Abstract
The dlt operon in Gram-positive bacteria encodes proteins that are necessary for the addition of d-alanine to teichoic acids of the cell wall. The addition of d-alanine to the cell wall results in a net positive charge on the bacterial cell surface and, as a consequence, can decrease the effectiveness of antimicrobials, such as cationic antimicrobial peptides (CAMPs). Although the roles of the dlt genes have been studied for some Gram-positive organisms, the arrangement of these genes in Clostridium difficile and the life cycle of the bacterium in the host are markedly different from those of other pathogens. In the current work, we determined the contribution of the putative C. difficile dlt operon to CAMP resistance. Our data indicate that the dlt operon is necessary for full resistance of C. difficile to nisin, gallidermin, polymyxin B and vancomycin. We propose that the d-alanylation of teichoic acids provides protection against antimicrobial peptides that may be essential for growth of C. difficile in the host.
Introduction
Clostridium difficile causes a potentially fatal intestinal disease that is increasing in incidence and severity (Dubberke et al., 2010). Infection is often chronic and difficult to eradicate (O’Brien et al., 2007). C. difficile is transmitted as dormant endospores that are highly resistant to killing by heat, antibiotics, oxidizing agents, UV radiation and many other treatments that are usually effective for killing bacteria (Vonberg et al., 2008; Setlow, 1995). C. difficile spores germinate in the intestine to produce vegetatively growing cells capable of producing toxins A and B, which have been implicated in disease progression and severity (Bartlett et al., 1977; McDonald et al., 2005).
Upon germination, actively growing C. difficile must contend with the onslaught of host defences present in the intestinal environment. The host produces a number of innate immune factors, such as cationic antimicrobial peptides (CAMPs) and bacteriolytic enzymes that protect the intestine from invading bacterial pathogens (Müller et al., 2005; Gudmundsson & Agerberth, 1999). Some of these CAMPs are products of host cells and others are produced by the intestinal microflora. These host effectors are attracted to the overall negative charge of bacterial cells and, as a consequence, bacterial resistance to these effectors can be achieved by charge modification of cell wall and membrane components (Nizet, 2006; Peschel, 2002). In many Gram-positive bacteria, this charge modification occurs by d-alanyl esterification of teichoic acid in the cell wall, which results in an increased positive charge on the cell (Neuhaus & Baddiley, 2003). The addition of d-alanine esters to teichoic acids is typically mediated by the products of the dlt operon, which encodes four proteins: DltA, a d-alanine : d-alanyl carrier protein ligase; DltB, a d-alanyl transfer protein; DltC, the d-alanyl carrier protein; and DltD, a d-alanine esterase. All four proteins are required for successful addition of d-alanine to the cell wall (Neuhaus & Baddiley, 2003). Incorporation of d-alanine into teichoic acid has been demonstrated to increase resistance of bacteria to CAMPs and bacteriolytic enzymes, as well as contribute to the virulence of pathogens such as Bacillus anthracis (Fisher et al., 2006), Listeria monocytogenes (Abachin et al., 2002), Streptococcus pneumoniae (Wartha et al., 2007) and Staphylococcus aureus (Weidenmaier et al., 2005).
We report that transcription of the C. difficile dlt operon is induced in response to CAMPs and is important for resistance to CAMPs during vegetative growth. In addition, we identified a putative regulatory gene as part of the dlt operon in C. difficile. This work represents what is believed to be the first report examining the role of the dlt operon in any Clostridium species.
Methods
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. C. difficile strains were grown in BHIS medium (Smith et al., 1981) supplemented with 0.1 % l-cysteine. Media for growth of C. difficile were supplemented with 250 µg d-cycloserine ml−1, 50 µg kanamycin ml−1 and 20 µg thiamphenicol ml−1; 5 µg erythromycin ml−1; or 5 µg erythromycin ml−1 and 50 µg kanamycin ml−1, as needed. C. difficile strains were maintained at 37 °C in an anaerobic chamber (Coy Laboratory Products) with an atmosphere of 10 % H2, 5 % CO2 and 85 % N2. Bacillus subtilis strains were routinely grown at 37 °C in L broth (Luria & Burrous, 1957) or BHIS medium supplemented with 1 µg erythromycin ml−1 when needed. For propagation of B. subtilis in the anaerobic chamber, the growth medium was supplemented with 5 mM KNO3. Escherichia coli strains were grown at 37 °C in L or BHIS medium supplemented with 20 µg chloramphenicol ml−1 or 100 µg ampicillin ml−1, as needed.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant genotype or features | Source or reference |
| E. coli strains | ||
| HB101 | F− mcrB mrr hsdS20 () recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 | B. Dupuy* |
| MC116 | HB101 containing pRK24 and pMC111 | This study |
| C. difficile strains | ||
| 630 | Clinical isolate | Wüst & Hardegger (1983) |
| JIR8094 | ErmS derivative of strain 630 | O’Connor et al. (2006) |
| MC120 | JIR8094 dltD : : ermB | This study |
| MC129 | MC120 : : Tn916 (dltDABC); ErmR | This study |
| B. subtilis strains | ||
| BS49 | CU2189 : : Tn916 | P. Mullany† |
| MC128 | BS49 Tn916 : : pMC118 | This study |
| Plasmids | ||
| pRK24 | Tra+ Mob+; bla tet | Thomas & Smith (1987) |
| pCE240 | C. difficile TargeTron construct based on pJIR750ai (group II intron, ermB : : RAM, ltrA); catP | C. Ellermeier‡ |
| pSMB47 | Tn916 integrational vector; CmR ErmR | Manganelli et al. (1998) |
| pMC111 | pCE240 retargeted to dltD | This study |
| pMC118 | 4812 bp DNA sequence corresponding to dltDABC plus 500 bp upstream sequence cloned as BamHI/SphI fragment in pSMB47 | This study |
Institut Pasteur, Paris, France.
