2005).
Our previous microarray analysis revealed a consistent up-regulation of CSN5 gene in the early HCC implying that CSN5 may be one of the early markers of malignant conversion (Kaposi-Novak et al., 2009). CSN5, also known as JAB1 or COPS5, is the catalytic center of COP9 signalosome (CSN), an evolutionary conserved multi-protein complex involved in the control of proteolysis via the ubiquitin proteasome pathway, transcription, protein phosphorylation, and intracellular distribution (Wei et al., 2008; Kato and Yoneda-Kato, 2009; Chamovitz, 2009; Shackleford and Claret, 2010). Although the precise molecular mechanisms remain to be elucidated, the most studied function of the CSN is the regulation of protein degradation, which occurs primarily through the removal of NEDD8 (a ubiquitin-like modifier) from cullin-RING-E3 ligases (CRL), thereby regulating ligase activity. Among the 8 CSN subunits (CSN1 to CSN8), CSN5 plays a central role in this deneddylation activity and controls the stability of a great variety of key intracellular regulators of survival and/or apoptotic programs, such as MYC, TGFβ1, MIF, c-Jun, HIF1α, p27, E2F1, and p53, and may function either as a monomer or as the fifth component of CSN complex. In addition to controlling the stability of proteins, the CSN also mediates transcriptional regulation by mechanisms that are not yet clearly defined. In particular, CSN5 seems to act as a transcriptional coactivator for MYC, which is essential for the transcriptional activation of genes involved in cell proliferation, angiogenesis, and cell invasion (Claret et al., 1996; Bae et al., 2002; Adler et al., 2006). CSN5 activities positively and negatively affect a number of pathways, including integrin signaling, cell cycle control, and apoptosis. Given the diversity of functions and a large number of effector molecules controlled by CSN5, we hypothesized that the inactivation of the overexpressed CSN5 gene could result in reestablishment of numerous anti-oncogenic programs involved in the control of cell proliferation and apoptosis and whereby be effective as an anti-cancer strategy.
RNA interference (RNAi) is a cellular mechanism for silencing gene expression which triggers a sequence-specific degradation of target mRNA (Fire et al., 1998; Elbashir et al., 2001). The double-stranded RNA substrates are enzymatically cleaved into siRNA and mediate the suppression of target gene expression through a phylogenetically conserved intrinsic pathway of multi-step processes (Sijen et al., 2001; Zamore, 2001). The mechanistic insight raised the hope that RNAi could be harnessed for the development of new drugs targeting all classes of pathological proteins (Hannon and Rossi, 2004; Bumcrot et al., 2006). We have previously described the development of SNALP as an effective systemic delivery vehicle for targeting siRNA to murine and primate liver as well as solid tumors and have demonstrated robust therapeutic silencing of endogenous hepatocyte, tumor or viral gene transcripts in the absence of any measurable immune response (Jeffs et al., 2005; Morrissey and Blanchard et al., 2005; Zimmermann et al., 2006; Judge et al., 2009).
Here we report that siRNA-mediated CSN5 knockdown inhibits cell cycle progression and greatly increases the rate of apoptosis in HCC cells in vitro. High-throughput transcriptomic analysis of CSN5 knockdown signature revealed a common subset of molecular alterations underlying the therapeutic responses, including down regulation of TGFβ1 and MYC-target genes, such as integrin β1 (ITGB1) and cyclin-dependent kinase 6 (CDK6). At the protein level, CSN5 depletion in HCC cells increased the phosphorylation of Smad2/3 and expression of Bak with a concomitant down-regulation of NF-κB, CDK6, cyclin D1 and Bcl-2, leading to apoptotic cell death. Finally, systemic delivery of the modified CSN5siRNA encapsulated in SNALP remarkably inhibited hepatic tumor growth in an orthotopic xenograft model of human liver cancer. These results demonstrate the important role for CSN5 in liver cancer progression and suggest targeting CSN5 as a promising novel tool for anti-cancer therapy.
