Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an incurable, highly metastatic disease that is largely resistant to existing treatments. A better understanding of the genetic basis of PDAC metastasis should facilitate development of improved therapies. To that end, we developed a novel mouse xenograft model of PDAC metastasis to expedite testing of candidate genes associated with the disease. Human PDAC cell lines BxPC-3, MiaPaCa-2 and Panc-1 stably expressing luciferase were generated and introduced by intracardiac injections into immunodeficient mice to model hematogenous dissemination of cancer cells. Tumor development was monitored by bioluminescence imaging (BLI). Bioluminescent MiaPaCa-2 cells most effectively recapitulated PDAC tumor development and metastatic distribution in vivo. Tumors formed in nearly 90% of mice and in multiple tissues, including normal sites of PDAC metastasis. Effects of p14ARF, a known suppressor of PDAC, were tested to validate the model. In vitro, p14ARF acted through a CtBP2-dependent, p53-independent pathway to inhibit MiaPaCa-2 invasive phenotypes, which correlated with reduced tumor cell colonization in vivo. These findings establish a new bioluminescent mouse tumor model for rapidly assessing the biological significance of suspected PDAC metastasis genes. This system may also provide a valuable platform for testing innovative therapies.
Keywords: Bioluminescence imaging, pancreatic ductal adenocarcinoma, ARF, CtBP2, xenograft model of metastasis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an incurable malignancy with a bleak median survival of 6 months and 5-year survival rate of less than 5 percent (1, 2). It claims the lives of over 37,000 Americans annually, typically between the ages of 40 and 80, and is the fourth leading cause of cancer deaths in this country (3). PDAC survival is so poor because it typically eludes diagnosis until patients have advanced, inoperable metastatic disease that is profoundly resistant to current therapies. Conventional approaches of surgical resection, radiation and/or chemotherapy fail to prevent local recurrence and metastasis, most likely because of pre-existing micro-metastases at initial diagnosis (4–6). Improved strategies for detecting and treating early and micro-metastatic stages of the disease are urgently needed.
A better understanding of molecular events controlling PDAC metastasis could provide new targets for detection and treatment of the disease. Several genetic alterations typify PDAC, namely early activating Kras mutations and subsequent sequential inactivation of the INK4a/ARF (a locus that encodes two unrelated proteins, p16INK4a and the Alternative Reading Frame protein), p53 and/or Smad4 tumor suppressors (1, 7). Genetically engineered mice with conditional expression of mutant KrasG12D in the pancreas, combined with inactivated p53, p16INK4a or p16INK4a/ARF, effectively mimic the progressive and metastatic nature of the human disease (8, 9). Nonetheless, tumors arise without mutations in the other principal targets commonly altered in human PDAC, suggesting that other genetic alterations are required for PDAC progression (8, 9). Indeed, high resolution genomic analyses have identified hundreds of candidate genes located on frequently altered regions of the PDAC genome (10–12).
It will be important to evaluate the role of candidate PDAC genes on disease progression and therapeutic interventions, however new experimental models are needed to do so in a rapid, cost-effective manner. While genetically engineered mouse models of PDAC are tremendously valuable and informative, they are expensive and time-consuming to generate. Moreover, variable tumor onset and inability to monitor tumor development at internal organ sites diminishes their usefulness in testing novel therapies. Similar limitations are associated with orthotopic models of PDAC metastasis that rely on implantation of tumor cells or tumor sections into the pancreas of recipient mice. In the absence of tumor tracking by ultrasound imaging or exploratory laparotomies, animals are euthanized at defined endpoints, requiring large cohorts of mice for statistical reasons. There are also surgical and survival considerations for the mice that can reduce the general utility of the approach.
Bioluminescent imaging (BLI) of cancer cells ectopically expressing bioluminescent proteins is a powerful approach that allows non-invasive serial assessment of tumor development and metastasis over time in live animals (13, 14). Photon emission from the luciferase-expressing cancer cells correlates well with tumor size and allows the sensitive detection of small numbers of cells (as few as 100) within deep tissue locations, a key advantage over fluorescence imaging (13, 15, 16). Recently, several groups generated bioluminescent mouse models of spontaneous pancreatic cancer (β-cell and acinar carcinoma) and demonstrated their value for pre-clinical therapeutic testing (17, 18). These transgenic models will also be useful for future genetic experimentation via crosses with other genetically modified mice. However, a bioluminescent model of PDAC, the predominant type of pancreatic cancer (>90%), is still lacking and time and cost issues associated with genetically engineered mice remain significant impediments to the rapid evaluation of genes with a suspected role in PDAC pathogenesis.
To expedite and simplify the biological testing of putative PDAC genes, we developed a new mouse xenograft model of PDAC metastasis that utilizes BLI to longitudinally track tumor formation and dissemination. We show this model is a fast and feasible method of studying genetic events that control pancreatic cancer metastasis, and directly demonstrate a p53-independent role for the p14ARF tumor suppressor in reducing PDAC metastatic colonization and tumor formation.
Materials and Methods
Cell culture
Panc-1 cells were maintained in standard Dulbecco’s Modified Eagles Medium (DMEM) containing 10% fetal bovine serum, 4 mM glutamine, and 100 ug/ml penicillin/streptomycin. MiaPaCa-2 cells were grown in the same media containing 2.5% equine serum. BxPC-3 cells were maintained in RPMI-1640 medium, 10% fetal bovine serum, 2 mM glutamine, 100 ug/ml penicillin/streptomycin, 2.5 g/liter D-glucose, 10 mM HEPES and 1 mM sodium pyruvate. Cells were purchased from and authenticated by ATCC, and used within 6 months after resuscitation.