Eastman Dental Institute, London, UK.
University of Iowa.
Strain and plasmid construction.
Oligonucleotides used in this study are listed in Table 2. C. difficile strain 630 DNA, GenBank accession number AM180355 (Sebaihia et al., 2006), was used as a template for PCR amplification unless otherwise noted. Sequencing of cloned DNA fragments was performed by the Tufts University Core Facility using an ABI 3130XL DNA sequencer. A null mutation in dltD was created in several steps. First, plasmid pMC111 was created by retargeting of the group II intron from pCE240 (kindly supplied by C. Ellermeier, University of Iowa) using primers oMC74, oMC75, oMC76 and the EBS Universal primer as outlined in the TargeTron users’ manual (Sigma-Aldrich), followed by initial cloning of the retargeted fragment in pCE240 digested with BsrGI/HindIII. Plasmid pCE240 is a derivative of pJIR750ai (Sigma-Aldrich) and is similar to pMTL007 (Heap et al., 2007). pMC111 was then introduced by transformation into E. coli strain HB101(pRK24), resulting in strain MC116. pRK24 is a derivative of the broad-host-range plasmid RP4, which mobilizes IncP oriT plasmids. MC116 was then mated with C. difficile strain JIR8094, resulting in transfer of pMC111 by conjugation, as previously described (Dineen et al., 2007), except that transconjugants were selected on BHIS plates supplemented with d-cycloserine, kanamycin and thiamphenicol. Inactivation of dltD was selected for by screening transconjugants for erythromycin resistance and thiamphenicol sensitivity, resulting in strain MC120. Insertional disruption of dltD was confirmed using primers oMC21 and oMC22, which are located outside the region of insertion in the dltD locus.
Table 2. Oligonucleotides used in this study.
Underlined bases denote retargeted sequences.
| Primer | Sequence | Use and location, or source |
| oMC21 | 5′-GCGAATTCGGTGAATTTATTTGGTGCTCC-3′ | dltD sequencing |
| oMC22 | 5′-GCGGATCCCATATAAGAATGCTCCTTTAAAATCC-3′ | dltD sequencing |
| oMC38 | 5′-AAAGACGGAGTCACAAGTCACC-3′ | dltD qPCR |
| oMC39 | 5′-CTGCTTTATACTCGTCACTTCCC-3′ | dltD qPCR |
| oMC44 | 5′-CTAGCTGCTCCTATGTCTCACATC-3′ | rpoC qPCR |
| oMC45 | 5′-CCAGTCTCTCCTGGATCAACTA-3′ | rpoC qPCR |
| oMC48 | 5′-CTTCAGGAAGTACAGGAAAGCC-3′ | dltA qPCR |
| oMC49 | 5′-TCCATGGATAAGACCAGGGTA-3′ | dltA qPCR |
| oMC74 | 5′-AAAAGCTTTTGCAACCCACGTCGATCGTGAAAAAGTTGTCTTGGTGCGCCCAGATAGGGTG-3′ | dltD intron retargeting |
| oMC75 | 5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCGTCTTGTTTAACTTACCTTTCTTTGT-3′ | dltD intron retargeting |
| oMC76 | 5′-CGCAAGTTTCTAATTTCGGTTACTTTTCGATAGAGGAAAGTGTCT-3′ | dltD intron retargeting |
| EBS Universal | 5′-CGAAATTAGAAACTTGCGTTCAGTAAAC-3′ | Sigma-Aldrich |
| oMC96 | 5′-CGTTCAGGTCAATTCTCTCTAGGC-3′ | cprA qPCR |
| oMC97 | 5′-GGTCAAGACCATTTGTAGGCTC-3′ | cprA qPCR |
| oMC98 | 5′-GCGGATCCCAGAGATGGAAGAGAAGAAGA-3′ | PdltDABC cloning |
| oMC100 | 5′-GCGCATGCGTCCATCAAATATTAGTATCAGATGG-3′ | PdltDABC cloning |
| oMC101 | 5′-GCCATCTTGTCCAGAACTC-3′ | dltB sequencing |
| oMC102 | 5′-GAGTTCTGGACAAGATGGC-3′ | dltB sequencing |
| oMC103 | 5′-GCCCATTAGACCTAAAATTACTG-3′ | dltB sequencing |
| oMC104 | 5′-CCTCGGTTGGTCCATATC-3′ | dltA sequencing |
| oMC105 | 5′-GCTCCTATCATACATGCCAT-3′ | dltA sequencing |
| oMC106 | 5′-CATCTTTGTCTCTTCCTTTGT-3′ | dltD sequencing |
| oMC107 | 5′-GCTGTAACCCCTTTATCTTT-3′ | dltD sequencing |
| oMC197 | 5′-CTTCCCTCTTCTCCCTGTATTTC-3′ | CD2855 qPCR |
| oMC198 | 5′-CTTCAAACTGGAGTGGATGGAG-3′ | CD2855 qPCR |
| oMC199 | 5′-GATATTGGACATGCTGTCCTTTGG-3′ | CD2850 qPCR |
| oMC200 | 5′-CGAGAATCAAGGTTTGTTTGATGG-3′ | CD2850 qPCR |
To complement the dltD disruption, a 4812 bp fragment containing the dltDABC operon and its upstream region was amplified using oMC98 and oMC100 as primers and cloned between the BamHI and SphI sites of pSMB47, generating pMC118. MC128 was created by integration of pMC118 into the chromosomal Tn916 locus of B. subtilis strain BS49. MC129 is a transconjugant from the mating of B. subtilis strain MC128 and C. difficile strain MC120, resulting in the integration of the plasmid : : Tn916 fusion of the donor strains into the C. difficile chromosome, as previously described (Haraldsen & Sonenshein, 2003).
MIC determination.
