Abstract
In TNF treated cells, TNFR1, TRADD, FADD, and RIP proteins form the signaling complex via modular interaction within their C-terminal death domains. Here we report that the death domain SXXE/D motifs, i.e., S381DHE motif of TNFR1-death domain, S215LKD and S296LAE motifs of TRADD-death domain are phosphorylated and this is required for stable TNFR1-TRADD complex formation and subsequent activation of NF-κB. Phospho-S215LKD and phospho-S296LAE motifs are also critical to TRADD for recruiting FADD and RIP. IKKβ plays a critical role in TNFR1 phosphorylation of S381, which leads to subsequent T cell migration and accumulation. Consistently, we observed in IBD specimens that TNFR1 was constitutively phosphorylated on S381 in those inflammatory T cells, which had accumulated in high numbers in the inflamed mucosa. Therefore, SXXE/D motifs found in the cytoplasmic domains of many TNFR family members and their adaptor proteins may serve to function as a specific interaction module for the α-helical death domain signal transduction.
Introduction
Tumor necrosis factor-α (TNF) is a proinflammatory cytokine, which can target its two cognate receptors and initiate the activation of NF-κB, caspase and the JNK pathways, leading to immune cell gene regulation, apoptosis and/or their immune cell activation. TNF bound TNF receptor 1 (TNFR1) recruits TNF receptor associated death domain protein (TRADD), an adaptor protein which serves as the platform for a further recruitment of receptor interacting protein kinase (RIP) and Fas associated death domain protein (FADD), initiating both NF-κB activation and apoptosis induction (1–3). The C-terminals of TNFR1, TRADD, FADD, and RIP all carry a discrete region termed the “death domain”, which is composed of six continuous α-helical bundles and responsible for homotypic interactions among these four proteins. The death domain has about 80~100 amino acids, similar in size to the SH2 domain, both of which can be packed into a similar globular structure. While the phosphotyrosine within the YXXL/I/V/Q/M sequence can be recognized by the SH2 domain during protein-protein interaction, it has been shown that the hydrophobic residue at the p+3 position is also important for SH2 domain recognition as revealed by synthesized peptide binding analysis (4).
For SH2 domain bearing proteins, such as growth factor receptor adaptor proteins or transcription factors like STATs, interactions between them and their substrates are cytokine or growth factor stimulation dependent. In the same way, for death domain proteins, the formation of the death domain complex is TNF stimulation dependent. One possible mechanism controlling this modular interaction is death domain phosphorylation, which depend on its nature/target may change the local conformation to favor its association with various different proteins. TNFR1 tyrosine phosphorylation has been reported and phosphotyrosine residue within the YXXL/V sequence has been identified within the death domain of TNFR1 or other death receptors (5, 6). Initial reports suggested that TNFR1 tyrosine phosphorylation had a negative effect on TNF induced NF-κB activation and growth modulation (6, 7). However, no consensus death domain recognition motif with phosphotyrosine has been established from sequence alignment or binding analysis among these death domain factors. Also the significance of TNFR1 serine phosphorylation on death domain signaling remains elusive, despite the reports of TNFR1 and FADD serine phosphorylation (7–9).
In this work, we have analyzed TNFR1 and TRADD serine phosphorylation and investigated the role of the SXXE/D motif-dependent TNF signal transduction. Conserved SXXE/D motifs have been identified in the death domains of TNFR1, TRADD, FADD, and RIP. We show here that the phospho-SXXE/D motifs of TNFR1 and TRADD death domains play a critical role in the death domain-death domain protein interaction and death domain signal transduction. TNF can also induce NF-κB activation or other cellular responses in a variety of cell types. We show in this study that TNF-induced and IKKβ-dependent TNFR1 death domain S381 phosphorylation plays an essential role in TNFR1-TRADD interaction, leading to NF-κB activation, T cell proliferation, apoptosis and migration. TNF plays acentral role in the pathophysiology of IBD as well as rheumatoid arthritis. Anti-TNF therapy has been documented to have clinical efficacy in treating these inflammatory diseases. Here we provide evidence that in IBD accumulating T cells in the inflammed intestinal tissues, may do soin response to constitutive TNFR1 S381 phosphorylation in these inflammatory lymphocytes. Together these findings provide insights into the determinants of specificity for death domain complex formation during signal transduction, which leads to inflammatory T cells activation, proliferation, and migration. Inasmuch; we suggest that SXXE/D motifs of death domain receptors or adaptors may provide novel targets for future therapeutic intervention.
Materials and Methods
Cell lines and Reagents
Jurkat, Daudi, H9, and SKW6.4 obtained from ATCC and thymocytes or splenocytes prepared from mice were grown in RPMI medium supplemented with 10% FBS at densities between 1×105~106 cells/ml. HeLa and 293T cells from ATCC were grown in DMEM medium supplemented with 10% FBS. Y→F substituted human TNFR1 (TNFR1-Y360F, TNFR1-Y401F, and TNFR1-Y360F/Y401F), S381→A substituted TNFR1 (TNFR1-S381A) and E384→A substituted TNFR1 (TNFR1-E384A), S→A substituted human TRADD (TRADD-S215A, TRADD-S296A and TRADD-S215A/S296A) were constructed by performing site-directed mutagenesis according to our published protocol (10). TNFR1 with S381DHE motif deletion mutation was constructed by designing two primers with two-step PCR amplification. TNFR1 and TRADD mutants were subcloned into pcDNA3.1 vector and pGK5 vector respectively. TNFR1 was also subcloned with a 6xc-Myc tag at its C-terminal end in pcDNA3.1 vector. GST-TNFR1 cDNA construction and protein preparation were described previously (11). Human IKKα and IKKβ were subcloned with Flag-tag in pcDNA3.1 vector. All these mutations were confirmed by DNA sequencing. Transfections were performed with lipofectamine 2000 by following the standard protocol (Invitrogen).
The polyclonal antibody against phospho-S381-TNFR1 was raised in rabbit against a synthetic phospho-peptide “GLpSDHEIDRLE”. Antibodies against TNFR1 (sc-7895), RIP (sc-7881), TRADD (sc-7868), Myc (sc-40), IκBα (sc-203), SOCS3 (sc-154), Rel-A (sc-7151), FADD (sc-271748), actin (sc-130301), GAPDH (sc-166545), protein A/G-agarose beads (sc-2003), and κB-site DNA oligo-beads (sc-372X) were purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA). Monoclonal anti-Flag was purchased from Sigma-Aldrich Corp (St. Louis, MO). IKKβ inhibitor PS-1145 was obtained from Millennia Cop. Both human and mouse TNF and human IL-15 were purchased from R&D Systems (Minneapolis, MN); LPS was from Sigma.
