Abstract
Salmonella enterica serovar Enteritidis (S. Enteritidis) is a major cause of food-borne gastroenteritis in humans worldwide. Poultry and poultry products are considered the major vehicles of transmission to humans. Using cell invasiveness as a surrogate marker for pathogenicity, we tested the invasiveness of 53 poultry-associated isolates of S. Enteritidis in a well-differentiated intestinal epithelial cell model (Caco-2). The method allowed classification of the isolates into low (n = 7), medium (n = 18) and high (n = 30) invasiveness categories. Cell invasiveness of the isolates did not correlate with the presence of the virulence-associated gene spvB or the ability of the isolates to form biofilms. Testing of representative isolates with high and low invasiveness in a mouse model revealed that the former were more invasive in vivo and caused more and earlier mortalities, whereas the latter were significantly less invasive in vivo, causing few or no mortalities. Further characterization of representative isolates with low and high invasiveness showed that most of the isolates with low invasiveness had impaired motility and impaired secretion of either flagella-associated proteins (FlgK, FljB and FlgL) or type III secretion system (TTSS)-secreted proteins (SipA and SipD) encoded on Salmonella pathogenicity island-1. In addition, isolates with low invasiveness had impaired ability to invade and/or survive within chicken macrophages. These data suggest that not all isolates of S. Enteritidis recovered from poultry may be equally pathogenic, and that the pathogenicity of S. Enteritidis isolates is associated, in part, with both motility and secretion of TTSS effector proteins.
Introduction
Salmonella enterica serovar Enteritidis (S. Enteritidis) is the leading cause of food-borne salmonellosis (WHO, 2008). S. Enteritidis-induced salmonellosis in humans is characterized by diarrhoea, fever, headache, abdominal pain, nausea and vomiting (CDC, 2007). S. Enteritidis is also increasingly reported from cases of invasive and extra-intestinal infections such as septicaemia, arthritis, endocarditis, meningitis and urinary tract infections (Ghosh & Vogt, 2006; Gordon et al., 2008; Katsenos et al., 2008; Kobayashi et al., 2009; Morpeth et al., 2009; Mutlu et al., 2009; Tena et al., 2007). Between 1990 and 2001, the US state and territorial health departments reported 677 S. Enteritidis outbreaks, which accounted for 23 366 illnesses, 1988 hospitalizations and 33 deaths (CDC, 2003). In 2006, countries within the European Union reported 1729 outbreaks caused by S. Enteritidis leading to 13 853 illnesses, 2714 hospitalizations and 14 deaths (EFSA, 2007). The Health Protection Agency of the UK reported 4194 cases of food-borne S. Enteritidis infection in 2008 (HPA, 2008). Poultry is considered the single largest reservoir of S. Enteritidis and most risk attribution studies have identified poultry and poultry products as the major source of human infection. S. Enteritidis is passed to humans mainly via handling and consumption of contaminated poultry meat and eggs (De Buck et al., 2004; Kimura et al., 2004; Little et al., 2008; Marcus et al., 2004; Patrick et al., 2004; Snow et al., 2007). For instance, eggs and egg-containing products were implicated as primary vehicles of S. Enteritidis infection in 298 (80 %) of the 371 known-source outbreaks reported to the Centers for Disease Control and Prevention from 1985 to 1999 (Patrick et al., 2004).
On-farm investigations have revealed that once chickens are exposed to S. Enteritidis, the entire poultry flock can become colonized quickly (Altekruse et al., 2006; Berrang et al., 2009; Foley et al., 2008). Infected birds shed bacteria in their faeces, leading to carcass contamination during slaughter house processing. S. Enteritidis is also invasive in both young and adult chickens. Young chickens less than 2 weeks of age often develop systemic disease with varying degrees of mortality (Duchet-Suchaux et al., 1995; Suzuki, 1994; Velge et al., 2005) whereas most adult hens colonized with S. Enteritidis become intestinal carriers and typically remain asymptomatic (Golden et al., 2008; Lister, 1988; Velge et al., 2005). S. Enteritidis can invade internal organs of laying hens and infect reproductive tissues leading to internal contamination of newly formed eggs (De Buck et al., 2004; Guard-Petter, 2001; Velge et al., 2005). It is believed that the specialized ability of S. Enteritidis to contaminate developing eggs acts as an ecological amplifier that facilitates dissemination of S. Enteritidis in the food chain and its eventual transmission to humans.
S. Enteritidis is the most genetically homogeneous serotype of Salmonella (Hudson et al., 2001; Liebana et al., 2001; Morales et al., 2005; Porwollik et al., 2005; Swaminathan et al., 2001). Despite limited genomic diversity, variation in phenotypic traits, including the ability to form biofilm, grow to high cell density, produce high-molecular-mass LPS and survive within egg albumin, has been commonly observed among field isolates of S. Enteritidis (Clavijo et al., 2006; Gast & Holt, 1995, 2001; Guard-Petter, 1998; Guard-Petter et al., 1999; Jain & Chen, 2007; Lock & Board, 1992; Pan et al., 2009; Parker et al., 2001; Solano et al., 1998; Yim et al., 2010). S. Enteritidis strains also vary in their virulence potential in poultry, irrespective of phage types (Poppe et al., 1993) and even within clonal lineages (Olsen et al., 1999; Yim et al., 2010). Dhillon et al. (1999) tested six strains of S. Enteritidis and showed that mortalities in experimentally challenged 1-day-old chickens ranged from 0 to 23 %, whereas Barrow (1991) reported that the mortalities caused by six strains of S. Enteritidis varied from 20 to 96 %. Similarly, Gast & Benson (1995, 1996) showed that S. Enteritidis strains differed significantly in not only their ability to cause mortality but also their ability to colonize intestinal tracts and invade the spleen and liver of 1-day-old chickens.
Although limited numbers of S. Enteritidis strains were tested, the above studies have shown that isolates of S. Enteritidis vary in their virulence potential, at least in poultry. Most other studies to characterize the pathogenic potential of S. Enteritidis strains have primarily focused on isolates recovered from either human clinical cases or non-poultry-associated environmental sources (Betancor et al., 2009; Lu et al., 1999; Pan et al., 2009; Solano et al., 1998, 2001; Yim et al., 2010). Humphrey et al. (1996) showed that human isolates of S. Enteritidis PT4 with enhanced heat and acid tolerance were more virulent in mice and more invasive in chickens compared with poultry-associated strains of S. Enteritidis. However, only two strains from each source were tested (Humphrey et al., 1996). Similarly, Pang et al. (2006) showed that poultry-associated strains of S. Enteritidis differed in their invasion potential in cultured Int-407 cells, but only eight strains were tested. Despite poultry being a reservoir host for S. Enteritidis and a major source of human infection worldwide, it is still unclear if differential pathogenicity translates into differential risk for human. In addition, members of this serotype of Salmonella harbour limited genetic variance and it is not clear what genetic mechanisms underlie differential virulence.
We hypothesized that not all S. Enteritidis isolates recovered specifically from poultry and poultry environments are equally pathogenic. By using invasion as a surrogate marker of virulence, we tested the invasion potential of 53 poultry-associated S. Enteritidis isolates in a well-differentiated human intestinal cell culture model (Caco-2). The results confirmed the identity of a low invasiveness group of isolates that were also less virulent in a mouse model. Lower virulence was associated, in part, with impaired motility and reduced secretion of Salmonella pathogenicity island-1 (SPI-1) type-three secretion (TTSS) secreted proteins. In addition, we found that isolates with low invasiveness were impaired in their ability to invade and/or survive within chicken macrophages, indicating that S. Enteritidis isolates that are less invasive in chickens are likely to be less pathogenic in mammalian hosts such as humans.
Methods
Bacterial strains.
S. Enteritidis isolates (Table 1) analysed in this study were stored at −80 °C in 15 % (v/v) phosphate-buffered glycerol. All isolates were grown in Luria–Bertani (LB) broth with shaking at 150 r.p.m. or on LB agar plates at 37 °C, except when indicated. All isolates were tested for their reactivity with Difco Salmonella O antiserum Group D1 by the plate agglutination test according to the manufacturer’s instructions. The MIC of gentamicin was determined for all isolates by the microdilution method described by Andrews (2001).
Table 1. Source, genotype and phenotypic characteristics of S. Enteritidis strains isolated from poultry and poultry environments.
| Strain (phage type) | Supplier designation | Source | Supplier* | MLVA type | Caco-2 cell invasion assay | Biofilm production | PCR screening | ||||
| Mean±sem log10(c.f.u. ml−1) | Invasion phenotype | Agar plate assay | Microtitre assay (OD490) | spvB | sefA | sdfI | |||||
| C1 | LK5 | Egg | SGSC | 4a | 5.84±0.06 | High | bf− | 0.09 | + | + | + |
| C19 (PT8) | SA5985 | Meat | SGSC | 11 | 4.46±0.06 | Low | bf− | 0.06 | + | + | + |
| C20 (PT13) | SA5986 | Heart blood | SGSC | 10c | 5.69±0.05 | High | bf+ | 0.16 | + | + | + |
| C44 | SE310 | Heart blood | SGSC | 10c | 5.69±0.04 | High | bf− | 0.10 | + | + | + |
| C45 | SE154 | Ovary | SGSC | 11 | 4.40±0.04 | Low | bf− | 0.06 | + | + | + |
| BC1 (PT8) | 2007-2700 | Yolk sac | AHC | 4b | 5.69±0.25 | High | bf− | 0.09 | + | + | + |
| BC2 | 2005-3637 | Turkey lung | AHC | 12 | 5.75±0.01 | High | bf+i | 0.14 | + | + | + |
| BC3 | 2005-3918 | Heart | AHC | 10c | 5.57±0.22 | High | bf+m | 0.11 | + | + | + |
| BC4 (PT8) | 2008-0455 | Heart | AHC | 10c | 5.41±0.28 | High | bf+m | 0.13 | + | + | + |
| BC5 | 2007-0734-52-1 | Farm environment | AHC | 6 | 5.39±0.07 | High | bf− | 0.10 | + | + | + |
| BC6 (PT13) | 2008-0742-1723F-8 | Fluff | AHC | 8a | 5.77±0.08 | High | bf+m | 0.16 | + | + | + |
| BC7 (PT8) | 2007-0275F-12 | Fluff | AHC | 10c | 5.66±0.03 | High | bf− | 0.11 | + | + | + |
| BC8 | 2005-2949 | Fluff | AHC | 9a | 5.81±0.14 | High | bf+m | 0.12 | + | + | + |
| A3 | E2006001733 | Chicken | MDH-PHLD | 1 | 5.37±0.17 | High | bf− | 0.11 | + | + | + |
| A5 | E2004001561 | Chicken swab | MDH-PHLD | 10a | 5.74±0.07 | High | bf− | 0.10 | + | + | + |
| A7 | I2008001102 | Litter | MDH-PHLD | 10b | 5.01±0.09 | Medium | bf+ | 0.14 | + | + | + |
| A8 | E2007001234 | Chicken | MDH-PHLD | 11 | 5.05±0.14 | Medium | bf+ | 0.15 | + | + | + |
| A11 | E2006001732 | Litter | MDH-PHLD | 10c | 4.65±0.12 | Low | bf− | 0.11 | + | + | + |
| P3 | 00-983-2 | Broiler carcass rinse | WADDL | 3 | 4.71±0.11 | Low | bf− | 0.11 | + | + | + |
| P12 | 90-734378(10) | Chicken | WADDL | 11 | 5.46±0.10 | High | bf+ | 0.19 | + | + | + |
| P13 | 388(9) | Layer chicken | WADDL | 11 | 5.48±0.07 | High | bf+m | 0.13 | + | + | + |
| P14 | 90-855307 | Chicken | WADDL | 11 | 5.42±0.05 | High | bf+i | 0.12 | + | + | + |
| P15 | 4/11/91210 | Chicken | WADDL | 11 | 5.35±0.05 | High | bf− | 0.11 | + | + | + |
| P16 | 9-1222W622-7 | Chicken | WADDL | 11 | 5.46±0.15 | High | bf− | 0.07 | + | + | + |
| P21 | 4/11/91560 | Chicken | WADDL | 9a | 5.76±0.23 | High | bf− | 0.06 | + | + | + |
| P27 | 9-1144 W4 22-10 | Chicken | WADDL | 11 | 5.64±0.02 | High | bf+ | 0.14 | + | + | + |
| Pu35 | 4534 | Chicken | FDIU | 12 | 5.11±0.19 | Medium | bf+ | 0.18 | + | + | + |
| Pu40 | 8879 | Chicken | FDIU | 12 | 5.14±0.30 | Medium | bf+ | 0.15 | + | + | + |
| Pu43 | 10520 | Chicken | FDIU | 11 | 5.10±0.15 | Medium | bf+m | 0.15 | + | + | + |
| Pu45 | 10610 | Chicken | FDIU | 2 | 4.64±0.12 | Low | bf+i | 0.13 | + | + | + |
| Pu48 | 12454 | Chicken | FDIU | 7 | 5.38±0.10 | High | bf+ | 0.15 | + | + | + |
| Pu50 | 15898 | Chicken | FDIU | 11 | 5.17±0.01 | Medium | bf+ | 0.17 | + | + | + |
| Pu51 | 15899 | Chicken | FDIU | 11 | 5.54±0.05 | High | bf− | 0.08 | + | + | + |
| G1† (PT4) | 22079 | Chicken | ESQRU | 13a | 4.93±0.14 | Medium | bf+ | 0.21 | + | + | + |
| G2† (PT13a) | 21046 | Chicken | ESQRU | 9b | 5.69±0.10 | High | bf− | 0.08 | + | + | + |
| G3† (PT13a) | 21027 | Chicken | ESQRU | 5 | 5.29±0.10 | Medium | bf+ | 0.16 | + | + | + |
| G9 (PT13a) | 21101 | Faecal | ESQRU | 4a | 5.42±0.24 | High | bf− | 0.05 | − | + | + |
| G10 (PT4) | 22080 | Farm environment | ESQRU | 13b | 5.12±0.00 | Medium | bf+ | 0.18 | + | + | + |
| G11 (PT13a) | 27003 | Chicken | ESQRU | 4a | 5.38±0.28 | High | bf+ | 0.24 | − | + | + |
| G12 (PT13a) | 21052 | Egg yolk | ESQRU | 4a | 5.44±0.24 | High | bf− | 0.08 | − | + | + |
| G14 (PT13a) | 21100 | Faecal | ESQRU | 4a | 5.13±0.07 | Medium | bf− | 0.06 | − | + | + |
| G15 (PT13) | 26017 | Chicken | ESQRU | 8b | 4.97±0.23 | Medium | bf+ | 0.15 | + | + | + |
| G21 (PT13a) | 21056 | Egg yolk | ESQRU | 4a | 5.13±0.03 | Medium | bf− | 0.11 | − | + | + |
| G22† | 21028 | Farm environment | ESQRU | 5 | 4.50±0.06 | Low | bf+ | 0.18 | + | + | + |
| G29† (PT4) | 22077 | Farm environment | ESQRU | 13a | 5.74±0.06 | High | bf− | 0.08 | + | + | + |
| G31 | 22150 | Egg | ESQRU | 4a | 5.08±0.03 | Medium | bf− | 0.07 | − | + | + |
| G33 (PT8) | 22083 | Farm environment | ESQRU | 10c | 4.86±0.27 | Medium | bf+m | 0.17 | + | + | + |
| G34 (PT13a) | 27035 | Oviduct | ESQRU | 4a | 5.17±0.17 | Medium | bf+ | 0.18 | − | + | + |
| G36† | 21030 | Farm environment | ESQRU | 5 | 4.89±0.06 | Medium | bf+ | 0.14 | + | + | + |
| G45 | 21063 | Spleen | ESQRU | 4a | 4.18±0.18 | Low | bf− | 0.09 | − | + | + |
| G46† | 21025 | Farm environment | ESQRU | 5 | 5.37±0.35 | High | bf+ | 0.19 | + | + | + |
| G47 (PT13a) | 27020 | Caeca | ESQRU | 4a | 5.67±0.04 | High | bf+ | 0.17 | − | + | + |
| UK (PT4) | P125109 | Chicken | WTSA | 13a | 5.91±0.06 | High | bf+ | 0.17 | + | + | + |
SGSC, Salmonella genetic stock centre, University of Calgary, BC, Canada; AHC, Animal Health Centre, Abbotsford, BC, Canada; MDH-PHLD, Minnesota Department of Health Public Health Laboratory Division, St Paul, MN, USA; WADDL, Washington Animal Disease Diagnostic Laboratory, Puyallup, WA, USA; FDIU, Field Disease Investigation Unit, Washington State University, Pullman, WA, USA; ESQRU, Egg Safety and Quality Research Unit, Athens, GA, USA; WTSA; Wellcome Trust Sanger Institute, London, UK.