Susceptibility tests were performed anaerobically in BHIS broth as follows. C. difficile strains were grown to OD600 0.5 and then diluted 1 : 100 in 1 ml fresh medium to give an initial concentration of approximately 5×105 c.f.u. ml−1. The medium was supplemented with a range of concentrations of nisin (MP Biomedicals), polymyxin B (Sigma-Aldrich, ≥6000 USP units mg−1), gallidermin (Alexis Biochemicals), LL-37 (cathelicidin) (AnaSpec), Magainin II (Phoenix Pharmaceuticals) or vancomycin (Sigma-Aldrich) and cultures were incubated for 18 h at 37 °C. The MIC was defined as the lowest concentration of antimicrobial peptide that prevented visible turbidity after 18 h. MICs were determined a minimum of three times for each isolate and represent the highest concentration of antimicrobial at which growth was consistently observed.
CAMP growth assays.
To assess the effects of CAMPs on growth, C. difficile strains were grown to late-exponential phase in BHIS and diluted 1 : 50 into fresh BHIS medium, BHIS with 10 µg nisin ml−1 or BHIS with 75 µg polymyxin B ml−1 to OD600 ~0.025. Samples were taken at hourly intervals to assess growth by optical density.
Quantitative reverse-transcription PCR (qPCR) analysis.
Exponential phase cultures of C. difficile grown in BHIS medium or BHIS supplemented with nisin, gallidermin or polymyxin B were harvested at OD600 0.2 (active growth), and RNA was prepared as previously described (McBride & Sonenshein, 2011; Dineen et al., 2007). RNA quantity and quality were measured by absorbance (A260 and A260/A280 ratio, respectively) on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). Primers for qPCR were designed using the online PrimerQuest tool from Integrated DNA Technologies (http://www.idtdna.com/Scitools/Applications/Primerquest), and amplification efficiencies for each primer set were determined prior to use. To control for chromosomal DNA contamination, mock cDNA synthesis reactions containing no reverse transcriptase were used as negative controls in subsequent amplifications. cDNA samples were diluted fourfold and used as templates for qPCR of rpoC (primers oMC44/oMC45), dltD (oMC38/oMC39), dltA (oMC48/oMC49), CD2855 (oMC197/oMC198) and CD2850 (oMC199/oMC200), using Roche SYBR Green I PCR mix and a Roche LightCycler 480 II thermocycler. Reactions were performed in a final volume of 20 µl using 4 µl diluted cDNA and 1 µM of each primer. Reactions were performed in triplicate using cDNA extracted from each of a minimum of three biological replicates, and results are presented as the means and sds of the data obtained. Amplification included 40 cycles of the following steps: 10 s at 95 °C, 10 s at 53 °C, 15 s at 72 °C. Results were calculated using the comparative cycle threshold method (Schmittgen & Livak, 2008), in which the amount of target mRNA is normalized to that of an internal control transcript (rpoC). The two-tailed Student’s t test was used to analyse the data.
Quantification of d-alanine ester content in teichoic acids.
Analysis of ester-linked d-alanine incorporation into cell walls was performed as described elsewhere, with minor modifications (Hyyrylainen et al., 2000; Fisher et al., 2006). Cultures (50 ml) of wild-type (JIR8094), dltD mutant (MC120) and the complemented strain (MC129) were grown to OD600 0.2–0.25 in BHIS anaerobically at 37 °C and harvested by centrifugation, and the cell pellets were stored at −80 °C prior to analysis. Cell pellets were thawed at room temperature and washed three times with 1 ml 0.1 M MES, pH 6.0 (Sigma-Aldrich). To partially purify the cell walls, cell pellets were boiled for 15 min in 0.5 ml 0.2 % SDS, 0.1 M MES, pH 6.0. Walls were then pelleted by centrifugation and washed five times with 1 ml 0.1 M MES, pH 6.0, and subsequently dried under vacuum in a heated tabletop vacuum centrifuge. Dried pellets were weighed to determine the amount of cell wall content for each sample. The cell wall material was then resuspended in 0.5 ml sodium pyrophosphate, pH 8.3 (EMD Chemicals), and incubated for 3 h at 60 °C to release ester-linked d-alanine residues. After incubation, the samples were centrifuged and the supernatants removed to a fresh tube. The assay for d-alanine ester content was performed as follows. A 0.1 ml volume of sample supernatant was mixed with 0.25 ml assay reagent [four volumes 0.1 M sodium pyrophosphate, pH 8.3, two volumes FAD (Sigma-Aldrich) at 0.2 mg ml−1 in 0.1 M sodium pyrophosphate, pH 8.3, one volume horseradish peroxidase (1000 units mg−1, Sigma-Aldrich) at 10 mg ml−1, one volume o-dianisidine dihydrochloride (Sigma-Aldrich) at 5 mg ml−1, and 0.1 volume d-amino acid oxidase (5.7 units mg−1, Sigma-Aldrich) at 13.15 µg ml−1]. The assay reactions were incubated at 37 °C for 15 min, at which time 1 ml 0.1 % SDS was added to stop the reaction. Samples were then centrifuged to remove any trace of precipitate and for each the A460 of the supernatant fluid was determined. The absorbance values were compared with a standard curve performed with d-alanine (Sigma-Aldrich) to determine the total amount of d-alanine esters present in each sample. The amount of d-alanine esters was divided by the weight of cell wall for each sample to determine the relative amount of d-alanine present. Results were calculated as the means and sds of three biological replicates, each tested in duplicate. The two-tailed Student’s t test was used to analyse the data.