T and B cells preparation and migration and immunostaining in situ
Human primary T cells from healthy blood donors were prepared by centrifugation of fresh blood through Histopaque of Sigma-Aldrich (St. Louis, MO) and taking the buffy coat. Cells were prepared by adsorbing the monocytes to gelatin coated plates and culturing non-adherent lymphocytes in RPMI medium supplemented with 10% FBS and PHA (1 μg/ml) for 3 days, followed by culturing in 20 ng/ml IL-15 for 3~6 days. Flow cytometric analysis demonstrated that these cells were >97% CD3 positive and CD56 negative. B cells were prepared from mouse spleen or human blood of healthy donors according to the published protocol (12). Flow cytometric analysis demonstrated that these enriched B cells were >97% positive for CD19 expression and lacked expression of CD4 and CD8. Real time migration of human primary T cells on cover glass with ICAM-1 and with or without TNF (50~200 ng/ml) treatment were studied by time-lapse video microscopy. Time-lapse movies were taken every 5 seconds. T cells were maintained at 37 °C with a FCS2 live cell imaging chamber (Bioptechs) in IL-15 (20 ng/ml) contained glucose medium. Intestinal tissue samples obtained from non-IBD (normal), Crohn’s disease, and ulcerative colitis were immunostained with anti-pS381-TNFR1, anti-CD3 and anti-CD20 according to the published protocol (13).
Coimmunoprecipitation, protein phosphorylation assay, DNA binding, luciferase reporter assay, and apoptosis analysis
(i) For coimmunoprecipitation, equal amount of cell lysates prepared in RIPA lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.5%Nonidet P-40) containing protease inhibitor cocktail, were incubated with the protein A/G-agarose bead-conjugated antibody overnight at 4°C. The antibody-beads were extensively washed and suspended in Laemmli Sample Buffer (Bio-Rad), boiled for 3–5 min. Such prepared immunoprecipitates or the whole cell lysates (5~10 μg) were analyzed by loading into 10% SDS-PAGE for electrophoresis, transferred to nitrocellulose membrane, and followed by Western blotting analysis. (ii) For TNFR1 or TRADD phosphorylation analysis in 293T cells, TNFR1 was transiently transfected in 293T cells for 24 hrs followed by 32P-orthophosphate metabolic labeling for additional 6 hrs. Immunoprecipitated TNFR1 proteins were analyzed in 10% SDS-PAGE. Radiolabeled proteins transferred to the nitrocellulose membrane were exposed to X-ray film for autoradiography. TNFR1 expression level was analyzed with anti-TNFR1 blot. For TNFR1 phosphorylation in vitro, immunopurified Flag-IKKα, Flag-IKKβ, or Flag-RIP (500 ng) was incubated in kinase reaction buffer containing 10μCi of γ-32P-ATP, 1 mM ATP, 10 mM MgCl2, 1 mM dithiothreitol, 100 mM NaCl, and 50 mM Tris-HCl (pH 7.8) at 30°C for 15 min. The substrate in these in vitro reactions was either glutathione-S-transferase(GST)-TNFR1 (1 μg). In a separate experiment, GST-TNFR1 wild-type (WT) or GST-TNFR1-S381A was incubated with purified Flag-IKKβ in kinase reaction buffer without isotope and TNFR1 phosphorylation was examined by blotting with anti-pS381-TNFR1. (iii) For DNA binding analysis, nuclear fractions were incubated with κB-site DNA oligo-beads overnight at 4°C. After extensive washes, the DNA beads binding NF-κB were analyzed in Western blot with anti-Rel A. (iv) For luciferase activity analysis, 293T cells (1×105) were transfected with lipofectamine 2000. A dual-luciferase reporter assay system was applied according to the instructions of the manufacturer (Promega). pRL vector (30 ng) was transfected along with 2xκB-luciferase reporter (300 ng) and 2 μg of either empty vector or TNFR1 or TRADD in WT or mutant forms. Firefly luciferase activity was measured in a luminometer and normalized on the basis of Renilla luciferase activity. (v) For cell viability analysis, cells were either stained with propidium iodide (PI) (0.5 mL of PI staining reagent: 0.1% NP-40, 0.1% Sodium Citrate and 50mg/mL PI; Sigma-Aldrich Corp) for flow cytometric analysis or stained with trypan blue for exclusion counting (14).
Results
TNF induced T cell morphological change is serine phosphorylation dependent
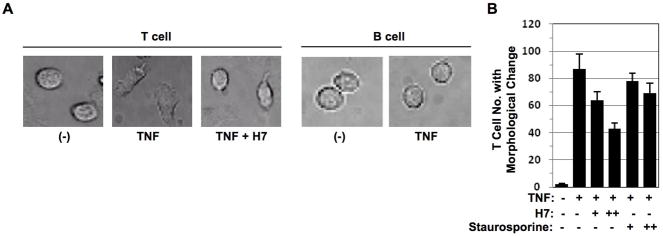
T cells but not B cells responded to TNF stimulation by a morphological change during cell migration (Fig. 1A). However, this morphological change in T cells was inhibited by treatment of serine kinase inhibitor H7 (Fig. 1A). TNF-induced T cell numbers with morphological changes were reduced by treatment of staurosporine (Sigma) another serine kinase inhibitor, to a lesser extent when compared with H7 (Fig. 1B). These data indicated that T cell morphological change induced by TNF was regulated by serine phosphorylation.
Figure 1. TNF induced T cell morphological change is serine phosphorylation dependent.
(A) Peripheral T cells and peripheral B cells obtained from healthy blood donors were cultured in the chambers coated with ICAM-1 in the presence or absence of TNF (200 ng/ml) with or without serine kinase inhibitor H7 pretreatment for 1hr and immediately subjected to time-lapse fluorescence imaging at 37°C. Cell morphological changes were recorded at time 0 or 30 min after. (B) Peripheral T cells and peripheral B cells obtained from healthy blood donors were cultured in the chambers coated with ICAM-1 in the presence or absence of TNF (50 ng/ml) with or without different dose of serine kinase inhibitors H7 or staurosporine pretreatment for 1 hr and immediately subjected to time-lapse fluorescence imaging at 37°C. Cell numbers with morphological changes were counted after 30 min. Data presented are mean ± SD. of triplicate determinations.
TNFR1 and TRADD death domains are phosphorylated on the SXXE/D motif
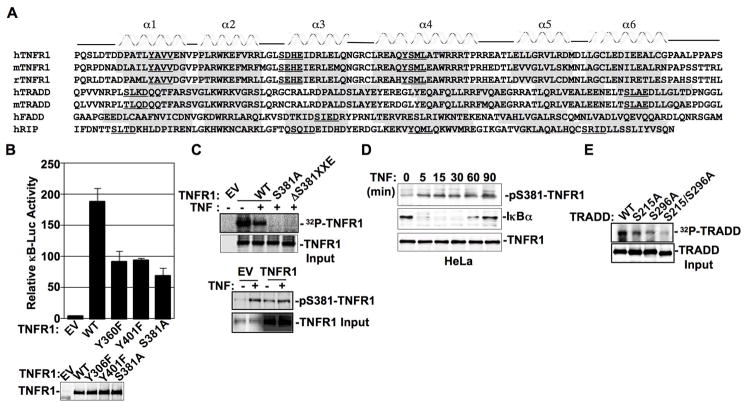
Secondary structure-based alignment indicates that TNFR1, TRADD, FADD, and RIP bear YXXL/V motifs and/or SXXE/D motifs in different α-helical segments of their death domains (Fig. 2A). TNFR1 death domain carries two YXXV/L motifs (Y360AVV and Y401SML) and one SXXE motif (S381DHE) conserved in all species (5–7). Y401XXL motif of α-helix-4 is conserved on all death receptors as well as on RIP whose phosphorylation has been reported to be necessary for recruiting the SH2 domain bearing-tyrosine phosphatase SHP-1 (6).