Results
The biological effects of CSN5 knockdown in human HCC cells in vitro
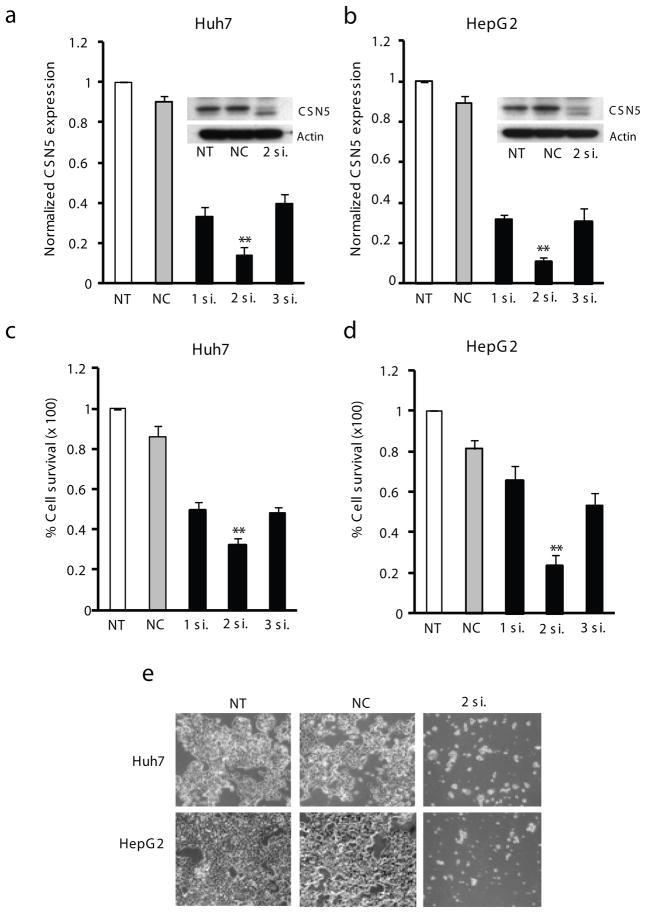
To address the functions of CSN5, the HCC cell lines Huh7 and HepG2 cells were transfected with three variants of CSN5-specific siRNA (CSN5-1-3) for 2 d, and efficiency of CSN5 silencing was determined by analysis of the steady-state levels of CSN5 mRNA and protein. The CSN5-2siRNA was the most effective in reducing CSN5 mRNA, and the downregulation was correlated with a remarkable decrease in target protein (Figures 1a and b). Consistent with these changes, CSN5-2siRNA caused a greater suppression of HCC cell growth and therefore was selected for further studies (Figure 1c–e). In two examined cell lines, Huh7 and HepG2, treatment with 15 nM CSN5-2siRNA resulted in 68% and 77% growth inhibition, respectively. Transfection with negative control (NC) siRNA did not affect cell growth. The growth inhibition in CSN5-depeleted HCC cells could be attributed in part to the defective cell cycle progression. FACS analysis revealed accumulation of G0/G1-phase cells and a concomitant decrease in the S-phase cells in both examined HCC cell lines two days after treatment with CSN5-2siRNA (Figure 2a). However, the most significant effect of CSN5 knockdown was a strong induction of apoptosis as measured by an assay detecting the denatured single-stranded DNA formed in apoptotic cells but not in the necrotic cells or cells with DNA breaks (Figure 2b). The result was reproduced in two additional HCC cell lines Huh1 and PLC/PRF/5 which showed a similar reduction in growth rate as a consequence of CSN5 silencing (Figure 2c). These data demonstrate that siRNA-mediated knockdown of CSN5 in human HCC cells inhibit cell cycle progression and increases predisposition to undergo apoptotic cell death.
Figure 1.
siRNA knockdown of CSN5 inhibits growth of HCC cells. (a–b) Downregulation of CSN5 mRNA and protein (insets) in Huh7 (a) or HepG2 (b) cells treated with CSN5-specific siRNA. Total RNAs and whole cell lysates were extracted at 48 h after treatment with 15 nM of siRNAs. CSN5 mRNA expression levels were calculated relative to GAPDH and normalized to untreated control. Actin protein was used as a loading control. The data are shown as means ± S.D. of triplicate experiments (**P < 0.01; Bootstrap Test). (c–e) Growth inhibition of Huh7 (c) or HepG2 (d) cells as measured by an MTT assay or microscopic observation (e) (X100 original magnification) 4 d after transfection. The cell survival was calculated relative to untreated cells. The data are shown as means ± S.D. of triplicate experiments (**P < 0.01, Bootstrap t-test). NT, no treatment; NC, negative control siRNA; 1si., CSN5-1siRNA; 2si., CSN5-2siRNA; 3si., CSN5-3siRNA.
Figure 2.
Biological effects of CSN5 siRNA knockdown in HCC cell lines. (a) Cell cycle analysis of HCC cells treated with CSN5-2siRNA for 2 d. (b) Detection of apoptosis 3 d after transfection with CSN5-2siRNA. (c) Effect of CSN5-2siRNA on the proliferation in various HCC cells. The cell survival was calculated relative to NCsiRNA treatment. All statistical analysis was performed using Bootstrap t-test. The data shown are means ± S.D. of triplicate experiments. **, P < 0.01.
The molecular mechanisms underlying the growth inhibitory effects of CSN5 knockdown
To identify the molecular targets and common mechanisms underlying the growth inhibitory effects caused by CSN5 silencing, Huh7 and HepG2 cells were treated with either NCsiRNA or CSN5-2siRNA for 48 h and subjected to transcriptomic analysis using gene expression microarrays. In both cell lines, we achieved a strong (> 8-fold) CSN5-specific inhibition (Figure 3a). The number of differentially expressed genes which displayed more than a 2-fold change was 768 (402 up- and 366 down-regulated genes) and 349 (127 up- and 222 down-regulated genes) in Huh7 and HepG2 cells, respectively, using permuted Bootstrap t-test (P < 0.05) (Supplementary Figures 1a and 2a). Given involvement of CSN5 gene in liver cancer progression, we then focused on the expressions of genes functionally interconnected with its principal regulators, MYC and TGFβ1. Consistent with the reported functional relationships (Wei et al., 2008; Kato and Yoneda-Kato, 2009), numerous genes affected by CSN5 inactivation in HCC cells were known/putative targets of MYC or TGFβ1 (Supplementary Figures 1b and c and Supplementary Figures 2b and c).