Originally isolated from mice captured from poultry farm premises and subsequently tested in a chicken infection model.
PCR screening of S. Enteritidis strains.
PCR amplification of the sefA gene, encoding SEF14 fimbrial antigen, was used for preliminary confirmation of the identity of Salmonella isolates (Doran et al., 1996). A chromosomally located sdfI gene reported to be unique to the serovar S. Enteritidis was targeted for the serotype-specific identification of S. Enteritidis (Agron et al., 2001; Alvarez et al., 2004; Clavijo et al., 2006; Trafny et al., 2006). A 60 kb virulence-plasmid-associated spvB gene was used as a marker for determination of the presence or absence of a typical virulence plasmid (pSEV) among S. Enteritidis isolates (Cho et al., 2007; Rotger & Casadesús, 1999; Rychlík et al., 2006, 2008). PCR amplification of complete ORFs of the sipA, sipD, flgK, fljB and flgL genes was performed to detect the presence or absence of these genes among selected S. Enteritidis isolates. All PCRs were run in 30 µl volumes containing 5 µl boiled lysates as template DNA and 21 µl Cougar PCR master mix (Custom Genome Services). Each primer was added to 200 nM (Table 2). The cycling conditions consisted of denaturation at 95 °C for 3 min followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C (spvB), 53 °C (sipA, sipD, flgK, fljB and flgL), 58 °C (sdf) or 59 °C (sefA) for 30 s and extension at 72 °C for 40 s. Samples were fractionated by using 1 % (w/v) agarose gel and visualized by ethidium bromide staining.
Table 2. Oligonucleotide sequences for PCR-based assays and MLVA typing.
| Target gene/locus | Primer | Sequence (5′–3′) | Annealing temperature (°C) | Amplicon size (bp) | Reference |
| sefA | SefA_F | GTGGTTCAGGCAGCAGTTACT | 59 | 334 | O’Regan et al. (2008) |
| SefA_R | CAGGGACATTTAGCGTTTCTTGAG | ||||
| sdfI | SdfI_F | TGTGTTTTATCTGATGCAAGAGG | 58 | 333 | Agron et al. (2001) |
| SdfI_R | CGTTCTTCTGGTACTTACGATGAC | ||||
| spvB | SpvB_F | CGGTTATAGAAGAGCTCCTGT | 55 | 349 | Rychlík et al. (2008) |
| SpvB_R | CCGGTATACGACTCTGTGATC | ||||
| SENTR1 | SENTR1-F | 6FAM-GCAACAGCAGCAGCAACAG | 55 | 440* (415)† | Malorny et al. (2008) |
| SENTR1-R | CCGAGCTGAGATCGCCAAG | ||||
| SENTR2 | SENTR2-F | VIC-CACTGGACGATCTGGATTTCTC | 55 | 519* (510)† | Malorny et al. (2008) |
| SENTR2-R | GTCGCCGTTACGCATCAAC | ||||
| SENTR3 | SENTR3-F | NED-CTAAACAAGCCGCTCATCCG | 55 | 494* (480)† | Malorny et al. (2008) |
| SENTR3-R | ACAACCTGCTGCTGTGCTG | ||||
| SENTR4 | SENTR4-F | 6FAM-GACCAACACTCTATGAACCAATG | 55 | 120* (120)† | Malorny et al. (2008) |
| SENTR4-R | ACCAGGCAACTATTCGCTATC | ||||
| SENTR5 | SENTR5-F | PET-CACCGCACAATCAGTGGAAC | 55 | 271* (270)† | Malorny et al. (2008) |
| SENTR5-R | GCGTTGAATATCGGCAGCATG | ||||
| SENTR6 | SENTR6-F | NED-ATGGACGGAGGCGATAGAC | 55 | 180* (177)† | Malorny et al. (2008) |
| SENTR6-R | AGCTTCACAATTTGCGTATTCG | ||||
| SENTR7 | SENTR7-F | VIC-ACGATCACCACGGTCACTTC | 55 | 135* (133)† | Malorny et al. (2008) |
| SENTR7-R | CGGATAACAACAGGACGCTTC | ||||
| SE-3 | SE-3F | 6FAM-CAACAAAACAACAGCAGCAT | 55 | 308* (307)† | Malorny et al. (2008) |
| SE-3R | GGGAAACGGTAATCAGAAAGT | ||||
| SE-7 | SE-7F | 6FAM-GATAATGCTGCCGTTGGTAA | 55 | 717* (691)† | Malorny et al. (2008) |
| SE-7R | ACTGCGTTTGGTTTCTTTTCT | ||||
| sipA | SipA_F | ATGGTTACAAGTGTAAGGACTCAG | 53 | 2055 | This study |
| SipA_R | ACGCTGCATGTGCAAGCCATC | ||||
| sipD | SipD_F | ATGCTTAATATTCAAAATTATTCCG | 53 | 1029 | This study |
| SipD_R | TCCTTGCAGGAAGCTTTTG | ||||
| flgK | FlgK_F | ATGTCCAGCTTGATTAATCAC | 53 | 1659 | This study |
| FlgK_R | GCGAATATTCAATAACGCATC | ||||
| fljB | FljB_F | ATGGCACAAGTCATTAATACAAAC | 53 | 1515 | This study |
| FljB_R | ACGCAGTAAAGAGAGGAC | ||||
| flgL | FlgL_F | ATGCGTATCAGTACCCAGATG | 53 | 951 | This study |
| FlgL_R | CCGGTTCAACTGGAAAAGC | ||||
| sipA | SipA_F_RT | TTTTAACGCCTCAGCGTCTT | 55 | 130 | This study |
| SipA_R_RT | CAGAGAAAGTGCCACAACGA | ||||
| sipD | SipD_F_RT | CCTCCCATTTTGGAAAGAAT | 55 | 181 | This study |
| SipD_R_RT | TATTTAGCGCTTCGCCTATG | ||||
| rpoD | RpoD_F | ACATGGGTATTCAGGTAATGGAAGA | 55 | 74 | Botteldoorn et al. (2006) |
| RpoD_R | CGGTGCTGGTGGTATTTTCA |
Based on S. Enteritidis NCTC13349.
Size analysed by using an ABI310 capillary sequence detection system (Applied Biosystems).
Multi-locus variable-tandem repeat analysis (MLVA) typing.
Primers including forward dye-labelled primers for MLVA were purchased from Applied Biosystems (Table 2). The multiplex-PCR procedure was performed as previously described with few modifications (Malorny et al., 2008). Briefly, a single colony of each strain was suspended in 200 µl 5 % Chelex 100 solution (Bio-Rad) with 100 µg proteinase-K, incubated at 56 °C for 45 min, and boiled for 20 min followed by centrifugation at 14 000 r.p.m. for 1 min. For all PCRs, 2 µl supernatant from boiled Chelex lysate was used as template DNA. Multiplex-PCRs were carried out in an iCycler thermal cycler (Bio-Rad) by using the Qiagen multiplex PCR kit, according to the manufacturer’s instructions, in 12.5 µl volumes with 1.25 µl 10× PCR buffer and 1.0 pmol of primer pairs SENTR1, SENTR2, SENTR3 and SE-7, and 0.25 pmol each of primers SENTR4, SENTR5, SENTR6, SENTR7 and SE-3 (Table 2). Results for the SENTR2 locus were difficult to interpret because of multiple peaks and therefore this locus was excluded from the analysis. Cycling conditions included an initial denaturation at 94 °C for 15 min, followed by 28 cycles of 94 °C for 30 s, 55 °C for 90 s and 72 °C for 90 s, with a final extension step at 72 °C for 10 min. Each multiplex reaction was diluted 1 : 10 with PCR-grade water. An aliquot (0.5 µl) of diluted multiplex PCR product was mixed with 0.5 µl GS1200LIZ size standard (Applied Biosystems) and 19 µl Hi-Di formamide (Applied Biosystems) (total volume 20 µl for capillary electrophoresis). Capillary electrophoresis was carried out by the Genomics Core Laboratory at Washington State University by using a 3730 DNA analyser with Pop-7 polymer (Applied Biosystems). The resulting electropherograms were analysed by using GeneMarker software (Softgenetics LLC). Fragment sizes for each VNTR locus were converted into numbers of predicted repeats and analysed with Bionumerics software (Applied Maths). Cluster analysis of the categorical coefficient was performed by using the unweighted pair group method with arithmetic average (UPGMA) algorithm (Sneath & Sokal, 1973).
In vitro invasion assays with differentiated Caco-2 cells.