DNA constructs and retrovirus-mediated protein expression
Retroviral constructs expressing firefly (Photinus pyralis) luciferase (pQClucIN) and untagged human p14ARF protein (bicistronic pMSCV-ARF-IRES-GFP) have been described previously (13, 19), as have expression constructs and siRNAs for CtBP2 (20, 21). To generate amphotropic retroviruses expressing luciferase, pQClucIN was transfected by calcium phosphate precipitation (22) with the VSV-G envelope plasmid (BD Biosciences) into GP-293 packaging cells. Retroviral supernatants were collected every 12 hours between 24–60 hours post-transfection, stored on ice, supplemented with 8 ug/ml polybrene and passed through sterile 0.45 um filters. Sequential infections (three rounds, each ≥ 4 hours in length) of low density pancreatic cancer cell lines (1.5 × 105 cells per 100 mm dish) were performed using 3–4 ml of virus per infection. Two days later, stable cell lines expressing luciferase were selected by addition of G418 (0.8 mg/ml for MiaPaCa-2, 0.5 mg/ml for Panc-1 and BxPC-3) to growth media for 2 weeks. Similar methods were used for the production and infection of GFP or p14ARF plus GFP from pMSCV-IRES-GFP and pMSCV-ARF-IRES-GFP plasmids, respectively. Sterile cell sorting was performed using a FACSDiva instrument (BD Biosciences) to enrich for successfully infected, GFP-positive cells in each population.
Cellular luciferase activity assay
A two-fold dilution series of cells (from 10,000 to 20 cells per well in 0.1 ml) was plated in duplicate in black 96-well plates, mixed with an equal volume of D-luciferin substrate (0.3 mg/ml), incubated for 5 minutes and imaged on a Xenogen IVIS100 imaging system (Caliper Life Sciences). Controls included wells with no cells or no D-luciferin.
Intracardiac injections
Luciferase-expressing cells were prepared at final concentrations of 1 × 106 viable cells per ml in PBS. All procedures involving animals were performed according to The University of Iowa Animal Care and Use Committee policies. Intracardiac injections and subsequent analyses were performed on 5–8 week old female scid mice (NCI, 01S11 SCID/NCr) that were anesthetized in a chamber with 3% isoflurane (Abbott Laboratories) (13). After removal from the chamber, mice were put in a ventral position and 100 ul (1 × 105 cells) of each cell suspension was slowly delivered into the left ventricle of the heart using a 30-gauge needle. To examine cell distribution following injection, animals were injected intraperitoneally with 200 ul of 15 mg/ml D-luciferin substrate, incubated for 5 minutes and imaged on the IVIS100 system.
BLI analyses of tumor growth rates and distribution
Metastatic colonization and tumor formation was monitored by serial BLI on the IVIS100 at weekly intervals beginning two weeks post-injections (13). Mice were anesthetized by isoflurane inhalation maintained at 2.5% through a nose cone and images obtained in dorsal and ventral presentations, 5 and 10 minutes after D-luciferin injection, respectively. Subsequently, a rectangular region of interest was placed around the dorsal and ventral images for each mouse, and total photon flux quantified using Living Image software v2.50 (Caliper Life Sciences) with the units of photons per second. Dorsal and ventral values were summed and plotted weekly per animal to obtain whole body tumor growth rates. Animals were observed twice weekly for adverse events associated with tumor growth (e.g., >15% body weight loss, immobility, loss of grooming, etc), at which time mice were euthanized. Statistical analyses comparing rates of tumor formation between the groups were determined using two-way ANOVA followed by Bonferroni post tests.
Post-mortem ex-vivo BLI was performed to identify precise anatomic locations of the tumors. Mice were injected with D-luciferin intraperitoneally followed by euthanasia after 5 minute incubation. Individual organs were excised and examined by IVIS100 imaging (bioluminescence signals are detectable up to 30 minutes after euthanasia). A significant number of tumors detected by BLI were collected for histology. Samples were fixed in 4% paraformaldehyde prior to hematoxylin and eosin staining at the University of Iowa Comparative Pathology Laboratory. Other tumors were processed under sterile conditions to generate tumor-derived cell lines for protein expression analyses.
Western blot analyses
Cells were lysed and total cellular protein (100 ug per sample) immunoblotted as described (23) with the exception that 1% Triton X-100, 0.1% SDS and 0.5% deoxycholate detergents were used in the lysis buffer. Proteins were detected by enhanced chemiluminescence (GE Biosciences) with antibodies to: GFP (Abcam, Ab290 rabbit polyclonal, 1:4,000), p14ARF (Novus, rabbit polyclonal, 1 ug/ml), CtBP2 (BD Transduction Laboratories), V5 tag (Invitrogen), ULF (kindly provided by Dr. Wei Gu, Columbia University), GAPDH (Abcam, Ab8245 mouse monoclonal, 1:20,000), and gamma-tubulin (Sigma, T6557 mouse monoclonal, 1:10,000).
Cell proliferation assays
Growth curves of MiaPaCa-2-luc cells expressing vector or human p14ARF were generated by plating cells at 1 × 104 cells per well (12-well dishes) in triplicate and counting cells every 2 days on a hemacytometer. The statistical significance of data was determined by a two-tailed, unpaired equal variance Student’s t-test.
In vitro migration and invasion assays
For wound assays measuring cell migration, equal numbers of cells were plated at 50% confluency in 6-well plates, and 24 hours after plating the cells were transfected with either siRNA or expression plasmids. Twenty-four hours after transfection, the confluent monolayer of cells was scraped to introduce a wound and cells were incubated for another 20 hours. Captured images were used to compare and quantify the percent wound closure by measuring the migration distance of cells found in the cleared area from the point the cells were scraped to the migration front in 5 independent × 100 fields per experiment.
For trans-well migration assays, cells were starved for 48 hours in serum-free DMEM containing 1% bovine serum albumin, harvested with trypsin, washed three times with PBS after trypsin inactivation, and 2 × 104 cells plated in triplicate or quadruplicate into fibronectin-coated 24-well trans-well inserts (Fisher Scientific, 8 um pore size) within 24-well plates. DMEM media in the bottom of the wells contained either 0% FBS (control) or 3% FBS as chemoattractant. Cells were incubated at 37oC, 5% CO2 for 24 hours, inserts removed from the original plates and the top of the membranes wiped with PBS-saturated Q-tips to remove non-migrated cells. Inserts were placed in new 24-well plates containing DMEM, followed by D-luciferin (0.15 mg/ml final concentration) addition. Plates were imaged after 5 minutes using the IVIS100 system to quantify migrated cells on the underside of the trans-well membrane. The percentage of migrated cells was calculated by dividing the luminescence levels of each cell line under the different conditions by the total luminescence levels (determined from cells plated directly into well without trans-well inserts).