Figure 3.
Transcriptomic analysis in gene expression following CSN5 knockdown. Huh7 and HepG2 cells were transfected with 15 nM of CSN5-2siRNA and analyzed for changes in gene expression at 48 h following treatment by illumina microarray. The means of the intensity log ratios in CSN5 siRNA-treated cells were calculated relative to the NCsiRNA-treated cells. (a) Fold-changes in expressions of CSN isoforms. (b) Heat-map overview of 127 genes commonly dysregulated in both Huh7 and HepG2. The scale bar (log2-transformed scale) indicate high (red) and low (green) expression levels. **, P <0.05 by Bootstrap t-test. (c–d) Expression targets of MYC (c) or TGFβ1 (d) in CSN5 knockdown signature. Up- and downregulated genes are indicated in red and green, respectively. Genes in gray are associated with the regulated genes.
To further explore the molecular mechanisms of CSN5 response, we have generated a common CSN5 knockdown signature consisting of 127 deregulated genes (Figure 3b and Supplementary Table 1). Using the Ingenuity Pathway Analysis (IPA) software, we identified common statistically significant pathway networks (score >22), suggesting that molecular alterations in the diverse oncogenic pathways may cooperatively result in the growth inhibition of HCC cells in response to CSN5 inactivation. The pattern of gene expression and the top biological functions in each network are shown in Supplementary Table 2. A PathwayStudio analysis illustrates that despite a limited gene to gene overlap, the expressions of twelve genes, known to be associated with MYC and/or TGFβ1 pathway, such as CDK6, BRCA1, ATF3, EIF2S1, SLC2A1, ITGB1, MMP11, GSN, TIMP2, PDGFβ, FSTL3 and LPL, were commonly deregulated (Figure 3c and d). Consisting with phenotypic changes, expressions of genes that are functionally involved in pro-apoptotic activity and tumor suppression (ATF3, MPM11, GSN, TIMP2, and FSTL3) were upregulated. Conversely, genes involved in anti-apoptosis, cell cycle progression, survival and metastasis (CDK6, EIF2S1, SLC2A1, ITGB1, and LPL) were downregulated. In particular, the cell cycle promoting function of cyclin-dependent kinase 6 (CDK6) which associates with cyclin D and targets the retinoblastoma protein to allow passage through the G1/S phase restriction point has been shown to have functional relationships with TGFβ1 and MYC (Mateyak et al., 1999; Tsubari et al., 1999).
To gain a better understanding of the molecular mechanisms linking growth suppression in HCC cells to the CSN5 deletion, we performed an extensive Western blot analysis of CSN components and selected CSN-controlled substrates. Supporting CSN5-specific inhibition on mRNA levels (Figure 3a), we found a significant reduction only in CSN5 protein levels while other CSN subunits (e.g. CSN1, CSN3 and CSN8) showed little or no change (Figure 4a). siRNA-mediated CSN5 silencing was sufficient to disrupt the CSN complex, and induced Cullin 1 hyperneddylation which was paralleled by a decrease in free NEDD8 (Figure 4a). Concomitant with a loss of CSN5 function, both examined HCC cell lines showed a decrease in the protein level of SKP2, a substrate-targeting subunit of the ubiquitin ligase complex that regulates entry into S phase by inducing the degradation of the cyclin-dependent kinase inhibitors p21 and p27 (Bashir et al., 2004). Interestingly, MYC, another candidate substrate of CSN-controlled ubiquitin ligases which could account for CSN5 knockdown phenotype, showed no obvious changes in the protein levels while accumulation of p53 as well as p27, a target of SKP2, was found only in HepG2 cell line characterized by wild type p53, but not in the p53 mutant Huh7 cells, reflecting cell type-specific differences (Figure 4a). However, in agreement with microarray data, and consistent with inhibition of cell cycle progression by CSN5 inactivation (Figure 1 and 2), both cell lines showed reduction in protein levels of cyclin D1, CDK6, and ITGB1 involved in the control of cell cycle progression although the expression of cyclin E, another regulator of G1 phase, as well as CDK2 and cyclin A, implicated in S and G2/M transition, were unaffected (Supplementary Figure 3). In support of the microarray analysis, CSN5 silencing caused increased phosphorylation of Smad 2 and 3, key mediators in TGFβ1 signaling, involved in the control of apoptotic cell death (Figure 4b). In addition, we found a significantly reduction in the NF-kB p65 level in both examined cell lines and a concomitant downregulation of anti-apoptotic Bcl-2 reflecting defective NF-kB activation in agreement with recent data linking CSN function to IkB-α regulation (Panattoni et al., 2008). In contrast, the levels of proapoptotic Bak were increased suggesting that activation of TGFβ1 signaling and/or inhibition of NF-κB pathway may be common mechanisms of growth inhibition and apoptotic induction following CSN5 inactivation.
Figure 4.