Figure 2. The death domains of TNFR1 and TRADD bear phospho-SXXE/D motifs.
(A) The death domains of TNFR1, TRADD, FADD, and RIP from different species were analyzed with the 3D-PSSM web tool version 2.5.6 (http://www.sbg.bio.ic.ac.uk/~3dpssm/) for α-helix and β-sheet prediction. (B) Empty vector (EV), wild-type (WT) TNFR1, TNFR1-Y360F, TNFR1-Y401F, or TNFR1-S381A was cotransfected with 2xκB-luciferase reporter in 293T cells for 24 hrs followed by luciferase reporter activity measurement. Data presented are mean ± S.D. of triplicate determinations. One representative anti-TNFR1 Western blot of indicated TNFR1 forms in 293T transfectants was shown in low panel. (C) EV, or indicated form of TNFR1 was transiently transfected in 293T cells followed by 32P-orthophosphate metabolic labeling. Immunoprecipitated TNFR1 proteins transferred to the nitrocellulose membrane were exposed to x-ray film for autoradiography. TNFR1 expression level was analyzed with anti-TNFR1 blot. In low panel, EV or TNFR1 transiently transfected 293T cells were treated with TNF for 30 min followed by Western blot analysis with anti-pS381-TNFR1 or anti-TNFR1. (D) Whole cell extracts, prepared from HeLa cells treated with TNF for indicated times, were subjected to Western blot analysis with anti-pS381-TNFR1, anti-IκBα, or anti-TNFR1. (E) TRADD of indicated forms were transiently transfected in 293T cells for 24 hrs followed by 32P-orthophosphate metabolic labeling for additional 6 hrs. The autoradiography of the immunoprecipitated TRADD were analyzed as described above. TRADD expression level was analyzed with anti-TRADD blot.
In 293T cells, while wild type TNFR1 was shown to recruit the SH2 domain bearing SOCS3, TNFR1 with Y401→F substitution attenuated TNFR1:SOCS3 interaction (data not shown). To determine the importance of these YXXL/V motifs in TNFR1 death domain signaling, we examined TNFR1 and TRADD interaction, in which single or doubly Y→F mutated TNFR1, i.e., TNFR1-Y360F, TNFR1-Y401F, and TNFR1-Y360F/Y401F, were produced. These mutants showed no apparent defect either in TNFR1:TRADD interaction or in their capacity to induce apoptosis (Figs. S1, S2), even though partially inhibiting NF-κB activation (Fig. 2B). By contrast, TNFR1 with S381→A substitution (TNFR1-S381A) inhibited both NF-κB activation and apoptosis induction significantly (Fig. 2B, Fig. S3). With E384→A substitution, TNFR1-E384A also affected NF-κB activation (Fig. S3), suggesting SXXE/D motif as a whole is important for phospho-SXXE/D motif modulatory signaling. These data suggest a more prominent role of serine phosphorylation than tyrosine phosphorylation in TNFR1 death domain dependent intracellular signal transduction.
We subsequently examined phosphorylation of the TNFR1 death domain SXXE motif in greater detail. TNFR1-S381A mutant or TNFR1 with “S381DHE” motif deletion mutation (TNFR1-ΔS381XXE) largely abolished TNFR1 phosphorylation in 293T cells, demonstrating that TNFR1 mainly undergoes serine phosphorylation rather than tyrosine phosphorylation (Fig. 2C). To specifically detect TNFR1 phosphorylation on S381, we prepared a polyclonal antibody against phospho-S381 of TNFR1. Utilizing this antibody, TNF-dependent TNFR1 phosphorylation on S381 was clearly detected in 293T cells (Fig. 2C lower panel) or in HeLa cells (Fig. 2D). It is worth noting that when overexpressed in 293T cells, TNFR1 became constitutively phosphorylated on S381 (Fig. 2C, lower panel).
TRADD, FADD and RIP death domains all bear SXXE/D motifs and TRADD death domain bears two SXXE/D motifs, i.e., S215XXD ofα-helix1 and S296XXE of α-helix6 (Fig. 2A). The fact that two bands of TRADD were often visible in different cell types (Fig. S4) suggests that TRADD was posttranslationally modified. In 293T cells, 32P-orthophosphate labeling analysis confirmed TRADD phosphorylation (Fig. 2E), While TRADD with S215→A substitution (TRADD-S215A) or S296→A substitution (TRADD-S296A) attenuated p32-phosphorylation intensity, TRADD with S215→A and S296→A double mutations (TRADD-S215A/S296A), which migrated faster in polyacrylamide gel, nearly swept away TRADD phosphorylation completely (Fig. 2E). Therefore, TRADD is phosphorylated on both SXXE/D motifs within the death domain.
The phospho-SXXE/D motif intermediates the death domain modular interaction
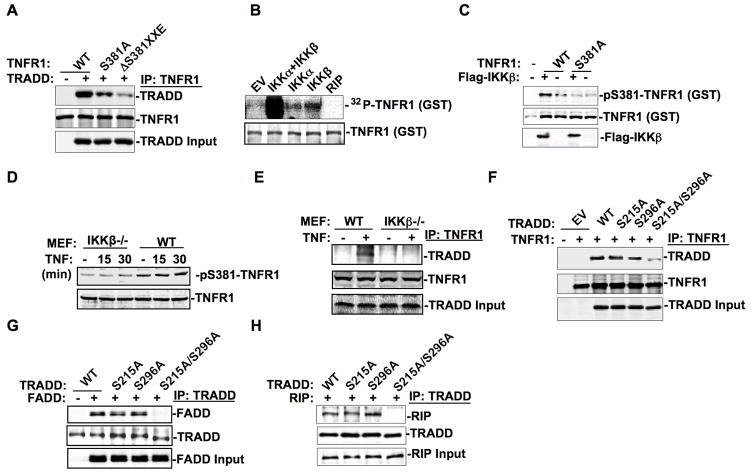
To investigate the role of phospho-SXXE/D motifs in death domain-death domain protein interaction, TRADD was cotransfected with wild type TNFR1 (TNFR1-WT) or TNFR1 mutant i.e., TNFR1-S381A, or TNFR1-ΔS381XXE in 293T cells. TNFR1-TRADD interaction was inhibited by TNFR1 with S381A mutation and worsened by TNFR1-with S381XXE deletion as revealed by coimmunoprecipitation analysis (Fig. 3A). We also examined TNFR1-TRADD complex stability by carrying out a peptide competition experiment, in which the synthesized unphosphorylated or phosphorylated peptide containing S381XXE motif (position 375–395) was incubated with the immuno-purified TNFR1-TRADD complex respectively. At a concentration of 300 nM, the phospho-peptide but not the control peptide disrupted the TNFR1-TRADD complex, indicating that phospho-S381XXE motif of TNFR1 death domain is involved in the formation of a stable TNFR1-TRADD complex (Fig. S5).