Trans-endothelial migration assays, which measure invasion of cells through a monolayer of primary HMVEC-Ls endothelial cells, were performed using serum-starved cells (see above) and BLI, as described (24). All cell migration assays (wound, trans-well and trans-endothelial) were performed with replicate samples (duplicate or triplicate) in at least three separate experiments and subjected to statistical analyses (ANOVA or Student’s t-test) to assess significance of the data.
Results
Generation of Bioluminescent PDAC Cell Lines
To facilitate rapid biological testing of putative PDAC genes, we sought to develop a novel mouse xenograft model of PDAC metastasis that utilized non-invasive, bioluminescence imaging (BLI) of tumor growth. The goals and general timeline of the study are outlined in Figure 1A. Three human pancreatic cancer-derived cell lines (Panc-1, MiaPaCa-2 and BxPC-3) were first transduced with retroviruses expressing the firefly luciferase gene and selected with antibiotic to generate stable luciferase-expressing populations (Figure 1A). The bioluminescence intensity of the new cell lines was measured by an in vitro luciferase activity assay in which cells were serially diluted, exposed to D-luciferin substrate, and the amount of light or photons emitted captured by BLI (Figures 1B and 1C). Moderate bioluminescence intensity was observed in all three cell lines (~20 to 30 photons/s/cell), which was highly stable and remained unchanged even after 3 months of culturing the cells without antibiotic selection (data not shown). Effective maintenance of the luciferase activity indicated these cell lines were suitable for long-term tumor studies in vivo.
Figure 1.
Establishment of bioluminescent pancreatic cancer cell lines. (A) Schematic of the time-frame for initiating the study, including months (Mos) needed for generating and testing stable luciferase-expressing cell lines. (B) In vitro luciferase activity assay of serially diluted Panc-1 (Panc), MiaPaCa-2 (MP2) and BxPC-3 (BxPC) cells infected with retroviruses encoding luciferase immediately after G418 selection. Heat map of the photon flux (photons per second) indicates levels of bioluminescent intensity within the cells. (C) Calibration curves of the data obtained in (B).
Tumor Formation Efficiency and Rates in Vivo
Each bioluminescent PDAC cell line was introduced into the arterial circulation of scid mice by injections into the left ventricle. This approach initially bypasses the lung capillary bed, unlike tail vein injections, and allows hematogenous dissemination of cancer cells via the arterial circulation (25). As seen previously, minimal mortality is associated with the procedure with only two out of 49 mice (~4%) dying from the injection process (Table 1).
Table 1.
Characteristics of the PDAC cell lines in the metastasis model
| Panc-1 | MiaPaCa-2 | BxPC-3 | ||||
|---|---|---|---|---|---|---|
| Classifications | Value | Percent | Value | Percent | Value | Percent |
| Mice injected | 17 | 17 | 15 (13)a | |||
| Mice that formed tumorsb | 12 | 71% | 15 | 88% | 12 | 92% |
| Mice autopsied | 12 | 71% | 13 | 76% | 9 | 69% |
| Mice autopsied with tumors | 9 | 75% | 11 | 85% | 9 | 100% |
| TO tumorsc | 2 | 22% | 1 | 8% | 6 | 67% |
| T+ tumorsc | 1 | 11% | 2 | 18% | 0 | 0% |
| ET tumorsc | 6 | 67% | 8 | 73% | 3 | 33% |
Two mice died shortly after injection, likely due to embolism, therefore they are excluded from subsequent calculations
As determined by BLI
Numbers and percentages for these tumors are based on analyses of the autopsied mice with tumors
TO = Thoracic only tumors
T+ = Thoracic + systemic tumors
ET = Extrathoracic (systemic) tumors
General performance in the model varied for each cell line, as assessed by BLI and ex vivo analyses (Table 1). The highest efficiency of tumor formation was observed for BxPC-3 (92%) and MiaPaCa-2 (88%) cells whereas Panc-1 cells produced tumors in only 71% of mice. Most animals that formed tumors underwent necropsy to verify if tumor location was systemic (extrathoracic, ET), restricted to the thoracic cavity (thoracic only, TO), or thoracic plus systemic tumors (T+). TO tumors included growths in the lungs, on the thoracic wall or on the outside of the heart and could represent aggressively colonizing cells or failed injections into the right ventricle, myo- or pericardium, or thoracic cavity. We found that mice injected with Panc-1 and MiaPaCa-2 cells mainly presented with systemic tumors (ET and T+) outside of the thoracic cavity (78% and 91%, respectively). By comparison, two-thirds of mice (67%) injected with BxPC-3 cells developed thoracic tumors.