Molecular mechanisms of therapeutic response to CSN5 targeting. (a) Western blot analysis of CSN components, neddylated cullins and CRL substrates in Huh7 and HepG2 cells treated with the indicated siRNA for 48 h. (b) Western blot analysis of phosphorylated Smad2/3 and the indicated proteins functionally involved in the cell proliferation or apoptosis. Actin was included as a loading control. (c) Growth inhibition of Huh7 or HepG2 cells as measured by an MTT assay 4 d after transfection of PDGFβ siRNA. (d,e) Induction of apoptosis (d) and caspase-3 (e) after transfection with PDGFβ siRNA. All data were calculated relative to NCsiRNA-treated cells. The data are shown as means ± S.D. of triplicate experiments (**P < 0.01, *P < 0.05, Bootstrap t-test). Pβsi., PDGFβ siRNA
Importantly, CSN5 knockdown also caused a compensatory upregulation of platelet-derived growth factor beta (PDGFβ), and decreased the expression of tumor suppressor BRCA1, reflecting a complex molecular dynamics caused by targeted inactivation of a single gene. When we blocked PDGFβ expression by siRNA, we found a significant reduction in survival and increased caspase-3-mediated apoptosis (Figure 4d and e) suggesting that a combined inactivation of several putative targets can amplify the therapeutic effect of CSN5 deletion in HCC cells. Taken together, these results show that siRNA depletion of a single gene may coordinately impact diverse oncogenic pathways acting in concert to drive liver cancer progression and provide a rationale for therapeutic targeting of CSN5.
Systemic delivery of CSN5 siRNA by SNALP suppresses orthotopic liver tumor growth
The next step was to validate if systemic silencing of CSN5 could suppress liver tumor growth. To prevent immune activation and enhance siRNA stability in vivo, native CSN5-2siRNA was chemically modified by selective incorporation of 2′-O-methyl (2′OMe) uridine or guanosine nucleosides into one strand of the siRNA duplex, and encapsulated into SNALP. A modified siRNA was then screened for in vivo application in terms of Huh7-luc+ cell growth and cytokine induction using murine Flt3L dendricytes isolated from mouse bone marrow. Among the variants tested, CSN5 3/8 sequence was the most effective in inhibiting tumor cell growth (about 80%) (Figure 5a). In addition, the modified sequence caused a minimal induction of IL-6 as compared to the treatment with native CSN5-2siRNA while injection of empty SNALP had little effect on IL-6 levels (Figure 5b).
Figure 5.
Selection of CSN5 3/8 for in vivo application based on the inhibition of tumor cell growth and minimal cytokine induction. (a) Inhibition of Huh7-luc+ cell growth after transfection with 15 nM of SNALP-formulated CSN5-2 (native) or its modified variants (CSN5-3/6~9, CSN5-4/6~9, CSN5-5/6~9). The siRNA transfectants were examined by MTT assay at 4 d after treatment. **, P < 0.01 (n=3) by Bootstrap t-test. SNALP-Luc, SNALP-fromulated siRNA targeting luciferase. (b) Quantification of IL-6 level after CSN5 targeting. Culture supernatants of Flt3L-derived dendrocytes were assayed for IL-6 using ELISA at 24 h after siRNA treatment. The data are shown as the means ± S.D. of triplicate experiments.
To test the therapeutic efficacy of CSN5 gene targeting we used an orthotopic mouse model of hepatocarcinoma and bioluminescence imaging (BLI) as a method of monitoring the kinetics of tumor growth. Mice with liver tumors derived from Huh7-luc+ cells were randomly assigned either to treatment or control group before starting siRNA therapy (Figure 6). The results showed that in control group treated with SNALP-βgal478 targeting β-galactosidase, tumors grew very rapidly and at the end point of the study (28 days) occupied a significant portion of liver parenchyma. In addition, the majority of mice developed ascites reflecting impairment of liver function characteristic for the end stage of liver cancer disease. In striking contrast, SNALP-CSN5 3/8 effectively inhibited the hepatic tumor growth formed by Huh7-luc+ cells and significantly improved well being of animals (Figure 6a and b). Consistent with this, mice receiving siRNA therapy showed a better histology with single and much smaller tumors as well as reduction in liver-to-body ratios as reflection of the reduced tumor burden (Figure 6c and d). These results indicate that CSN5 is an important regulator of HCC growth and may represent a promising target for systemic therapy of human HCC.
Figure 6.
Systemic delivery of CSN5 3/8siRNA by SNALP suppresses human HCC growth in a mouse model of orthotopic transplantation. SCID/beige mice received 0.5×106 Huh7-luc+ cells through intrasplenic injection resulting in tumorous growth in the liver. Mice were randomly assigned either to control (SNALP-βgal478) or treatment (SNALP-CSN5 3/8) group based on the intensity of bioluminescence before initiation of siRNA therapy. Two mg/kg of SNALP-βgal478 and SNALP-CSN5 3/8 were injected into tail vein at 8, 11, 14 and 18 days. (a) Representative in vivo bioluminescence images of Huh7-xenografts. Images were set at the same pseudocolor scale to show the relative bioluminescence changes over time. (b) Quantification of bioluminescence. The total flux is plotted as photon/second. **, P < 0.01, (n=8 vs. n=6) by Mann-Whitney U-test. (c) Histopathological evaluation. Shown are representative photos of gross liver morphology and histopathology at 28 days after transplantation. H&E staining, original magnification X50. (d) Liver-to-body weight ratios. The data are shown as the means ± S.D. in each treatment group. **P < 0.01, Mann-Whitney U-test.