Figure 3. The phospho-SXXE/D motif intermediates the death domain modular interaction.
(A) In 293T cells, TNFR1 of indicated forms were cotransfected with TRADD followed by coimmunoprecipitation analysis. Anti-TNFR1 immunoprecipitates were subjected to Western blot analysis with anti-TRADD or anti-TNFR1. (B) Flag-tagged IKKα, IKKβ, and RIP were immunoprecipitated from 293T transfectants and were incubated with GST-TNFR1 purified from bacteria in phosphorylation reaction buffer containing γ-32P-ATP for 15 min at 30°C. Radiolabeled proteins were separated in 10% SDS-PAGE, transferred to the nitrocellulose membrane, and exposed to X-ray film for autoradiography. (C) Purified GST-TNFR1 was incubated with purified Flag-IKKβ in reaction buffer followed by Western blot analysis with anti-pS381-TNFR1 or indicated antibodies. (D) Whole cell extracts, prepared from wild-type or IKKβ−/−MEFs treated with TNF for indicated times, were subjected to Western blot analysis with anti-pS381-TNFR1 or anti-TNFR1. (E) Whole cell extracts were prepared from wild-type or IKKβ deficient MEFs treated with or without TNF (50 ng/ml) for 15min. Anti-TNFR1 immunoprecipitates were analyzed with anti-TRADD in Western blot. (F–H) In 293T cells, TRADD of indicated forms were transfected along with TNFR1 (F), FADD (G), or RIP (H) followed by coimmunoprecipitation with anti-TNFR1 (F) or anti-TRADD (G, H) and blotted with indicated antibodies in Western blot analysis.
We next sought to identify the protein kinase responsible for TNFR1 phosphorylation on S381. Coimmunoprecipitation analysis revealed that both IKKα and IKKβ could form complex with TNFR1 in 293T cells with IKKα or IKKβ transfected along with TNFR1 (Fig. S6, S7). Bacteria purified GST-TNFR1 was applied to an in vitro kinase assay in the presence ofγ-32P-ATP, revealing a synergistic effect between IKKα and IKKβ on TNFR1 phosphorylation, despite that IKKβ alone was more efficient than IKKα in TNFR1 phosphorylation induction (Fig. 3B). When purified TNFR1, WT but not S381A form, was incubated with purified Flag-IKKβ, in an in vitro reaction, TNFR1-S381 phosphorylation was clearly detected with the antibody against pS381-TNFR1(Fig. 3C). This supports the conclusion that IKKβ is able to phosphorylate TNFR1 on S381 in 293T cells (Fig. S8). However, in IKKβ−/− MEFs, there was a basal level of TNFR1 S381 phosphorylation (Fig. 3D), which was most likely contributed by endogenous IKKα. Importantly, TNFR1-TRADD complex formation was induced by TNF treatment of wild-type MEFs but not in IKKβ−/−MEFs (Fig. 3E), suggesting that IKKβ may play an important role in both TNFR1 and TRADD activation.
Since TNFR1-TRADD complex formation was not completely abolished when TNFR1 was introduced with S381A mutation or S381XXE deletion mutation (Fig.3A), we suspected that TRADD serine phosphorylation was also involved in TNFR1-TRADD complex formation. TNFR1-TRADD complex formation was largely abolished when both S215A and S296A mutations were introduced (Fig. 3F), suggesting that both pS215XXD and pS296XXE motifs of TRADD were involved in TNFR1-TRADD interaction (2, 15). Moreover, TRADD phosphorylation on S215XXD and S296XXE motifs performed an important role in TRADD-FADD or TRADD-RIP complex formation, as TRADD-S215A/S216A but not TRADD-S215A or TRADD-S296A failed to recruit FADD or RIP in 293T cells (Fig. 3, G and H). These results strongly indicate that the two phospho-SXXE/D motifs of the TRADD death domain play a central role in this death domain-tetramer complex formation.
NF-κB dependent gene regulation requires the SXXE/D motif phosphorylation
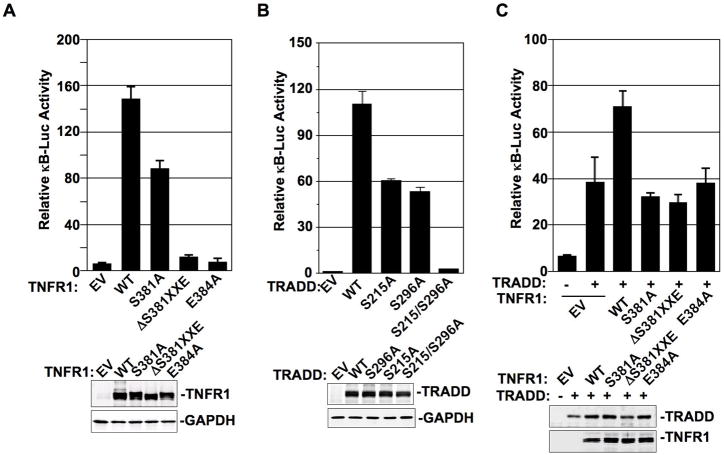
Given that TNFR1-TRADD complex formation led to IκB degradation and NF-κB nuclear translocation, a potentiating effect of the SXXE/D motif phosphorylation in their death domains was expected to initiate NF-κB activation. In 293T cells, transient transfection of either TNFR1 or TRADD alone could trigger NF-κB transcriptional activation, as reflected by the results of NF-κB dependent luciferase reporter activation assays of Fig.4, A and B. While TNFR1-S381A, TRADD-S215A, and TRADD-S296A mutants reduced NF-κB activation, TNFR1-ΔS381XXE, TNFR1-E384A, and TRADD-S215A/S296A mutants almost completely abrogated NF-κB activation (Fig.4, A and B). Along these lines, we found that TRADD-dependent NF-κB activation was further enhanced by co-transfecting TRADD with TNFR1-WT but not with TNFR1 mutants (Fig. 4C). These results strongly support our model that both the phosphorylated serine and the negatively charged p+1 residue of the “SXXE/D motif” in the death domains are important in NF-κB activation.
Figure 4. NF-κB-dependent gene regulation requires the SXXE/D motif phosphorylation.