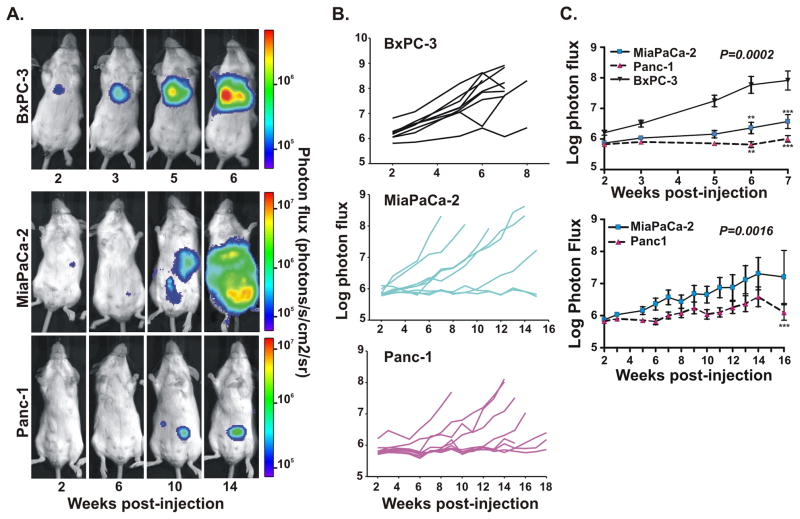
The high percentage of systemic tumors for Panc-1 and MiaPaCa-2 cell types suggested that the predominance of TO tumors for BxPC-3 cells was not likely caused by failed injections but rather due to aggressive colonization. Consistent with that notion, weekly BLI imaging of tumor growth post-injection showed a significantly faster rate of tumor formation for BxPC-3 cells than the other two cell lines (Figure 2). Serial bioluminescence images (Figure 2A) and quantification of whole body tumor growth (Figure 2B) in individual BxPC-3-injected mice revealed discrete tumor foci by 3-4 weeks and lethal tumor growth by 5–7 weeks. In contrast, MiaPaCa-2 and Panc-1 groups of mice displayed delayed rates of tumor development overall (most animals surviving 14–18 weeks after injection), and Panc-1 tumors were often small in size (Figures 2A and 2B). Analyses of the average tumor growth rates for each group demonstrated statistically significant differences between all three groups, with tumor onset fastest in BxPC-3 mice and slowest in Panc-1 animals (Figure 2C).
Figure 2.
BLI of serially imaged tumors reflect variable rates and efficiency of tumor formation for the different PDAC cell lines. (A) Bioluminescence images at the indicated times (weeks) post-injection with luciferase-expressing BxPC-3, MiaPaCa-2 or Panc-1 cells. Each set of images was taken from the same mouse. Note the different time scales and corresponding heat maps for BxPC-3 mice versus MiaPaCa-2 and Panc-2 mice. (B) Representative experiment showing individual tumor growth rates (n=9 mice per group) injected with the specified cell type, as reflected by total photon flux (photons per second) per animal over the indicated time periods. (C) Average tumor growth rates (mean ± standard error) quantified from (B). P values obtained from two-way ANOVA analyses show statistically significant differences occurred between BxPC-3 with Panc-1 or MiaPaCa-2 (top panel, p=0.0002) or between Panc-1 and MiaPaCa-2 (bottom panel, p=0.0016) over the 7 week or 16 week time courses, respectively. ** p<0.01, *** p<0.001 values are indicated for individual time points, as calculated by two-way ANOVA followed by Bonferroni post tests.
Tumor Distribution Widest and Most Clinically-Relevant for Mice Injected with Miapaca-2 and Panc-1 Cells
BLI does not pinpoint the exact tumor site, therefore ex vivo imaging of the PDAC mice was performed to assess precise anatomical location and distribution frequency of the tumors. Notably, the average number of tumor sites/organs colonized per animal, as determined from BLI and necropsied animals, was similar for all three cell types (1.36 for Panc-1, 1.33 for BxPC-3, and 1.31 for MiaPaCa-2) with a range of 1-3 tumor sites per animal. The total number of tumors formed per animal ranged from 1 to 5, with average number of tumors per mouse equaling 2.1 for Panc-1, 2.3 for MiaPaCa-2, and 1.6 for BxPC-3. Consistent with BLI data in Table 1 and as shown in Figures 3A and 3B, the majority of tumors (56%) isolated from BxPC-3 injected mice resided in the thoracic region. These included tumors around the heart or within the thoracic wall, as well as two neoplasms in the lungs. The small percentage of BxPC-3 mice with systemic tumors had growths at multiple organ sites per animal, including the lower jaw region of the head and neck, liver, and celiac plexus. Importantly, the combination of BLI and histological analyses verified the host organ site and human epithelial origin of the tumors that developed from each of the cell lines (Figure 3C).
Figure 3.
Bioluminescent MiaPaCa-2 and Panc-1 cells displayed greatest tumor dissemination and clinically-relevant distribution in the intracardiac metastasis model. (A) Graphical and (B) diagrammatic representations of the different organ sites colonized by luciferase-expressing BxPC-3, MiaPaCa-2 or Panc-1 cells, as determined by ex vivo BLI analyses of necropsied mice. Total number of tumors analyzed is indicated in (A). (C) Ex vivo histological analyses of tissues containing tumors (T) derived from each cell line by hematoxylin and eosin staining. In the adrenal gland, the MiaPaCa-2 tumor effaced the entire tissue and showed significant necrosis (N). Corresponding whole body bioluminescent images for the excised tumors are shown with yellow boxes denoting the signal location of each tumor processed for histology.
Ex vivo analyses confirmed a wider range of organs was colonized by MiaPaCa-2 and Panc-1 cells compared to BxPC-3 cells, with drastically fewer tumors (only 12% to 25%, respectively) located in the thoracic area (Figures 3A and 3B). Systemic Panc-1 tumors mainly grew in the adrenal glands, liver and bone (femur). Likewise, the adrenal glands were a predominant organ site for MiaPaCa-2 tumors, as well as the lower jaw area of the head and neck (lymph nodes and/or bone), fat pads of the breast, and the celiac plexus. Single tumors in the liver and pancreas were also seen among the tumors isolated from MiaPaCa-2 mice. These results suggest some preferences in tumor site colonization between the cell types (e.g., lack of Panc-1 in the celiac plexus or lack of BxPC-3 in the adrenal glands). More significantly, higher percentages of MiaPaCa-2 (35%) and Panc-1 (42.5%) tumors formed in clinically-relevant organ sites compared to BxPC-3 tumors (22%). Those organs included the adrenal glands, liver, and celiac plexus; all normal sites of PDAC metastases in patients with the disease (26).