Discussion
Hepatocarcinogenesis is a multi-step process driven by the accumulation of multiple genetic and epigenetic alterations (Thorgeirsson and Grisham, 2002). Previously, we have identified MYC as one of the driver genes for malignant conversion from the dysplastic stage to early HCC (Kaposi-Novak et al., 2009). Notably, the presence of the MYC signature significantly correlated with increased expression of CSN5, as well as with a higher overall transcription rate of genes located in the 8q chromosome region. However, the significance of CSN5 upregulation in liver cancer has remained undetermined. In this study, we demonstrate that siRNA-mediated knockdown of CSN5 caused a consistent and strong induction of apoptotic cell death and delayed cell cycle progression in the examined human HCC cells. These results suggest that overexpression of CSN5 may contribute to both cell survival and proliferation and thus represent a prognostic marker for malignant conversion in liver cancer. CSN5 overexpression has been also found in breast, thyroid, skin, ovarian, lung and pancreatic cancer (Sui et al., 2001; Esteva et al., 2003; Ito et al., 2003; Kouvaraki et al., 2003; Fukumoto et al., 2004; Goto et al., 2004; Ivan et al., 2004; Adler et al., 2008), implying that CSN5 targeting may be an effective therapeutic strategy against various cancer types. Consistent with this, depletion of CSN5 has been shown to result in growth inhibition of pancreatic cell line (Fukumoto et al., 2006).
The massive apoptosis and defective cell cycle progression in CSN5-depleted HCC cells was paralleled by coordinated dysregulation of numerous gene expression programs acting downstream of CSN5-regulated transcription factors. As shown in Figure 3, at transcriptional level, CSN5 knockdown-mediated downregulation of CDK6, a key regulator in G1/S phase transition, was functionally associated with both MYC and TGFβ1, and correlated with the increased expression of proapoptotic effectors (e.g. ATF3, activating transcription factor 3) (Kashiwakura et al., 2008), or activator of metastasis (e.g. MMP11, matrix metalloproteinase 11) (Brasse et al., 2010). In this study we also found that the decreased levels of CDK6 and cyclin D1 proteins were correlated with the activation of TGFβ1 and inhibition of NF-κB pathways (Figure 4b). These observations may provide the molecular mechanism(s) underlying the phenotypic changes triggered by siRNA silencing of CSN5 expression, such as inhibition of G1/S transition and induction of apoptosis.
Among the important findings of the study is the demonstration of high efficiency of CSN5 targeting in a mouse xenograft model of human liver cancer. The primary obstacle for therapeutic application of RNAi is a successful delivery to targeted cell type in vivo (Pai et al., 2006; Hokaiwado et al., 2008). The SNALP-based technology has unique technical advantages for a systemic siRNA delivery to liver (Rossi, 2006; Whitehead et al., 2009). First, target siRNA modified and incorporated into SNALP is protected from both glomerular filtration and serum nucleases whereby increasing the half-life of the circulating siRNA. In particular, 2′OMe-modification of siRNA utilized in this study mediated potent silencing of target mRNA without cytokine induction, toxicity or off-target effects associated with unmodified siRNA (Judge and Robbins et al., 2005; Judge and Sood et al., 2005). Second, the use of SNALP technology allowed reducing the dosing regimen up to 3 mg/kg, which is 10-fold less as compared to the standard dose of siRNA without a carrier (Morrissey and Blanchard et al., 2005). In our study, treatment with a moderate dose of 2 mg/kg was sufficient for the effective suppression of liver tumor growth in vivo (Figure 6). Finally, SNALP formulated target siRNA injected intravenously primarily traffics to liver and spleen with little accumulation in other organs. Although more work is needed to optimize the therapeutic dose of SNALP-target siRNA for different stages of HCC disease, our work demonstrates the potency of systemic RNAi targeting of CSN5 without an overt toxicity in a mouse model of orthotopic transplantation of human HCC cells and suggests its clinical utility for treatment of HCC disease.