(A) EV and different forms of TNFR1 were transiently transfected with 2xκB-site luciferase reporter in 293T cells for 24 hrs followed by luciferase reporter activity measurement. Data presented are mean ± S.D. of triplicate determinations. Experiments were repeated three times. One representative anti-TNFR1 Western blot of indicated TNFR1 forms in 293T transfectants was shown in low panel. (B) EV and different forms of TRADD were transiently transfected with 2xκB-site luciferase reporter in 293T cells for 24 hrs followed by luciferase reporter activity measurement and data process as described in (A). (C) EV or TRADD WT was transfected with 2xκB-site luciferase reporter alone or with different forms of TNFR1 cotransfection in 293T cells for 24 hrs followed by luciferase reporter activity measurement and data process as described in (A).
TNF activates NF-κB in T cells via TNFR1 S381 phosphorylation and TNFR1-TRADD complex formation
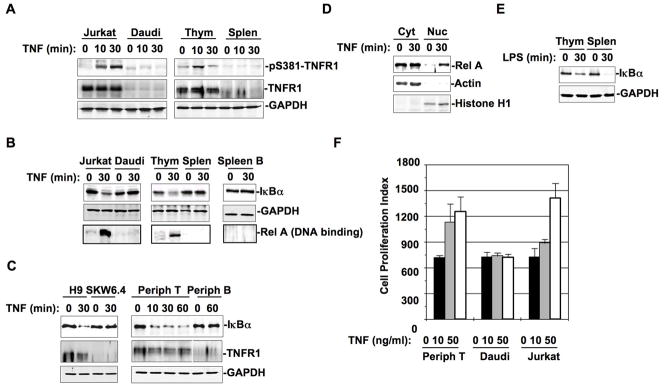
It has been reported that both T and B cells respond to TNF for activation during inflammatory processes. However, TNF treatment for 10 or 30 min induced TNFR1 S381 phosphorylation in Jurkat T lymphoma line and mouse thymocytes that were primarily T cells, but not in Daudi cells, a B lymphoma line, or mouse splenocytes that contained 40% B cells (Fig. 5A). TNF treatment of Jurkat cells induced the formation of a complex between TNFR1 and TRADD (Fig. S9), supporting the conclusion that serine phosphorylated TNFR1 and serine phosphorylated TRADD form modulatory protein-protein interaction upon TNF treatment. TNF treatment induced IκB degradation and NF-κB activation for DNA binding in Jurkat cells and mouse thymocytes but not in Daudi cells, mixed mouse splenocytes or purified mouse spleen B cells (Fig. 5B). When another pair of human T and B lymphoma lines, H9 and SKW6.4, or purified peripheral T and B cells from normal human blood donors were tested, IκB (IκBα) degradation and NF-κB (Rel A) nuclear translocation were only detected in TNF-treated T cells and not in TNF-treated B cells (Fig. 5, C and D).
Figure 5. TNF activates NF-κB in T cells via TNFR1-S381 phosphorylation and TNFR1-TRADD complex formation.
(A) Jurkat cells, Daudi cells, mouse thymocytes, and mouse splenocytes were treated with TNF (50 ng/ml) for different times as indicated. Whole cell extracts (equal amount) were subjected to Western blot analysis with anti-pS381-TNFR1 or anti-TNFR1. (B) Jurkat cells, Daudi cells, mouse thymocytes, mouse splenocytes, and purified mouse spleen B cells were treated with or without TNF for 30 min. While the cytosolic fractions prepared from these cells were analyzed with anti-IκBα in Western blot (upper panel), the nuclear fractions were incubated with κB-site oligo-agarose beads overnight at 4°C. DNA precipitated proteins were subjected to Western blot analysis with anti-RelA (low panel). (C) Whole extracts from H9 and SKW6.4 cells treated with or without TNF (50 ng/ml) for 30 min were analyzed with anti-IκBα or anti-TNFR1 in Western blot. Human peripheral T and B cells from normal blood donors were treated with TNF for different times as indicated. Whole cell lysates prepared from these peripheral T or B cells were analyzed with anti-IκBα or anti-TNFR1 in Western blot. (D) Cytosolic and nuclear fractions from above human peripheral T cells treated with or without TNF for 30 min were analyzed for Rel-A nuclear translocation in Western blot. (E) Whole cell extracts of mouse thymocytes or splenocytes treated with or without LPS (10 ng/ml) for 30 mins were analyzed with anti-IκBα in Western blot. (F) Equal amount (4×105) of peripheral T cells, Daudi cells, and Jurkat cells were incubated with or without TNF (10 or 50 ng/ml) for 24 hrs in 96-well plate. Cell proliferation was determined by applying CyQUANT cell proliferation assay kit from Invitrogen (Carlsbad, CA) according to the protocol. Cells were lysed by addition of 100 μl CyQUANT GR dye buffer and the fluorescence intensity was measured with a microplate fluorescence reader FLx800 from Bio-Tek Instruments (Winooski, VT) with excitation at 485nm and emission detection at 530 nm. The fluorescence intensity is presented as cell proliferation index.
The significantly lower expression level of TNFR1 by B cells has been suggested as an explanation for the failure of B cells to response to TNF treatment by NF-κB activation (Fig. 5, A and C). However, TNFR1 associated death domain factors including TRADD, FADD, and RIP are all expressed at comparable levels in both B and T cells, while TNFR2 expression level was low overall in both T and B cells (data not shown). The TNFR1 associated inhibitor A20 has been reported to induce RIP poly-ubiquitination and inhibit TRADD downstream signal transduction in T cells (15, 16). Utilizing mass spectrometry for proteomic analysis, no apparent difference was detected for proteins associated with TNFR1 in B cells with or without treatment of A20 or other established inhibitors (data not shown). LPS induced IκB degradation in both thymocytes and splenocytes (Fig. 5E), suggesting that a similar level of IKK activity is present in T and B cells.
We then examined the effect of TNF stimulation on T and B cell viability. TNF treatment for 12–24 hrs stimulated proliferation in peripheral T cells and Jurkat cells, but not in Daudi cells (Fig. 5F). We also compared different sources of T and B cells for their apoptotic response to TNF treatment. In this regard, we found that TNF mediated apoptosis could be detected in Jurkat cells or thymocytes, whereas only a spontaneous or background level of apoptosis was observed in Daudi cells or splenocytes (Fig. S10). These results indicate that TNF can selectively activate T cells over B cells via NF-κB activation and this appears to require serine phosphorylation-dependent TNFR1-TRADD complex formation.