The ARF Tumor Suppressor Inhibits PDAC Migration and Invasion in Vitro
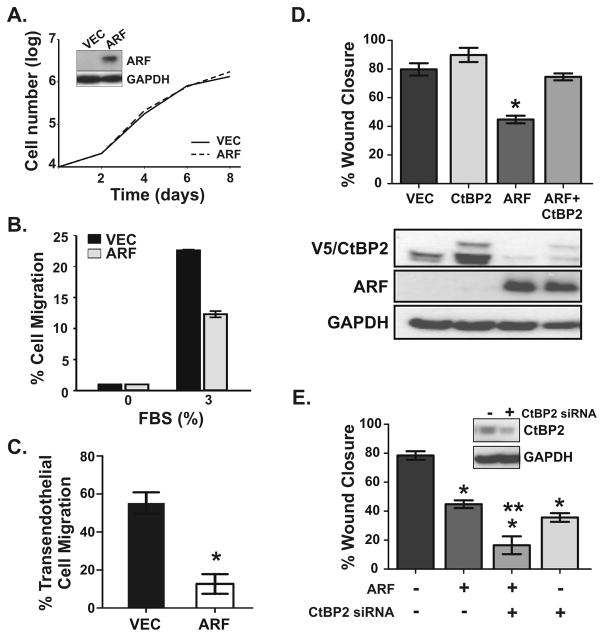
Through a p53-Independent Mechanism We used the ARF tumor suppressor (p14ARF in humans), an established inhibitor of PDAC, to validate the utility of the model for testing putative PDAC metastasis genes. ARF is inactivated in approximately 40% of human PDAC (1, 27) and its genetic deletion accelerates tumor formation and metastasis in mouse models of the disease (9). Since ARF is not expressed in the cell lines used in this study, we reintroduced human ARF by retroviral infection into our bioluminescent MiaPaCa-2 cells. These cells were chosen because they formed systemic tumors with high efficiency and optimal rate of onset. Cells were well-transduced (60–75% efficiency), but sterile cell sorting for GFP-positive cells was performed to obtain higher percentages of successfully infected cells in both vector control (90%) and ARF (88%) populations. Western blotting showed selective expression of ARF in the ARF-infected cells (inset, Figure 4A), and luciferase activity assays confirmed identical bioluminescence was maintained in both cell populations (data not shown).
Figure 4.
The ARF tumor suppressor inhibits MiaPaCa-2 cell motility in vitro through a p53-independent, CtBP2-dependent mechanism. (A) Representative growth curves of cells stably expressing vector (VEC, solid line) or ARF (dashed line). Western blots (inset) confirm ectopic ARF expression in these ARF-null cells. GAPDH is the loading control. (B) Relative migration of vector (V) and ARF (A) expressing cells induced by 3% FBS, normalized to samples plated in 0% FBS, in a trans-well chemotaxis assay. Data shown are the average of duplicate experiments (each sample in quadruplicate). Findings were reproduced in four separate analyses. (C) Representative transendothelial migration analysis of vector (VEC) and ARF expressing cells. The mean and SD were calculated from triplicate samples (*, p=0.00016). (D) VEC and ARF expressing cells were transfected with control pcDNA3 vector or pcDNA3-V5/CtBP2 and subjected to wound healing migration assays (*, p < 0.0001 for ARF alone compared to all other samples). Western blots of cell lysates from the same cells show levels of ARF, CtBP2 (endogenous plus slower migrating V5-tagged form) and GAPDH. (E) VEC and ARF expressing cells were transfected with control or CtBP2 siRNA (20 nmol/L each siRNA) and wound healing migration assays conducted. Statistically significant differences (p < 0.0001) were observed between each condition compared to control cells (*), as well as between ARF expressing cells transfected with control versus CtBP2 siRNA (**). Western blots (inset) confirm CtBP2 knockdown. For panels D and E, data were averaged from three independent experiments with each experiment conducted in triplicate.
Effects of ARF on the in vitro tumorigenic phenotype of MiaPaCa-2 cells were first analyzed. Growth curves showed no inhibitory effect of ARF on the survival or proliferative rate of the cells (Figure 4A). By comparison, three separate analyses of cell migration (4B&D) and invasion (Fig 4C) showed that ARF suppressed the migratory and invasive capability of the cells two- to four-fold. ARF’s selective ability to impair the migration and invasion of the MiaPaCa-2 cells, but not their proliferation, has similarly been observed in hepatocellular carcinoma cell lines (28). Both cancer cell types express a mutated, non-functional version of p53, the primary mediator of ARF-induced cell cycle arrest or death (29). These data provide new, direct evidence that ARF inhibits the migratory behavior and invasiveness of pancreatic cancer cells and that this activity is independent of p53.
Previous studies demonstrated that ARF prevents cancer cell migration by binding to the transcriptional corepressor C-terminal binding protein 2 (CtBP2) and promoting its degradation (20, 21, 28). In accordance with those findings, Western blot analyses showed decreased expression of endogenous CtBP2 in MiaPaCa-2 cells expressing ARF compared to empty vector (Fig 4D, bottom panel, lanes 3 versus 1). We then tested the functional role of CtBP2 by altering its expression levels, and as predicted, overexpression of V5-tagged CtBP2 in MiaPaCa-2 cells completely interfered with ARF’s anti-migratory activity (Fig 4D). Conversely, siRNA-mediated knockdown of CtBP2 inhibited cell motility to a similar degree as overexpression of ARF, and its loss accentuated the anti-migratory activity of ARF in a synergistic manner (Fig 4E). These results demonstrate that a primary mechanism by which ARF inhibits pancreatic cancer cell motility is by blocking CtBP2 expression and function.