Materials and methods
siRNA
All native CSN5 siRNA duplexes used for in vitro studies were chemically synthesized by Ambion (Austin, TX, USA) (CSN5-1: sense, 5′-CCAUUACUUUAAGUACUGCtt-3′; antisense, 5′-GCAGUACUUAAAGUAAUGGtg-3′, CSN5-2: sense, 5′-GGAUCACC AUUACUUUAAGtt-3′; antisense, 5′-CUUAAAGUAAUGGUGAUCCtt-3′, CSN5-3: sense, 5′-CCGAAAAUCAGAAGACAAAtt-3′; antisense, 5′-UUUGUCUUCUGAUU UUCGGtc-3′, PDGFβsi: sense, 5′-GCACCGAGGUGUUCGAGAUtt-3′; antisense, 5′-AUCUCGAACACCUCGGUGCgc-3′). For in vivo application, CSN5-2 siRNA was synthesized by Integrated DNA Technologies (Coralville, IA, USA) in a large quantity and modified by 2′-OMe method (Judge and Bola et al., 2005), and then encalpsulated into SNALP using a published method (Jeffs et al., 2005) (CSN5 3/8: sense, 5′-GGAmUCACCAUmUACmUUmUAAGUU-3′; antisense, 5′-CUUAAAmGUAAUGmG UmGAUCCUU-3′, mU, 2′O-methyl uridine; and mG, 2′O-methyl guanosine). Negative control siRNA molecules that do not target any endogenous transcript were used for control experiments. Silencer Negative Control #1 siRNA (Ambion) and SNALP-formulated βgal478siRNA (Judge and Sood et al., 2005) were used for in vitro and in vivo studies, respectively.
Cell culture and transfection of siRNA in vitro
PLC and HepG2 were obtained from the American Type Culture Collection (Rockville, MD, USA), Huh7 from Riken Cell Bank (Tsukuba, Ibaraki, Japan) deposited by Dr. Nam-Ho Huh and Huh1 from Health Science Research Resource Bank (Osaka, Japan). The cells were maintained in DMEM/F-12 media (Mediatech, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Norcross, GA, USA) at 37°Cin the presence of 5% CO2. For the measurements of cell proliferation and apoptosis, cells were seeded at 25% confluence in 96-well plates one day before transfection in 100 μl of culture media without antibiotics. Lipofectamine 2000 was mixed with siRNA molecules in a volume of 50 μl Opti-MEM I (both from Invitrogen, Carlsbad, CA, USA) and added to HCC cells. The medium was replaced 24 h after transfection. The negative control siRNAs (NCsiRNA) were used in the same quantity and transfected to the cells simultaneously.
Measurement of cell proliferation and apoptotic cell death
The growth inhibitory effects of control and target siRNA were studied using the Vybrant MTT Cell Proliferation Assay (Invitrogen) as recommended by the manufacturer. Absorbance was measured at 540 nm using an ELISA reader SpectraMAX 190 (Molecular Devices, Sunnyvale, CA, USA). The percentage of viable cells was calculated by comparing the optical density using the following formula: (1 - absorbance of an experimental well)/absorbance of an untreated control well × 100). The induction of apoptosis was measured using ApoStrand ELISA Apoptosis Detection Kit (Biomol International, Plymouth Meeting, PA, USA).
Quantitative real-time RT-PCR
The changes in target gene expression on mRNA level were detected using real-time quantitative RT-PCR. Total RNA was isolated using Tri reagent (Molecular Research Center, Cincinnati, OH, USA) according to the protocol recommended by the manufacturer. One μg of RNA was reverse transcribed using random primers supplied in the High-Capacity cDNA Archieve Kit (Applied Biosystems, Carlsbad, CA, USA). cDNA of CSN5 gene was amplified using corresponding pair of primers (forward 5′-TCTGCTGAAGATGGTGATGC-3′; reverse, 5′-GCCAACCTGTTTTGCATTTT-3′) synthesized by Operon Technology (Valencia, CA, USA), Power SYBR Green PCR Master Mix and ABI 7700HT PCR Machine (both from Applied Biosystems). The mRNA levels of GAPDH were used for normalization. All reactions were performed in triplicate.
Western blot analysis
The amount of total protein was determined by BCA Protein Assay Kit (Pierce, Rockford, IL, USA). 100 μg of total protein were run using 4–20% SDS-polyacrylamide gel and transferred onto PVDF membrane (Invitrogen). The membrane was blocked with 5% milk/Tris-buffered saline plus Tween20 (TBST) and incubated with primary antibodies against human CSN3 (RR12), CSN5 (FL-334), CSN8 (C-19), Cullin 1 (H-213), p53 (FL-393), p27 (F-8), Myc (C-19), SKP2 (H-435), pSmad2/3 (SC-11769), NFκB p65 (C-20), ITGB1 (M-106), CDK2 (M2), CDK6 (H-230), cyclin D1 (DCS-6), cyclin E (M-20), cyclin A (BF683), Bak (G-23), Bcl-2 (N-19) and Bcl-xL (H-5) (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA). CSN1 (#S0055) antibody was purchased from Epitomics (Burlingame, CA, USA) and Novus Biologics (Littleton, CO, USA), respectively. The antibodies against NEDD8 (#2745S) and caspase-3 (#9661) were available from Cell Signaling (Danvers, MA, USA). Immunoreactive bands were visualized using ECL Plus Western Blotting Detection System (GE Healthcare, Piscataway, NJ, USA). The equal loading was assessed by probing the same membrane with Actin antibody (A1978, Sigma, St. Louis, MO, USA).