Results
Identification and expression of the C. difficile dltDABC operon
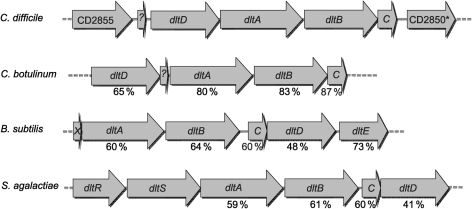
The dlt genes were identified as CD2851–CD2854 on the chromosome of the sequenced strain 630 (Sebaihia et al., 2006), based on sequence similarity to the dlt genes of B. subtilis. C. difficile has homologues of all four required dlt genes arranged in the order dltDABC (Fig. 1). The dltDABC genes are flanked upstream by a putative oxidoreductase, CD2855. The B. subtilis dlt operon also has a fifth gene, dltE, which encodes a putative oxidoreductase that does not have a known role in d-alanine incorporation into teichoic acid. In addition, a putative regulatory gene, CD2850, which encodes a protein similar to the DeoR family of transcriptional regulators, is located downstream of the dltC gene in C. difficile. We also searched for homologues of dltX, a small (144 bp) gene of unknown function that accompanies the dlt genes of many Gram-positive bacteria (Abi Khattar et al., 2009; Palumbo et al., 2006; Allen, 2008), but were unable to find any sequences with significant similarity. However, a 123 bp ORF was present between CD2855 and dltD. Based on the location and size of this ORF, this sequence may have a similar, albeit unknown, function in C. difficile (Fig. 1).
Fig. 1.
Comparison of the dlt regions of C. difficile and of other Gram-positive bacteria. Shown is the organization of the dlt operon and accessory genes of C. difficile (GenBank accession no. AM180355), Clostridium botulinum (CP001078), B. subtilis (AL009126) and Streptococcus agalactiae (CP000114). Numbers below the arrows indicate percentage similarity of proteins to the corresponding C. difficile sequences.
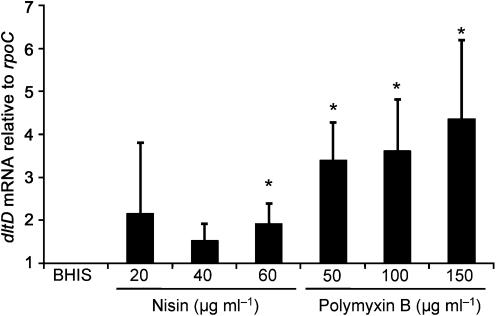
The dlt operons characterized to date add d-alanine esters to teichoic acid and, by changing the charge on the cell wall, provide resistance to CAMPs (Neuhaus & Baddiley, 2003). In order to determine whether the annotated dlt genes of C. difficile play a similar role, we analysed expression of these genes during growth in CAMPs. We chose to use the bacteria-derived model CAMPs nisin and polymyxin B because these CAMPs have been used extensively to study antimicrobial peptide resistance in other Gram-positive and Gram-negative bacteria, and because CAMPs are made by many bacterial species in the gut (Lactobacillus, Lactococcus, Streptococcus, Enterococcus, etc.). We used qPCR to examine the expression of the operon, specifically the dltD transcript, during early exponential growth in standard BHIS medium and medium supplemented with a range of concentrations of the CAMPs nisin and polymyxin B. As shown in Fig. 2, transcription of dltD was induced when wild-type C. difficile was grown in the presence of either CAMP. Expression of dltD increased with the concentration of CAMPs in the medium and was higher in cells grown in polymyxin B than in nisin. These results are consistent with the expression profiles of dlt genes noted for other Gram-positive bacteria grown in media containing CAMPs (Li et al., 2007a, b).
Fig. 2.
qPCR analysis of dltD expression in C. difficile during growth in polymyxin B and nisin. C. difficile wild-type (JIR8094) was grown to OD600 0.2 in BHIS alone or BHIS supplemented with 20, 40 or 60 µg nisin ml−1, or 50, 100 or 150 µg polymyxin B ml−1, as indicated. RNA was harvested, cDNA synthesized and qPCR performed using gene-specific primers for dltD. Results were normalized to an internal control gene (rpoC). The means and sds of biological replicates are shown. Asterisks, P≤0.05 by Student’s t test.
Effects of the disruption of dltD on CAMP resistance
To test whether the dltDABC locus has a direct role in CAMP resistance, we introduced an insertion mutation into the first gene of the operon, dltD. To do so, we used a TargeTron-based group II intron (Karberg et al., 2001) that was retargeted for integration into dltD at nucleotide 353 of the coding sequence (see Methods). The resulting strain, MC120, was then tested for the MICs of nisin, polymyxin B, gallidermin and vancomycin. The MC120 mutant had decreased MICs for all four of these compounds (Table 3). We also tested the resistance of the wild-type and MC120 strains to two animal-derived CAMPs, LL-37 and Magainin II. We observed no difference in the LL-37 MIC values of the wild-type, dlt mutant and complemented strains (see below). In the case of Magainin II, all of the strains studied were resistant to the highest concentration of the compound tested (>100 µg ml−1). As a result, these CAMPs were not used in further analyses.
Table 3. MICs (μg ml−1) for C. difficile strains with different CAMPs.
| CAMP | Strain | ||
| 630 | MC120 | MC129 | |
| Nisin | 90 | 50 | 110 |
| Polymyxin B | 300 | 150 | 400 |
| Gallidermin | 0.5 | 0.3 | 0.4 |
| Vancomycin | 1 | 0.75 | 1 |
| LL-37 | 4 | 4 | 4 |
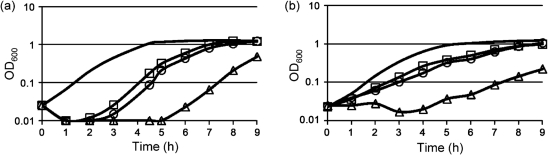
The dltD mutant also demonstrated significant growth delays when exposed to sublethal concentrations of either nisin (10 µg ml−1) (Fig. 3a) or polymyxin B (75 µg ml−1) (Fig. 3b). Though the growth rate of the MC120 mutant grown in CAMPs eventually caught up with that of the parental strain, dilution of the growing mutant culture into fresh medium containing CAMPs again produced a delay in active growth (data not shown), indicating that the mutant strain neither adapted to growth in CAMPs nor acquired a resistance mutation.