Caco-2 human colon adenocarcinoma cells were obtained from the American Type Culture Collection (ATCCHTB-37) and were routinely grown in Dulbecco’s modified Eagle medium (DMEM) (Invitrogen) supplemented with 10 % (v/v) fetal bovine serum (FBS; Invitrogen) in 95 % air/5 % CO2 at 37 °C. Propagated cells were seeded at approximately 106 cm–2 into 75 cm2 flasks (BD falcon). After 5 days of culture, cells were harvested by trypsinization and resuspended in DMEM with 10 % FBS. Subsequently, cells were plated onto 12-well plates (Cellstar) at ~1×106 cells per well. The medium was changed daily until confluency and thereafter every second day. Confluency occurred after 5–6 days of incubation and well-differentiated monolayers of Caco-2 cells were obtained 14 days after seeding for the assays. The state of cell differentiation was monitored by estimating the production of intestinal alkaline phosphatase by using a SensoLyte pNPP alkaline phosphatase assay kit (AnaSpec) following the manufacturer’s instructions. Intestinal alkaline phosphatase is a brush border enzyme expressed exclusively in villus-associated enterocytes, and expression indicates the development of digestive and absorptive function (Goldberg et al., 2008; Hara et al., 1993). Cell differentiation was also confirmed by monitoring the development of well-developed apical brush borders by using transmission electron microscopy (TEM; data not shown). Bacterial inocula were prepared from overnight (~16 h) cultures in LB broth, and 14-day-old monolayers of well-differentiated Caco-2 cells were infected at an m.o.i. ratio of 10 : 1 (S. Enteritidis : Caco-2 cells). Culture plates were centrifuged at 450 g at room temperature for 10 min to bring the bacteria into contact with cells. This centrifugation step also eliminated the possible effects of variations in the motility among the strains, which could otherwise influence the outcome of the assay. Inoculated monolayers were incubated for 2 h at 37 °C to allow bacteria to invade the cells. Subsequently, cells were washed three times with PBS. Next, 1 ml DMEM containing 100 µg gentamicin ml–1 was placed in each well and incubated for 2 h to kill extracellular bacteria. The MIC of gentamicin for all the isolates used in this study was <0.125 µg. The supernatants from representative gentamicin-treated samples revealed no extracellular bacteria (data not shown), indicating that the exposure of gentamicin used for the invasion assay was sufficient to kill extracellular bacteria. Following incubation, the monolayers were washed three times with PBS and lysed with 0.5 % (v/v) Triton X-100 (Fisher Biotech) in PBS for 10 min at 37 °C. Serial dilutions of suspensions were made in PBS and inoculated onto LB plates to determine the c.f.u. that survived gentamicin treatment and hence had invaded the Caco-2 cells. Each isolate was tested in triplicate in two or more independent experiments.
Detection of biofilm phenotypes.
Two different assays were used to determine biofilm phenotypes of S. Enteritidis strains. Qualitative determination of biofilm formation was based on colony morphology on brilliant green agar (BGA) plates (Acumedia) as described previously with a few modifications (Guard-Bouldin et al., 2004; Guard-Petter et al., 1996; Humphrey et al., 1996). Briefly, 100 colonies of each S. Enteritidis isolate were spot-transferred onto BGA at 20 colonies per plate (150 mm diameter) at least 20–25 mm apart. The plates were incubated at 37 °C for 16 h followed by incubation for an additional 5 days at 25±2 °C (mean±sd). Colony morphology was observed every 24 h. The proportion of biofilm-producing colonies was recorded as a percentage of the total colonies counted 5 days post-inoculation. Strains were classified as follows: biofilm positive (bf+; 100 % of colonies are positive for organic matrix that covers the entire colony, giving a wrinkled appearance), biofilm negative (bf−; 100 % of colonies, including their edges, appear smooth), intermediately positive (bf+i; ≥80 % colonies with wrinkled edges and smooth centre or vice versa giving a semi-wrinkled appearance) or mixed positive (bf+m; both bf+ and bf− colony types are present within the same culture).
Quantitative determination of biofilm production was performed by using a microtitre assay as described previously with minor modifications (de Silva et al., 2002; Eftekhar & Speert, 2009). Briefly, 100 µl overnight cultures (diluted 1 : 100 in trypticase soy broth, TSB) were inoculated into 96-well flat-bottomed polystyrene microtitre plates (Evergreen) and incubated for 3 days at room temperature (25 ±2 °C). Well contents were decanted, washed three times with 200 µl sterile PBS (pH 7.2), air-dried and stained with 250 µl 0.1 % (w/v) safranin for 15 min followed by a single wash with PBS. Bound safranin was dissolved by adding 250 µl 30 % (v/v) acetic acid solution. OD490 readings were recorded by using an ELISA plate reader (BioTek). Biofilm-positive (G3) and biofilm-negative (G2) control isolates were included in each plate, as was the background control of TSB without bacteria. Four wells were tested per strain in two independent experiments. The mean OD values of the test isolates were used to generate a receiver operating characteristic curve. An OD490 of ≥0.12 was chosen to distinguish biofilm-positive from biofilm-negative strains (92 and 96 % predicted sensitivity and specificity, respectively).
In vivo invasion assay in mouse model.
Mouse challenge studies were performed according to protocols approved by the WSU-Institutional Animal Care and Use Committee. Six- to eight-week-old female BALBc mice, weighing 20–24 g, were obtained from a local breeding colony maintained at the Animal Resource unit, WSU. The mice were raised in static micro-isolator cages at 25 °C with 12 h light cycle and fed on a Salmonella-free commercial diet (Lab Diet 5001; Purina). Food and water were supplied ad libitum until the end of the experiment. Eight S. Enteritidis isolates with different cell-invasion potential in cultured Caco-2 cells were used for the mouse invasiveness assay. Bacterial isolates were grown overnight in LB medium as described above and serially diluted in PBS to obtain desired cell counts. Mice were inoculated intragastrically with 200 µl PBS containing ~1×106 cells. Control mice were inoculated with 200 µl PBS. Each isolate was inoculated in three animals and each animal was raised in a separate isolator cage to prevent cross contamination. To exclude observer bias in interpretation of the results, all clinical and bacteriological assessments were conducted by personnel blind to the identity of the challenge isolates. Faecal pellets were collected from each mouse at 3, 6, 9, 15 and 21 days post-infection (PI) and processed for isolation and enumeration of challenge isolates as described below. Challenged mice were observed, at least three times daily, for mortality and clinical signs such as ruffled fur, hunched backs and lethargy for up to 21 days PI. Mice showing all of the above symptoms were considered moribund and were killed by carbon dioxide asphyxiation. Samples of liver, spleen and intestine were collected by using aseptic techniques, homogenized in cold PBS and bacterial c.f.u. number was determined by direct plating of serially diluted organ homogenates on xylose-lysine dulcitol (XLD) agar (Criterion) plates. Samples that were negative after direct plating on XLD agar were enriched by inoculating into standard tetrathionate enrichment broth (Difco) and incubated at 37 °C for 24 h followed by plating onto XLD agar. The results for enrichment cultures were recorded as either positive or negative. Experiments were terminated after 3 weeks when all the surviving mice were killed and organ samples were processed as described above.
Secretory protein profiling.
Secreted proteins were recovered as described previously (Amy et al., 2004; Arricau et al., 1998; Kaniga et al., 1995). S. Enteritidis isolates were grown overnight (16 h) at 37 °C in 10 ml LB broth supplemented with 300 mM NaCl in a 50 ml conical tube with shaking at 200 r.p.m. and adjusted to an OD600 of 1. Culturing of Salmonella in high osmolarity conditions has been shown to induce expression of TTSS encoded by SPI-1 and flagellar proteins. Bacteria were harvested by centrifugation at 8000 g for 10 min, and culture supernatant containing secretory proteins was collected and passed through a 0.2 µm filter. Proteins present in 2 ml supernatant were precipitated by adding 10 % (v/v) trichloroacetic acid and incubated on ice for 1 h. Precipitated proteins were then recovered by centrifugation at 10 000 r.p.m. for 30 min. The pellets were washed once with ice-cold acetone and air-dried. The protein pellets were resuspended in 50 µl Laemmli buffer. Subsequently, 10 µl of this suspension was separated on 12 % (w/v) acrylamide gels by SDS-PAGE. After electrophoresis, proteins were stained by using a silver staining kit (Pierce) as per the manufacturer’s recommendations. Each isolate was tested independently in three or more experiments. The differentially expressed protein bands were excised from the gel and sequenced by using MALDI MS/MS.
Motility assay and TEM.
To test whether variation in cell invasion corresponded to variation in motility, the motility of representative isolates from low and high invasiveness groups was assessed at 37 °C on semi-solid LB agar (0.3 %). Briefly, the procedure involved growing bacterial isolates overnight (16 h) in LB broth at 37 °C. Subsequently, 1 µl overnight culture was stab-inoculated into semi-solid media and the plates were incubated at either 37 °C (Experiment 1) or 42 °C (Experiment 2). The diameter of the halo of growth was measured for each isolate after 7 h of incubation. Each strain was tested in duplicate and each experiment was repeated at least twice. For TEM, culture samples were taken from the edge of the motile colony grown at 42 °C. Carbon–Formvar-coated grids (200 mesh) were floated on a drop of cells suspended in distilled water and then allowed to dry. The grids were stained in 2 % uranyl acetate for 2.5 min and viewed on a Philips model CM200 TEM at an accelerating voltage of 200 kV (Franceschi Microscopy and Imaging Center, WSU). At least 10 fields were viewed for each isolate.
In vitro invasion assays with chicken macrophage cells.
Chicken macrophage cells (HD11) were routinely maintained in Iscove’s modified Dulbecco’s medium (IMDM; Invitrogen) supplemented with 10 % (v/v) FBS (Invitrogen) in 95 % air/5 % CO2 at 37 °C (Bohez et al., 2006; He et al., 2003). Propagated cells were seeded at approximately 106 cm–2 into 75 cm2 flasks (BD falcon). After 48 h of culture, cells were harvested by trypsinization and resuspended in IMDM with 10 % FBS. Subsequently, cells were plated onto 12-well plates (Cellstar) at ~1×106 cells per well. After 24 h incubation in 95 % air/5 % CO2 at 37 °C, monolayers of HD11 were achieved. Bacterial inocula were prepared from overnight (~16 h) cultures in LB broth and monolayers were infected at an m.o.i. ratio of approximately 50 : 1 (S. Enteritidis : HD11 cells) in DMEM. Culture plates were centrifuged at 1000 g at room temperature for 5 min to bring the bacteria into contact with cells. Inoculated monolayers were incubated for 2 h at 42 °C to allow cells to phagocytose the bacteria. Subsequently, cells were washed three times with PBS to remove non-adherent extracellular bacteria. Next, 1 ml DMEM containing 100 µg gentamicin ml–1 was placed in each well and incubated for 2 h at 42 °C to kill cell-adherent extracellular bacteria. Following treatment with gentamicin, the monolayers were washed three times with PBS and lysed with 0.5 % (v/v) Triton X-100 (Fisher Biotech) in PBS for 10 min at 37 °C. Serial dilutions of suspensions were made in PBS and inoculated onto LB agar plates to determine the number of organisms that survived gentamicin treatment and hence had invaded and/or survived within the HD11 cells. Each isolate was tested in duplicate and at least three independent experiments were performed.
Real-time RT-PCR.
Representative high invasive (BC8 and P21) and low invasive strains (C19 and C45) were cultured overnight (16 h) in LB broth at 42 °C under known SPI-1-inducing conditions as described above (Amy et al., 2004; Arricau et al., 1998; Kaniga et al., 1995). Total bacterial RNA was extracted by using the Ambion Ribopure kit as per the manufacturer’s protocol followed by DNase I (Ambion) treatment to remove residual DNA. The purified RNA was quantified by using a Nanodrop ND-1000 spectrophotometer and quality was assessed by gel electrophoresis and ethidium bromide staining. The absence of DNA contamination was checked by PCR amplification by using total RNA as a template and primers specific for a housekeeping gene, rpoD (Botteldoorn et al., 2006). Real-time RT-PCR was performed to measure the transcriptional levels of sipA, sipD and the housekeeping gene rpoD, by using an iQ5 iCycler (Bio-Rad) and SYBR green I mastermix (Bio-Rad). One microgram of DNase-treated total RNA from two independent cultures was reverse transcribed to cDNA by using Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. Amplifications were performed by using Ssofast Evagreen Supermix (Bio-Rad). Optimal PCR conditions and primer efficiencies were determined for each primer pair (Table 2) by reference to a standard curve generated from a series of fourfold dilutions of the positive control pGEM-T Easy (Promega) plasmid containing the amplicon of interest. Following amplification, melting curves were generated to verify PCR product identity. Expression levels were normalized to that of the housekeeping gene rpoD and the resultant cycle threshold (Ct) values were analysed by using the relative expression software tool (REST 2009) for determination of relative expression levels of genes of interest (Pfaffl et al., 2002).
Statistical analysis.
Differences in the invasiveness of S. Enteritidis isolates in Caco-2 and HD11 cells were analysed by means of one-way anova followed by multiple comparisons with a Tukey–Kramer test. Subsequently, k-means clustering was used to classify the isolates by their level of Caco-2 cell invasion into three major groups: high, medium and low invasiveness. The difference in the virulence potential of selected isolates in a mouse model was evaluated by Kaplan–Meier survival analysis and log-rank test. Results of biofilm formation from quantitative microtitre plate assays and qualitative agar plate assays were compared by one-way anova or Student’s t-test. A Spearman rank-correlation coefficient was used to examine the correlation between S. Enteritidis phenotypes such as Caco-2 cell invasiveness, biofilm formation, motility and mouse virulence. All statistical tests were performed by using NCSS 2007.