ARF Inhibits Pancreatic Metastatic Tumor Colonization in Vivo
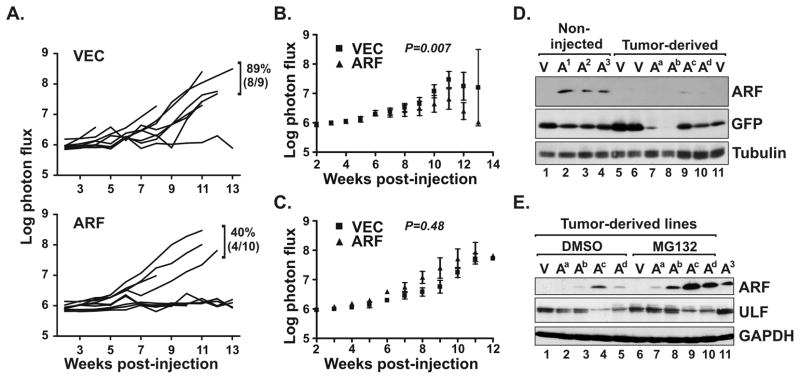
We next tested if ARF would suppress tumor colonization and development in vivo in the metastasis model. Quantification of whole body tumor growth showed that only 40% of mice receiving ARF expressing cells developed tumors whereas 89% of vector control animals formed tumors (Figure 5A). This marked disparity in tumor formation efficiency resulted in statistically significant differences in the average tumor growth rates between the two groups (Figure 5B). Notably, exclusive analysis of the tumor-forming animals from both groups revealed overlapping rates of tumor growth (Figure 5C). Therefore, we tested if ARF expression was maintained in the tumors that developed by measuring ARF levels in tumor-derived cell lines. As a comparative control, Western blot analyses of in vitro cultured, non-injected MiaPaCa-2 cells showed sustained, strong expression of ARF during maintenance in culture (Figure 5D, lanes 2-4). In the tumors, however, exogenous ARF expression was either lost entirely or dramatically down-regulated (lanes 7–10).
FIGURE 5.
ARF expression reduces the efficiency of PDAC tumor cell colonization in vivo. (A) Tumor growth rates, as measured by total photon flux (photons per second) over a 13-week period, following intracardiac injections with bioluminescent MiaPaCa-2 cells expressing vector control (VEC) or ARF. Frequency of tumor formation and number of animals tested are indicated and were significantly different (p=0.013 from normal approximation of proportions). (B) Average tumor growth rates for all mice, including those that did not form tumors, injected with MiaPaCa-2 cells expressing vector (squares) or ARF (triangles). Two-way ANOVA analysis shows significant differences between the two groups over the 13-week time course (p=0.007). (C) Selective analysis of average tumor growth rates from animals in (B) that formed tumors showed no difference (p=0.43) between vector and ARF groups. (D) Immunoblot analysis of ARF and GFP levels in cultured MiaPaCa-2 cells expressing GFP (V, vector) or ARF plus GFP (A), before (non-injected) or after (tumor-derived) introduction into mice. Note dramatic loss of ARF expression in all four tumor-derived lines (designated Aa-d). Non-injected cells were maintained in culture for 0 (A1), 3 (A2) or 5 (A3) weeks. (E) Western blot analyses of ARF and ULF expression levels (with GAPDH loading control) in the tumor-derived vector (V) and ARF (Aa-d) cells following treatment for 4 hours with vehicle control (DMSO) or MG132 (50 uM). Non-injected, untreated ARF expressing cells (A3) are included for comparison. .
ARF is known to be negatively regulated by N-terminal ubiquitination and proteasomal degradation (30). ULF, a recently identified ubiquitin ligase for ARF (also called TRIP12) (31, 32), destabilizes ARF and is predicted to be upregulated in human pancreatic cancers (32). We speculated that ULF expression may be increased in tumor-derived versus non-injected cells, but this was not observed in Western analyses (Fig 5E, lanes 1-5 versus lane 11). Interestingly, a clear inverse correlation between ARF and ULF levels was seen within the tumor-derived cells, with lower ULF levels corresponding to higher ARF expression and vice versa (Fig 5E). Consistent with that finding, inhibition of the proteasome with MG132 led to a marked increase in ARF stability and expression in the tumor-derived lines (lanes 7–10 versus 1–4). These data indicate that ARF expression is greatly restricted in pancreatic tumor cells through a proteasome-dependent mechanism, possibly through ULF and/or other as yet unidentified ubiquitin ligases for ARF.
Together, the findings herein verify the utility of our novel model for assaying candidate PDAC metastasis genes and demonstrate a direct, p53-independent role for ARF in suppressing PDAC metastatic colonization and tumor growth in vivo.
Discussion
The goal of this study was to develop a bioluminescent mouse model of PDAC metastasis that would facilitate rapid and relatively inexpensive in vivo analyses of putative PDAC metastasis genes. There is significant need for such a model given that PDAC is a highly metastatic, deadly malignancy for which there are currently no cures or effective treatments (1, 3). Although a number of elegant, genetically engineered mouse models of PDAC exist that effectively mimic the human disease (33), they are expensive and time consuming to generate. They are also unsuitable for efficiently evaluating large numbers of genes with suspected roles in PDAC tumor formation and metastasis. Here, we describe the development and characterization of a novel mouse xenograft model of PDAC that employs BLI to longitudinally track metastatic colonization and tumor growth in vivo.
Three frequently studied human PDAC cell lines (Panc-1, MiaPaCa-2 and BxPC-3) expressing luciferase were generated and tested in this model. This allowed for non-invasive, serial imaging of tumor growth in vivo following intracardiac injection and hematogenous distribution of the cells in scid mice. BxPC-3 cells displayed the fastest rates of tumor onset and lethality, and the highest efficiency of tumor formation. Their rapid rate of tumor growth would be amenable for testing genetic alterations predicted to slow tumor metastasis, but not as useful for those that would accelerate the process. However, the use of BxPC-3 cells in the model was complicated by their tendency to form non-systemic tumors restricted to the thoracic region. Our data suggested this was due to aggressive tumor growth rather than failed injections into the heart, nonetheless it diminishes their utility.
Of the three cell types, MiaPaCa-2 cells behaved optimally in the metastasis model. They displayed efficient metastatic colonization (~90% of mice injected formed tumors), a moderate rate of tumor onset experimentally suitable for in vivo studies, and a wide distribution of systemic tumors. Tumors developed at organ sites commonly colonized by human PDAC metastases, including the liver, adrenal glands and celiac plexus. MiaPaCa-2 tumors also formed in the mammary fat pads and craniofacial region, including lower jaw bone and lymph nodes, and one was even found invading the pancreas and spleen. While these overall patterns of tumor distribution do not exactly mimic the clinical spectrum of human PDAC metastasis, more than one-third of tumors developed at metastatic sites normally seen in PDAC patients.