Fig. 3.
Growth of C. difficile wild-type and mutant isolates in nisin and polymyxin B. C. difficile wild-type (JIR8094, □), MC120 (dltD : : intron : : ermB, △) and MC129 (dltD : : intron : : ermB, dltDABC+, ○) strains were grown in BHIS medium or BHIS medium supplemented with 10 µg nisin B ml−1 (a) or 75 µg polymyxin B ml−1 (b). The growth rates of strains in BHIS alone were indistinguishable and are thus shown as a single solid line for clarity.
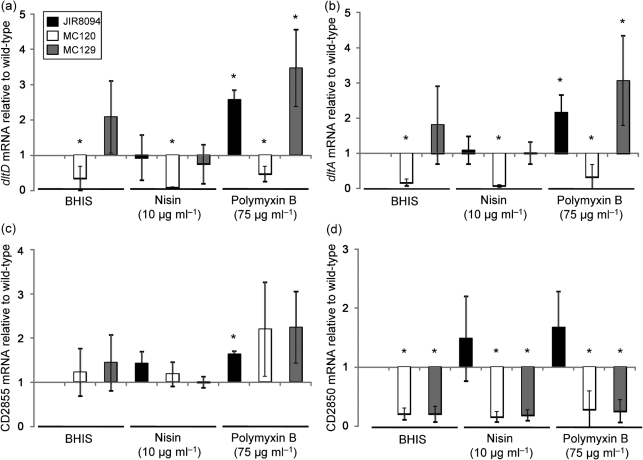
The dlt genes in other bacteria are known to form a transcriptional unit and, as a result, disruption of the first gene in the operon results in a polar effect on transcription of the downstream genes. Because the dlt genes of C. difficile are arranged in a different orientation from that in most other species [dltDABC vs dlt(X)ABCD], it was not clear whether the dltDABC genes and the upstream oxidoreductase (CD2855) constitute a single transcriptional unit. For this reason, we performed qPCR analysis of the dltD, dltA and CD2855 transcripts during growth in the presence and absence of the CAMPs polymyxin B and nisin to determine the effects on transcription of the insertional disruption in dltD. As Fig. 4 illustrates, expression of dltD and the downstream dltA transcript was severely impaired in a dltD null mutant, as compared with the wild-type parent strain. These results confirm that targeted insertion in the dltD gene not only disrupts transcription of that gene but also results in markedly decreased expression of the downstream gene, providing evidence that these genes are part of a transcriptional unit. In contrast, expression of the CD2855 oxidoreductase was not induced in either the wild-type or the dltDABC null mutant under any condition tested (Fig. 4c), indicating that it is not part of the same transcriptional unit as dltDABC. Because CD2855 is not co-regulated with the dlt genes, it is unlikely to play a direct role in d-alanylation.
Fig. 4.
Expression of the dlt genes of wild-type and mutant strains in the presence of the CAMPs nisin and polymyxin B. C. difficile wild-type (JIR8094), MC120 (dltD : : intron : : ermB) and MC129 (dltD : : intron : : ermB, dltDABC+) strains were grown to OD600 0.2 in BHIS or BHIS supplemented with 10 µg nisin ml−1 or 75 µg polymyxin B ml−1. RNA was harvested, cDNA synthesized and qPCR performed using gene-specific primers for (a) dltD, (b) dltA, (c) CD2855 or (d) CD2850. Results were normalized to an internal control gene (rpoC) and are presented as the ratio of each transcript level to that of wild-type cells grown in BHIS. The means and sds of biological replicates are shown. Asterisks, P≤0.05 by Student’s t test.
To confirm that the defect in CAMP resistance of MC120 was due to disruption of the dlt operon, we complemented MC120 with the native version of the dltDABC region, including a 500 bp upstream region predicted to contain the potential dltX ORF and the promoter for these genes. Complementation of MC120 with the dltDABC genes restored the MICs and growth properties in CAMPs to those of the wild-type (Table 3, Fig. 3). Similar to the wild-type, expression of dltDABC in MC129 was also induced during growth in nisin and polymyxin B (Fig. 4a, b).
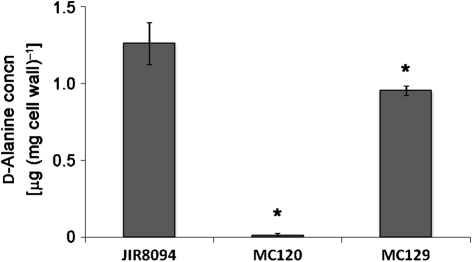
To determine whether the decreased CAMP resistance of the dltD mutant was due to a decrease in the incorporation of d-alanine esters into teichoic acids of the cell wall, we isolated and partially purified cell walls from the wild-type, dltD mutant (MC120) and dltDABC complemented mutant (MC129) to assay for ester-linked d-alanine. As shown in Fig. 5, the d-alanine content of the dltD mutant was approximately 100-fold less than that of wild-type C. difficile, while the d-alanine content of the dltDABC complemented strain was restored to near wild-type levels. Taken together, the MIC values, transcriptional analyses and d-alanine content of the dltD mutant indicate that the dltDABC gene products are responsible for incorporation of d-alanine esters into the teichoic acids of C. difficile and contribute to resistance to CAMPs.
Fig. 5.
Analysis of ester-linked d-alanine content in C. difficile cell walls. Cell walls partially purified from C. difficile wild-type (JIR8094), MC120 (dltD : : intron : : ermB) and MC129 (dltD : : intron : : ermB, dltDABC+) strains harvested after growth to OD600 0.2 in BHIS were assayed for incorporation of d-alanine into teichoic acids of the cell wall. Results represent the means and sds of three biological replicates, each replicate tested in duplicate samples. Asterisks, P≤0.05 by Student’s t test.