Microarray analysis
Biotin-labeled cRNA was linearly amplified according to manufacturer’s specification (AMIL1791; Ambion, Austin, TX, USA). As input, 200 ng total RNA were used for in vitro transcription (IVT) reactions for 16 h at 37 °C. The efficiency of this single round amplification was measured by NanoDrop (ND1000, Thermo Scientific, Barrington, IL, USA). Hybridization, washing, detection (Cy3-streptavidin, Amersham Biosciences, GE Healthcare) and scanning were performed on an Illumina iScan system (Illumina, San Diego, CA, USA) using reagents and following protocols supplied by the manufacturer. Briefly, the biotinylated cRNA (750 ng/sample)was hybridized on Sentrix whole genome beadchips human Ref-8v3 for 18 h at 58°C while rocking (5 rpm). The beadchip covers ~ 24,000 RefSeq transcripts. Image analysis and data extraction were performed automatically using Illumina GenomeScan Software. All microarray data was submitted to Gene Expression Omnibus database with the accession number of GSE26485.
Functional network analysis
To explore the functional relationships among the genes with altered expression in HCC cells treated with CSN5 siRNA, a pathway analysis was carried out using the Ingenuity Pathway Analysis tool (Ingenuity Systems, Redwood City, CA, USA). The significance of each network was estimated by scoring system provided by Ingenuity. The scores were determined by the number of differentially expressed genes within each network and the strength of the associations among the network members. Once over-represented and functionally relevant genes were identified, their functional association was validated by independent pathway analysis tool PathwayStudio (Ariadne Genomics, Rockville, MD, USA).
Generation of HCC reporter cell line expressing luciferase
Huh7 cells were transfected with pGL4.17 vector (Promega, Madison, WI, USA) expressing firefly luciferase and neomycin resistance gene using Lipofectamine 2000. To enhance the expression of luciferase gene, β-actin promoter from pCAGEN plasmid (Addgene, Cambridge, MA, USA) was subcloned into multicloning site of pGL4.17. Cells were selected for antibiotic resistance with Geneticin (Invitrogen), and surviving colonies were amplified and screened for bioluminescence in complete media supplemented with 150 μg/ml D-luciferin (Biosynth International, Itasca, IL, USA) by in vitro imaging (IVIS Imaging System, Xenogen, Alameda, CA, USA). The Huh7-1H6 clone with a highest expression was selected and used throughout the study.
Systemic administration of SNALP-formulated siRNA and BLI in vivo
Six-week-old male SCID/Beige mice (Charles River Laboratories, Wilmington, MA, USA) were anesthetized by inhalation of 5% isoflorane in oxygen. Cells (5 × 105 in 50 μL phosphate-buffered saline) were injected into the splenic pulp using a 27-gauge needle. Spleen was removed 30 sec after injection and wound was closed in two layers using 3-0 silk suture and surgical clips. Body and liver weights were recorded at death. Animals were housed in an AAALAC facility and cared for in accordance with the guidelines from the Animal Care and Use Committee at the US National Cancer Institute, NIH. Mice with liver tumors derived from Huh7-luc+ cells were randomly assigned to treatment or control groups based on bioluminescence imaging before initiation of siRNA-therapy. SNALP-formulated siRNAs (2 mg/kg) were injected into the lateral tail vein four times with a 3-day interval. Tumor growth was monitored by BLI for 4-weeks with 3–4 day intervals using an IVIS Imaging System. Images and measurements of luciferase signals were analyzed using Living Image Software (Xenogen). Ten minutes prior to in vivo imaging, mice were anesthesized with 1–3% isoflurane (Abbott Laboratories) and given an i.p. injection luciferin (Biosynth International) at 150 mg/kg in DPBS. Regions of interest (ROI) from displayed images were drawn around the tumor sites and quantified as photons/second using the software.
Supplementary Material
Acknowledgments
This project was supported by the Intramural Research Program of the Center for Cancer Research, NCI.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler AS, Littlepage LE, Lin M, Kawahara TL, Wong DJ, Werb Z, et al. CSN5 isopeptidase activity links COP9 signalosome activation to breast cancer progression. Cancer Res. 2008;68:506–515. doi: 10.1158/0008-5472.CAN-07-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsura M, Cavin LG. Nuclear factor-kappaB and liver carcinogenesis. Cancer Lett. 2005;229:157–169. doi: 10.1016/j.canlet.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Avila MA, Berasain C, Torres L, Martín-Duce A, Corrales FJ, Yang H, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM, Bae SK, et al. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem. 2002;277:9–12. doi: 10.1074/jbc.C100442200. [DOI] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Berasain C, Herrero JI, García-Trevijano ER, Avila MA, Esteban JI, Mato JM, et al. Expression of Wilms’ tumor suppressor in the liver with cirrhosis: relation to hepatocyte nuclear factor 4 and hepatocellular function. Hepatology. 2003;38:148–157. doi: 10.1053/jhep.2003.50269. [DOI] [PubMed] [Google Scholar]