A key element of our model is the use of intracardiac injections into the left ventricle to introduce the bioluminescent pancreatic cancer cells directly into the arterial circulation. This approach circumvents early steps of metastasis, namely invasion and intravasation, but it does evaluate the extravasation, metastatic colonization and growth of tumor cells at clinically relevant sites of metastasis. Indeed, it has been used to successfully model metastasis of retinoblastoma, breast and prostate cancers, among others (13, 34–37). Importantly, our experiments are the first to show that the ARF tumor suppressor effectively inhibits the process of pancreatic tumor cell colonization in vivo.
The fact that ARF had no suppressive effect on in vitro proliferation of MiaPaCa-2 cells is interesting and indicates it is interfering with other aspects of tumor colonization. In that regard, in vitro assays showed that ARF significantly impairs the migration and invasion of MiaPaCa-2 cells. The decreased invasiveness of ARF-expressing cells through endothelial cell monolayers in vitro would be expected to greatly reduce the ability of those cells to extravasate in vivo, thereby reducing their ability to form tumors. Such results are consistent with findings that a high percentage of human pancreatic ductal adenocarcinomas, which are highly motile and metastatic, have inactivated ARF (1, 27).
The anti-migratory and anti-tumor effects of ARF observed in this study were independent of the p53 tumor suppressor, a primary effector of ARF (29), since MiaPaCa-2 cells express a mutated form of the protein. This supports in vitro evidence in other cancer cell types that ARF functions independent of p53 to inhibit tumor cell migration. Specifically, ARF suppresses the in vitro migration and invasion of lung, colon and hepatocellular carcinoma (HCC) cell lines that lack functional p53 (21, 28). In the HCC study, ARF selectively impaired tumor cell migration and invasion without affecting their proliferation (28), as seen here in MiaPaCa-2 cells. Notably, these p53-independent effects of ARF required its ability to bind and inhibit CtBP2. Our studies show that ARF employs the same mechanism here to limit pancreatic cancer cell migration in vitro, suggesting that the interplay between CtBP2 and ARF may play a critical and significant role in controlling PDAC metastasis in vivo.
The robust selective pressure against ARF expression in vivo, which was not seen in adherent cultures in vitro, was remarkable. Specifically, ARF expression was dramatically reduced in all tumors derived from the ARF group of mice. In two of the four tumors analyzed, a nearly complete lack of both ARF and GFP was observed suggesting that non-infected (~12%) or poorly infected cells in the population had a discernable tumorigenic advantage. By comparison, two other tumors from the ARF group of mice retained GFP yet lost ARF expression. Given that ARF and GFP were ectopically expressed from a bicistronic retroviral construct, this implied a specific mechanism for ARF destabilization is activated in vivo. Indeed, we found ARF is down-regulated by the proteasome in these cells and there is an inverse correlation between levels of ARF and ULF, its recently discovered ubiquitin ligase (32). Since ULF levels appear unchanged in the tumor-derived versus non-injected adherent cells, ULF activity (not levels) and/or other ubiquitin ligases may be turned on by paracrine factors in vivo, thereby facilitating ARF destruction. Simple loss of cell adherence may also contribute to ARF down-regulation since soft agar plating likewise led to loss of exogenous ARF expression (supplemental Fig S1). Interestingly, no correlation between ULF and ARF expression was seen in soft agar colonies, unlike in the tumor-derived cells, suggesting different factors may be involved.
In summary, although much progress has been made with genetic modeling in defining key alterations that promote the pancreatic ductal adenocarcinoma, this cancer remains one of the deadliest and most difficult to treat in humans. This necessitates a more comprehensive understanding of molecular mechanisms required for PDAC development and metastasis. To address that need, we created and validated a novel mouse model of PDAC metastasis that will facilitate economically feasible and rapid in vivo evaluation of candidate genes that control PDAC metastasis. This system should also provide a powerful platform for pre-clinical testing of unique chemotherapies that can effectively treat PDAC.
Supplementary Material
Acknowledgments
The authors are grateful for ULF antibodies from Wei Gu (Columbia University), assistance from the University of Iowa Flow Cytometry facility and helpful discussion of the work from Frederick Quelle and members of the M.H. and D.Q. laboratories.
Grant Support: NIH grants R01-CA090367 (D.E.Q.), R21-CA127031 (D.E.Q., M.D.H.) and R03-CA143763 (S.R.G.), ACS Research Scholar Grant (S.R.G.) and the University of Iowa Biological Sciences Funding Program (D.E.Q., M.D.H.).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Devel. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M. Pancreatic Cancer. New Eng J Med. 362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao YP, Xu JQ, Thun MJ. Cancer Statistics, 2009. Ca-A Cancer J for Clinicians. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Allison DC, Piantadosi S, Hruban RH, Dooley WC, Fishman EK, Yeo CJ, et al. DNA content and other factors associated with ten-year survival after resection of pancreatic carcinoma. J Surg Oncol. 1998;67:151–9. doi: 10.1002/(sici)1096-9098(199803)67:3<151::aid-jso2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Burris HA. Recent updates on the role of chemotherapy in pancreatic cancer. Sem Oncol. 2005;32:S1–S3. doi: 10.1053/j.seminoncol.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Yeo CJ. Pancreatic cancer - In brief. Curr Prob Cancer. 2002;26:170. [Google Scholar]