Identification of a regulatory gene within the dlt operon
The regulation of the dlt operon and d-alanylation of teichoic acid are not known to have been studied in any Clostridium species. However, in the spore-forming bacterium B. anthracis, the dlt genes are turned on during spore germination in the host and are necessary for resistance to innate host defences (Fisher et al., 2006). Because a potential regulatory protein is located immediately adjacent to the dlt operon, we examined the regulation of its expression. The gene for this regulatory protein, CD2850, is situated downstream of the operon and separated from dltC by 217 bp. CD2850 has homology to the DeoR family of transcriptional regulators, which often function as repressors of gene transcription and are expressed in an inverse manner relative to the genes that they regulate (i.e. regulator transcription decreases when target transcription increases; Hammer-Jespersen & Munch-Petersen, 1975), although in some circumstances they may behave as positive regulators (Haghjoo & Galán, 2007). Because of the close proximity of CD2850 to the dlt genes, we investigated whether expression of CD2850 coincides with expression of the dlt operon. Using qPCR, we found that the CD2850 regulator was expressed in response to either nisin or polymyxin B in wild-type cells (Fig. 4d). However, expression of CD2850 was drastically reduced in both the dltD insertional mutant (MC120) and the dltDABC-complemented strain (MC129). These data indicate that CD2850 expression is dependent on transcription of dltDABC, and provide evidence that dltDABC and CD2850 constitute a transcriptional unit. Considering that an increase in CD2850 expression accompanies the increase in dltDABC expression, it is unlikely that the CD2850 regulatory protein is a negative regulator of the dlt operon. However, because the dltDABC-complemented strain (MC129) does not express CD2850, we can also infer that this putative regulator is not required for expression of the dlt operon under the conditions tested.
Discussion
The dlt operon of Gram-positive bacteria is responsible for the addition of d-alanine to teichoic acids, which results in a decreased negative charge on the bacterial cell surface. This change in surface charge directly diminishes the interaction of host innate immune effectors, such as CAMPs, with the bacterial surface. As a consequence, the addition of d-alanine can lead to decreased effectiveness of these molecules in protecting against Gram-positive infection. Although these genes have been studied for some organisms, the arrangement of otherwise similar genes in C. difficile and the life cycle of the bacterium in the host are markedly different from those of other pathogens. In the current work, we found that the expression of the dltDABC genes increased when wild-type C. difficile was grown in the presence of CAMPs, and that disruption of the dlt operon by insertional mutation resulted in a decrease in resistance to multiple CAMPs, including polymyxin B, nisin, gallidermin and vancomycin. These results imply strongly that the C. difficile dlt operon plays a role in protecting the bacteria against host defences, especially against bacterial CAMPs that may be produced by the indigenous microbiota.
Many regulatory schemes have been found to control transcription of the dlt genes of Gram-positive bacteria (Neuhaus & Baddiley, 2003). Some bacteria use a two-component system [e.g. Streptococcus agalactiae (Poyart et al., 2001)], some use a single regulator [e.g. L. monocytogenes (Mandin et al., 2005)] and others use multiple regulators [e.g. S. aureus (Li et al., 2007a, b; Dunman et al., 2001; Saïd-Salim et al., 2003; Kraus et al., 2008; Koprivnjak et al., 2006)] to control transcription of the dlt genes. Many of the dlt transcriptional regulators also control the expression of virulence genes (Mandin et al., 2005; Dunman et al., 2001; Saïd-Salim et al., 2003; Kraus et al., 2008; Li et al., 2007a, b). Our investigation of dlt transcription revealed a co-expressed regulatory protein encoded downstream of the dltDABC ORFs, but we could find no evidence that the putative regulator plays either a positive or a negative role in dlt expression. In Streptococcus agalactiae, the two-component regulatory system that is transcribed with the dlt genes is also unnecessary for dltABCD transcription under the conditions tested, leading to the hypothesis that these regulators are involved in regulating the level of d-alanine esters under unknown conditions (Poyart et al., 2001). This may be the case for the CD2850 gene in C. difficile as well.
In B. subtilis, the dlt operon is regulated by an assortment of sigma factors and sporulation regulatory proteins, depending on the growth stage of the bacterium (Perego et al., 1995; Cao & Helmann, 2004). The principal regulator in B. subtilis is SigX, a sigma factor that regulates cell envelope metabolism and resistance to antimicrobial peptides (Cao & Helmann, 2004). Secondary transcription factors for dlt include SigD and Spo0A (Perego et al., 1995; Estacio et al., 1998). C. difficile does not have a SigX homologue, but does have homologues of SigD and Spo0A (Sonenshein et al., 2005). However, no obvious SigD-binding sites are present in the dlt region of C. difficile. Little is known about the expression characteristics of any of these regulatory factors during growth of C. difficile in the host. Given that C. difficile in the intestinal tract encounters a very different environment from that of B. subtilis in the soil and has a more limited repertoire of sigma factors (Sonenshein et al., 2005), it is likely that regulation of d-alanine incorporation into the cell wall of C. difficile will be distinct from that in the other sporulating bacteria characterized to date.
In conclusion, we have demonstrated that the dlt operon of C. difficile is expressed as a consequence of exposure to CAMPs, and that this operon is directly involved in the addition of d-alanine residues to teichoic acids as well as resistance to antimicrobial peptides. The production of CAMPs in the host represents a critical component of innate immunity. Thus, the decreased viability in the presence of any of these compounds may predicate decreased fitness in the host. Though we did not observe a decreased MIC for the dlt mutant with the human CAMP LL-37, the decreased MIC of the mutant for a variety of other CAMPs suggests that this strain has a general defect in growth in the presence of cationic peptides that may affect its survival in vivo. In B. anthracis, transcription of the dlt operon is initiated during germination and outgrowth, and mutants that do not express these genes are attenuated during outgrowth in vitro and for virulence in an animal model (Fisher et al., 2006). Because C. difficile undergoes an analogous developmental evolution from the dormant spore form to a vegetative cell in vivo, it too must perform this task in the presence of host innate effectors, and therefore could be similarly susceptible during the outgrowth phase and periods of rapid growth. We are currently examining the role of dltDABC during outgrowth and vegetative growth in in vivo models of disease. Based on the roles of the C. difficile dltDABC genes in survival in the presence of CAMPs, we hypothesize that analogous genes could also affect the viability of related pathogens, such as Clostridium botulinum.