Results
PCR-based screening and MLVA typing of S. Enteritidis isolates
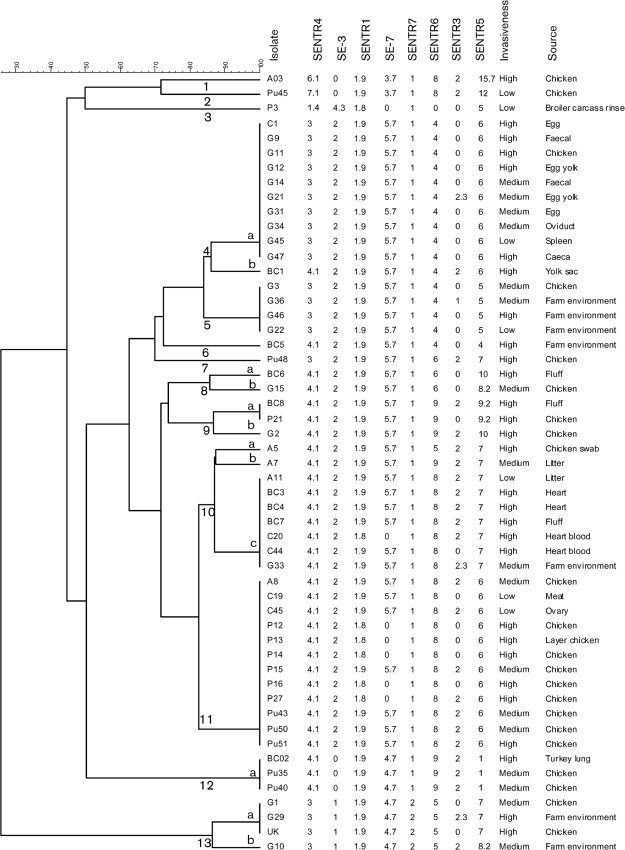
PCR screening was conducted to confirm the identity of each isolate. All isolates tested positive for the presence of serogroup D-specific sefA and S. Enteritidis-specific sdfI (Table 2). In addition, all strains were confirmed as being of smooth type based on their seroreactivity with Salmonella O antiserum Group D1 (data not shown). MLVA distinguished 53 S. Enteritidis isolates from diverse sources into 13 main clusters sharing on average ~85 % allelic congruence (Fig. 1). These were classified into five unique and 14 cluster-associated MLVA types. There was considerable phenotypic diversity in terms of cell invasion potential and biofilm formation among the isolates within each cluster. We found no correlation between MLVA type and cell invasiveness (Spearman’s rank-correlation coefficient, 0.07; P = 0.6) or the ability of isolates to form biofilm (Spearman’s rank-correlation coefficient, 0.26; P = 0.06).
Fig. 1.
Cluster analysis of 53 S. Enteritidis isolates recovered from diverse poultry sources. Eight major clusters (4, 5 and 8–13) were defined as groups of closely related strains sharing on average ≥85 % allelic congruence, as resolved by MLVA genotypes.
S. Enteritidis isolates differ in their invasiveness in differentiated Caco-2 cells
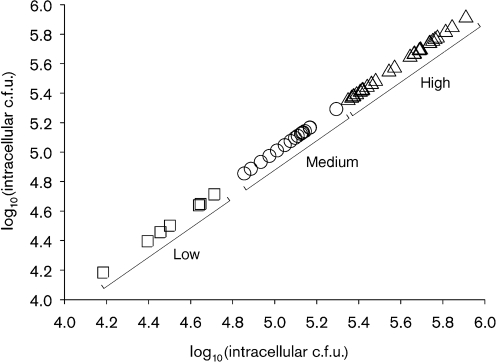
A standard gentamicin protection assay was used to test the invasiveness of S. Enteritidis isolates in human colon cancer cell line Caco-2. Initially, we screened several isolates of S. Enteritidis by using undifferentiated Caco-2 cultures that were grown for 48 h, but we observed a low degree of invasion and a high degree of variance between the replicates (data not shown). We therefore tested Caco-2 cells for their state of cell differentiation and the effect of this state on invasion by S. Enteritidis. Alkaline phosphatase activity was significantly higher (P<0.01, Student’s t-test) in 14-day-old cells compared with 5-day-old cells (Table 3) (Burns et al., 2001; Hara et al., 1993). In addition, 14-day-old cells showed well-developed apical brush borders, as visualized by TEM, indicating that 14-day-old cells were well differentiated (data not shown). Subsequently, we tested the invasive ability of representative S. Enteritidis isolates along with a clinical isolate of S. Typhimurium (positive control) and a laboratory Escherichia coli strain (negative control) in 5-day-old (undifferentiated cells) and 14-day-old (differentiated cells) Caco-2 confluent cultures. The invasion rates of both S. Enteritidis G2 and S. Typhimurium were 13- and 2-fold higher in differentiated cells than in undifferentiated cells (Table 3). Consequently, 14-day-old well-differentiated cultures of Caco-2 cells were used to screen the invasiveness of all 53 S. Enteritidis isolates. There were significant differences (P<0.05) in invasiveness [mean±sem log10(c.f.u. ml−1)] between different isolates (Table 1) with G45 being minimally invasive (4.18±0.18) and UK being highly invasive (5.91±0.06). When a statistical cluster analysis via a k-means clustering algorithm was applied to cell invasiveness data for all the isolates tested, a kink in the sum of squares located at k = 3 indicated that three was the optimum number of clusters (Hastie et al., 2001). As a result, three groups with cluster centres of 5.58±0.03 (high invasiveness, n = 30), 5.07±0.06 (medium invasiveness, n = 16) and 4.50±0.04 (low invasiveness, n = 7) log10 (c.f.u. ml−1) were identified (Fig. 2). These differences in the mean log10(c.f.u.) between the three groups were significant (Tukey–Kramer test; P<0.05).
Table 3. Differences in invasion rates of S. Enteritidis between differentiated and undifferentiated Caco-2 cells.
| Culture length (days) | Invasion (%)* | Mean±sem alkaline phosphatase (µg ml−1)† | ||
| S. Enteritidis G2 | S. Typhimurium | E. coli K-12 | ||
| 5 | 0.05 | 0.06 | 0 | 0.5±0.11 |
| 14 | 0.68 | 0.13 | 0 | 1.1±0.04 |
Percentage invasion = [organisms recovered (c.f.u. ml−1)/organisms inoculated (c.f.u. ml−1)]×100.
The difference in alkaline phosphatase activity between 5- and 14-day-old cells was significant (P<0.001).
Fig. 2.
A k-means cluster plot of the variable Caco-2 cell invasiveness of 53 S. Enteritidis isolates recovered from diverse poultry sources. Three groups of isolates with low (n = 7), medium (n = 18) and high (n = 30) invasiveness were identified based on their cell invasion potential.
Invasiveness of S. Enteritidis isolates is not associated with the presence of a virulence plasmid
Nine (17 %) of 53 isolates used in this study tested negative for the spvB gene, indicating that these isolates lacked a ‘typical’ virulence-associated plasmid (pSEV). Based on the invasiveness of these plasmid-negative isolates in Caco-2 cells, three (G11, G12 and G47) isolates belong to the high invasiveness category, four (G14, G21, G31 and G34) to the medium invasiveness category and one (G45) to the low invasiveness category (Table 1). There was no association (Fisher’s exact test, P = 0.4) between the presence or absence of the spvB gene and the invasiveness of isolates.
Cell invasion potential of S. Enteritidis isolates is not related to biofilm formation
By using a BGA plate assay, 20 (37.7 %), 23 (43.4 %), three (5.7 %) and seven (13.2 %) of the 53 S. Enteritidis isolates were identified as bf+, bf−, bf+i and bf+m, respectively (Table 1). The proportion with intermediate (semi-wrinkled) colony morphology among isolates showing bf+i phenotype was ≥80 % whereas the proportion of typical biofilm-positive colonies among isolates showing bf+m phenotype ranged from 10 to 93 % (data not shown). Because of the presence of several isolates with bf+m and bf+i phenotypes it was difficult to classify these isolates either as bf+ or as bf−. Consequently, we confirmed the results of the BGA plate assay by using a quantitative microtitre plate assay. The OD490 of the bf+i phenotype ranged from 0.12 to 0.14 whereas that for the bf+m phenotype ranged from 0.11 to 0.17, indicating that according to the quantitative microtitre assay, all of these isolates except BC3 (OD490 = 0.11) were biofilm formers. Therefore, for data analysis purposes, all isolates with bf+, bf+i or bf+m phenotypes were merged together as bf+ and compared with the isolates showing typical bf− colony types. Quantitative biofilm production, as determined by microtitre plate assay, was significantly higher in isolates that were found to be biofilm producers by the BGA plate assay [geometric mean (95 % confidence interval) OD490 = 0.15 (0.14–0.16) versus 0.08 (0.07–0.09); P<0.01] (Table 1). In general, all biofilm-positive isolates except BC3 were identified as biofilm formers by microtitre plate assay whereas all biofilm-negative isolates were identified as biofilm negative, indicating strong concordance between the results of the two assays. The results indicated that a higher proportion of isolates with medium (81.2 %, 13/16) and high (50 %, 15/30) invasiveness formed a biofilm compared with isolates with low invasiveness (28.5 %, 2/7). However, this association was not statistically significant (Spearman’s rank-correlation coefficient, −0.12; P = 0.35).
Caco-2 cell invasion correlates with virulence potential of S. Enteritidis isolates in a mouse model
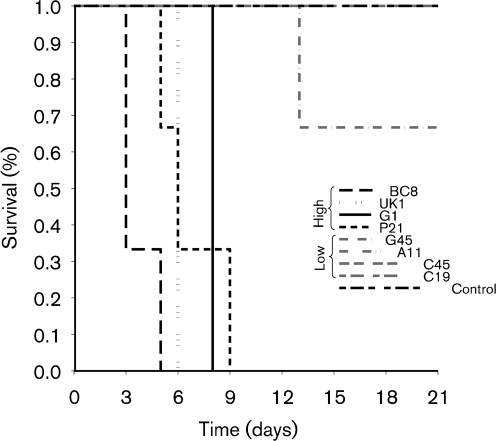
To determine whether cell invasiveness correlates with in vivo virulence, representative S. Enteritidis isolates with low (C19, C45, A11 and G45), medium (G1) and high (BC8, P21 and UK) invasiveness were tested by using an in vivo mouse challenge model. There were no differences between the in vitro growth kinetics of low and high invasiveness isolates (data not shown). S. Enteritidis isolates with high or medium invasiveness were more virulent and caused more and earlier mortality compared with isolates with low invasiveness (Kaplan–Meier survival analysis–log-rank test, P<0.01; Fig. 3). The mean survival time for mice inoculated with the high/medium invasiveness isolates P21, BC8, G1 and UK was 6 days (range = 3–9 days). All mice inoculated with high/medium invasiveness isolates died or were killed by 9 days PI. In contrast, no clinical signs or mortality were recorded in mice challenged with low invasiveness isolates C45, C19 and A11. Isolate G45 caused limited and delayed mortality (one of three mice died at 13 days PI). The mean survival duration of mice challenged with low invasiveness isolates and negative control was 20.3 (range = 13–23 days) and 21 days, respectively. There was a significant correlation between high invasiveness of S. Enteritidis isolates and virulence in the mouse model (Spearman’s rank-correlation coefficient, 0.95; P = 0.0002).
Fig. 3.
Kaplan–Meier survival curve showing percentage survival rates in mice challenged with representative S. Enteritidis isolates with high and low invasiveness in cultured Caco-2 cells.
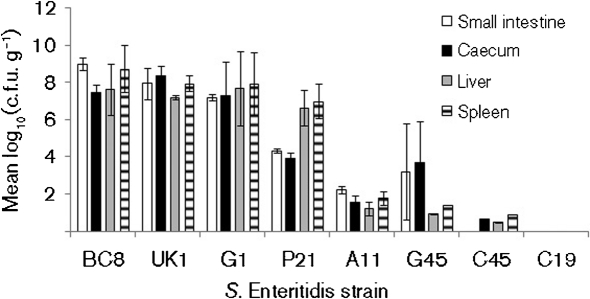
The number of bacteria per gram of tissue from the small intestine, caecum, liver and spleen from all animals that died during the experiment as well as from those that survived the challenge until the end of experiment at 21 days PI was determined. The mean (±sem) number [log10(c.f.u. g−1)] for isolates with high invasiveness was 7.10±0.99 (small intestine), 6.74±0.97 (caecum), 7.28±0.24 (liver) and 7.90±0.35 (spleen). In contrast, mean values for isolates with low invasiveness were 1.36±0.80 (small intestine), 1.47±0.80 (caecum), 0.66±0.26 (liver) and 1.03±0.40 (spleen). The numbers of bacteria recovered from different organs from animals challenged with high/medium invasiveness isolates were significantly higher (P<0.01) than those of low invasiveness isolates (Fig. 4), supporting the results of survival curve analysis. It is important to note, however, that colony counts were made at different time points with recoveries from highly invasive isolates mostly collected from mice that died between days 3 and 9 whereas for low virulence isolates bacteria were mostly recovered from mice that were killed at 21 days PI. In C19-challenged mice, no Salmonella was isolated from any of the organs tested for any of the mice. In C45-challenged mice, Salmonella was isolated from all the organs from only one of the three mice either by direct plating or by pre-enrichment procedures. In G45-challenged mice, Salmonella was isolated from all internal organs of the mouse that died at 13 days PI, and from intestine, but not the liver and spleen, of the two mice that survived until 21 days PI. Because of the variability in the recovery of the challenge isolates from mice challenged with C19, C45 and G45 isolates, the challenge experiment with these three isolates was repeated as described previously. The results from these repeat experiments were very similar to the first experiment (data not shown).