TNF induces T cells migration via TNFR1 S381 phosphorylation
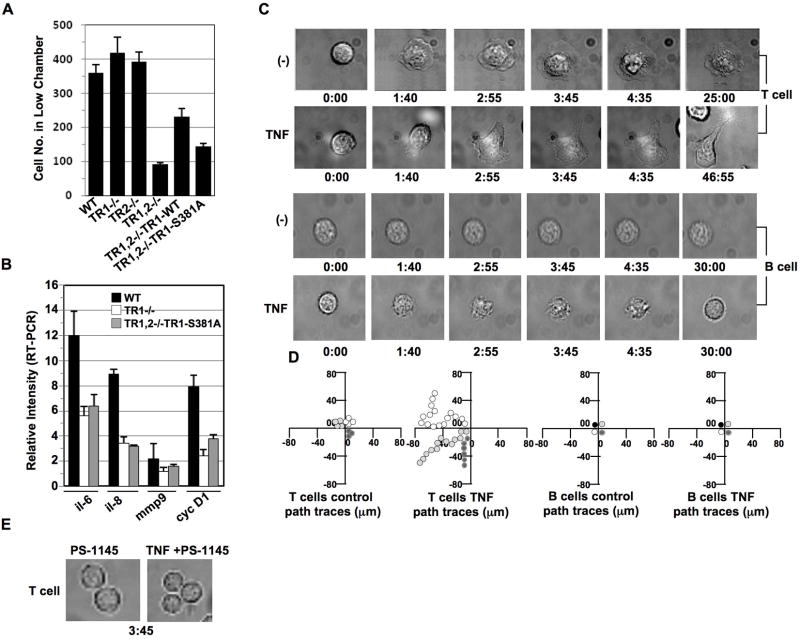
Among TNF’s variety of actions are the regulation of cell viability and the induction of cell migration. To investigate the role of TNFR1 in regulating cell migration, fibroblasts obtained from wild type, TNFR1−/−, TNFR2−/−, and TNFR1/TNFR2 double−/− mice were assessed for their comparative ability to migrate using a cell chamber migration assay. Interestingly, while TNFR2 has been previously shown to play a more critical role than TNFR1 in cell migration (17), we observed here that neither TNFR1−/− alone nor TNFR2−/− alone showed any negative effect on cell migration (Fig. 6A). However, cell migration was severely inhibited when both TNFR1 and TNFR2 were knock out (Fig. 6A); suggesting that TNFR1 and TNFR2 play a collaborative role in regulating cell migration. Furthermore, reconstitution of TNFR1-WT but not TNFR1-S381A mutant form into TNFR1/TNFR2 −/− partially restored cell migration (Fig. 6A), indicating that TNFR1 involvement in cell migration is mediated through S381 phosphorylation. We then performed RT-PCR to examine the expression of genes related to cell migration in these TNFR1−/− or TNFR1, TNFR2−/− MEFs. Among those genes assessed, IL-6, IL-8, Matrix metallopeptidase-9 (MMP-9), and cyclin D1 were apparently reduced in their expression in TNFR−/− MEFs when compared with wild type MEFs (Fig. 6B). The introduction of TNFR1-S381A mutant in these TNFR1/TNFR2−/− MEFs showed little or no rescue effect on regulation of the expression of those genes (Fig. 6B). Thus, we believe that TNFR1 phosphorylation on S381 plays a critical role in NF-κB-dependent gene regulation involved in cell migration and that the presence of both TNFR1 and TNFR2 on the cell is still critical for cell migration.
Figure 6. TNF induces T but not B cell migration.
(A) Equal amount (5×105) of mouse embryonic fibroblasts obtained from TNFR1−/−, TNFR2−/− or TNFR1 and TNFR2 double−/− (TNFR1,2−/−) mice, as well as the TNFR1-WT or TNFR1-S381A reconstituted fibroblasts were seeded in the upper chamber (polycarbonate membranes with 8.0-μm pore size) in DMEM medium supplied with 5% FBS. 12 hrs later, the cells migrated to the low chamber were fixed, stained with H&E, and visualized with a light microscope according to our published protocol (13). The above H&E stained cells in the lower chamber were counted from three vision areas with a light microscope. (B) In TNFR1−/− MEFs or TNFR1, TNFR2−/− MEFs with TNFR1-S381A mutant reintroduction, indicated genes were analyzed for their transcriptional regulation by performing RT-PCR. Data are expressed as mean ± S.D. of three experiments. (C) Peripheral T cells and peripheral B cells obtained from healthy blood donors were cultured in the chambers coated with ICAM-1, in the presence or absence of TNF (200 ng/ml) and immediately subjected to time-lapse fluorescence imaging at 37 °C. Cell morphological changes were recorded at different times (min) as indicated. Experiments were repeated on T lymphocyte preparations from three independent donors. (D) The migration of the above T and B cells in (C) was tracked over a 30-min period. Each line represents one cell. (E) TNF treatment at indicated time failed to induce cell morphological change in peripheral T cells pretreated with IKKβ inhibitor PS-1145 for 2 hrs.
We then compared T cell and B cell migration in response to TNF treatment. In the absence of TNF, T cells prepared from peripheral blood of normal donors, remained still in their ICAM-1 coated chamber without evidence of observable crawling or movement in a directed fashion (Fig. 6C). In the presence of TNF, T cells became polarized and migrated on the ICAM-1 coated surface with an approximate steady state migration velocity of 10 μm/min (Fig. 6D). These migrating T cells exhibited rapid shape change, formation of constriction rings, and concomitant cytoplasmic streaming. The migrating T cells treated with TNF were comparable with those exposed to treatment for integrin activation (18). Under the same condition, peripheral B cells failed to respond to TNF treatment with respect to the development of directed migration or migration related morphological change (Fig. 6C and D). Pretreatment of T cells with IKKβ inhibitor PS1145 abolished T cell migration in response to TNF stimulation (Fig. 6E). Above results demonstrated that TNFR1 S381 phosphorylation by IKKβ leads to NF-κB activation and downstream gene expression is required for TNF to induce a T cell migration response.
T cells with constitutive expression of TNFR1-S381 phosphorylation accumulate in the inflamed intestines of IBD patients
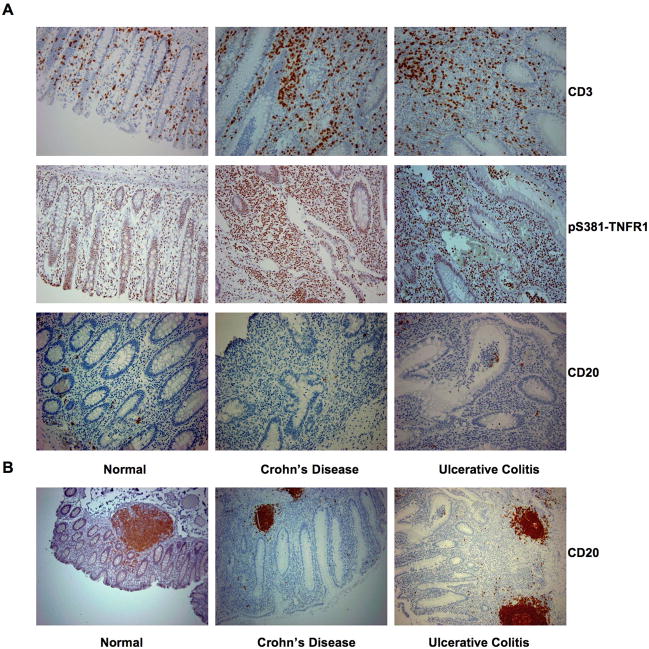
In IBD, TNF plays an essential role in recruiting immune cells to inflammatory sites (19). Intestinal tissue samples obtained from patients of Crohn’s disease or ulcerative colitis were compared with normal intestinal tissues for T cell accumulation. In all the intestinal tissue samples analyzed, T cells became most apparently in the inflamed mucosa in both Crohn’s disease and ulcerative colitis specimens (Fig. 7A). These accumulating T cells were exclusively phospho-S381-TNFR1 positive (Fig. 7A). In contrast, B cells, localized mainly in the intestinal B cell follicles or in the germinal centers, were sparsely visible (Fig. 7B). These results suggest that TNFR1 S381 constitutive phosphorylation is likely involved in activating and/or driving the influx of T cells into the areas of inflammation seen in the bowel (19, 20). Further supporting the requirement of SXXE/D motif phosphorylation during signal transduction via TNFR1-TRADD centered death domain complex formation.