Acknowledgements
We thank J. Sorg, L. Bouillaut and A. Camilli for helpful suggestions and discussions during the course of this work and for criticism of the manuscript, C. Ellermeier (University of Iowa) for pCE240, Gayatri Vedantam for the help with LL-37 MICs, and J. P. van Pijkeren and the Britton laboratory (Michigan State University) for the group II intron algorithm. This work was supported by the US National Institutes of Health via a research grant (AI057637 to A. L. S.) and a National Research Service Award (DK082156 to S. M. M), a Natalie V. Zucker Research grant to S. M. M., and by a core facility grant (NS047243) to the Tufts University Center for Neuroscience Research.
Abbreviations:
- CAMP
cationic antimicrobial peptide
- qPCR
quantitative reverse-transcription PCR
Footnotes
Edited by: A. J. O'Neill
References
- Abachin E., Poyart C., Pellegrini E., Milohanic E., Fiedler F., Berche P., Trieu-Cuot P. (2002). Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 43, 1–14. 10.1046/j.1365-2958.2002.02723.x [DOI] [PubMed] [Google Scholar]
- Abi Khattar Z., Rejasse A., Destoumieux-Garzón D., Escoubas J. M., Sanchis V., Lereclus D., Givaudan A., Kallassy M., Nielsen-Leroux C., Gaudriault S. (2009). The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J Bacteriol 191, 7063–7073. 10.1128/JB.00892-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, D. (2008) Transcriptional regulation of the dlt operon in Enterococcus faecalis and further characterization of a dltA mutant. MS thesis, Kansas State University, Manhattan, KS. [Google Scholar]
- Bartlett J. G., Onderdonk A. B., Cisneros R. L., Kasper D. L. (1977). Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis 136, 701–705. [DOI] [PubMed] [Google Scholar]
- Cao M., Helmann J. D. (2004). The Bacillus subtilis extracytoplasmic-function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol 186, 1136–1146. 10.1128/JB.186.4.1136-1146.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen S. S., Villapakkam A. C., Nordman J. T., Sonenshein A. L. (2007). Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66, 206–219. 10.1111/j.1365-2958.2007.05906.x [DOI] [PubMed] [Google Scholar]
- Dubberke E. R., Butler A. M., Yokoe D. S., Mayer J., Hota B., Mangino J. E., Khan Y. M., Popovich K. J., Fraser V. J. (2010). Multicenter study of Clostridium difficile infection rates from 2000 to 2006. Infect Control Hosp Epidemiol 10, 1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunman P. M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., Brown E. L., Zagursky R. J., Shlaes D., Projan S. J. (2001). Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183, 7341–7353. 10.1128/JB.183.24.7341-7353.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estacio W., Anna-Arriola S. S., Adedipe M., Márquez-Magaña L. M. (1998). Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis. J Bacteriol 180, 3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher N., Shetron-Rama L., Herring-Palmer A., Heffernan B., Bergman N., Hanna P. (2006). The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J Bacteriol 188, 1301–1309. 10.1128/JB.188.4.1301-1309.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson G. H., Agerberth B. (1999). Neutrophil antibacterial peptides, multifunctional effector molecules in the mammalian immune system. J Immunol Methods 232, 45–54. 10.1016/S0022-1759(99)00152-0 [DOI] [PubMed] [Google Scholar]
- Haghjoo E., Galán J. E. (2007). Identification of a transcriptional regulator that controls intracellular gene expression in Salmonella typhi. Mol Microbiol 64, 1549–1561. 10.1111/j.1365-2958.2007.05754.x [DOI] [PubMed] [Google Scholar]
- Hammer-Jespersen K., Munch-Petersen A. (1975). Multiple regulation of nucleoside catabolizing enzymes: regulation of the deo operon by the cytR and deoR gene products. Mol Gen Genet 137, 327–335. 10.1007/BF00703258 [DOI] [PubMed] [Google Scholar]
- Haraldsen J. D., Sonenshein A. L. (2003). Efficient sporulation in Clostridium difficile requires disruption of the σK gene. Mol Microbiol 48, 811–821. 10.1046/j.1365-2958.2003.03471.x [DOI] [PubMed] [Google Scholar]
- Heap J. T., Pennington O. J., Cartman S. T., Carter G. P., Minton N. P. (2007). The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70, 452–464. 10.1016/j.mimet.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Hyyrylainen H. L., Vitikainen M., Thwaite J., Wu H., Sarvas M., Harwood C. R., Kontinen V. P., Stephenson K. (2000). d-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J Biol Chem 275, 26696–26703. [DOI] [PubMed] [Google Scholar]
- Karberg M., Guo H., Zhong J., Coon R., Perutka J., Lambowitz A. M. (2001). Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol 19, 1162–1167. 10.1038/nbt1201-1162 [DOI] [PubMed] [Google Scholar]
- Koprivnjak T., Mlakar V., Swanson L., Fournier B., Peschel A., Weiss J. P. (2006). Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J Bacteriol 188, 3622–3630. 10.1128/JB.188.10.3622-3630.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D., Herbert S., Kristian S. A., Khosravi A., Nizet V., Götz F., Peschel A. (2008). The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol 8, 85. 10.1186/1471-2180-8-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Cha D. J., Lai Y., Villaruz A. E., Sturdevant D. E., Otto M. (2007a). The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 66, 1136–1147. 10.1111/j.1365-2958.2007.05986.x [DOI] [PubMed] [Google Scholar]