- 7.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 8.Hingorani SR, Wang LF, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53(R172H) and KraS(G12D) cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Bardeesy N, Aguirre AJ, Chu GC, Cheng Kh, Lopez LV, Hezel AF, et al. From the Cover: Both p16Ink4a and the p19Arf-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci U S A. 2004;101:9067–72. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidenblad M, Lindgren D, Veltman JA, Jonson T, Mahlamaki EH, Gorunova L, et al. Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic implications. Oncogene. 2005;24:1794–801. doi: 10.1038/sj.onc.1208383. [DOI] [PubMed] [Google Scholar]
- 12.Heidenblad M, Schoenmakers EFPM, Jonson T, Gorunova L, Veltman JA, van Kessel AG, et al. Genome-Wide Array-Based Comparative Genomic Hybridization Reveals Multiple Amplification Targets and Novel Homozygous Deletions in Pancreatic Carcinoma Cell Lines. Cancer Res. 2004;64:3052–9. doi: 10.1158/0008-5472.can-03-3159. [DOI] [PubMed] [Google Scholar]
- 13.Drake JM, Gabriel CL, Henry MD. Assessing tumor growth and distribution in a model of prostate cancer metastasis using bioluminescence imaging. Clin Exper Metastasis. 2005;22:674–84. doi: 10.1007/s10585-006-9011-4. [DOI] [PubMed] [Google Scholar]
- 14.Contag CH, Spilman SD, Contag PR, Oshiro M, Eames B, Dennery P, et al. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–31. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 15.Lyons SK, Lim E, Clermont AO, Dusich J, Zhu LY, Campbell KD, et al. Noninvasive bioluminescence imaging of normal and spontaneously transformed prostate tissue in mice. Cancer Res. 2006;66:4701–7. doi: 10.1158/0008-5472.CAN-05-3598. [DOI] [PubMed] [Google Scholar]
- 16.Edinger M, Sweeney TJ, Tucker AA, Olomu AB, Negrin RS, Contag CH. Noninvasive assessment of tumor cell proliferation in animal models. Neoplasia. 1999;1:303–10. doi: 10.1038/sj.neo.7900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang N, Lyons S, Lim E, Lassota P. A Spontaneous Acinar Cell Carcinoma Model for Monitoring Progression of Pancreatic Lesions and Response to Treatment Through Noninvasive Bioluminescence Imaging. Clin Cancer Res. 2009;15:4915–24. doi: 10.1158/1078-0432.CCR-08-2256. [DOI] [PubMed] [Google Scholar]
- 18.Zumsteg A, Strittmatter K, Klewe-Nebenius D, Antoniadis H, Christofori G. A bioluminescent mouse model of pancreatic beta-cell carcinogenesis. Carcinogenesis. 2010;31:1465–74. doi: 10.1093/carcin/bgq109. [DOI] [PubMed] [Google Scholar]
- 19.di Tommaso A, Hagen J, Tompkins V, Muniz V, Dudakovic A, Kitzis A, et al. Residues in the alternative reading frame tumor suppressor that influence its stability and p53-independent activities. Exper Cell Res. 2009;315:1326–35. doi: 10.1016/j.yexcr.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paliwal S, Pande S, Kovi RC, Sharpless NE, Bardeesy N, Grossman SR. Targeting of C-Terminal Binding Protein (CtBP) by ARF Results in p53-Independent Apoptosis. Mol Cell Biol. 2006;26:2360–72. doi: 10.1128/MCB.26.6.2360-2372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paliwal S, Kovi RC, Nath B, Chen YW, Lewis BC, Grossman SR. The Alternative Reading Frame Tumor Suppressor Antagonizes Hypoxia-Induced Cancer Cell Migration via Interaction with the COOH-Terminal Binding Protein Corepressor. Cancer Res. 2007;67:9322–9. doi: 10.1158/0008-5472.CAN-07-1743. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–52. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tompkins VS, Hagen J, Frazier AA, Lushnikova T, Fitzgerald MP, di Tommaso A, et al. A Novel Nuclear Interactor of ARF and MDM2 (NIAM) That Maintains Chromosomal Stability. J Biol Chem. 2007;282:1322–33. doi: 10.1074/jbc.M609612200. [DOI] [PubMed] [Google Scholar]
- 24.Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 Enhances Transendothelial Migration and Represses the Epithelial Phenotype of Prostate Cancer Cells. Mol Biol Cell. 2009;20:2207–17. doi: 10.1091/mbc.E08-10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arguello F, Baggs RB, Frantz CN. A murine model of experimental metastasis to bone and bone-marrow. Cancer Res. 1988;48:6876–81. [PubMed] [Google Scholar]
- 26.Goulart BH, Clark JW, Lauwers GY, Ryan DP, Grenon N, Muzikansky A, et al. Long term survivors with metastatic pancreatic adenocarcinoma treated with gemcitabine: a retrospective analysis. J Hematol Oncol. 2009:2. doi: 10.1186/1756-8722-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hustinx SR, Leoni LM, Yeo CJ, Brown PN, Goggins M, Kern SE, et al. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Modern Path. 2005;18:959–63. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 28.Chen YW, Paliwal S, Draheim K, Grossman SR, Lewis BC. p19Arf Inhibits the Invasion of Hepatocellular Carcinoma Cells by Binding to C-terminal Binding Protein. Cancer Res. 2008;68:476–82. doi: 10.1158/0008-5472.CAN-07-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 30.Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Devel. 2004;18:1862–74. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collado M, Serrano M. The TRIP from ULF to ARF. Cancer Cell. 2010;17:317–8. doi: 10.1016/j.ccr.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Chen DL, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–U193. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: Consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 34.Weydert CJ, Esser AK, Mejia RA, Drake JM, Barnes JM, Henry MD. Endothelin-1 inhibits prostate cancer growth in vivo through vasoconstriction of tumor-feeding arterioles. Cancer Biol Ther. 2009;8:720–9. doi: 10.4161/cbt.8.8.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drake JM, Strohbehn G, Moreland JG, Henry MD. Transendothelial migration selects for prostate cancer cells with elevated Zeb1 expression and loss of epithelial identity. Clin Exper Metastasis. 2009;26:860–1. [Google Scholar]
- 36.Song HT, Jordan EK, Lewis BK, Liu W, Ganjei J, Klaunberg B, et al. Rat model of metastatic breast cancer monitored by MRI at 3 tesla and bioluminescence imaging with histological correlation. J Translational Med. 2009:7. doi: 10.1186/1479-5876-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji XD, Cheng L, Wei F, Li HM, Wang MY, Tian YH, et al. Noninvasive Visualization of Retinoblastoma Growth and Metastasis via Bioluminescence Imaging. Invest Ophthal Visual Sci. 2009;50:5544–51. doi: 10.1167/iovs.08-3258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.