- Brasse D, Mathelin C, Leroux K, Chenard MP, Blaise S, et al. Matrix metalloproteinase 11/stromelysin-3 exerts both activator and repressor functions during the hematogenous metastatic process in mice. Int J Cancer. 2010;127:1347–1355. doi: 10.1002/ijc.25309. [DOI] [PubMed] [Google Scholar]
- Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz DA. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009;10:352–358. doi: 10.1038/embor.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret FX, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Esteva FJ, Sahin AA, Rassidakis GZ, Yuan LX, Smith TL, Yang Y, et al. Jun activation domain binding protein 1 expression is associated with low p27(Kip1) levels in node-negative breast cancer. Clin Cancer Res. 2003;9:5652–5659. [PubMed] [Google Scholar]
- Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z, et al. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–2604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fukumoto A, Ikeda N, Sho M, Tomoda K, Kanehiro H, Hisanaga M, et al. Prognostic significance of localized p27Kip1 and potential role of Jab1/CSN5 in pancreatic cancer. Oncol Rep. 2004;11:277–284. [PubMed] [Google Scholar]
- Fukumoto A, Tomoda K, Yoneda-Kato N, Nakajima Y, Kato JY. Depletion of Jab1 inhibits proliferation of pancreatic cancer cell lines. FEBS Lett. 2006;580:5836–5844. doi: 10.1016/j.febslet.2006.09.042. [DOI] [PubMed] [Google Scholar]
- Goto A, Niki T, Moriyama S, Funata N, Moriyama H, Nishimura Y, et al. Immunohistochemical study of Skp2 and Jab1, two key molecules in the degradation of P27, in lung adenocarcinoma. Pathol Int. 2004;54:675–681. doi: 10.1111/j.1440-1827.2004.01679.x. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Hokaiwado N, Takeshita F, Banas A, Ochiya T. RNAi-based drug discovery and its application to therapeutics. IDrugs. 2008;11:274–278. [PubMed] [Google Scholar]
- Ito Y, Yoshida H, Matsuzuka F, Matsuura N, Nakamura Y, Nakamine H, et al. Jun activation domain-binding protein 1 expression in malignant lymphoma of the thyroid: its linkage to degree of malignancy and p27 expression. Anticancer Res. 2003;23:4121–4125. [PubMed] [Google Scholar]
- Ivan D, Diwan AH, Esteva FJ, Prieto VG. Expression of cell cycle inhibitor p27(Kip1) and its inactivator Jab1 in melanocytic lesions. Mod Pathol. 2004;17:811–818. doi: 10.1038/modpathol.3800123. [DOI] [PubMed] [Google Scholar]
- Jeffs LB, Palmer LR, Ambegia EG, Giesbrecht C, Ewanick S, MacLachlan I. A scalable, extrusion free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2005;13:494–504. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence- dependent stimulation of the mammalian innate immune response. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Coulouarn C, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–2782. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwakura Y, Ochiai K, Watanabe M, Abarzua F, Sakaguchi M, Takaoka M, et al. Down-regulation of inhibition of differentiation-1 via activation of activating transcription factor 3 and Smad regulates REIC/Dickkopf-3-induced apoptosis. Cancer Res. 2008;68:8333–8341. doi: 10.1158/0008-5472.CAN-08-0080. [DOI] [PubMed] [Google Scholar]
- Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14:1209–1225. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- Kouvaraki MA, Korapati AL, Rassidakis GZ, Tian L, Zhang Q, Chiao P, et al. Jun activation domain-binding protein 1 expression in breast cancer inversely correlates with the cell cycle inhibitor p27(Kip1) Cancer Res. 2003;63:2977–2981. [PubMed] [Google Scholar]
- Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–S55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, et al. Activity of stabilized siRNAs in a mouse model of HBV replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, et al. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464–477. doi: 10.1038/sj.gt.3302694. [DOI] [PubMed] [Google Scholar]
- Panattoni M, Sanvito F, Basso V, Doglioni C, Casorati G, Montini E, et al. Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J Exp Med. 2008;205:465–477. doi: 10.1084/jem.20070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- Rossi JJ. SNALPing siRNAs in vivo. Gene Ther. 2006;13:583–584. doi: 10.1038/sj.gt.3302661. [DOI] [PubMed] [Google Scholar]
- Rust C, Gores GJ. Apoptosis and liver disease. Am J Med. 2000;108:567–574. doi: 10.1016/s0002-9343(00)00370-3. [DOI] [PubMed] [Google Scholar]
- Shackleford TJ, Claret FX. JAB1/CSN5: a new player in cell cycle control and cancer. Cell Div. 2010;5:26–39. doi: 10.1186/1747-1028-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sui L, Dong Y, Ohno M, Watanabe Y, Sugimoto K, Tai Y, et al. Jab1 expression is associated with inverse expression of p27(kip1) and poor prognosis in epithelial ovarian tumors. Clin Cancer Res. 2001;7:4130–4135. [PubMed] [Google Scholar]
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Tsubari M, Taipale J, Tiihonen E, Keski-Oja J, Laiho M. Hepatocyte growth factor releases mink epithelial cells from transforming growth factor beta1-induced growth arrest by restoring Cdk6 expression and cyclin E-associated Cdk2 activity. Mol Cell Biol. 1999;19:3654–3663. doi: 10.1128/mcb.19.5.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Andersen DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD. RNA interference: listening to the sound of silence. Nat Struct Biol. 2001;8:746–751. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.