Fig. 4.
Numbers of Salmonella recovered from small intestine, caecum, liver and spleen of mice challenged with S. Enteritidis isolates with high/medium and low invasiveness in cultured Caco-2 cells.
Virulence of S. Enteritidis is associated with secretion of TTSS secreted proteins and motility
We determined the secretory protein profiles from the supernatant of representative isolates with low and high invasiveness cultured in LB broth containing 300 mM NaCl, corresponding to conditions allowing SPI-1 gene expression induction. Five major bands of secretory proteins with molecular masses of 74 (p1), 59 (p2), 52 (p3), 36 (p4) and 34 (p5) kDa were differentially expressed, based on the intensity of bands on silver-stained polyacrylamide gels, among isolates with low and high invasiveness (Fig. 5). These differences were consistently observed in at least three independent experiments and therefore all five protein bands were excised from the gel for protein identification by MALDI MS/MS. The deduced amino acid sequences from the above five proteins corresponded to SipA (p1), FlgK (p2), FljB (p3), SipD (p4) and FlgL (p5) (Fig. 5). The bands corresponding to FlgK, FljB and FlgL were not detected in isolate G45 (Fig. 5). Two isolates with low invasiveness (C19 and C45) showed lower band intensity for SipA (SPI-1 TTSS effector protein) and SipD (SPI-1 translocon assembly protein) when compared with the secretory protein profiles of isolates with high/medium invasiveness (Fig. 5). Consequently, we tested the levels of expression of mRNA transcripts for sipA and sipD in low invasiveness isolates C19 and C45 and compared these with the representative high invasiveness isolates P21 and BC8. Interestingly, expression levels of sipA and sipD were 16- and 11-fold higher, respectively, in the high invasiveness isolates compared with the low invasiveness isolates. This difference was statistically significant (P<0.05) and was consistent with impaired transcription of sipA and sipD being responsible for reduced secretion of these proteins.
Fig. 5.
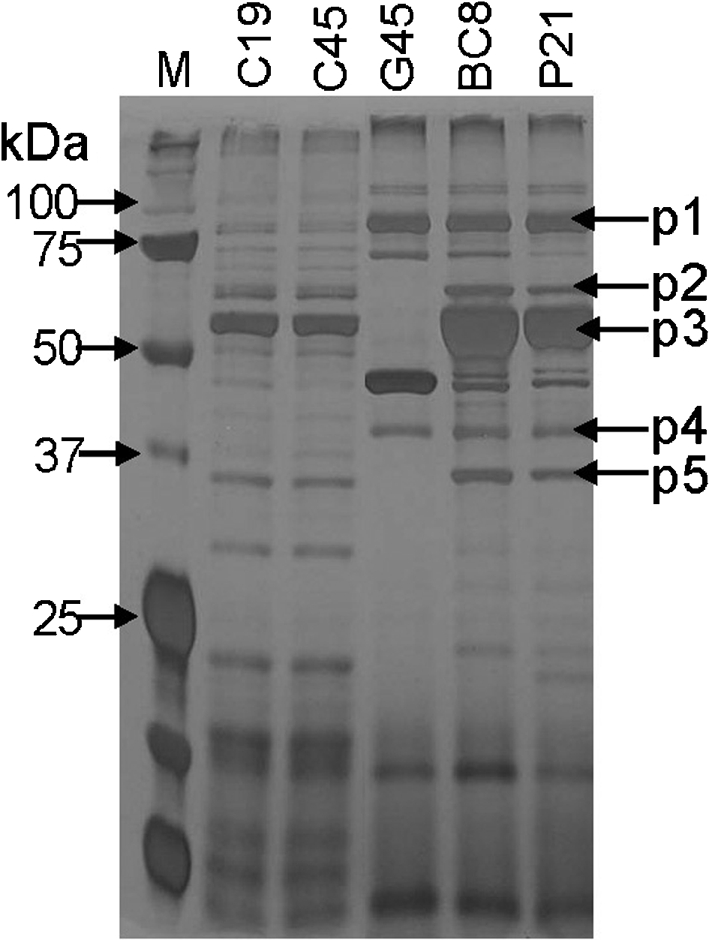
Secretory protein profile of representative S. Enteritidis isolates with low and high invasiveness in cultured Caco-2 cells. Lane M, protein molecular mass standard. The deduced amino acid sequences from the five major protein bands (indicated by arrows) corresponded to SipA (p1, 74 kDa), FlgK (p2, 59 kDa), FljB (p3, 52 kDa), SipD (p4, 36 kDa) and FlgL (p5, 34 kDa).
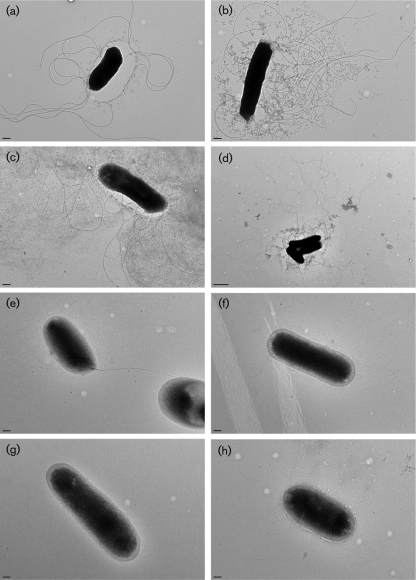
Because three flagella-associated proteins, FlgK (flagellar filament protein), FljB (flagellin) and FlgL (flagellar hook-associated protein), were absent from the supernatant of isolate G45, we postulated that the motility of this isolate was impaired. Consequently, we determined the motility of representative isolates with low and high invasiveness on a semi-solid motility medium at two temperatures (25 and 42 °C). At both temperatures, the mean diameter of the halo of growth of low invasiveness isolates was significantly smaller (Tukey–Kramer test, P<0.01) compared with high/medium invasiveness isolates (Table 4), indicating that, apart from the impaired secretion of SPI-1 TTSS proteins SipA and SipD, isolates with low invasiveness (C19, C45, A11 and G45) also had impaired motility. TEM of cultures grown at 42 °C revealed that isolates with high invasiveness produced long, thin, peritrichous, flagellar appendages (Fig. 6a–d). In contrast, flagellar appendages were not visible among isolates with low invasiveness, with the exception of isolate C19 which occasionally showed the presence of a unipolar flagellum (Fig. 6e–h).
Table 4. Intra-macrophage survival and motility of representative low- and high-cell invasiveness isolates of S. Enteritidis.
| Strain | Caco-2 cell invasion phenotype | Motility (cm) at:* | Intramacrophage survival in HD-11 cells [log10(c.f.u. ml–1)]† | |
| 37 °C | 42 °C | |||
| C19 | Low | 0.35±0.03 | 0.10±0.00 | 4.46±0.16 |
| C45 | Low | 0.37±0.02 | 0.20±0.04 | 4.08±0.16 |
| A11 | Low | 0.37±0.06 | 0.20±0.04 | 5.87±0.09 |
| G45 | Low | 0.10±0.00 | 0.10±0.00 | 5.12±0.12 |
| UK | High | 3.65±0.09 | 3.60±0.07 | 6.23±0.04 |
| BC8 | High | 3.15±0.10 | 1.40±0.04 | 6.01±0.05 |
| P21 | High | 2.97±0.14 | 1.82±0.14 | 6.04±0.17 |
| G1 | Medium | 3.82±0.16 | 0.67±0.04 | 5.99±0.05 |
Values are mean±sem diameter of the halo of growth on motility agar from measurements conducted on at least two colonies each from two independent experiments.
Each isolate was tested in duplicate. Values for c.f.u. ml−1 are mean±sem numbers of bacteria that survived inside the avian macrophages (HD11 monolayers) from three independent experiments.
Fig. 6.
Transmission electron micrographs of S. Enteritidis strains UK (a), BC8 (b), P21 (c), G1 (d), C19 (e), C45 (f), G45 (g) and A11 (h). Samples were taken from the edge of a motile colony. Bars, 0.2 µm.
S. Enteritidis isolates with impaired motility and/or secretion of TTSS proteins have impaired ability to invade and/or survive in chicken macrophages
We hypothesized that the low invasiveness isolates (C19, C45, A11 and G45) showing impaired motility and altered secretion of one or more SPI-1 TTSS effector proteins are also less invasive in the chicken host. Consequently, we used chicken macrophage cells (HD11) to test the ability of representative isolates with low and high invasiveness to invade and/or survive within the chicken cells. Overall, the mean (±sem) intra-macrophage c.f.u. of low invasiveness isolates [log10(c.f.u.) = 4.88±0.13] was significantly lower than that of high invasiveness isolates [log10(c.f.u.) = 6.07±0.08] (Tukey–Kramer test; P<0.01, Table 4).
Discussion
Invasion in cultured epithelial cells is routinely used as a measure of pathogenicity of Salmonella isolates (Dibb-Fuller et al., 1999; van Asten et al., 2000, 2004). Fewer studies have been conducted to determine differences in invasiveness of field isolates of S. Enteritidis (Pan et al., 2009; Pang et al., 2006; Solano et al., 1998). Although these studies have shown that not all isolates of S. Enteritidis have similar cell invasive potential, only limited numbers of isolates recovered mostly from human, animal, food and the environment were tested. This is the first report in which more than 50 wild-type S. Enteritidis isolates recovered specifically from diverse poultry sources have been tested for their invasiveness by using well-differentiated Caco-2 cells as a model for infection. Given enough time, Caco-2 cells spontaneously differentiate to closely mimic a small-bowel-like cellular phenotype, as indicated by dome formation, presence of microvilli and expression of brush border enzymes (e.g. alkaline phosphatase) after confluency (Hara et al., 1993). In addition, differentiation of Caco-2 cells has been shown to be an important factor for invasion by Salmonella and for expression of surface receptors for enterotoxigenic E. coli (Finlay & Falkow, 1990; Kernéis et al., 1992). We observed that the invasion rate of stationary phase cultures of S. Enteritidis was 13 times higher for differentiated cells than for undifferentiated cells (Table 3). When this model was used to screen stationary phase cultures of all the field isolates, seven (13 %) isolates were identified with significantly lower invasiveness (Table 1). These data indicate that although poultry and poultry products are major sources of infection for humans worldwide, not all isolates of S. Enteritidis recovered from poultry may be equally pathogenic.
The majority (50–90 %) of field isolates of S. Enteritidis are known to produce biofilm, an extracellular matrix composed of high-molecular-mass LPS, flagella and fimbriae (Guard-Petter et al., 1996; Guard-Petter, 2001; Marin et al., 2009; Solano et al., 2001, 2002). Similar to data from these reports, 56.6 % (30/53) of S. Enteritidis isolates tested here produced biofilm. Biofilm formation in S. Enteritidis isolates has also been reported to be associated with the enhanced disruption of cultured Caco-2 cells and increased virulence in chickens (Solano et al., 1998, 2001). In this study, however, there was no significant correlation (P = 0.35) between invasiveness and the ability of S. Enteritidis isolates to form biofilms. Nevertheless, the majority (71 %) of isolates with low invasiveness tested bf− whereas lower proportions of isolates with medium (19 %) and high (50 %) invasiveness were bf−, indicating that biofilm may, in part, play a role in the pathogenesis of S. Enteritidis infection. Some isogenic strains of S. Enteritidis (bf+m) show both bf+ and bf− colonies in varying proportions (Guard-Petter et al., 1996; Humphrey et al., 1996). Subcultures from isogenic single bf+ or bf− colonies result in similar mixed phenotypes (J. Guard, unpublished data). Using a BGA plate assay, seven (13 %) isolates in this study were identified as isogenic, with the proportion of bf+ colonies ranging from 10 to 90 % (data not shown). When biofilm production in these isogenic strains was confirmed by quantitative microtitre assay, all strains were classified as bf+ (Table 1). Interestingly, when chickens are infected with a mixture of highly invasive bf+ and less invasive bf− S. Enteritidis isolates, the bf+ S. Enteritidis acts as a ‘helper’ phenotype and aids access of the less orally invasive bf− strain to the parenteral (post-mucosal) environment in chickens (Gast et al., 2002; Guard-Petter, 2001; Parker & Guard-Petter, 2001). It is possible that a similar phenomenon may have contributed to the higher cell invasion potential of the isogenic strains (bf+m) identified in this study.