- Li M., Lai Y., Villaruz A. E., Cha D. J., Sturdevant D. E., Otto M. (2007b). Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A 104, 9469–9474. 10.1073/pnas.0702159104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Burrous J. W. (1957). Hybridization between Escherichia coli and Shigella. J Bacteriol 74, 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P., Fsihi H., Dussurget O., Vergassola M., Milohanic E., Toledo-Arana A., Lasa I., Johansson J., Cossart P. (2005). VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol 57, 1367–1380. 10.1111/j.1365-2958.2005.04776.x [DOI] [PubMed] [Google Scholar]
- Manganelli R., Provvedi R., Berneri C., Oggioni M. R., Pozzi G. (1998). Insertion vectors for construction of recombinant conjugative transposons in Bacillus subtilis and Enterococcus faecalis. FEMS Microbiol Lett 168, 259–268. 10.1111/j.1574-6968.1998.tb13282.x [DOI] [PubMed] [Google Scholar]
- McBride S. M., Sonenshein A. L. (2011). Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79, 167–176. 10.1128/IAI.00731-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L. C., Killgore G. E., Thompson A., Owens R. C., Jr, Kazakova S. V., Sambol S. P., Johnson S., Gerding D. N. (2005). An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353, 2433–2441. 10.1056/NEJMoa051590 [DOI] [PubMed] [Google Scholar]
- Müller C. A., Autenrieth I. B., Peschel A. (2005). Intestinal epithelial barrier and mucosal immunity. Innate defenses of the intestinal epithelial barrier. Cell Mol Life Sci 62, 1297–1307. 10.1007/s00018-005-5034-2 [DOI] [PubMed] [Google Scholar]
- Neuhaus F. C., Baddiley J. (2003). A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev 67, 686–723. 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V. (2006). Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol 8, 11–26. [PubMed] [Google Scholar]
- O’Brien J. A., Lahue B. J., Caro J. J., Davidson D. M. (2007). The emerging infectious challenge of Clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect Control Hosp Epidemiol 28, 1219–1227. 10.1086/522676 [DOI] [PubMed] [Google Scholar]
- O’Connor J. R., Lyras D., Farrow K. A., Adams V., Powell D. R., Hinds J., Cheung J. K., Rood J. I. (2006). Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol 61, 1335–1351. 10.1111/j.1365-2958.2006.05315.x [DOI] [PubMed] [Google Scholar]
- Palumbo E., Deghorain M., Cocconcelli P. S., Kleerebezem M., Geyer A., Hartung T., Morath S., Hols P. (2006). d-Alanyl ester depletion of teichoic acids in Lactobacillus plantarum results in a major modification of lipoteichoic acid composition and cell wall perforations at the septum mediated by the Acm2 autolysin. J Bacteriol 188, 3709–3715. 10.1128/JB.188.10.3709-3715.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Glaser P., Minutello A., Strauch M. A., Leopold K., Fischer W. (1995). Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 270, 15598–15606. [DOI] [PubMed] [Google Scholar]
- Peschel A. (2002). How do bacteria resist human antimicrobial peptides? Trends Microbiol 10, 179–186. 10.1016/S0966-842X(02)02333-8 [DOI] [PubMed] [Google Scholar]
- Poyart C., Lamy M. C., Boumaila C., Fiedler F., Trieu-Cuot P. (2001). Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J Bacteriol 183, 6324–6334. 10.1128/JB.183.21.6324-6334.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saïd-Salim B., Dunman P. M., McAleese F. M., Macapagal D., Murphy E., McNamara P. J., Arvidson S., Foster T. J., Projan S. J., Kreiswirth B. N. (2003). Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185, 610–619. 10.1128/JB.185.2.610-619.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Sebaihia M., Wren B. W., Mullany P., Fairweather N. F., Minton N., Stabler R., Thomson N. R., Roberts A. P., Cerdeño-Tárraga A. M., et al. (2006). The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38, 779–786. 10.1038/ng1830 [DOI] [PubMed] [Google Scholar]
- Setlow P. (1995). Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu Rev Microbiol 49, 29–54. 10.1146/annurev.mi.49.100195.000333 [DOI] [PubMed] [Google Scholar]
- Smith C. J., Markowitz S. M., Macrina F. L. (1981). Transferable tetracycline resistance in Clostridium difficile. Antimicrob Agents Chemother 19, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L., Haraldsen J. D., Dupuy B. (2005). RNA polymerase and alternative sigma factors. In Handbook on Clostridia, pp. 659–669. Edited by Durre P. Boca Raton, FL: Taylor and Francis. [Google Scholar]
- Thomas C. M., Smith C. A. (1987). Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol 41, 77–101. 10.1146/annurev.mi.41.100187.000453 [DOI] [PubMed] [Google Scholar]
- Vonberg R. P., Kuijper E. J., Wilcox M. H., Barbut F., Tüll P., Gastmeier P., van den Broek P. J., Colville A., Coignard B., et al. (2008). Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect 14 Suppl 52–20. 10.1111/j.1469-0691.2008.01992.x [DOI] [PubMed] [Google Scholar]
- Wartha F., Beiter K., Albiger B., Fernebro J., Zychlinsky A., Normark S., Henriques-Normark B. (2007). Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol 9, 1162–1171. 10.1111/j.1462-5822.2006.00857.x [DOI] [PubMed] [Google Scholar]
- Weidenmaier C., Peschel A., Kempf V. A., Lucindo N., Yeaman M. R., Bayer A. S. (2005). DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect Immun 73, 8033–8038. 10.1128/IAI.73.12.8033-8038.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst J., Hardegger U. (1983). Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob Agents Chemother 23, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]