Figure 7. T cells accumulated in the inflammatory intestines of IBD patients are TNFR1-S381 constitutively phosphorylated.
(A) Local intestinal mucosa from control non-IBD, inflamed Crohn’s disease, or inflamed ulcerative colitis were immunostained with anti-CD3, anti-pS381-TNFR1, and anti-CD20 respectively. (B) Intestinal tissue samples obtained from non-IBD, Crohn’s disease, and ulcerative colitis were immunostained with anti-CD20 to show the B cell follicles
Discussion
Like tyrosine phosphorylation, serine phosphorylation plays a critical role in regulating signaling for cell survival, apoptosis, or cell migration (21, 22). SXXE/D motifs, found in the cytoplasmic domains of many TNFR family members as well as their adaptor proteins, are involved in serine signal transduction. However, unlike PXSP or PS/SP motifs that are phosphorylated by MAPK/cdc2PK type kinases for β-stranded WW domain (ββ) interaction (23), SXXE/D motifs are phosphorylated by CK-II/IKK type kinases and have been shown to associate with TNFR1 (24). We demonstrated here that IKKβ and, to a lesser extent, IKKα can phosphorylate the “SXXE” motif of TNFR1 death domain.
In the helical death domain (αααααα) of Fas, the α-helix3 is involved in direct contact with FADD death domain within theα-helix2-α-helix3 region, which bears the SXXE/D motif (25). Besides, S215 of TRADD death domain and positive charge residues of both TRADD and TNFR1 death domains have been previously noted to play a role in the interaction between these two molecules (26, 27). Both SXXE/D motifs of the TRADD death domain were required for upstream TNFR1 and downstream FADD/RIP binding simultaneously, suggesting that TRADD may either form dimer prior to mediating the TNFR1 and FADD/RIP complex formation or sequentially recruit TNFR1, FADD, and RIP. Furthermore, the TNFR1 SXXE motif has also been demonstrated to be important for TNFR1-TRADD complex formation.
These observations are reminiscent of those made for theα-helix sandwiched and β-strand centered SH2 domains of Src (αββββα) and STAT (αβββαα) that bear phosphorylated tyrosine residues (28, 29). For two STAT protein molecules, positively charged residues of αA and βB of their SH2 domains recognize reciprocal phospho-tyrosine residue located at the extended C-terminus of the SH2 domain during dimerization (28). Serine phosphorylation of the SXXE/D motif is important for death domain mediated modular interaction. S381 phosphorylation of TNFR1 death domain SXXE motif may enhance the protein-protein interaction by juxtaposing another negatively charged group adjacent to D382 and E384 for stabilizing its association with the positive groove in TRADD. Comparing phosphotyrosine with, phosphoserine indicates phosphoserine is less bulky in structure. Thus the negatively charged residue at the p+3 position in the death domain SXXE/D motif presumably plays a more important role in strengthening the modular interaction than the hydrophobic residue of YXXL//I/V/M/Q motif does in interacting with the SH2 domain.
The effect of the phospho-SXXE/D motif may not only be limited to the the homologous domain interaction. TNFR2 bears at least three species-conserved SXXE/D motifs within the C-terminus. The helical ankyrin repeats (ANK) of ASB3 and the TRAF-C domain of TRAF2 were found to dock on those TNFR2 SXXE motifs that are most likely phosphorylated (10, 30). Phospho-SXXE/D motifs have also been identified in other helical structures such as ANK of IκBα. Ubiquitin-protein ligases are responsible for phospho-SXXE/D recognition during IκB degradation and the release of NF-κB. Positively charged residue of R47 within the helical ANK repeats of p16INK4α interacts with a negatively charged residue (i.e., Glu) of the N-terminal SXXE motif of cdk4 (31).
The observation that TNF preferentially activates T cells via SXXE motif phosphorylation within TNFR1 death domain and this signaling, may explain why T cells, but not B cells accumulated in areas of inflammation; thus, playing a dominant role in inflammatory process. Although TNF may activate other signaling pathways such as JNK in both T and B cells, SXXE and/or SXXD motif-mediated NF-κB activation appears to play a more critical role in cell growth regulation, apoptosis, cell migration and the accumulation of T cell at sites of inflammation (11, 32). It is tempting to think that TNFR1-S381 may become constitutively phosphorylated in T cells in inflamed intestinal mucosa; presumably due to increased TNF level in the diseased lamina propria as opposed to adjacent un-inflamed bowel. Further we suggest that constitutive phosphorylation of TNFR1 should promote inflammation by regulating T cells proliferation and migration to the inflamed gut of IBD patients (33). Thus, we feel that the disruption of the modular death domain-death domain interaction in T cells, by targeting phospho-SXXE/D motifs, may represent a potentially novel therapeutic approach for the treatment of inflammatory conditions, like IBD.
Supplementary Material
Acknowledgments
We thank for D. V. Goeddel for human TNFR1 and human TRADD plasmids; V. Dixit for human FADD plasmid; B. Seed for human RIP plasmid; S. Ghosh for 2xκB-luciferase reporter construct; G. S. Hotamisligil for wild type, TNFR1−/−, TNFR2−/−, and TNFR1, TNFR2 −/− MEFs; and I. Verma for wild type, IKKα−/−, and IKKβ−/− MEFs.
This research was supported by NIH RO1 grant to Y.E.C. Z.Z. was partially supported by the fellowship from China Scholarship Council (CSC). C.Y. was supported by Shanghai Pujiang Program 09PJ1408600.