Regardless of phage types, geographical origins or source of isolation, S. Enteritidis is one of the most genetically homogeneous serotypes of Salmonella and as a result is poorly discriminated by the commonly used genetic subtyping methods, such as phage typing, pulsed field gel electrophoresis or comparative genomic hybridization microarrays (Betancor et al., 2009; Botteldoorn et al., 2010; Hudson et al., 2001; Liebana et al., 2001; Malorny et al., 2008; Morales et al., 2005; Olsen et al., 1999; Pan et al., 2009; Pang et al., 2006; Porwollik et al., 2005; Saeed et al., 2006; Swaminathan et al., 2001). Because of its greater discriminatory capacity, MLVA was used to assess the genetic similarity among S. Enteritidis isolates tested in this study (Boxrud et al., 2007; Cho et al., 2007; Malorny et al., 2008). The majority (90 %) of isolates were grouped into eight clusters (clusters 4, 5 and 8–13) with a within-cluster allelic congruence of ≥85 % (Fig. 1). Some isolates that were recovered from different sources occurred within the same cluster, suggesting a considerable degree of genetic similarity. This was not unexpected as several studies using different genotyping methods have shown that isolates of S. Enteritidis are genetically closely related. Despite the clonal nature of S. Enteritidis, field isolates vary in their phenotypes. For instance, differences in the virulence potential between S. Enteritidis isolates within the same phage types and even within the same clonal lineages have been reported (Olsen et al., 1999; Yim et al., 2010). When we compared invasiveness or biofilm-forming ability of the isolates with their MLVA clusters, it was clear that each cluster included isolates with more than one phenotype (Table 1), indicating extensive phenotypic diversity among genetically closely related isolates of S. Enteritidis. Our data confirm the genetic homogeneity of S. Enteritidis and support the existence of phenotypic diversity within each genetic cluster.
Field isolates of S. Enteritidis contain a serovar-specific high-molecular-mass plasmid (pSEV) that encodes spvRABCD (Salmonella plasmid virulence) genes (Rychlík et al., 2006; Rotger & Casadesús, 1999). pSEV contributes to systemic infection of mice but not chickens (Bakshi et al., 2003; Nakamura et al., 1985), and spvB, in particular, plays a role in the pathogenesis by ribosylating actin of the macrophages and destabilizing the cytoskeleton (Lesnick et al., 2001; Otto et al., 2000; Tezcan-Merdol et al., 2001). Thus, identification of individual genes of the spv operon from S. Enteritidis is commonly used as a marker to assess the status of pSEV in field isolates (Rodríguez et al., 1998; Rychlík et al., 2008; Soto et al., 2003, 2006). Nine (17 %) isolates from our collection tested negative for the presence of the spvB gene, indicating that these isolates lacked the typical virulence plasmid. Our results are in agreement with those of Bakshi et al. (2003), who reported that 16 % (5/24) of field isolates lacked pSEV. Additional studies have reported that the proportion of plasmid-free isolates of S. Enteritidis ranged from 3 % (2/65) to 8 % (5/60) (Nakamura et al., 1985; Ridley et al., 1998). Overall, there was no statistically significant correlation between the presence of the spvB virulence gene and the invasiveness of S. Enteritidis isolates. Interestingly, all the plasmid-free isolates formed a single MLVA cluster 4b (Fig. 1), indicating a possible clonal relationship among these isolates. Nevertheless, isolates within cluster 4a with 100 % similarity by MLVA showed variable phenotypes. For instance, strain G47 was bf+, highly invasive in Caco-2 cells (Table 1) and motile, and survival rates within chicken macrophages were similar to those of other highly invasive strains (data not shown). In contrast, strain G45 was non-motile, did not form biofilm and showed low invasiveness in both cell lines (Tables 1 and 4). These results further suggest that genetically homogeneous isolates of S. Enteritidis may vary in their virulence potential and warrant further investigations on the mechanism underlying this differential virulence.
To determine whether the invasiveness of isolates correlated with their virulence, we compared the virulence of representative isolates in mice. All the isolates with low invasiveness caused no mortality or significantly lower mortality than high invasiveness isolates (Fig. 3). These data support our hypothesis that not all isolates of S. Enteritidis recovered from poultry are equally pathogenic. Although the molecular mechanisms underlying the differences in the virulence of S. Enteritidis remain poorly understood, both flagella and TTSS proteins encoded by virulence-associated genes located on SPI-1 and SPI-2 are known to play a major role in the pathogenesis of S. Enteritidis infection in cultured epithelial cells and in chicken and mouse challenge models (Allen-Vercoe & Woodward, 1999; Cogan et al., 2004; Dibb-Fuller et al., 1999; Methner & Barrow, 1997; Parker & Guard-Petter, 2001; van Asten et al., 2000, 2004; Yim et al., 2010). We observed that isolates with low invasiveness also had impaired secretion of SPI-1-encoded TTSS proteins (SipA and SipD) or motility-associated proteins (FlgK, FljB and FlgL) encoded by flagellar TTSS (Fig. 5). PCR amplification of the complete ORFs of sipA, sipD, flgK, fljB and flgL revealed that these genes were uniformly present in all isolates with low invasiveness and in representative isolates with high invasiveness (data not shown), indicating that the differential protein expression among these isolates was not due to the differences in gene content. Interestingly, TEM revealed that isolates with low invasiveness lacked flagellar appendages (Fig. 6e–h), which corroborates the impaired motility of these isolates and also partly explains the absence of bands corresponding to FlgK, FljB and FlgL proteins in isolate G45 (Fig. 5). It has also been shown that non-flagellar mutants of S. Enteritidis are strongly attenuated in their ability to invade cultured epithelial cells (Dibb-Fuller et al., 1999; van Asten et al., 2000), further suggesting that lower virulence of low invasiveness isolates is at least in part associated with their impaired motility (Table 4). It has been proposed that in S. Typhi, a host-adapted serotype, repression of flagellin may facilitate evasion from TLR5-mediated innate responses of host mucosal epithelium, thereby enhancing systemic bacterial dissemination from the intestine (Winter et al., 2008, 2010). Repression of flagellin was also shown to enable increased dissemination of S. Typhimurium to the spleen in a chicken model (Winter et al., 2010). In contrast, deletion of FliC in S. Enteritidis caused virulence attenuation leading to significantly reduced counts in the spleen of orally infected chickens (Allen-Vercoe & Woodward, 1999; Parker & Guard-Petter, 2001). Because S. Enteritidis primarily causes gastroenteritis in immunocompetent individuals, we surmise that S. Enteritidis strains with impaired motility identified in our study are less likely to utilize the proposed pathway to cause systemic disease. Nevertheless, it would be interesting to test this hypothesis by using a non-murine model in which mucosal barriers against Salmonella infection mimic human mucosal barriers.
Previous studies have also revealed that genes encoded by SPI-1 TTSS are involved in intestinal invasion whereas genes encoded on SPI-2 are required for systemic infection and survival within macrophages (Desin et al., 2009; Dieye et al., 2009; Jones et al., 2007; Li et al., 2009; Rychlík et al., 2009; Wisner et al., 2010). Mutations in the genes encoding secretory effector proteins or genes that regulate their expression lead to virulence attenuation in S. Enteritidis (Amy et al., 2004; Fardini et al., 2007; Parker & Guard-Petter, 2001). At least two low invasive isolates, C19 and C45, tested here showed impaired secretion of SipA and SipD proteins (Fig. 5). It is possible that the differential transcriptional or translational regulation of TTSS accounts for the observed differences in virulence and other phenotypes observed in this study. With real-time quantitative RT-PCR we confirmed that the mRNA expression levels of sipA and sipD in these low invasiveness isolates were 16- and 11-fold lower, respectively, than the representative high invasiveness isolates BC8 and P21. Therefore, our results not only corroborate published reports indicating the roles of TTSS or flagellar secreted proteins in the pathogenesis of S. Enteritidis infection but also provide the first evidence of impaired motility and secretion of TTSS proteins among wild-type isolates of S. Enteritidis. Further investigations that compare global transcriptional or proteome signatures of strains with low and high invasiveness should provide more insights into the detailed molecular mechanisms that regulate the differences in virulence potential of this important food-borne pathogen.
Finally, because poultry is the major source of infection for humans, it may be argued that poultry isolates that are more invasive or pathogenic in chickens may carry or express virulence factors that enhance their fitness and virulence in humans. This argument is consistent with reports showing that clinical isolates of S. Enteritidis are more virulent in mice and chickens compared with S. Enteritidis isolates recovered from chickens or environmental sources (Humphrey et al., 1995, 1996; Solano et al., 1998). Moreover, S. Enteritidis isolates recovered from eggs or from the human clinical cases showed greater adherence to and invasiveness of chicken ovarian granulosa cells compared with the isolates recovered from chicken caeca (Saeed et al., 2006). Our data show that isolates with high invasiveness in cultured Caco-2 cells are able to invade and/or survive within cultured chicken macrophages in significantly higher numbers than isolates with low invasiveness (Table 4), suggesting that isolates with low invasiveness are likely to invade and/or survive less well in chickens. Determination of oral invasiveness of representative high and low invasiveness isolates in a 1-day-old chicken model may be required to confirm these results.
In conclusion, our study shows that although poultry is the major source of infection for humans, not all poultry-associated S. Enteritidis isolates are equally pathogenic, as measured by invasion assays using well-differentiated human intestinal epithelial cells (Caco-2) and live BALB/c mice. Further investigations on the detailed molecular mechanism underlying this differential virulence are required in order to understand the pathogenesis of this important food-borne pathogen.
Acknowledgements
Carol Casavant provided technical assistance. This project was funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health and Department of Health and Human Services, under contract numbers N01-A1-30055. We gratefully acknowledge the technical assistance of Salma Al-Adwani and Swathi Kotla at various stages of this work.
Abbreviations:
- MLVA
multi-locus variable-number tandem repeat analysis
- PI
post-infection
- SPI
Salmonella pathogenicity island
- TEM
transmission electron microscopy
- TTSS
type III secretion system
Footnotes
Edited by: D. L. Gally
References
- Agron P. G., Walker R. L., Kinde H., Sawyer S. J., Hayes D. C., Wollard J., Andersen G. L. (2001). Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl Environ Microbiol 67, 4984–4991. 10.1128/AEM.67.11.4984-4991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Vercoe E., Woodward M. J. (1999). The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J Med Microbiol 48, 771–780. 10.1099/00222615-48-8-771. [DOI] [PubMed] [Google Scholar]
- Altekruse S. F., Bauer N., Chanlongbutra A., DeSagun R., Naugle A., Schlosser W., Umholtz R., White P. (2006). Salmonella enteritidis in broiler chickens, United States, 2000–2005. Emerg Infect Dis 12, 1848–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J., Sota M., Vivanco A. B., Perales I., Cisterna R., Rementeria A., Garaizar J. (2004). Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J Clin Microbiol 42, 1734–1738. 10.1128/JCM.42.4.1734-1738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy M., Velge P., Senocq D., Bottreau E., Mompart F., Virlogeux-Payant I. (2004). Identification of a new Salmonella enterica serovar Enteritidis locus involved in cell invasion and in the colonisation of chicks. Res Microbiol 155, 543–552. 10.1016/j.resmic.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Andrews J. M. (2001). Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48 Suppl 15–16. [DOI] [PubMed] [Google Scholar]
- Arricau N., Hermant D., Waxin H., Ecobichon C., Duffey P. S., Popoff M. Y. (1998). The RcsB–RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol 29, 835–850. 10.1046/j.1365-2958.1998.00976.x. [DOI] [PubMed] [Google Scholar]
- Bakshi C. S., Singh V. P., Malik M., Singh R. K., Sharma B. (2003). 55 kb plasmid and virulence-associated genes are positively correlated with Salmonella enteritidis pathogenicity in mice and chickens. Vet Res Commun 27, 425–432. 10.1023/A:1025720306045. [DOI] [PubMed] [Google Scholar]
- Barrow P. A. (1991). Experimental infection of chickens with Salmonella enteritidis. Avian Pathol 20, 145–153. 10.1080/03079459108418749. [DOI] [PubMed] [Google Scholar]
- Berrang M. E., Bailey J. S., Altekruse S. F., Shaw W. K., Jr, Patel B. L., Meinersmann R. J., Fedorka-Cray P. J. (2009). Prevalence, serotype, and antimicrobial resistance of Salmonella on broiler carcasses postpick and postchill in 20 U.S. processing plants. J Food Prot 72, 1610–1615. [DOI] [PubMed] [Google Scholar]
- Betancor L., Yim L., Fookes M., Martinez A., Thomson N. R., Ivens A., Peters S., Bryant C., Algorta G., et al. (2009). Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates. BMC Microbiol 9, 237. 10.1186/1471-2180-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohez L., Ducatelle R., Pasmans F., Botteldoorn N., Haesebrouck F., Van Immerseel F. (2006). Salmonella enterica serovar Enteritidis colonization of the chicken caecum requires the HilA regulatory protein. Vet Microbiol 116, 202–210. 10.1016/j.vetmic.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Botteldoorn N., Van Coillie E., Grijspeerdt K., Werbrouck H., Haesebrouck F., Donné E., D’Haese E., Heyndrickx M., Pasmans F., Herman L. (2006). Real-time reverse transcription PCR for the quantification of the mntH expression of Salmonella enterica as a function of growth phase and phagosome-like conditions. J Microbiol Methods 66, 125–135. 10.1016/j.mimet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Botteldoorn N., Van Coillie E., Goris J., Werbrouck H., Piessens V., Godard C., Scheldeman P., Herman L., Heyndrickx M. (2010). Limited genetic diversity and gene expression differences between egg- and non-egg-related Salmonella Enteritidis strains. Zoonoses Public Health 57, 345–357. [DOI] [PubMed] [Google Scholar]
- Boxrud D., Pederson-Gulrud K., Wotton J., Medus C., Lyszkowicz E., Besser J., Bartkus J. M. (2007). Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J Clin Microbiol 45, 536–543. 10.1128/JCM.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. L., Griffith A., Barry J. J., Jonas M., Chi E. Y. (2001). Transcytosis of gastrointestinal epithelial cells by Escherichia coli K1. Pediatr Res 49, 30–37. 10.1203/00006450-200101000-00010. [DOI] [PubMed] [Google Scholar]
- CDC (2003). Outbreaks of Salmonella serotype Enteritidis infection associated with eating shell eggs – United States, 1999–2001. MMWR Morb Mortal Wkly Rep 51, 1149–1152. [PubMed] [Google Scholar]
- CDC (2007). Salmonella serotype Enteritidis infections among workers producing poultry vaccine, Maine, November–December 2006. MMWR Morb Mortal Wkly Rep 56, 877–879. [PubMed] [Google Scholar]
- Cho S., Boxrud D. J., Bartkus J. M., Whittam T. S., Saeed M. (2007). Multiple-locus variable-number tandem repeat analysis of Salmonella Enteritidis isolates from human and non-human sources using a single multiplex PCR. FEMS Microbiol Lett 275, 16–23. 10.1111/j.1574-6968.2007.00875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo R. I., Loui C., Andersen G. L., Riley L. W., Lu S. (2006). Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl Environ Microbiol 72, 1055–1064. 10.1128/AEM.72.2.1055-1064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogan T. A., Jørgensen F., Lappin-Scott H. M., Benson C. E., Woodward M. J., Humphrey T. J. (2004). Flagella and curli fimbriae are important for the growth of Salmonella enterica serovars in hen eggs. Microbiology 150, 1063–1071. 10.1099/mic.0.26791-0. [DOI] [PubMed] [Google Scholar]
- De Buck J., Van Immerseel F., Haesebrouck F., Ducatelle R. (2004). Colonization of the chicken reproductive tract and egg contamination by Salmonella. J Appl Microbiol 97, 233–245. 10.1111/j.1365-2672.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- de Silva G. D., Kantzanou M., Justice A., Massey R. C., Wilkinson A. R., Day N. P., Peacock S. J. (2002). The ica operon and biofilm production in coagulase-negative Staphylococci associated with carriage and disease in a neonatal intensive care unit. J Clin Microbiol 40, 382–388. 10.1128/JCM.40.02.382-388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desin T. S., Lam P. K., Koch B., Mickael C., Berberov E., Wisner A. L., Townsend H. G., Potter A. A., Köster W. (2009). Salmonella enterica serovarEnteritidis pathogenicity island 1 is not essential for but facilitates rapid systemic spread in chickens. Infect Immun 77, 2866–2875. 10.1128/IAI.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon A. S., Alisantosa B., Shivaprasad H. L., Jack O., Schaberg D., Bandli D. (1999). Pathogenicity of Salmonella enteritidis phage types 4, 8, and 23 in broiler chicks. Avian Dis 43, 506–515. 10.2307/1592649. [DOI] [PubMed] [Google Scholar]
- Dibb-Fuller M. P., Allen-Vercoe E., Thorns C. J., Woodward M. J. (1999). Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145, 1023–1031. 10.1099/13500872-145-5-1023. [DOI] [PubMed] [Google Scholar]
- Dieye Y., Ameiss K., Mellata M., Curtiss R., III (2009). The Salmonella Pathogenicity Island (SPI) 1 contributes more than SPI2 to the colonization of the chicken by Salmonella enterica serovar Typhimurium. BMC Microbiol 9, 3. 10.1186/1471-2180-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran J. L., Collinson S. K., Clouthier S. C., Cebula T. A., Koch W. H., Burian J., Banser P. A., Todd E. C., Kay W. W. (1996). Diagnostic potential of sefA DNA probes to Salmonella enteritidis and certain other O-serogroup D1 Salmonella serovars. Mol Cell Probes 10, 233–246. 10.1006/mcpr.1996.0033. [DOI] [PubMed] [Google Scholar]
- Duchet-Suchaux M., Léchopier P., Marly J., Bernardet P., Delaunay R., Pardon P. (1995). Quantification of experimental Salmonella enteritidis carrier state in B13 leghorn chicks. Avian Dis 39, 796–803. 10.2307/1592416. [DOI] [PubMed] [Google Scholar]
- EFSA (2007). The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA J 130, 1–352. [Google Scholar]
- Eftekhar F., Speert D. P. (2009). Biofilm formation by persistent and non-persistent isolates of Staphylococcus epidermidis from a neonatal intensive care unit. J Hosp Infect 71, 112–116. 10.1016/j.jhin.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Fardini Y., Chettab K., Grépinet O., Rochereau S., Trotereau J., Harvey P., Amy M., Bottreau E., Bumstead N., et al. (2007). The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica serovar Enteritidis. Infect Immun 75, 358–370. 10.1128/IAI.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., Falkow S. (1990). Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J Infect Dis 162, 1096–1106. [DOI] [PubMed] [Google Scholar]
- Foley S. L., Lynne A. M., Nayak R. (2008). Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J Anim Sci 86 Suppl. 14E149–E162. 10.2527/jas.2007-0464. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Benson S. T. (1995). The comparative virulence for chicks of Salmonella enteritidis phage type 4 isolates and isolates of phage types commonly found in poultry in the United States. Avian Dis 39, 567–574. 10.2307/1591810. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Benson S. T. (1996). Intestinal colonization and organ invasion in chicks experimentally infected with Salmonella enteritidis phage type 4 and other phage types isolated from poultry in the United States. Avian Dis 40, 853–857. 10.2307/1592309. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Holt P. S. (1995). Differences in the multiplication of Salmonella enteritidis strains in liquid whole egg: implications for detecting contaminated eggs from commercial laying flocks. Poult Sci 74, 893–897. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Holt P. S. (2001). Multiplication in egg yolk and survival in egg albumen of Salmonella enterica serotype Enteritidis strains of phage types 4, 8, 13a, and 14b. J Food Prot 64, 865–868. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Guard-Petter J., Holt P. S. (2002). Characteristics of Salmonella enteritidis contamination in eggs after oral, aerosol, and intravenous inoculation of laying hens. Avian Dis 46, 629–635. 10.1637/0005-2086(2002)046[0629:COSECI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ghosh T. S., Vogt R. L. (2006). Cluster of invasive salmonellosis cases in a federal prison in Colorado. Am J Infect Control 34, 348–350. 10.1016/j.ajic.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Goldberg R. F., Austen W. G., Jr, Zhang X., Munene G., Mostafa G., Biswas S., McCormack M., Eberlin K. R., Nguyen J. T., et al. (2008). Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc Natl Acad Sci U S A 105, 3551–3556. 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden N. J., Marks H. H., Coleman M. E., Schroeder C. M., Bauer N. E., Jr, Schlosser W. D. (2008). Review of induced molting by feed removal and contamination of eggs with Salmonella enterica serovar Enteritidis. Vet Microbiol 131, 215–228. 10.1016/j.vetmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Gordon M. A., Graham S. M., Walsh A. L., Wilson L., Phiri A., Molyneux E., Zijlstra E. E., Heyderman R. S., Hart C. A., Molyneux M. E. (2008). Epidemics of invasive Salmonella enterica serovar Enteritidis and S. enterica serovarTyphimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis 46, 963–969. 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- Guard-Bouldin J., Gast R. K., Humphrey T. J., Henzler D. J., Morales C., Coles K. (2004). Subpopulation characteristics of egg-contaminating Salmonella enterica serovar Enteritidis as defined by the lipopolysaccharide O chain. Appl Environ Microbiol 70, 2756–2763. 10.1128/AEM.70.5.2756-2763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard-Petter J. (1998). Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl Environ Microbiol 64, 2166–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard-Petter J. (2001). The chicken, the egg and Salmonella enteritidis. Environ Microbiol 3, 421–430. 10.1046/j.1462-2920.2001.00213.x. [DOI] [PubMed] [Google Scholar]
- Guard-Petter J., Keller L. H., Rahman M. M., Carlson R. W., Silvers S. (1996). A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol Infect 117, 219–231. 10.1017/S0950268800001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard-Petter J., Parker C. T., Asokan K., Carlson R. W. (1999). Clinical and veterinary isolates of Salmonella enterica serovar Enteritidis defective in lipopolysaccharide O-chain polymerization. Appl Environ Microbiol 65, 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara A., Hibi T., Yoshioka M., Toda K., Watanabe N., Hayashi A., Iwao Y., Saito H., Watanabe T., Tsuchiya M. (1993). Changes of proliferative activity and phenotypes in spontaneous differentiation of a colon cancer cell line. Jpn J Cancer Res 84, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. H. (2001). The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer. [Google Scholar]
- He H., Crippen T. L., Farnell M. B., Kogut M. H. (2003). Identification of CpG oligodeoxynucleotide motifs that stimulate nitric oxide and cytokine production in avian macrophage and peripheral blood mononuclear cells. Dev Comp Immunol 27, 621–627. 10.1016/S0145-305X(03)00013-2. [DOI] [PubMed] [Google Scholar]
- HPA (2008). Salmonella in humans (excluding S. Typhi & S. Paratyphi). http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/Salmonella/EpidemiologicalData/salmDataHumanse/ Accessed 31 March 2010.