Abbreviations used in this article
- TNF
tumor necrosis factor-α
- TNFR1
TNF receptor 1
- TRADD
TNF receptor-associated death domain protein
- RIP
receptor interacting protein kinase
- FADD
Fas associated death domain protein
- ANK
ankyrin repeats
- WT
wild type
- EV
empty vector
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 2.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 3.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 4.Domchek SM, Auger KR, Chatterjee S, Burke TR, Jr, Shoelson SE. Inhibition of SH2 domain/phosphoprotein association by a nonhydrolyzable phosphonopeptide. Biochemistry. 1992;31:9865–9870. doi: 10.1021/bi00156a002. [DOI] [PubMed] [Google Scholar]
- 5.Darnay BG, Aggarwal BB. The p80 TNF receptor-associated kinase (p80TRAK) associates with residues 354–397 of the p80 cytoplasmic domain: similarity to casein kinase. FEBS Lett. 1997;406:101–105. doi: 10.1016/s0014-5793(97)00251-2. [DOI] [PubMed] [Google Scholar]
- 6.Daigle I, Yousefi S, Colonna M, Green DR, Simon HU. Death receptors bind SHP-1 and block cytokine-induced anti-apoptotic signaling in neutrophils. Nat Med. 2002;8:61–67. doi: 10.1038/nm0102-61. [DOI] [PubMed] [Google Scholar]
- 7.Gambelli F, Di P, Niu X, Friedman M, Hammond T, Riches DW, Ortiz LA. Phosphorylation of tumor necrosis factor receptor 1 (p55) protects macrophages from silica-induced apoptosis. J Biol Chem. 2004;279:2020–2029. doi: 10.1074/jbc.M309763200. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy NJ, Budd RC. Phosphorylation of FADD/MORT1 and Fas by kinases that associate with the membrane-proximal cytoplasmic domain of Fas. J Immunol. 1998;160:4881–4888. [PubMed] [Google Scholar]
- 9.Van Linder AA, Cottin V, Leu C, Riches DW. Phosphorylation of the membrane proximal region of tumor necrosis factor receptor CD120a (p55) at ERK consensus sites. J Biol Chem. 2000;275:6996–7003. doi: 10.1074/jbc.275.10.6996. [DOI] [PubMed] [Google Scholar]
- 10.Chung AS, Guan YJ, Yuan ZL, Albina JE, Chin YE. Ankyrin repeat and SOCS box 3 (ASB3) mediates ubiquitination and degradation of tumor necrosis factor receptor II. Mol Cell Biol. 2005;25:4716–4726. doi: 10.1128/MCB.25.11.4716-4726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Wu TR, Cai S, Welte T, Chin YE. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol. 2000;20:4505–4512. doi: 10.1128/mcb.20.13.4505-4512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li CK, Pender SL, Pickard KM, Chance V, Holloway JA, Huett A, Goncalves NS, Mudgett JS, Dougan G, Frankel G, MacDonald TT. Impaired immunity to intestinal bacterial infection in stromelysin-1 (matrix metalloproteinase-3)-deficient mice. J Immunol. 2004;173:5171–5179. doi: 10.4049/jimmunol.173.8.5171. [DOI] [PubMed] [Google Scholar]
- 13.Yuan ZL, Guan YJ, Wang L, Wei W, Kane AB, Chin YE. Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol Cell Biol. 2004;24:9390–9400. doi: 10.1128/MCB.24.21.9390-9400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi AR, Chung CS, Song GY, Lomas J, Priester RA, Ayala A. F-kappaB activation has tissue-specific effects on immune cell apoptosis during polymicrobial sepsis. Shock. 2002;18:380–386. doi: 10.1097/00024382-200210000-00015. [DOI] [PubMed] [Google Scholar]
- 15.He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22:6034–6045. doi: 10.1128/MCB.22.17.6034-6045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Kondo S, Shivji GM, Fujisawa H, Mak TW, Sauder DN. Tumour necrosis factor receptor II (p75) signalling is required for the migration of Langerhans’ cells. Immunology. 1996;88:284–288. doi: 10.1111/j.1365-2567.1996.tb00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woywodt A, Ludwig D, Neustock P, Kruse A, Schwarting K, Jantschek G, Kirchner H, Stange EF. Mucosal cytokine expression, cellular markers and adhesion molecules in inflammatory bowel disease. Eur J Gastroenterol Hepatol. 1999;11:267–276. doi: 10.1097/00042737-199903000-00010. [DOI] [PubMed] [Google Scholar]
- 19.Kim M, Carman CV, Yang W, Salas A, Springer TA. The primacy of affinity over clustering in regulation of adhesiveness of the integrin {alpha}L{beta}2. J Cell Biol. 2004;167:1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juffermans NP, Paxton WA, Dekkers PE, Verbon A, de Jonge E, Speelman P, van Deventer SJ, van der Poll T. Up-regulation of HIV coreceptors CXCR4 and CCR5 on CD4(+) T cells during human endotoxemia and after stimulation with (myco)bacterial antigens: the role of cytokines. Blood. 2000;96:2649–2654. [PubMed] [Google Scholar]
- 21.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 22.Parker D, Jhala US, Radhakrishnan I, Yaffe MB, Reyes C, Shulman AI, Cantley LC, Wright PE, Montminy M. Analysis of an activator: coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 23.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;28:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 24.Devin A, Lin Y, Yamaoka S, Li Z, Karin M, Liu Zg. The alpha and beta subunits of IkappaB kinase (IKK) mediate TRAF2-dependent IKK recruitment to tumor necrosis factor (TNF) receptor 1 in response to TNF. Mol Cell Biol. 2001;21:3986–3994. doi: 10.1128/MCB.21.12.3986-3994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 26.Park A, V, Baichwal R. Systematic mutational analysis of the death domain of the tumor necrosis factor receptor 1-associated protein TRADD. J Biol Chem. 1996;271:9858–9862. doi: 10.1074/jbc.271.16.9858. [DOI] [PubMed] [Google Scholar]
- 27.Telliez JB, Xu GY, Woronicz JD, Hsu S, Wu JL, Lin L, Sukits SF, Powers R, Lin LL. Mutational analysis and NMR studies of the death domain of the tumor necrosis factor receptor-1. J Mol Biol. 2000;300:1323–1333. doi: 10.1006/jmbi.2000.3899. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 29.Vadlamudi RK, Sahin AA, Adam L, Wang RA, Kumar R. Heregulin and HER2 signaling selectively activates c-Src phosphorylation at tyrosine 215. FEBS Lett. 2003;543:76–80. doi: 10.1016/s0014-5793(03)00404-6. [DOI] [PubMed] [Google Scholar]
- 30.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 31.Byeon IJ, Li J, Ericson K, Selby TL, Tevelev A, Kim HJ, O’Maille P, Tsai MD. Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 32.Delhase M, Li N, Karin M. Kinase regulation in inflammatory response. Nature. 2000;406:367–368. doi: 10.1038/35019154. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Fu YX. Tumor necrosis factor family members and inflammatory bowel disease. Immunol Rev. 2005;204:144–155. doi: 10.1111/j.0105-2896.2005.00218.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.