- Hudson C. R., Garcia M., Gast R. K., Maurer J. J. (2001). Determination of close genetic relatedness of the major Salmonella enteritidis phage types by pulsed-field gel electrophoresis and DNA sequence analysis of several Salmonella virulence genes. Avian Dis 45, 875–886. 10.2307/1592867. [DOI] [PubMed] [Google Scholar]
- Humphrey T. J., Slater E., McAlpine K., Rowbury R. J., Gilbert R. J. (1995). Salmonella enteritidis phage type 4 isolates more tolerant of heat, acid, or hydrogen peroxide also survive longer on surfaces. Appl Environ Microbiol 61, 3161–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T. J., Williams A., McAlpine K., Lever M. S., Guard-Petter J., Cox J. M. (1996). Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol Infect 117, 79–88. 10.1017/S0950268800001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Chen J. (2007). Attachment and biofilm formation by various serotypes of Salmonella as influenced by cellulose production and thin aggregative fimbriae biosynthesis. J Food Prot 70, 2473–2479. [DOI] [PubMed] [Google Scholar]
- Jones M. A., Hulme S. D., Barrow P. A., Wigley P. (2007). The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar Typhimurium in the chicken. Avian Pathol 36, 199–203. 10.1080/03079450701264118. [DOI] [PubMed] [Google Scholar]
- Kaniga K., Trollinger D., Galán J. E. (1995). Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol 177, 7078–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsenos C., Anastasopoulos N., Patrani M., Mandragos C. (2008). Salmonella enteritidis meningitis in a first time diagnosed AIDS patient: Case report. Cases J 1, 5. 10.1186/1757-1626-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernéis S., Chauvière G., Darfeuille-Michaud A., Aubel D., Coconnier M. H., Joly B., Servin A. L. (1992). Expression of receptors for enterotoxigenic Escherichia coli during enterocytic differentiation of human polarized intestinal epithelial cells in culture. Infect Immun 60, 2572–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A. C., Reddy V., Marcus R., Cieslak P. R., Mohle-Boetani J. C., Kassenborg H. D., Segler S. D., Hardnett F. P., Barrett T., et al. (2004). Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin Infect Dis 38 Suppl. 3S244–S252. 10.1086/381576. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Hall G. S., Tuohy M. J., Knothe U., Procop G. W., Bauer T. W. (2009). Bilateral periprosthetic joint infection caused by Salmonella enterica serotype Enteritidis, and identification of Salmonella sp. using molecular techniques. Int J Infect Dis 13, e463–e466. 10.1016/j.ijid.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Lesnick M. L., Reiner N. E., Fierer J., Guiney D. G. (2001). The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol Microbiol 39, 1464–1470. 10.1046/j.1365-2958.2001.02360.x. [DOI] [PubMed] [Google Scholar]
- Li S., Zhang Z., Pace L., Lillehoj H., Zhang S. (2009). Functions exerted by the virulence-associated type-three secretion systems during Salmonella enterica serovar Enteritidis invasion into and survival within chicken oviduct epithelial cells and macrophages. Avian Pathol 38, 97–106. 10.1080/03079450902737771. [DOI] [PubMed] [Google Scholar]
- Liebana E., Garcia-Migura L., Breslin M. F., Davies R. H., Woodward M. J. (2001). Diversity of strains of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J Clin Microbiol 39, 154–161. 10.1128/JCM.39.1.154-161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister S. A. (1988). Salmonella enteritidis infection in broilers and broiler breeders. Vet Rec 123, 350. 10.1136/vr.123.13.350. [DOI] [PubMed] [Google Scholar]
- Little C. L., Rhoades J. R., Hucklesby L., Greenwood M., Surman-Lee S., Bolton F. J., Meldrum R., Wilson I., McDonald C., et al. (2008). Survey of Salmonella contamination of raw shell eggs used in food service premises in the United Kingdom, 2005 through 2006. J Food Prot 71, 19–26. [DOI] [PubMed] [Google Scholar]
- Lock J. L., Board R. G. (1992). Persistence of contamination of hens’ egg albumen in vitro with Salmonella serotypes. Epidemiol Infect 108, 389–396. 10.1017/S095026880004989X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Manges A. R., Xu Y., Fang F. C., Riley L. W. (1999). Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect Immun 67, 5651–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malorny B., Junker E., Helmuth R. (2008). Multi-locus variable-number tandem repeat analysis for outbreak studies of Salmonella enterica serotype Enteritidis. BMC Microbiol 8, 84. 10.1186/1471-2180-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus R., Rabatsky-Ehr T., Mohle-Boetani J. C., Farley M., Medus C., Shiferaw B., Carter M., Zansky S., Kennedy M., et al. (2004). Dramatic decrease in the incidence of Salmonella serotype Enteritidis infections in 5 FoodNet sites: 1996-1999. Clin Infect Dis 38 Suppl. 3S135–S141. 10.1086/381579. [DOI] [PubMed] [Google Scholar]
- Marin C., Hernandiz A., Lainez M. (2009). Biofilm development capacity of Salmonella strains isolated in poultry, risk factors and their resistance against disinfectants. Poult Sci 88, 424–431. 10.3382/ps.2008-00241. [DOI] [PubMed] [Google Scholar]
- Methner U., Barrow P. A. (1997). Significance of motility of Salmonella enteritidis and Salmonella typhimurium as a virulence factor and on the expression of the inhibition phenomenon in vitro and in vivo in SPF chickens. Berl Munch Tierarztl Wochenschr 110, 391–396. (in German). [PubMed] [Google Scholar]
- Morales C. A., Porwollik S., Frye J. G., Kinde H., McClelland M., Guard-Bouldin J. (2005). Correlation of phenotype with the genotype of egg-contaminating Salmonella enterica serovar Enteritidis. Appl Environ Microbiol 71, 4388–4399. 10.1128/AEM.71.8.4388-4399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morpeth S. C., Ramadhani H. O., Crump J. A. (2009). Invasive non-Typhi Salmonella disease in Africa. Clin Infect Dis 49, 606–611. 10.1086/603553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu H., Babar J., Maggiore P. R. (2009). Extensive Salmonella enteritidis endocarditis involving mitral, tricuspid valves, aortic root and right ventricular wall. J Am Soc Echocardiogr 22, 210.e1–210.e3. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Sato S., Ohya T., Suzuki S., Ikeda S. (1985). Possible relationship of a 36-megadalton Salmonella enteritidis plasmid to virulence in mice. Infect Immun 47, 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. E., Tiainen T., Brown D. J. (1999). Levels of virulence are not determined by genomic lineage of Salmonella enterica serotype Enteritidis strains. Epidemiol Infect 123, 423–430. 10.1017/S0950268899003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan E., McCabe E., Burgess C., McGuinness S., Barry T., Duffy G., Whyte P., Fanning S. (2008). Development of a real-time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples. BMC Microbiol 8, 156. 10.1186/1471-2180-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Tezcan-Merdol D., Girisch R., Haag F., Rhen M., Koch-Nolte F. (2000). The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol Microbiol 37, 1106–1115. 10.1046/j.1365-2958.2000.02064.x. [DOI] [PubMed] [Google Scholar]
- Pan Z., Carter B., Núñez-García J., Abuoun M., Fookes M., Ivens A., Woodward M. J., Anjum M. F. (2009). Identification of genetic and phenotypic differences associated with prevalent and non-prevalent Salmonella Enteritidis phage types: analysis of variation in amino acid transport. Microbiology 155, 3200–3213. 10.1099/mic.0.029405-0. [DOI] [PubMed] [Google Scholar]
- Pang J. C., Lin J. S., Tsai C. C., Tsen H. Y. (2006). The presence of major world-wide clones for phage type 4 and 8 Salmonella enterica serovar Enteritidis and the evaluation of their virulence levels by invasiveness assays in vitro and in vivo. FEMS Microbiol Lett 263, 148–154. 10.1111/j.1574-6968.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- Parker C. T., Guard-Petter J. (2001). Contribution of flagella and invasion proteins to pathogenesis of Salmonella enterica serovar Enteritidis in chicks. FEMS Microbiol Lett 204, 287–291. 10.1111/j.1574-6968.2001.tb10899.x. [DOI] [PubMed] [Google Scholar]
- Parker C. T., Liebana E., Henzler D. J., Guard-Petter J. (2001). Lipopolysaccharide O-chain microheterogeneity of Salmonella serotypes Enteritidis and Typhimurium. Environ Microbiol 3, 332–342. 10.1046/j.1462-2920.2001.00200.x. [DOI] [PubMed] [Google Scholar]
- Patrick M. E., Adcock P. M., Gomez T. M., Altekruse S. F., Holland B. H., Tauxe R. V., Swerdlow D. L. (2004). Salmonella enteritidis infections, United States, 1985–1999. Emerg Infect Dis 10, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30, e36. 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C., Demczuk W., McFadden K., Johnson R. P. (1993). Virulence of Salmonella enteritidis phagetypes 4, 8 and 13 and other Salmonella spp. for day-old chicks, hens and mice. Can J Vet Res 57, 281–287. [PMC free article] [PubMed] [Google Scholar]
- Porwollik S., Santiviago C. A., Cheng P., Florea L., Jackson S., McClelland M. (2005). Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J Bacteriol 187, 6545–6555. 10.1128/JB.187.18.6545-6555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. M., Threlfall E. J., Rowe B. (1998). Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol 36, 2314–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez M., de Diego I., Mendoza M. C. (1998). Extraintestinal salmonellosis in a general hospital (1991 to 1996): relationships between Salmonella genomic groups and clinical presentations. J Clin Microbiol 36, 3291–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotger R., Casadesús J. (1999). The virulence plasmids of Salmonella. Int Microbiol 2, 177–184. [PubMed] [Google Scholar]
- Rychlík I., Gregorova D., Hradecka H. (2006). Distribution and function of plasmids in Salmonella enterica. Vet Microbiol 112, 1–10. 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Rychlík I., Hradecka H., Malcova M. (2008). Salmonella enterica serovar Typhimurium typing by prophage-specific PCR. Microbiology 154, 1384–1389. 10.1099/mic.0.2007/015156-0. [DOI] [PubMed] [Google Scholar]
- Rychlík I., Karasova D., Sebkova A., Volf J., Sisak F., Havlickova H., Kummer V., Imre A., Szmolka A., Nagy B. (2009). Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens. BMC Microbiol 9, 268. 10.1186/1471-2180-9-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A. M., Walk S. T., Arshad M., Whittam T. S. (2006). Clonal structure and variation in virulence of Salmonella enteritidis isolated from mice, chickens, and humans. J AOAC Int 89, 504–511. [PubMed] [Google Scholar]
- Sneath P. H., Sokal R. R. (1973). Numeral Taxonomy. San Francisco: W. H. Freeman. [Google Scholar]
- Snow L. C., Davies R. H., Christiansen K. H., Carrique-Mas J. J., Wales A. D., O’Connor J. L., Cook A. J., Evans S. J. (2007). Survey of the prevalence of Salmonella species on commercial laying farms in the United Kingdom. Vet Rec 161, 471–476. 10.1136/vr.161.14.471. [DOI] [PubMed] [Google Scholar]
- Solano C., Sesma B., Alvarez M., Humphrey T. J., Thorns C. J., Gamazo C. (1998). Discrimination of strains of Salmonella enteritidis with differing levels of virulence by an in vitro glass adherence test. J Clin Microbiol 36, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C., Sesma B., Alvarez M., Urdaneta E., Garcia-Ros D., Calvo A., Gamazo C. (2001). Virulent strains of Salmonella enteritidis disrupt the epithelial barrier of Caco-2 and HEp-2 cells. Arch Microbiol 175, 46–51. 10.1007/s002030000236. [DOI] [PubMed] [Google Scholar]
- Solano C., García B., Valle J., Berasain C., Ghigo J. M., Gamazo C., Lasa I. (2002). Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol 43, 793–808. 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Soto S. M., González-Hevia M. A., Mendoza M. C. (2003). Antimicrobial resistance in clinical isolates of Salmonella enterica serotype Enteritidis: relationships between mutations conferring quinolone resistance, integrons, plasmids and genetic types. J Antimicrob Chemother 51, 1287–1291. 10.1093/jac/dkg193. [DOI] [PubMed] [Google Scholar]
- Soto S. M., Rodríguez I., Rodicio M. R., Vila J., Mendoza M. C. (2006). Detection of virulence determinants in clinical strains of Salmonella enterica serovar Enteritidis and mapping on macrorestriction profiles. J Med Microbiol 55, 365–373. 10.1099/jmm.0.46257-0. [DOI] [PubMed] [Google Scholar]
- Suzuki S. (1994). Pathogenicity of Salmonella enteritidis in poultry. Int J Food Microbiol 21, 89–105. 10.1016/0168-1605(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Swaminathan B., Barrett T. J., Hunter S. B., Tauxe R. V., CDC PulseNet Task Force (2001). PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7, 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena D., González-Praetorius A., Bisquert J. (2007). Urinary tract infection due to non-typhoidal Salmonella: report of 19 cases. J Infect 54, 245–249. 10.1016/j.jinf.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Tezcan-Merdol D., Nyman T., Lindberg U., Haag F., Koch-Nolte F., Rhen M. (2001). Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol Microbiol 39, 606–619. 10.1046/j.1365-2958.2001.02258.x. [DOI] [PubMed] [Google Scholar]
- Trafny E. A., Kozłowska K., Szpakowska M. (2006). A novel multiplex PCR assay for the detection of Salmonella enterica serovar Enteritidis in human faeces. Lett Appl Microbiol 43, 673–679. 10.1111/j.1472-765X.2006.02007.x. [DOI] [PubMed] [Google Scholar]
- van Asten F. J., Hendriks H. G., Koninkx J. F., Van der Zeijst B. A., Gaastra W. (2000). Inactivation of the flagellin gene of Salmonella enterica serotype Enteritidis strongly reduces invasion into differentiated Caco-2 cells. FEMS Microbiol Lett 185, 175–179. 10.1016/S0378-1097(00)00098-7. [DOI] [PubMed] [Google Scholar]
- van Asten F. J., Hendriks H. G., Koninkx J. F., van Dijk J. E. (2004). Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int J Med Microbiol 294, 395–399. 10.1016/j.ijmm.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Velge P., Cloeckaert A., Barrow P. (2005). Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet Res 36, 267–288. 10.1051/vetres:2005005. [DOI] [PubMed] [Google Scholar]
- WHO (2008). http://www.who.int/gfn/activities/CDB_poster_Sept09.pdf.
- Winter S. E., Raffatellu M., Wilson R. P., Rüssmann H., Bäumler A. J. (2008). The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol 10, 247–261. [DOI] [PubMed] [Google Scholar]
- Winter S. E., Winter M. G., Godinez I., Yang H. J., Rüssmann H., Andrews-Polymenis H. L., Bäumler A. J. (2010). A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog 6, e1001060. 10.1371/journal.ppat.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisner A. L., Desin T. S., Koch B., Lam P. K., Berberov E. M., Mickael C. S., Potter A. A., Koster W. (2010). Salmonella enterica subspecies enterica serovar Enteritidis Salmonella pathogenicity island 2 type III secretion system: role in intestinal colonization of chickens and systemic spread. Microbiology 156, 2770–2781. 10.1099/mic.0.038018-0. [DOI] [PubMed] [Google Scholar]
- Yim L., Betancor L., Martinez A., Giossa G., Bryant C., Maskell D., Chabalgoity J. A. (2010). Differential phenotypic diversity among epidemic-spanning Salmonella enterica serovar enteritidis isolates from humans or animals. Appl Environ Microbiol 76, 6812–6820. 10.1128/AEM.00497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]